Abstract
The combination of laser capture microdissection and mass spectrometry represents a powerful technology for studying spatially-resolved proteomes. Moreover, the compositions of integral membrane proteomes have rarely been studied in a spatially-resolved manner. In this study, ocular lens tissue was carefully dissected by laser capture microdissection and conditions for membrane protein enrichment, trypsin digestion, and mass spectrometry analysis were optimized. Proteomic analysis allowed the identification of 170 proteins, 136 of which were identified with more than one peptide match. Spatial differences in protein expression were observed between cortical and nuclear samples. In addition, the spatial distribution of posttranslational modifications to lens membrane proteins, such as the lens major intrinsic protein AQP0, were investigated and regional differences were measured for AQP0 C-terminal phosphorylation and truncation.
Introduction
Integral membrane proteins are targets for many pharmacological compounds and are crucial to many cellular functions, such as ion regulation and transport, molecular recognition and response, and energy transduction. About 30 to 40 percent of all human proteins are predicted to be integral membrane proteins. However, given the highly hydrophobic nature of these proteins and the analytical challenges they present, even basic characterization falls far behind soluble proteins. The hydrophobic nature of membrane proteins results in poor solubility, incomplete enzymatic digestion, and difficulties in protein separation and identification which hinders the study of membrane proteomics and posttranslational modifications of these proteins.
The spatial distribution of membrane proteins and their posttranslational modifications has rarely been studied not only because of the difficulties of working with membrane proteins, but also because of the lack of precise microdissection techniques. Laser capture microdissection (LCM) has emerged as an ideal tool for selectively extracting individual cells or area of tissue of interest with a microscopic region as small as 3–5 µm in diameter from their natural environment.1–3 The major disadvantage of LCM is that it isolates minute amounts of material which challenges detection limits of modern technologies. Recently, the combination of laser capture microdissection (LCM) and highly sensitive mass spectrometry has been applied to study the proteomics of specific areas of tissue;4–7 however, little work has been done on membrane proteins.
The ocular lens has a relatively simple cellular composition with an anterior monolayer of epithelial cells and the underlying fiber cells that form the bulk of the organ. Epithelial cells differentiate continuously in the equatorial region into lens fiber cells that lie on the top of the older fiber cells. Fiber cell differentiation is accompanied by rapid cellular elongation with a concomitant increase in plasma membrane surface area and loss of organelles.8 Unlike other tissues, there is no lenticular cell turnover resulting in newly differentiated cells being added to the lens mass on a continuous basis, in a concentric manner.9 Therefore, fiber cells in the different regions of the lens are in different stages of differentiation, are of different age, and are extensively modified in an age-dependent manner. Hence, although cellularly simplistic, the ocular lens serves as an excellent model tissue for spatially-resolved proteomics studies.
The most abundant integral membrane protein in the lens is the major intrinsic protein (MIP) now known to be a member of the aquaporin (AQP) family of membrane water channels, named AQP0. AQP0 is essential for establishing proper fiber cell structure and organization10, functions as a water-selective channel11, and has a role in fiber cell adhesion12. Posttranslational modifications of AQP0 have been studied previously and include: phosphorylation, deamidation, methionine oxidation and truncation. 13–19 When lenses were manually dissected into 3–4 concentric regions these modifications were found to be heterogeneously distributed in different lens regions15 and some may be associated with cataract20. Considering the dramatic biochemical, physiological, and morphologic changes involved in differentiation of lens epithelial cells into fiber cells as well as the subsequent aging of fiber cells, it is reasonable that posttranslational modifications of proteins are spatially distributed throughout the tissue corresponding to specific stages of lens fiber cell differentiation and age. Understanding the differences of protein expression and posttranslational modifications in these different aged fiber cells will help our understanding of lens development and aging.
In this study, specific regions of lens tissue were isolated using LCM and the lens membrane proteome was analyzed with particular attention to posttranslational modifications of the lens major intrinsic membrane protein, AQP0. The LCM conditions to capture lens tissue and subsequent membrane protein analysis by electrospray tandem mass spectrometry (ESI-MS/MS) were optimized. Differential protein expression and spatially-distinct AQP0 modifications were repeatedly found in different bovine lenses. These studies demonstrate a powerful new tool in the arsenal for investigating the spatially-resolved tissue distributions of integral membrane proteins and their modified forms.
Methods
Materials
Bovine lenses were obtained from PelFreez Biologicals (Rogers, AK). Ammonium bicarbonate, sodium fluoride, EDTA, urea, DTT, sodium hydroxide and trifluoroacetic acid (TFA), were purchased from Sigma (St. Louis, MO). All HPLC grade solvents were purchased from Fisher (New Jersey, NJ). Sequence-grade modified trypsin was obtained from Promega (Madison, WI). RapiGest detergent was purchased from Waters Corp. (Milford, MA).
Frozen tissue sectioning
Frozen bovine lenses were quickly decapsulated by briefly placing under running water and using forceps to pull off capsule. The decapsulated lenses were attached to a chuck with optimal cutting temperature (OCT) medium (Tissue-Tek®, Sakura Finetek, Torrance, CA) or encapsulated with OCT. Lenses were sectioned equatorially in a cryostat (HM505E Microm, Walldorf, Germany) at −20°C. 12 µm sections were cut through the whole lens, but certain sections were collected for LCM as indicated in Figure 1. Sections were picked up by Fisherbrand Superfrost/Plus slides (Fisher Scientific, Pittsburgh, PA) coated with a thin layer of anhydrous ethanol and frozen immediately in a chilled slide box on dry ice. After sectioning, sections were stored in a slide box at −80°C until needed.
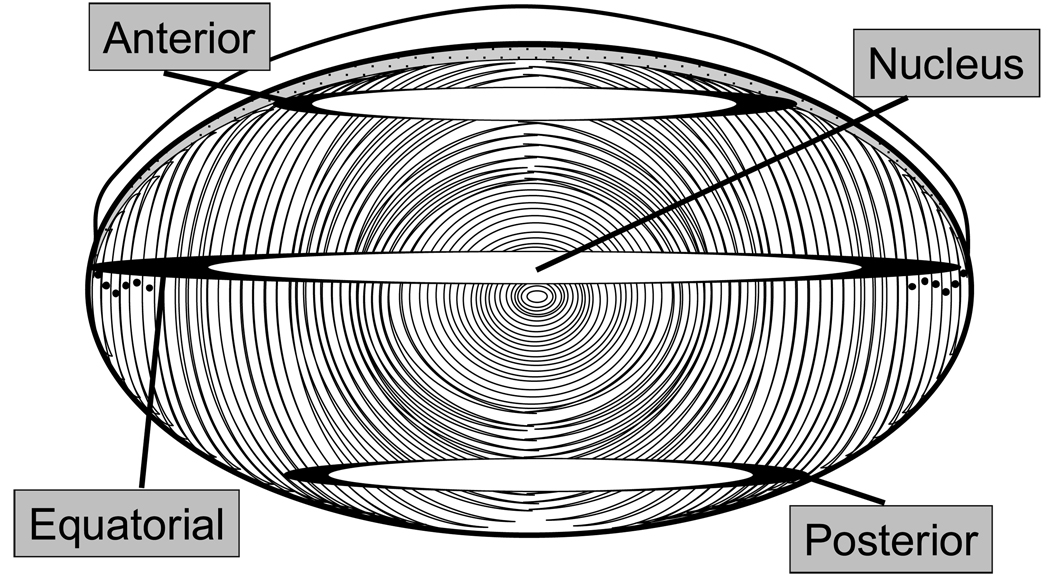
Figure 1.
A diagram of a lens that indicates the anterior, equatorial, and posterior sections used for laser capture microdissection. Four regions of the lens, shaded in black, were the captured and subjected to proteomic analysis.
Section staining, dehydration, and LCM
Frozen tissue sections were thawed and partially air dried. Slides were immediately fixed in 75% ethanol for 1 min. After fixation sections were placed in water for 1 min to remove any OCT remaining on the sections followed by dehydration through a series of graded ethanol and xylene washes: 75% ethanol for 1 min, 95% ethanol for 1 min, 95% ethanol for 1 min, 100% ethanol for 1 mins, 100% ethanol for 2 mins, xylene for 5 mins (2X). Following incubation in xylene, sections were dried for 15 mins in a dessicator. One section from each region of the lens was stained with hematoxylin and eosin to guide the capture.
LCM was performed using a PixCell® II instrument (Arcturus, Mountain View, CA). Specific tissue sections were captured using a 30µm diameter laser beam at 70–100 mW power with a pulse duration of 3 ms. In this experiment, the outer cortex from each section and the nucleus from the equator sections were captured (Figure 1). For outer cortex samples, 7 concentric cell layers (30 µm per layer) were captured from the very edge of each section. For some anterior sections, a thin dark region of tissue was apparent after dehydration which is believed to be the epithelium cell layer. Once this dark edge was found, the capture was performed adjacent to it to avoid collecting epithelial cells. For nuclear samples, a circular area with a radius of 15 laser beam diameters (450 µm) was captured from the centers of the equator sections. Tissues collected from homogeneous populations of cells were captured on three caps from four to eight serial sections of each region of the lens (a total of 3,000–4,000 laser shots) and films were pooled together.
Membrane protein preparation
After microdissection, the LCM caps were inserted into an Eppendorf tube containing buffer (200 µL 20 mM ammonium bicarbonate buffer containing 7 M urea, 5 mM EDTA, 10 mM NaF and 1 mM DTT, pH8 ). The tube was placed upside down and incubated at 4°C for 30 mins. The tube was centrifuged at 9, 0000 g for 10 min. and the supernatant was discarded. This washing process was repeated three times. The films on the caps were removed, combined and washed with the above buffer twice followed by separate washes with: 0.1 M NaOH, water, 95% ethanol and water (X5). A blank film was treated in the same way as above to identify any potential contamination from the film. To compare LCM results to results from manually dissected tissue a very thin layer of lens tissue was cut from the anterior region of the decapsulated frozen bovine lens using a razor blade. This tissue was homogenized in the same ammonium bicarbonate buffer as above. The homogenate was centrifuged at 90,000 g for 30 mins and the pellets were collected. The pellets were washed in the same way as above to collect the membrane proteins.
Trypsin digestion
100 µL of 0.1% RapiGest (in 50mM ammonium bicarbonate buffer) and 100 ng of trypsin were added to the Eppendorf tube containing LCM films. The tubes were incubated at 37 °C for 24 hours and digestion was stopped by adding TFA to a final concentration of 0.5%. The sample was incubated at 37°C for 30 mins and centrifuged at 2,0000 g for 5 mins to remove the by-products of the RapiGest detergent. The supernatant was collected and dried in a SpeedVac.
NanoLC-ESI/MS/MS
Tryptic peptides were reconstituted in H2O (0.1% TFA) and analyzed by HPLC-ESI-MS/MS. The peptides were separated on an Ultimate Nano-LC system equipped with a Switchos (Dionex, Germany). Samples were desalted on a trap column (PepMap, C18, 5µm particle size, 300µm i.d. × 5mm, 100 Å pore size) for 4 mins with a 20 µL/min flow rate and then eluted from an analytical column (PepMap C18, 150 mm × 75 µm, 3 µm, 100 Å, Dionex, Germany) using the following gradient: 0–80 mins: 2–50% ACN (0.02% HFBA), 80–100 mins: 50–95% ACN (0.02% HFBA) with a 0.18 µL/min flow rate. The flow was directly infused into a nano-ESI-LTQ mass spectrometer (Thermo Electron, San Jose, CA). The mass spectrometer was operated in data dependent mode with the top 5 most abundant ions in each mass spectrum being selected for subsequent MS/MS scans using a normalized collision energy of 35%. Mass spectra were acquired over a m/z range of 360–2000. Dynamic exclusion (repeat count 2, repeat duration 0.5 min., exclusion duration 3 min.) was enabled to allow detection of less abundant ions.
Data analysis
All MS/MS spectra were searched with Bioworks (version 3.3, Thermo Electron, San Jose, CA) using the Swiss-Prot bovine protein database. Peptide identifications were made according to the following criteria: cross-correlation (XCorr) values were at least 1.5 for +1 charged ions, 2.0 for +2 charged ions, and 2.5 for +3 charged ions; and protein probability scores less than 0.001. Proteins identified by only one peptide were excluded from the final analysis. Potential posttranslational modifications including deamidation, phosphorylation, methionine oxidation, and truncation were considered in the searches. All MS/MS spectra used to assign posttranslational modifications presented in this paper were manually verified. False positive rates were determined by comparing peptides and proteins identified using the criteria for matching described above and searching a reversed Swiss-Prot bovine database. The calculated rates were 6.3% at the peptide level and 0.66% at the protein level (requiring two peptide matches).
For quantitative analysis, selected ion chromatograms were generated by specifying the mass-to-charge ratio of a predicted peptide (+/− 1 m/z unit) and the chromatogram was subjected to three-point Boxcar smoothing using the Qual Browser program of the Xcalibur software. Peaks were detected using ICIS peak detection algorithm. Peak area integration was accomplished using Xcalibur’s default options.
Results
Optimization of LCM conditions
Sections were treated in different ways to optimize lenticular tissue capture. The cortical regions of the lens attached to the slides very tightly and resulted in poor laser-induced transfer to films, whereas the nuclear regions of the lens easily detached from the slides. A thin layer of anhydrous ethanol on the slides promoted tissue attachment to the glass slides while maintaining the quality of the sections. Sections were partially air dried before fixation by 75% ethanol to avoid detachment of the nucleus during fixation and water washes. In addition it was found that a 1 min. water wash after fixation was an essential step to promote efficient tissue transfer to LCM caps.
One section from each of the three lens regions (anterior, equator, and posterior) was stained using hematoxylin and eosin and showed that cortical tissue was more lightly stained than nuclear tissue. For outer cortex samples, 7 concentric layers (7 × 30 µm laser diameter) were captured corresponding to the outer region of the lightly stained outer cortex of the lens. The nucleus sample was captured from the heavily stained center of the equator section. Generally about 3000–4000 laser shots were taken per cap which took about 20 mins. Digital images were taken of tissue before and after microdissection (Figure 2).
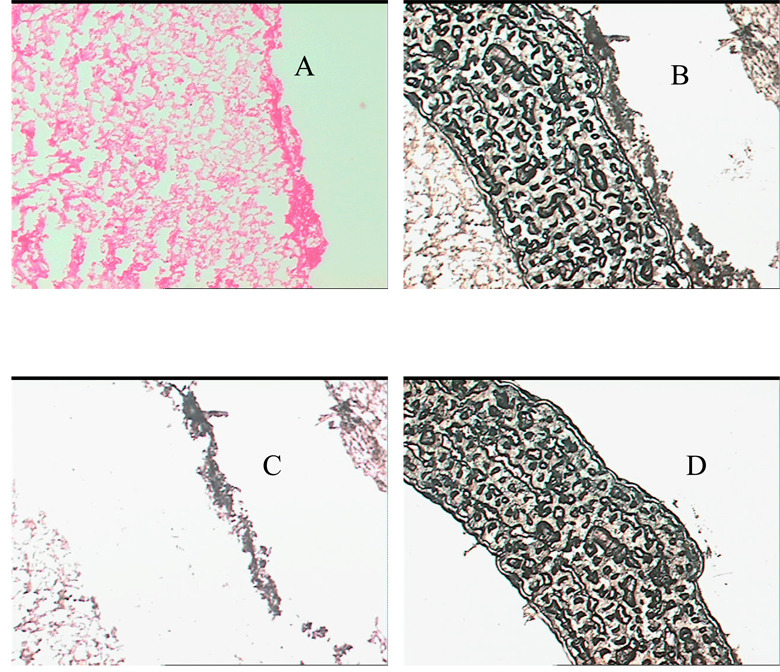
Figure 2.
Images of the laser capture microdissected bovine lens anterior cortical tissue. A) H&E stained tissue before LCM, B) optical image of tissue section after laser capture but before tissue removal, C) optical image of tissue section after LCM and tissue removal, and D) optical image of captured tissue on the LCM cap.
Protein identification and comparison of homogenized control and LCM samples
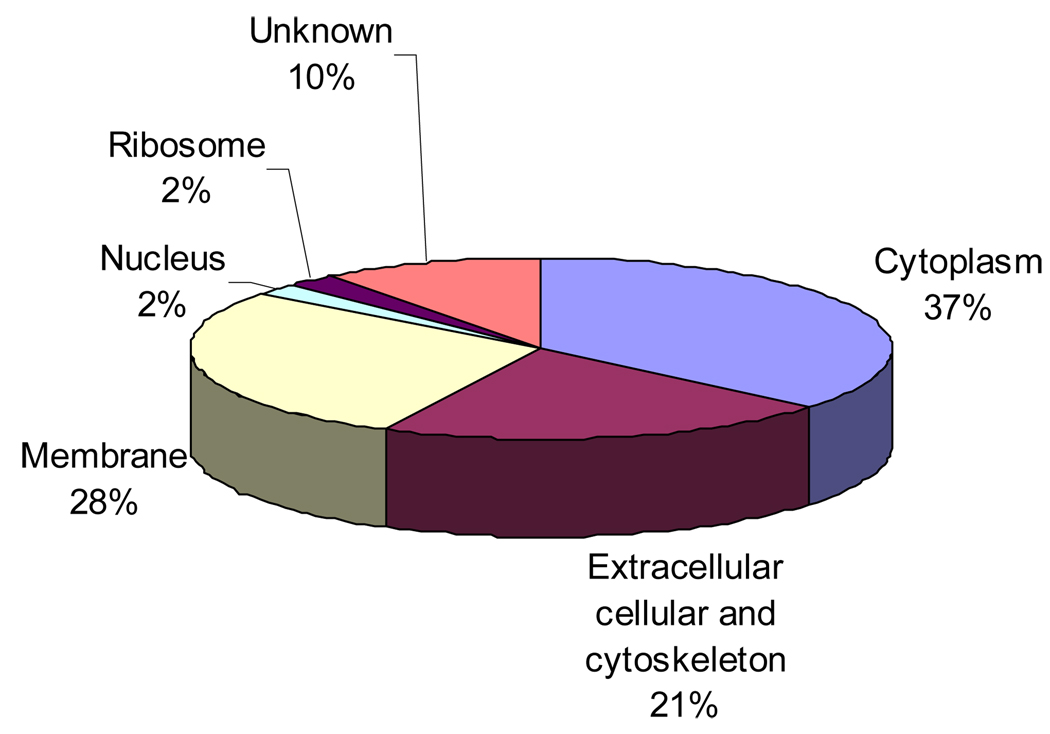
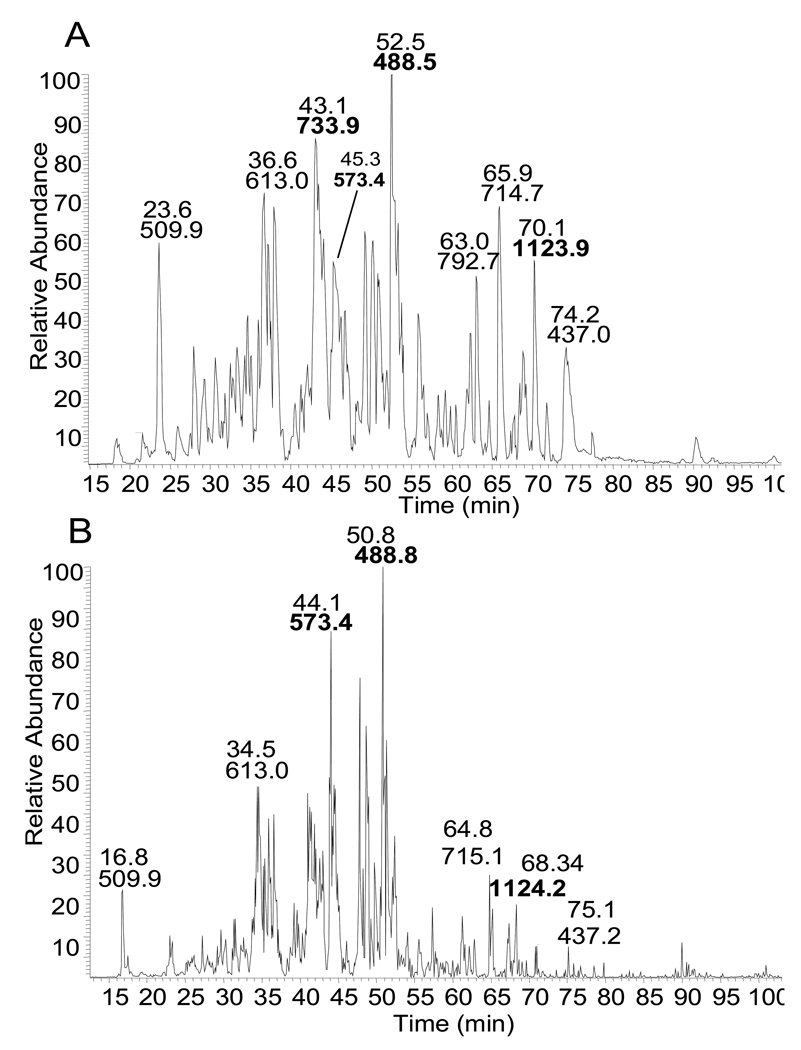
Typically 60–90 proteins were identified from each LCM sample. A summation of results from 12 LCM samples revealed a total 170 proteins identified of which 136 proteins were identified with more than one peptide match. These 136 proteins are listed in Table A of the supporting information. A general summary of protein identification frequency is provided in Table 1. Lens membrane proteins including AQP0, MP20 and gap junction proteins (connexin 50 and connexin 46) were present in all of the samples with sequence coverage ranging from 16% to 45%. AQP0 peptides identified in all samples include: 188–196, 239–259, 234–238, and 229–238phosphorylated. AQP0 229–233 was observed in 11 of the 12 experiments, 242–259 was observed in 10 of 12 experiments and 227–233 appeared in 8 of 12 experiments. Lens specific cytoskeletal proteins filensin and phakinin (CP49), often seen in lens membrane preparations, were also detected in every sample examined. Many other integral membrane and membrane associated proteins were also observed including transporters, receptors, and cell adhesion molecules. Figure 3 indicates the subcellular localization of identified proteins determined based on Swiss-Prot database annotations. The signal of many proteins was sufficient to identify posttranslational modifications such as phosphorylation, truncation, deamidation, and methionine oxidation. At least 7 phosphorylation sites in connexin 50 and one phosphorylation site in filensin were identified in the LCM samples (data not shown). Other phosphorylation sites are discussed below. Even though LCM samples were extensively washed, the highly abundant crystallin proteins remained in the samples in significant amounts; however, the crystallin signals have been dramatically decreased by washing. Predicted integral membrane proteins identified comprise 28% of all of the proteins identified in the sample and when combined with cytoskeletal proteins comprise 50% of proteins identified. Strong signals corresponding to AQP0 and connexin 50 peptides can be seen in the base peak chromatogram (Figure 4).
Table 1.
Summary of protein identification with multiple peptides hits from laser captured bovine lens cells:
| Number of proteinsa | Number of times identified |
|---|---|
| 21 | 12 |
| 9 | 11 |
| 1 | 10 |
| 11 | 9 |
| 6 | 8 |
| 2 | 7 |
| 3 | 6 |
| 6 | 5 |
| 6 | 4 |
| 12 | 3 |
| 21 | 2 |
| 38 | 1 |
| Total : 136 | |
Results from 12 LCM experiments: four regions per lens dissected and analyzed in triplicate.
Figure 3.
Summary of the cellular localization of the identified proteins. 136 proteins identified in the 12 samples were sorted based on the known cellular localization listed in the Swiss-Prot database annotation.
Figure 4.
Base peak chromatograms from LC-MS/MS analysis. Base peak chromatogram indicating major AQP0 tryptic peptide signals from: A) anterior cortex laser capture microdissected sample and B) lens homogenate from manually dissected anterior cortex. Major AQP0 and connexin 50 peptides are highlighted in bold font.
To investigate the effect of LCM on the observed protein profiles, a very thin layer of lens tissue was gently dissected from the anterior of the lens using a razor blade. Since the tissue was collected from the very anterior edge of the lens, it is thought that this sample has a very similar composition to the laser captured anterior outer cortex sample. All of the proteins identified in the manually dissected sample were also present in the LCM samples. The extent of protein sequence coverage of the major membrane proteins was very similar between the two samples; and a comparison of base peak chromatograms (Figure 4) indicates comparable signals for AQP0 and connexin 50 between LCM and manually dissected samples.
A comparison of proteins identified from different laser captured regions of the lens revealed similar profiles among cortical regions but some differences between cortical and nuclear regions. Proteins identified in all cortical samples but absent from the nuclear samples included: Desmin (O62654), Tubulin beta-2B chain (Q6B856), Catenin beta-1(Q0VCX4), Ezrin (P31976), Neuronal cell adhesion molecule (Q6Q146). All of these proteins are cell adhesion-related membrane proteins or cytoskeletal proteins. Other cytoskeletal protein such as vimentin and spectrin were positively identified but with very low sequence coverage in the nucleus which may indicate either low abundance of these proteins in the nucleus or high levels of posttranslational modification.
Identification of posttranslational modifications in the C-terminal of AQP0
The major posttranslational modifications in AQP0 reported previously were found in all LCM samples including: methionine oxidation, deamidation, phosphorylation, and truncation. In this study, we are mainly interested in the spatial distributions of these posttranslational modifications. Two major modifications (phosphorylation and C-terminal truncation) were carefully analyzed quantitatively to investigate spatial differences.
Phosphorylation
Several major phosphorylation sites of bovine lens AQP0 reported previously were found in all LCM samples including: Ser 235, Ser 243 and Ser 245. A very small amount of Ser 231 phosphorylation was also found in the nucleus sample. The relative amounts of phosphorylation at these major sites was investigated by calculating the ratio of phosphorylated peptides compared to the unphosphorylated counterparts assuming similar ionization efficiencies of the peptides used in the calculation21. The result is shown in Figure 5.
Figure 5.
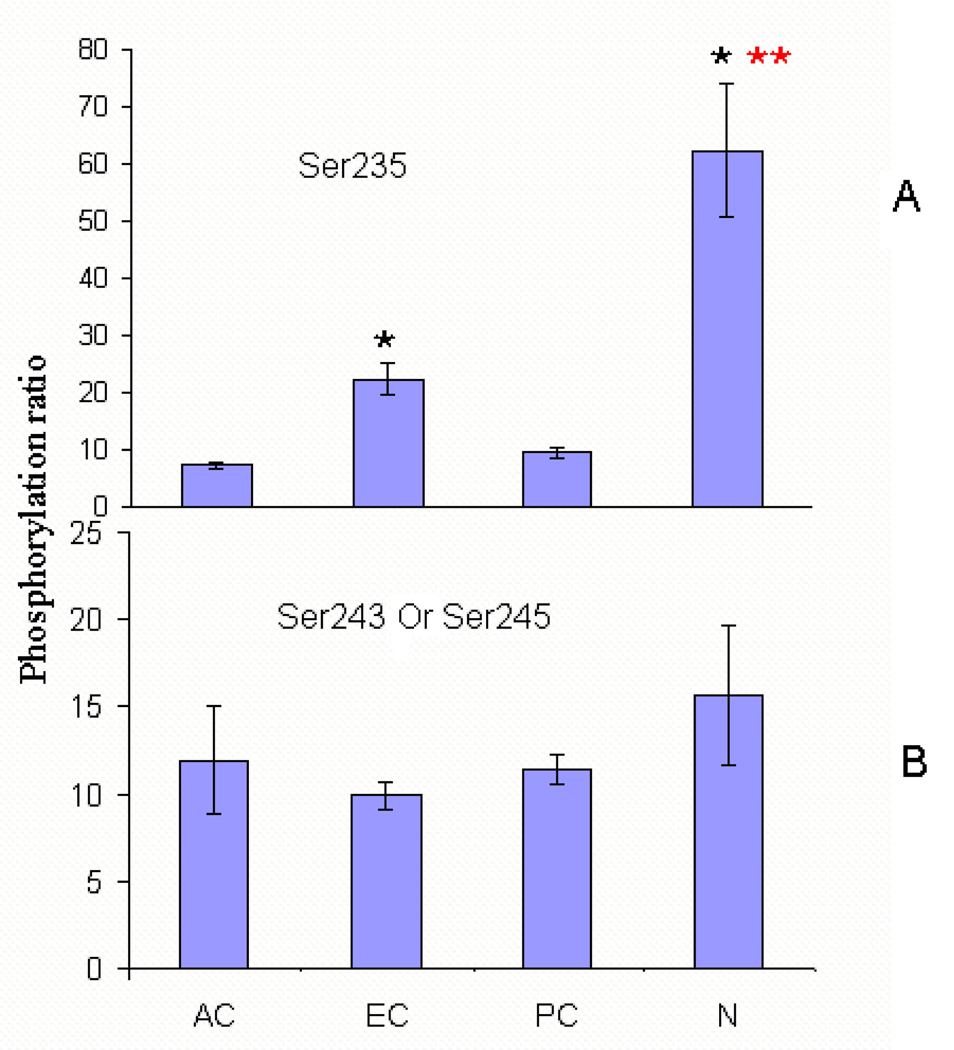
The spatial distribution of AQP0 C-terminal phosphorylation. The relative percentage of phosphorylation in the different regions of the lens are expressed by the ratio of phosphorylation. The ratio of S235 phosphorylation = peak area of phosphorylated 229–238/ peak area of unphosphorylated 234–238; the ratio of phosphorylation on S243 or S245= peak area of mono-phosphorylated 239–259/ peak area of unphosphorylated 239–259. The ratios of phosphorylation are plotted: (A) S235 phosphorylation and (B) S243 or S245 phosphorylation. A single asterisk (*) indicates significance compared to the anterior outer cortex. A double asterisk (**) indicates significance compared to the equator outer cortex (p< 0.01). AC, anterior cortex; EC, equatorial cortex; PC, posterior cortex; N, nucleus.
As reported previously18, when Ser 235 is phosphorylated, tryptic cleavage is blocked and the observed phosphorylated peptide includes residues 229–238, whereas in the unphosphorylated situation, peptide 229–238 is efficiently cleaved into two peptides (229–233 and 234–238). The ratio of peak area of peptide 229–238 with Ser 235 phosphorylated to the peak area of the unphosphorylated peptide 234–238 was used to compare the relative percentage of phosphorylation within the different regions of the lens. The mean percentage of Ser 235 phosphorylation in four different regions of the lens was 7.3±0.5 (n=3) in the anterior cortex, 9.5±0.8 (n=3) in the posterior cortex, 22.4±2.7 (n=3) in the equatorial cortex and 62.4±11.5 (n=3) in the nucleus. The levels of phosphorylation at Ser 235 in the anterior cortex and posterior cortex were not significantly different (p > 0.01). However, the levels of phosphorylation at Ser 235 were significantly different in the equatorial cortex and in the nucleus (p < 0.01), and both were significantly different from the anterior outer cortex and posterior outer cortex (p < 0.01) (See Figure 5(A)).
Careful examination of the MS/MS spectra of the peptide 239–259 with one Ser phosphorylated revealed that both Ser 243 and Ser 245 could be phosphorylated and the singly phosphorylated peptides coeluted. Therefore, calculation of the total percentage of phosphorylation includes both sites. The results are shown in Figure 5(B), which indicates that the levels of phosphorylation of Ser 243 and Ser 245 in the regions analyzed are very similar (approximately 10–15%) and the differences among regions are not statistically significant.
C-terminal truncation of AQP0
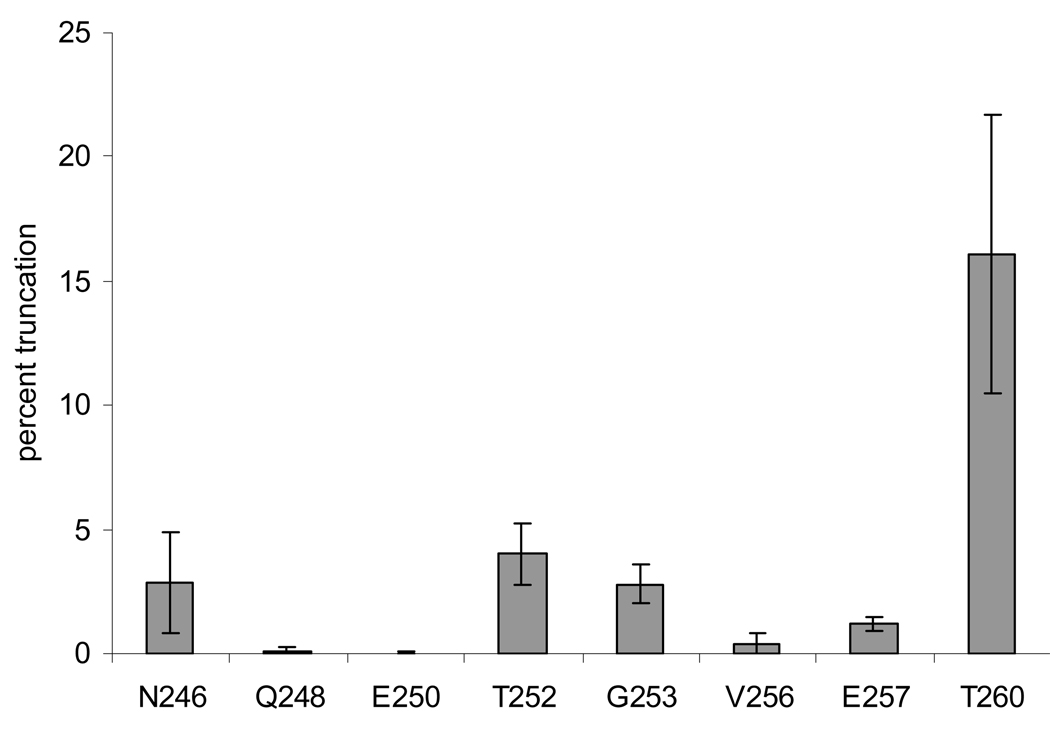
C-terminal truncation was detected in the nucleus of the bovine lens. All the MS/MS spectra of the truncated peptides were manually verified. The relative level of truncation at each site was calculated as the ratio of the peak area of each truncated peptide to the peak area of the expected tryptic peptide containing residues 239–259. The results are shown in Figure 6. Truncation was rarely detected or the levels of truncation were extremely low in all of the outer cortex samples. The major truncation sites in the nucleus include: N246, T252, G253, E257 and T260. Since the proteins were digested by trypsin (expected to produce a 239–259 peptide), only miscleaved 239–260 was detected for T260 truncation. If trypsin cleaved at K259 of the T260 truncated form of the protein, the resulting product would be a threonine residue that is undetectable in this experiment. Therefore, the level of truncation at T260 may be significantly under-estimated. Nevertheless, significant levels of truncation at T260 were observed in all of the nucleus samples while this truncation was not detected in any cortex sample.
Figure 6.
C-terminal truncation of AQP0 in bovine lens. C-terminal truncation of AQP0 was rarely detected in the outer cortex samples (not shown), but was detected in all of the nucleus samples (gray bars). The relative level of truncation at each site was calculated as the ratio of the peak area of each truncated peptide to the peak area of peptide 239–259.
Truncation of connexin 50 and connexin 46
Glutamic acid, E290, was previously reported to be a calpain cleavage site,22 In this study, truncation products of connexin 50 at E290 were detected in the nucleus samples and occasionally detected in the anterior outer cortex samples albeit at very low levels. Truncation products of connexin 46 at D124 and L255 were also detected in the nucleus samples. The selected ion chromatograms for these truncation products are shown in (Supplemental Data Figure A).
Discussion
The combination of LCM and mass spectrometry has been previously employed for proteome analysis and biomarker discovery; however, the present study is the first study to use this powerful approach to investigate the spatial distribution of analytically challenging integral membrane proteins and their posttranslational modifications. The difficulties of investigating membrane proteins are primarily due to their poor solubility; however, the LCM approach used here has advantages for sample preparation of insoluble proteins. After LCM, membrane proteins cannot be extracted from the films by mass spectrometry compatible buffers; therefore, samples can be extensively washed to remove soluble proteins and trypsin digestion can be carried out directly on the films. A blank film was treated in the same way as the samples and analyzed by LC-MS/MS revealing no significant contaminations that prevented protein analysis. Trypsin digestion was most efficient on the film when RapiGest was used. Compared to the homogenized sample, LCM samples had higher crystallin signal most likely because those proteins are buried in the films making them difficult to remove by simply washing protocols.
When all tandem mass spectra were searched against the Swiss-Protein bovine database, 136 proteins were identified with at least two peptide matches. 50% of these proteins corresponded to predicted membrane or cytoskeletal proteins, which indicates that levels of soluble proteins had been reduced during sample preparation. A comparison of the proteins expressed in the different lens regions indicated that several cell adhesion-related or cytoskeletal proteins were not found or found at very low abundance in the nucleus of the lens. These results are consistent with previous reports, for example, cadherin and catenin were reported to be present in the cortical fiber cells but absent in the nucleus23, 24. Moreover, neuronal cell adhesion molecule has been found only in the cortex of the lens23. Vimentin25 and spectrin26 were also reported to be absent in the nucleus. In the present experiments these proteins can be detected in the nucleus, but the sequence coverage was extremely low suggesting low abundance.
Sufficient signals were obtained from LCM samples to examine protein posttranslational modifications. Modifications to AQP0 play important roles in its function in the lens and may be linked to fiber cell differentiation. For example, phosphorylation affects calmodulin binding which regulates water permeability27. Also, truncation is predicted to affect the interaction of AQP0 with lens specific cytoskeletal proteins filensin and CP49.28 Therefore, spatial definition of AQP0 modifications is critical to understanding AQP0 function in lens development, aging, and cataractogenesis.
Serine 235 has been reported to be the major phosphorylation site in AQP0 and resides in a very conserved region in vertebrate AQP0s.17, 18 The results reported in this study indicate the interesting finding that the levels of phosphorylation of S243 and of S245 do not significantly change in different lens regions. The level of S235 phosphorylation is highest in the nucleus and lowest in the anterior and posterior regions. This result implies that the level of S235 phosphorylation may be spatially regulated. Further study of phosphorylation in other regions of the lens is needed to draw definitive conclusions.
Age and fiber cell specific C-terminal truncation of AQP0 has been reported previously.15–17 In the present study, truncation of AQP0 at several sites was detected from the limited LCM-derived samples. Although considerable variability was observed in the quantification of truncation products, the locations of major truncation sites in the bovine lens were consistent among samples. The level of truncation in the outer cortex was not detectable or very low, but high in the nucleus, which is consistent with previous published results.15–17 The major site of truncation in the bovine lens nucleus was found at T260, whereas, the major sites of truncation in human lens were reported at N246 and N259.16–17 This may simply result from the difference between species or from an artifact of the analysis. In the bovine lens, residue 259 is a lysine, which is a trypsin cleavage site; therefore, any in vivo K259 truncation cannot be detected in the present experiment.
Considering the structural and functional importance of integral membrane proteins and mechanisms for their regulation, it is extremely valuable to investigate the spatial distribution of these proteins and their posttranslational modifications. The alteration in these proteins in the lens, for example, could potentially lead to the development of cataract. The present study demonstrates that the combination of LCM and mass spectrometry make it possible to study integral membrane proteins and their posttranslational modifications in a spatially-resolved manner. The application of this approach to more complex tissues should prove highly productive in the search for disease mechanisms and therapeutic targets.
Supplementary Material
This material is available at http://pubs.acs.org.
Acknowledgements
Financial support for this project was provided by NIH grant EY-13462 (KLS) and the Kilpatrick Medical Research Fund. The authors thank Dr. Debra Hazen-Martin and the MUSC Histo-pathology core for access and training on the LCM instrument. The authors acknowledge use of the MUSC Mass Spectrometry Facility.
References
- 1.Zhou G, Li H, DeCamp D, Chen S, Shu H, Gong Y, Flaig M, Gillespie JW, Hu N, Taylor PR, Emmert-Buck MR, Liotta LA, Petricoin EF, Zhao Y. 2D differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Mol. Cell. Proteomics. 2002;1:117–124. doi: 10.1074/mcp.m100015-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Craven RA, Totty N, Harnden P, Selby PJ, Banks RE. Laser capture microdissection and two-dimensional polyacrylamide gel electrophoresis: evaluation of tissue preparation and sample limitations. Am. J. Pathol. 2002;160:815–822. doi: 10.1016/S0002-9440(10)64904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 4.Wang HY. Laser capture microdissection in comparative proteomic analysis of hepatocellular carcinoma. Methods Cell Biol. 2007;82:689–707. doi: 10.1016/S0091-679X(06)82025-X. [DOI] [PubMed] [Google Scholar]
- 5.Umar A, Luider TM, Foekens JA, Pasa-Tolić L. NanoLC-FT-ICR MS improves proteome coverage attainable for approximately 3000 laser-microdissected breast carcinoma cells. Proteomics. 2007;7:323–329. doi: 10.1002/pmic.200600293. [DOI] [PubMed] [Google Scholar]
- 6.Haqqani AS, Nesic M, Preston E, Baumann E, Kelly J, Stanimirovic D. Characterization of vascular protein expression patterns in cerebral ischemia/reperfusion using laser capture microdissection and ICAT-nanoLC-MS/MS. FASEB J. 2005;9:1809–1821. doi: 10.1096/fj.05-3793com. [DOI] [PubMed] [Google Scholar]
- 7.Baker H, Patel V, Molinolo AA, Shillitoe EJ, Ensley JF, Yoo GH, Meneses-García A, Myers JN, El-Naggar AK, Gutkind JS, Hancock WS. Proteome-wide analysis of head and neck squamous cell carcinomas using laser-capture microdissection and tandem mass spectrometry. Oral Oncol. 2005;41:183–199. doi: 10.1016/j.oraloncology.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Bassnett S, Winzenburger PA.Morphometric analysis of fibre cell growth in the developing chicken lens Exp. Eye Res 200376291–302 [DOI] [PubMed] [Google Scholar]
- 9.Taylor VL, Al-Ghoul KJ, Lane CW, Davis VA, Kuszak JR, Costello MJ. Morphology of the normal human lens. Invest. Ophthalmol. Vis. Sci. 1996;37:1396–1410. [PubMed] [Google Scholar]
- 10.Dunia IM, Recouvreur M. Assembly of connexins and MP26 in lens fiber plasma membranes studied by SDS-fracture immunolabeling. J Cell Sci. 1998;111:2109–2120. doi: 10.1242/jcs.111.15.2109. [DOI] [PubMed] [Google Scholar]
- 11.Gorin MS, Yancey SB, Cline J, Revel JP, Horwitz J. Cell. 1984;39:49–59. doi: 10.1016/0092-8674(84)90190-9. [DOI] [PubMed] [Google Scholar]
- 12.Michea LF, Andrinolo D, Ceppi H, Lagos N. Biochemical evidence for adhesion-promoting role of major intrinsic protein isolated from both normal and cataractous human lenses. Exp. Eye Res. 1995;61:293–301. doi: 10.1016/s0014-4835(05)80124-1. [DOI] [PubMed] [Google Scholar]
- 13.Han J, Schey KL. Proteolysis and mass spectrometric analysis of an integral membrane: aquaporin 0. J Proteome Res. 2004;3:807–812. doi: 10.1021/pr049945w. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Little M, David LL, Giblin FJ, Schey KL. Sequence and peptide map of guinea pig aquaporin 0. Mol. Vis. 2004;10:215–222. [PubMed] [Google Scholar]
- 15.Ball LE, Garland DL, Crouch RK, Schey KL. Post-translational modifications of aquaporin 0 (AQP0) in the normal human lens: spatial and temporal occurrence. Biochemistry. 2004;43:9856–9865. doi: 10.1021/bi0496034. [DOI] [PubMed] [Google Scholar]
- 16.Schey KL, Little M, Fowler JG, Crouch RK. Characterization of human lens major intrinsic protein structure Invest Ophthalmol. Vis Sci. 2000;41:175–182. [PubMed] [Google Scholar]
- 17.Schey KL, Fowler JG, Shearer TR, David L. Modifications to rat lens major intrinsic protein in selenite-induced cataract. Invest. Ophthalmol. Vis. Sci. 1999;40:657–667. [PubMed] [Google Scholar]
- 18.Schey KL, Fowler JG, Schwartz JC, Busman M, Dillon J, Crouch RK. Complete map and identification of the phosphorylation site of bovine lens major intrinsic protein. Invest. Ophthalmol. Vis. Sci. 1997;38:2508–2515. [PubMed] [Google Scholar]
- 19.Lampe PD, Johnson RG. Amino acid sequence of in vivo phosphorylation sites in the main intrinsic protein (MIP) of lens membranes. Eur. J Biochem. 1990;194:541–547. doi: 10.1111/j.1432-1033.1990.tb15650.x. [DOI] [PubMed] [Google Scholar]
- 20.Schey KL, Fowler JG, Shearer TR, David L. Modifications to rat lens major intrinsic protein in selenite-induced cataract. Invest. Ophthalmol. Vis. Sci. 1999;40:657–667. [PubMed] [Google Scholar]
- 21.Steen H, Jebanathirajah JA, Rush J, Morrice N, Kirschner MW. Phosphorylation Analysis by Mass Spectrometry: Myths, Facts, and the Consequences for Qualitative and Quantitative Measurements. Mol. Cell Proteomics. 2006;5:172–181. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Lin JS, Fitzgerald S, Dong Y, Knight C, Donaldson P, Kistler J. Processing of the gap junction protein connexin50 in the ocular lens is accomplished by calpain. Eur J Cell Biol. 1997;73:141–149. [PubMed] [Google Scholar]
- 23.Watanabe M, Kobayashi H, Yao R, Maisel H. Adhesion and junction molecules in embryonic and adult lens cell differentiation. Acta Ophthalmol Suppl. 1992;205:46–52. doi: 10.1111/j.1755-3768.1992.tb02180.x. [DOI] [PubMed] [Google Scholar]
- 24.Atreya PL, Barnes J, Katar M, Alcala J, Maisel H. N-cadherin of the human lens. Curr Eye Res. 1989;8:947–956. [PubMed] [Google Scholar]
- 25.Sandilands A, Prescott AR, Carter JM, Hutcheson AM, Quinlan RA, Richards J, FitzGerald PG. Vimentin and CP49/filensin form distinct networks in the lens which are independently modulated during lens fibre cell differentiation. J Cell Sci. 1995;108:1397–1406. doi: 10.1242/jcs.108.4.1397. [DOI] [PubMed] [Google Scholar]
- 26.Maisel H, Ellis M. Cytoskeletal proteins of the aging human lens. Curr Eye Res. 1984;3:369–381. doi: 10.3109/02713688408997222. [DOI] [PubMed] [Google Scholar]
- 27.Lindsey Rose KM, Wang Z, Magrath GN, Hazard ES, Hildebrandt JD, Schey KL. Aquaporin 0-Calmodulin Interaction and the Effect of AQP0 Phosphorylation. Biochemistry. doi: 10.1021/bi701980t. in press. [DOI] [PubMed] [Google Scholar]
- 28.Lindsey Rose KM, Gourdie RG, Prescott AR, Quinlan RA, Crouch RK, Schey KL. The C terminus of lens aquaporin 0 interacts with the cytoskeletal proteins filensin and CP49. Invest. Ophthalmol. Vis. Sci. 2006;47:1562–1570. doi: 10.1167/iovs.05-1313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This material is available at http://pubs.acs.org.