Abstract
Considering the potential role of interleukin-8 (IL-8) in inflammation, angiogenesis, tumorigenesis, and metastasis, we investigated the molecular mechanism involved in IL-8-mediated signaling. In this report we provide evidence that like TNF, an inducer of NF-κB and also a NF-κB-dependent gene product, IL-8 induces NF-κB in a unique pathway. IL-8 induces NF-κB activation in a dose-dependent manner in different cell types as detected by a DNA-protein binding assay. IL-8 induces NF-κB-dependent reporter gene expression as well as ICAM-1, VCAM-1, and Cox-2 expression. IL-8 also induces IκBα phosphorylation followed by degradation and p65 translocation. IL-8 induces c-Jun N-terminal kinase (JNK) and mitogen-activated protein kinase (MAPK) in a dose- and time-dependent manner. IL-8-induced NF-κB activation is for the most part unaltered when cells are transfected with dominant-negative TRADD, FADD, or TRAF2, but is inhibited with dominant-negative TRAF6-, NIK-, IKK-, or IκBα-transfected cells. The data suggest that IL-8-induced NF-κB activation proceeds through a TRAF2-independent but TRAF6-dependent pathway, followed by recruitment of IRAK and activation of IKK. IL-8-induced NF-κB activation is not observed in a cell-permeable peptide that has TRAF6 binding motif-treated cells or IRAK-deficient cells. IL-8-induced NF-κB activation proceeds mostly through interaction with TRAF6 and partially through the Rho-GTPase pathways. This is the first report that IL-8 induces NF-κB in a distinct pathway, and activation of NF-κB and its dependent genes may be one of the pathways of IL-8-induced inflammation and angiogenesis.
Chemokines regulate cell migration and leukocyte homing in the inflammatory and allergic sites. Beside its crucial role in inflammatory and allergic responses, chemokines play a vital role in tumor-associated angiogenesis and tumor progression. Interleukin-8 (IL-8),1 a CXC chemokine, is involved in these processes (1–4). In biological solution IL-8 is a dimer, although the monomer is also biologically active (5, 6). Biological functions of IL-8 include plasma exudation, neutrophil accumulation, and general granulocytophilia in vivo (7), chemotaxis for recruitment of leukocytes mainly neutrophils by diapedesis through endothelial spaces (8), degranulation of neutrophils (9), respiratory burst response (10), and mobilization of intra-cellular Ca2+ (11) in vitro. IL-8 specifically chemoattracts several cell types, which is the basis for inflammation. Neovascularization is a crucial step in tumor growth and metastasis (12, 13). Recent reports suggest that in addition to cell proliferation and migration, endothelial cell survival and death are also important components for tumor angiogenesis (14). Tumor angiogenesis is regulated by aberrant production of angiogenic and anti-angiogenic factors expressed both in malignant tumor and host cells (15, 16). The receptors for IL-8 are widely expressed on normal and various tumor cells (17–20). However the mechanisms regulating IL-8-mediated angiogenesis are not well understood.
Among a variety of transcription factors, NF-κB plays a critical role in regulation of genes involved in inflammation and tumorigenesis (21–24). NF-κB is activated by several endogenous inducers and at the same time regulates the expression of some of these inducers. For example, TNF-α is a potent inducer of NF-κB and also is regulated by NF-κB. TNF-α activates NF-κB by recruitment of NIK followed by IKK and thereby phosphorylates IκBα, which leads to the degradation of IκBα and subsequent release of NF-κB (22, 25). Once NF-κB is activated, it drives the expression of several genes having NF-κB responsive elements within their promoters. These include proinflammatory genes like TNF-α, IL-1, IFNγ, Cox-2, and genes involved in tumorigenesis such as adhesion molecules, matrix metalloproteases, and also chemokines like IL-8 (26, 27). Several reports suggest the regulation of IL-8 production, which is one of the key mediators of inflammation by NF-κB (28–30). As discussed earlier, IL-8 is a potent chemokine involved in inflammation and cancer. We were interested to determine whether IL-8 can also regulate NF-κB, similar to TNF, IL-1, and IL-10 (26, 27, 31). TRAF6 is the only TRAF family member that participates in signal transduction of both the TNF receptor superfamily and the IL-1 receptor (IL-1R)/Toll-like receptor (TLR) superfamily through the recruitment of IL-1 receptor-associated kinase (IRAK) (32). IL-1 interacts by sequential recruitment of MyD 88, IRAK, TRAF6, and NIK (33–37) and finally activate IκB kinases (IKKs) and mitogen-activated protein kinase (MAPK) kinase kinase (MEKK) 1, which in turn activates MAPK kinase (MEK), c-Jun N-terminal kinase (JNK), and p38 MAPK (38, 39). Chemoattractants (FMLP, PAF, C3a, and C5a) bind their respective seven transmembrane-spanning G-protein-coupled receptors, and subsequently activate the transcription factor NF-κB (40–42). Formyl methionyl leucyl phenylalanine (FMLP)-mediated activation of NF-κB is caused by stimulation of RhoA, a small GTPase activity (43). However a recent report suggests that Kaposi’s sarcoma-associated virus G protein-coupled receptor is homologous to the IL-8R and activates AP-1 and CREB via the Erk1/2 and PI3K/Akt pathways, whereas NF-κB activation is independent of PI3K/Akt (44).
In this study we clearly demonstrate that IL-8 acts as a potent activator of NF-κB. IL-8 also induces NF-κB-dependent reporter gene expression and NF-κB-dependent genes such as ICAM-1, VCAM-1, and Cox-2. IL-8 also induces JNK and MAPK in a time- and concentration-dependent manner. IL-8-mediated induction of NF-κB was not influenced by DN-TRADD, -FADD, and -TRAF2 but was inhibited by DN-TRAF6, -NIK, and -IKK, indicating involvement of these in IL-8-mediated activation of NF-κB. From the results we obtained for IL-8 using the DN constructs, IL-1 receptor-associated kinase (IRAK) knock-out cells, or TRAF6-binding peptide (TRAF6-BP), which inhibits TRAF6-mediated cell signaling (45) we demonstrate that IL-8 might function through recruitment of IRAK and TRAF6 followed by NIK, IKK, finally leading to the phosphorylation and degradation of IκBα thereby activating NF-κB. IL-8 mediates its action partially through induction of the GTPase pathway, like FMLP and induces NF-κB. With already existing data on the importance of IL-8 in inflammation and angiogenesis, and our finding that IL-8 activates NF-κB, we can say that one of the mechanisms of IL-8-mediated inflammation is through activation of NF-κB. In this report we are providing the data on IL-8-mediated NF-κB activation, unlike other chemoattractants, through the IRAK-TRAF6 pathway, which might be helpful in understanding its involvement for metastasis and new vascularization. Further our results make IL-8 an important therapeutic target in diseases involving the aberrant regulation of either IL-8 or NF-κB.
EXPERIMENTAL PROCEDURES
Materials
Glycine, dithiothreitol, leupeptin, FMLP, phenylmethyl-sulfonyl fluoride, aprotinin, 4-methyl umbelliferyl phosphate, bovine serum albumin, and anti-tubulin antibody were obtained from Sigma. Penicillin, streptomycin, RPMI 1640 medium, Dulbecco’s modified Eagle’s medium, and fetal bovine serum were obtained from Invitrogen Life Technologies (Grand Island, NY). IL-8 was obtained from Pharmingen (San Diego, CA). TNF was obtained from Peprotech (Rocky Hill, NJ). Antibodies against IL-1R1, IL-1R2, IL-8, IL-8R, TLR4, p65, IKKα, IκBα, c-Rel, JNK, ICAM-1, VCAM-1, ERK, CRM1, and Cox-2 and double-stranded Oct1 gel shift oligonucleotide were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against IRAK, phospho-IκBα, and phospho-MAPK were obtained from Cell Signaling Technologies (Beverly, MA). Mounting medium with DAPI and goat-anti-rabbit IgG (Alexa-Fluor-conjugated) were obtained from Molecular Probes (Eugene, OR). Cell-permeable TRAF6-binding peptide with Ka-posi fibroblast growth factor leader sequence (AAVALLPAVLLALLAP-RKIPTEDEYTDRPSQPST; leader sequence in italics), and the mutant TRAF6-binding peptide (AAVALLPAVLLALLAP-RKIATADEATDRPSQPST; leader sequence in italics and mutated amino acids underlined) were chemically synthesized and purified by high performance liquid chromatography. Recombinant C3 toxin was kindly provided by Prof. G. S. Chhatwal (German Center for Biotechnology, Braunschweig, Germany). The plasmid constructs for p65, NF-κB-SEAP, Cox-2-luciferase, and dominant-negative constructs for TRADD, FADD, TRAF2, TRAF6, NIK, IKK, and IκBα were kindly supplied by Prof. B. B. Aggarwal of the University of Texas M. D. Anderson Cancer Center, Houston, TX.
Cell Lines
The cell lines used in this study were as follows: U-937 (histiocytic lymphoma), HeLa (epithelial cells), THP1 (monocytic cells), H4 (glial cells), and RAW264.7 (mouse macrophages). These cells were obtained from American Type Culture Collection (Manassas, VA). The IRAK knock-out HEK293 cells (lacks IRAK protein and mRNA) and IRAK-reconstituted cells (restoration of IRAK in IRAK knock-out cells, which restored their responsiveness to IL-1) (46) were kindly provided by Dr. Xiaoxia Li, Dept. of Immunology, Lerner Research Institute, Cleveland, OH. Peripheral blood mononuclear cells (PBMC) and neutrophils were isolated from fresh blood by the Ficoll-Hipaque method (47). U-937, THP-1, and IRAK-reconstituted and knock-out HEK293 cells were cultured in RPMI 1640 medium whereas others were in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). All cells were free from mycoplasma.
NF-κB Activation Assay
To determine NF-κB activation, EMSA was conducted essentially as described (48). Briefly, 8 μg of nuclear extract proteins were incubated with 32P end-labeled 45-mer double-stranded NF-κB oligonucleotides from HIV-LTR, 5′-TTG TTA CAA GGG ACT TTC CGC TGG GGA CTT TCC AGG GAG GCG TGG-3′ (bold indicates NF-κB binding site) for 30 min at 37 °C, and the DNA-protein complex formed was separated from free oligonucleotide on 6.6% native polyacrylamide gels. The specificity of binding was examined by competition with unlabeled oligonucleotides. Visualization and quantitation of radioactive bands were done as indicated above.
NF-κB-dependent Reporter Gene Transcription Assay
The IL-8-induced NF-κB-dependent reporter gene transcription was measured as described previously (49). Briefly, U-937 cells were transiently transfected with Qiagen SuperFect transfection reagent (Hilden, Germany) with 1 ml of medium containing dominant-negative plasmid DNAs for TRADD, FADD, TRAF2, TRAF6, NIK, IKK, or IκBα (1 μg each) along with 0.5 μg of NF-κB promoter DNA linked to the heat-stable secretory alkaline phosphatase (SEAP) and green fluorescent peptide (GFP) constructs. The total amount of DNA was maintained at 3 μg by the addition of the control plasmid pCMVFLAG1 DNA. After 3 h, cells were washed, cultured for 12 h, and then treated with IL-8 or TNF for 24 h. Cell culture-conditioned medium was harvested, and 25 μl was analyzed for SEAP activity essentially as per the Clontech protocol (Palo Alto, CA). The average number (± S.D.) of relative fluorescent light units for each transfection was then determined and reported as fold activation with respect to control vector-transected cells. This reporter system was specific, because TNF-induced NF-κB SEAP activity was inhibited by overexpression of IκBα mutants lacking either Ser32 or Ser36 (49). For transfection control, cells were co-transfected with β-galactosidase, and the activity was assayed from each transfection. For transfection efficiency, the GFP-positive cells were counted under a fluorescent microscope.
Cox-2-dependent Reporter Gene Transcription
IL-8- and TNF-dependent luciferase reporter gene expression was carried out as described before (50). U-937 cells were transiently co-transfected with SuperFect transfection reagent with 1 μg of Cox-2-luciferase plasmid and 0.5 μg of β-galactosidase. After 3 h, cells were washed and stimulated with different concentrations of either IL-8 or TNF for 24 h. The cell pellets were extracted with lysis buffer (part of the luciferase assay kit from Promega). Cell extracts were incubated with the firefly luciferin (substrate, Promega). Light emission was monitored with a luminometer, and values were calculated as fold activation over vector-transfected value. The activity of β-galactosidase was assayed from the same extracts.
Immunocytochemistry
The effect of IL-8 on nuclear translocation of p65 was examined by the immunocytochemical method as described (50) with slight modifications. Briefly, after treatment cells were fixed, permeabilized, and incubated with rabbit polyclonal anti-human p65 antibody followed by goat anti-rabbit IgG-Alex-Fluor. Cells were mounted with mounting medium with DAPI (to counterstain the nucleus) and analyzed under a fluorescence microscope.
JNK Assay
JNK was assayed by a modified method as described earlier (49). Briefly, cells, stimulated with either different concentrations of IL-8 or different times with IL-8 (100 ng/ml) were extracted, and the cell extract (250 μg/treatment) was used to detect JNK activity using GST-Jun as substrate.
MAPK Kinase Assay
U-937 cells, stimulated with either different concentrations of IL-8 or at different time periods with IL-8 (100 ng/ml) at 37 °C and phosphorylated MAPK, were assayed using phosphospecific anti-p44/42 MAP kinase (Thr202/Tyr204) antibody (New England Biolabs) by Western blot (49).
RESULTS
In this study, we examined the mechanism of IL-8-mediated signal transduction. For most of the studies, U-937 cells were used, since these cells have been well characterized in our laboratory. The concentrations and duration of exposure of different inducers or chemicals employed for this study had no effect on cell viability.
IL-8 Induces NF-κB Activation
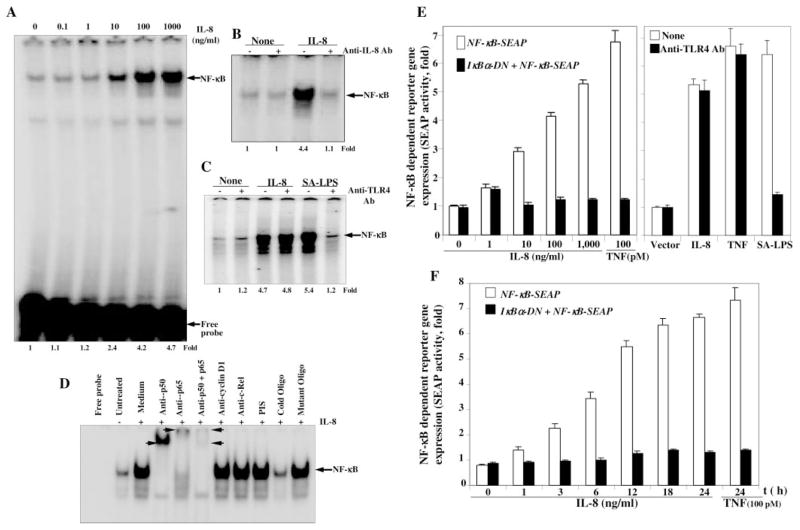
To determine the effect of IL-8 on activation of NF-κB, U-937 cells (2 × 106/ml) were incubated with different concentrations of IL-8 for 2 h and nuclear extracts assayed to detect NF-κB by EMSA. IL-8-induced NF-κB activation was shown in a dose-dependent manner in U-937 cells (Fig. 1A). These results suggest that IL-8 induces NF-κB potently in U-937 cells.
Fig. 1. Effect of IL-8 on NF-κB activation.
U-937 cells (2 × 106/ml) were stimulated with different concentrations of IL-8 for 2 h. Cytoplasmic and nuclear extracts were prepared. 8 μg of nuclear proteins were analyzed in 6.6% native gel and assayed for NF-κB (A). 100 ng of IL-8 was incubated with 1 μg of anti-IL-8 antibody for 1 h at 37 °C. U-937 cells were stimulated with this mixture or IL-8 (100 ng/ml) at 37 °C for 2 h. After these treatments, nuclear extracts were prepared and assayed for NF-κB (B). IL-8 (100 ng) or SA-LPS (100 ng of LPS incubated with 20 μl of serum for 1 h) was incubated with anti-TLR4 antibody (1 μg) for 1 h at 37 °C. U-937 cells were stimulated with IL-8 or SA-LPS and those mixtures for 2 h. Then nuclear extracts, prepared from cells were assayed for NF-κB (C). Nuclear extracts, prepared from untreated or IL-8-induced U-937 cells, were incubated for 15 min with different antibodies and cold or mutated NF-κB oligonucleotides, and then assayed for NF-κB (D). U-937 cells were transiently transfected with NF-κB plasmid linked to the SEAP gene with or without dominant-negative IκBα plasmid for 3 h. Cells were then cultured for 12 h followed by stimulation with different concentrations of IL-8 or 100 pM TNF for 24 h (E, left panel) or different times with 100 ng/ml IL-8 or 100 pM TNF for 24 h (F). In another set, transfected cells were stimulated without or with anti-TLR4 antibody mixture with IL-8, TNF, or SA-LPS for 24 h (E, right panel). Culture supernatants were assayed for SEAP activity as described under “Experimental Procedures.” Results are expressed as fold activity over the nontransfected control.
To determine whether the induced NF-κB band is due to IL-8-mediated cell activation, 100 ng of IL-8 was incubated with 1 μg of anti-IL-8 antibody for 1 h at 37 °C. U-937 cells were then incubated with this IL-8 and antibody mixture for 2 h at 37 °C. The nuclear extracts were prepared and assayed for NF-κB by EMSA. The results show in Fig. 1B that IL-8 induced NF-κB. Anti-IL-8 antibody alone or in combination with IL-8 did not induce NF-κB activation. This result suggests that IL-8-mediated NF-κB activation is specifically caused by IL-8-mediated signaling.
To determine whether NF-κB activation is caused by IL-8, and not by endotoxin, IL-8 was incubated with anti-TLR antibody for 1 h, and then U-937 cells were stimulated with IL-8 and anti-TLR4 antibody mixture. Nuclear extracts were then assayed for NF-κB. The results in Fig. 1C show that anti-TLR antibody-preincubated IL-8 activated NF-κB similar to IL-8 alone, suggesting that IL-8-mediated NF-κB activation is not caused by endotoxin.
To detect the composition and specificity of the induced band visualized by EMSA, nuclear extracts from IL-8-activated U-937 cells were incubated with Abs p50 (NF-κBI), p65 (Rel A), or in combination and 50-fold excess of cold or mutant NF-κB. Abs to either subunit of NF-κB shifted the band to a higher molecular size (Fig. 1D), thus suggesting that the IL-8-activated complex consisted of p50 and p65 subunits. Neither pre-immune serum nor irrelevant Abs such as anti-c-Rel or anti-cyclin D1 had any effect on the mobility of NF-κB. The complex completely disappeared in the presence of cold NF-κB indicating the specificity of NF-κB. Mutant NF-κB was unable to bind with the IL-8-induced protein complex, further suggesting its specificity.
IL-8 Induces NF-κB-dependent Reporter Gene Expression
We have shown that IL-8 induces NF-κB activation in different cell types. To detect the role of IL-8 on NF-κB-dependent reporter gene expression, U-937 cells were transiently transfected with NF-κB-containing plasmid linked to the SEAP gene GFP construct with or without IκBα -DN plasmids for 3 h followed by culture for 12 h. The GFP-positive U-937 cells were counted under a fluorescent microscope and 23 and 22% in SEAP + GFP- and SEAP + GFP + IκBα -DN-transfected pool of cells were found, respectively. Cells were stimulated with different concentrations of IL-8 or TNF (100 pM) for 24 h at 37 °C in a CO2 incubator. The SEAP activity was assayed in culture supernatant. Results are expressed as fold activity over the nontransfected control. The results in Fig. 1E show that IL-8 induced SEAP activity in a dose-dependent manner, and about 5-fold induction of SEAP activity was observed at 1000 ng/ml IL-8 or 100 pM TNF. IκBα -DN-transfected cells showed no induction of SEAP activity at any concentration of IL-8 or 100 pM TNF, suggesting IL-8 mediated induction of active NF-κB-dependent gene transcription. The SEAP-transfected cells were incubated with IL-8, TNF, or serum-activated LPS (SA-LPS) alone or in mixture with anti-TLR4 antibody (1 μg each for 1 h) for 24 h. Then SEAP activity was assayed from culture supernatant. The IL-8, TNF, or SA-LPS increased SEAP activities. The mixture of anti-TLR4 antibody with IL-8 or TNF, but not SA-LPS did not block induced SEAP activities (Fig. 1F1) indicating that IL-8- or TNF-induced NF-κB activation is not caused by endotoxin. When transfected cells were stimulated with 100 ng/ml IL-8 for different times, the time-dependent activation of NF-κB-dependent SEAP activity was observed (Fig. 1F), further suggesting NF-κB-dependent SEAP induction by IL-8. The β-galactosidase activity from cell extracts showed almost similar reduction of absorbance (as per the Promega protocol) at 420 nm, suggesting the transfection control for each treatment.
IL-8 Induces Phosphorylation and Degradation of IκBα
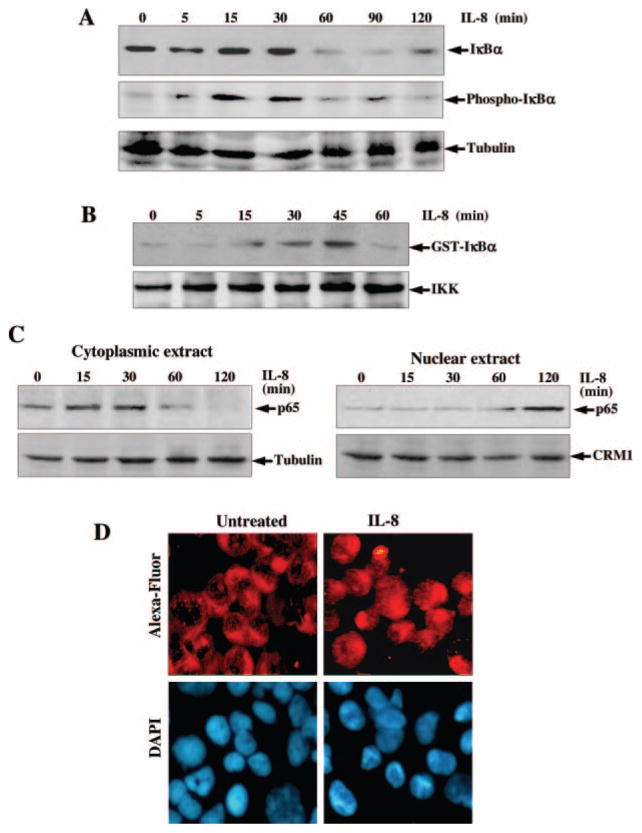
The translocation of NF-κB to the nucleus is preceded by the phosphorylation, ubiquitination, and proteolytic degradation of IκBα (51). To determine whether the IL-8-mediated NF-κB activation was caused by an effect on IκBα phosphorylation and subsequent degradation, the cytoplasmic level of IκBα proteins was examined by Western blot. IκBα degradation started at 60 min of IL-8 (100 ng/ml) treatment in U-937 cells and was completed within 90 min. The band reappeared by 120 min indicating NF-κB-dependent IκBα resynthesis (Fig. 2A). To determine whether IL-8 induces IκBα phosphorylation, Western blot with antibodies against the serine-phosphorylated form of IκBα was performed. As shown in Fig. 2A, middle panel, phosphorylated IκBα band appeared at 15 and 30 min of IL-8 stimulation. At 60 and 90 min of IL-8 incubation, the IκBα bands were not visible, indicating its degradation. Upon reprobing the membrane with anti-tubulin antibody, we found that the band intensities in all lanes were uniform, indicating equal loading of extracted protein.
Fig. 2.
A, effect of IL-8 on phosphorylation and degradation of IκBα. U-937 cells were stimulated with 100 ng/ml IL-8 for different times. Cytoplasmic extracts (50 μg/sample) were assayed for IκBα and phospho-IκBα by Western blot. The same blot was reprobed with anti-tubulin antibody. B, effect of IL-8 on IKK activation. U-937 cells were stimulated with 100 ng/ml IL-8 for different times, and whole cell extracts were prepared, 250 μg of protein were immunoprecipitated with anti-IKKα antibody, and IKK was assayed using GST-IκBα as substrate protein. 50 μg of protein were analyzed in 10% SDS-PAGE to detect IKKα by Western blot. C and D, effect of IL-8 on translocation of p65. Cytoplasmic and nuclear extracts were analyzed in 10% SDS-PAGE and assayed for p65 (C) by Western blot. The gel, which ran with cytoplasmic extracts, was reprobed with anti-tubulin antibody, and the gel with nuclear extracts was reprobed with anti-CRM1 antibody. U-937 cells were stimulated with 100 ng/ml IL-8 for 2 h. Cells were subjected to immunocytochemistry (D) as described under “Experimental Procedures.” Alexa-Fluor took on a red color to detect p65 (upper panel) and DAPI took on a blue color for the nucleus (lower panel).
IL-8 Induces IKK
As IL-8 induces IκBα phosphorylation and subsequent degradation, and these events occur through activation of an upstream kinase, IKK, we investigated whether IL-8 can induce IKK. U-937 cells were stimulated with IL-8 (100 ng/ml) for different times, and the IKK was assayed in vitro using GST-IκBα as substrate protein. As shown in Fig. 2B, the radiolabeled GST-IκBα bands were increased with increasing IL-8 incubation time until 45 min. At the later time, the band decreased dramatically. The results suggest that IL-8 induces IKK activation, which correlates with phosphorylation and degradation of IκBα.
IL-8 Induces Translocation of the p65 Subunit of NF-κB
Whether IL-8 induces nuclear translocation of the p65 subunit of NF-κB was also examined by Western blot analysis. U-937 cells were stimulated with 100 ng/ml IL-8 for different times and then cytoplasmic extracts (CE) and nuclear extracts (NE) were prepared. 60 μg of CE and 40 μg of NE proteins were analyzed in 10% SDS-PAGE, and p65 was detected by Western blot analysis. Upon IL-8 treatment, p65 levels were decreased in the cytoplasm with increasing time, and there was a corresponding increase in p65 levels in the nucleus (Fig. 2C). These results indicate that IL-8 induces the nuclear translocation of p65 NF-κB. Upon reprobing the same blot with anti-tubulin antibody all lanes showed equal loading of proteins. IL-8-induced translocation of p65 from cytoplasm to nucleus was also observed from immunofluorescence data where in unstimulated cells the p65 remained in the cytoplasm but in IL-8-stimulated cells it stayed mostly in the nucleus (Fig. 2D). These data further support IL-8-mediated activation of NF-κB.
IL-8 Induces ICAM-1, VCAM-1, and Cox-2 Expression
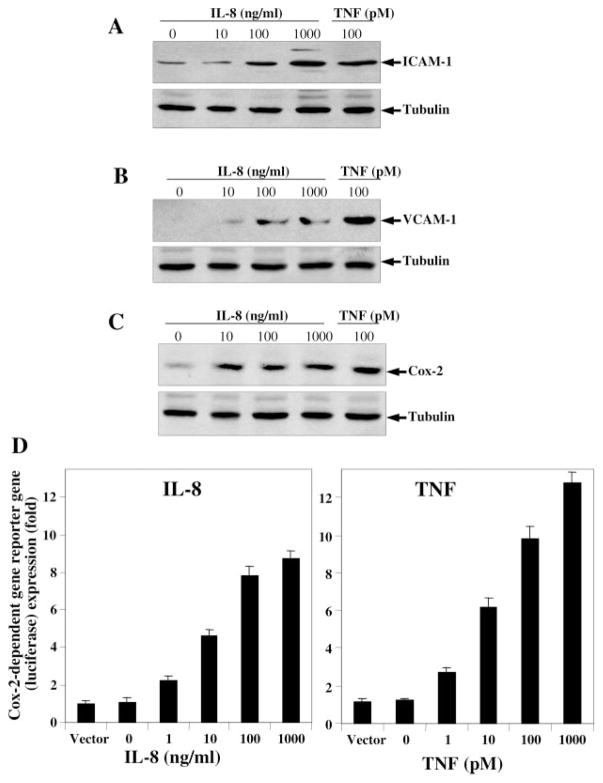
As IL-8 induces NF-κB and dependent reporter gene expression, the expression of NF-κB-regulated genes like adhesion molecules and Cox-2 were detected. U-937 cells were treated with different concentrations of IL-8 and 100 pM TNF for 24 h. Then cells were washed, and extracts were prepared. The 100-μg cell extract proteins were analyzed in 10% SDS-PAGE to detect ICAM-1, VCAM-1, and Cox-2 by Western blot. IL-8-induced ICAM-1 (Fig. 3A), VCAM-1 (Fig. 3B), and Cox-2 (Fig. 3C) expression was increased in a dose-dependent manner for IL-8 and also by TNF (100 pM). The results suggest that IL-8 not only induces NF-κB and NF-κB-dependent reporter gene but also induces NF-κB-dependent active transcription of the adhesion molecules and Cox-2 in cells. Upon reprobing the gels with anti-tubulin antibody, we found that the band intensities in all lanes were uniform, indicating equal loading of extracted protein in all lanes.
Fig. 3. Effect of IL-8 on induction of ICAM-1, VCAM-1, and Cox-2.
U-937 cells were stimulated with different concentrations of IL-8 and 100 pM of TNF for 24 h at 37 °C, CO2 incubator. 100 μg of extract proteins were analyzed in 10% SDS-PAGE, and ICAM-1 (A), VCAM-1 (B), or Cox-2 (C) were detected by Western blot. All these gels were reprobed with anti-tubulin monoclonal antibody. U-937 cells were co-transfected with the Cox-2-luciferase plasmid and β-galactosidase gene. After 3 h of transfection, cells were stimulated with different concentrations of IL-8 or TNF for 24 h. The luciferase (D) and β-galactosidase enzymes activity was measured from cell extracts.
IL-8 Induces Cox2-dependent Reporter Gene Expression
Although we have shown by Western blot that IL-8 induces Cox-2 expression, Cox-2-dependent gene transcription was also assayed. To determine the effect of IL-8-induced Cox-2-dependent reporter gene expression, we transiently transfected U-937 cells with the Cox-2-luciferase and β-galactosidase constructs for 3 h with SuperFect transfection reagent, cultured for 12 h, and then incubated with different concentrations of IL-8 or TNF for 24 h. The results show in Fig. 3D that IL-8- and TNF-induced luciferase activity in a dose-dependent manner. An almost 8.7-fold increase in luciferase activity over the vector control was noted upon stimulation with 1000 ng/ml IL-8 whereas almost 12.8-fold activity was observed with 1 nM TNF. These results demonstrate that both IL-8 and TNF potentially induce Cox-2-dependent reporter gene expression. Compared with IL-8, TNF is a more potent inducer of Cox-2. The β-galactosidase activity from cell extracts showed an almost similar reduction of absorbance (as per Promega protocol) at 420 nm, suggesting the transfection control for each treatment.
IL-8 Induces NF-κB Activation in Different Cell Types
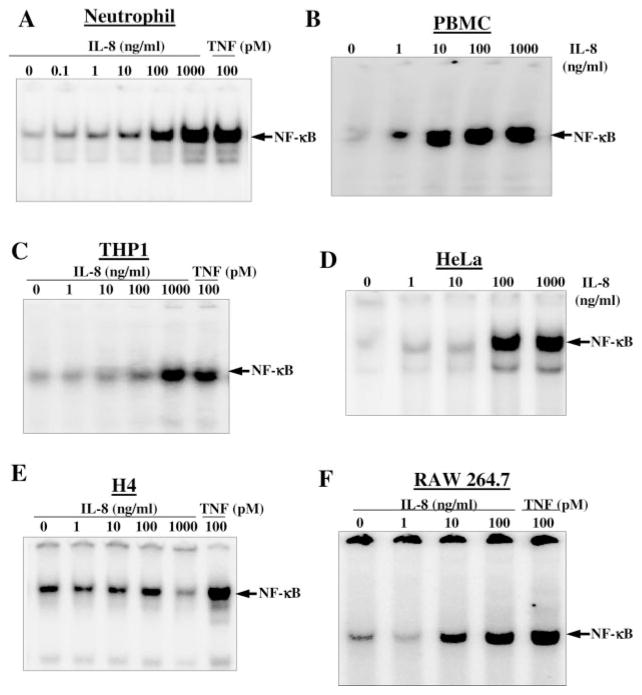
IL-8-mediated induction of NF-κB has been demonstrated in U-937 cells. We were also interested in detecting the effect of IL-8 on induction of NF-κB in other cell types. Neutrophils and PBMC, isolated from fresh blood and THP1 cells (2 × 106/ml), were incubated with different concentrations of IL-8 or 100 pM TNF for 2 h at 37 °C. Similarly, HeLa, H4, and RAW264.7 cells, at 60% confluent state were incubated with different concentrations of IL-8 for 2 h. Cells were washed with D-PBS, nuclear extracts were prepared, and then assayed for NF-κB by EMSA. In Fig. 4, IL-8-induced NF-κB was shown in a dose-dependent manner in neutrophils (Fig. 4A), PBMC (Fig. 4B), THP1 (Fig. 4C), HeLa (Fig. 4D), and RAW264.7 (Fig. 4F) cells. In H4 cells, the IL-8 did not induce NF-κB, whereas TNF potently induced NF-κB activation (Fig. 4E). The results suggest that IL-8 induces NF-κB in most of the cell types.
Fig. 4. Effect of IL-8 on NF-κB activation in different cells.
Neutrophils (A), PMBC (B), THP1 (C), HeLa (D), H4 (E), and RAW264.7 (F) cells, at 60% confluency were stimulated with different concentrations of IL-8 or 100 pM TNF for 2 h. Nuclear extracts were prepared, and 8 μg of nuclear proteins were used to detect NF-κB by EMSA.
IL-8 Induces MEK Activation
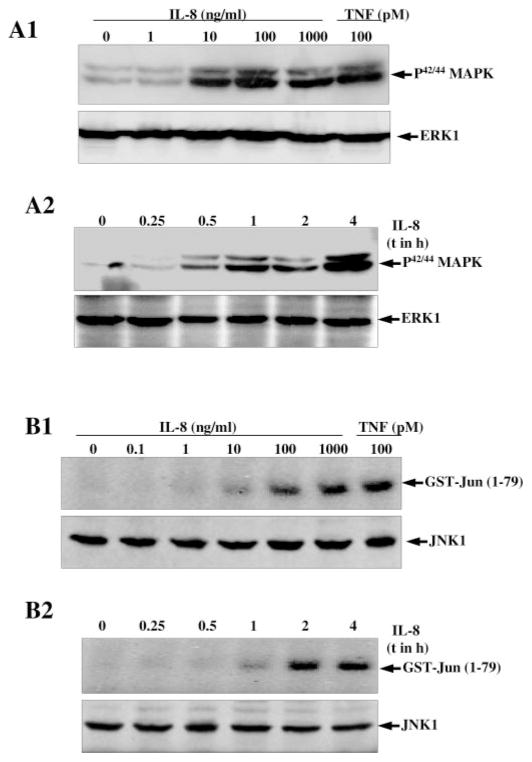
It has been reported that MEK is required for different inducer-mediated NF-κB activation, so the question of whether IL-8 can activate MAPK was investigated. U-937 cells were stimulated with either different concentrations of IL-8 for 2 h or 100 ng/ml IL-8 for different times. Then whole cell extract proteins (100 μg) were analyzed in 10% SDS-PAGE and the phosphorylated form of MAPK (MEK) was detected by Western blot. The results showed that IL-8 induced phosphorylation of MAPK in a dose- (Fig. 5, panel A1) and time- (Fig. 5, panel A2)-dependent manner. The activation of MAPK with 100 ng/ml IL-8 was almost similar to 100 pM TNF. The results suggest the IL-8-mediated phosphorylation of MAPK.
Fig. 5. Effect of IL-8 on MEK and JNK activation.
U-937 cells were stimulated with different concentrations of IL-8 and 100 pM TNF for 2 h (panel A1) or at different times with 100 ng/ml IL-8 as indicated (panel A2) at 37 °C. The cells were used to detect phosphorylated MAPK (MEK). Both blots were reprobed with anti-ERK1 antibody. For JNK, U-937 cells were stimulated with different concentrations of IL-8 and 100 pM TNF (panel B1) or for different times with 100 ng/ml IL-8 (panel B2) at 37 °C for 2 h. Then JNK was assayed from cell extracts as described under “Experimental Procedures.” 50 μg of extract proteins from the same treatments were detected JNK1 by Western blot.
IL-8 Induces JNK Activation
Whether IL-8 can induce JNK was also examined. U-937 cells were pretreated with different concentrations of IL-8 or 100 pM TNF for 2 h or at different times with 100 ng/ml IL-8; activation of JNK was then measured. IL-8 induced JNK in a dose-dependent manner and almost equivalently to 100 pM TNF and 1000 ng/ml IL-8 (Fig. 5, panel B1). IL-8-induced JNK activation was shown to occur in a time-dependent manner, and maximum activation was observed in 2 h with 100 ng/ml of IL-8 (Fig. 5, lane B2). The extracts from the different treatments when probed with anti-JNK1 antibody by Western blot showed equal intensities of JNK1 bands indicating equal levels of expression. The results suggest that IL-8 mediated JNK activation, similar to TNF, in a dose- and time-dependent manner.
IL-8-induced NF-κB Activation Is a TRAF2-independent Pathway, but a TRAF6-, NIK-, IKK-, and IκBα-dependent Pathway
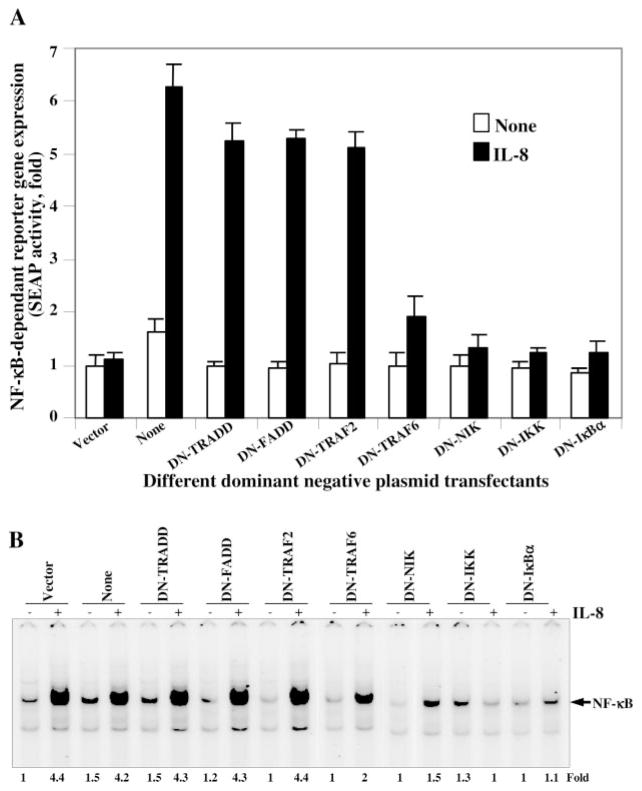
To detect the pathway involved in IL-8-induced NF-κB activation, U-937 cells were co-transfected with different dominant-negative constructs (TRADD, FADD, TRAF2, TRAF6, NIK, IKK, or IκBα), NF-κB-SEAP reporter gene, GFP, and β-galactosidase constructs. After 3 h of transfection, cells were washed and cultured for 12 h followed by 24 h of incubation with IL-8 (100 ng/ml). The GFP-positive cells were 22, 24, 25, 23, 22, 24, or 22% for dominant-negative constructs of TRADD, FADD, TRAF2, TRAF6, NIK, IKK, or IκBα, respectively. The culture supernatant was assayed for SEAP activity. In Fig. 6A, the IL-8-induced SEAP activity was increased 6.28-fold over vector-transfected cells. In cells transfected with dominant-negative TRADD, FADD, or TRAF2, the IL-8-induced SEAP activity was increased 5.24-, 5.28-, or 5.12-fold over vector control. However, cells transfected with dominant-negative TRAF6, NIK, IKK, or IκBα almost completely inhibited IL-8-induced SEAP activity. The data suggest that IL-8-mediated NF-κB activation is independent of TRADD, FADD, or TRAF2 whereas it is dependent on TRAF6, NIK, IKK, or IκBα. The β-galactosidase activity from whole cell extracts showed almost similar reduction of absorbance (as per Promega protocol) at 420 nm, suggesting the transfection control for each treatment.
Fig. 6. Effect of different dominant-negative constructs on IL-8-induced NF-κB activation.
U-937 cells were transiently co-transfected with dominant-negative constructs of TRADD, FADD, TRAF2, TRAF6, NIK, IKK, and IκBα; NF-κB-containing plasmid linked to the SEAP gene; GFP construct; and β-galactosidase plasmid for 3 h. Cells, cultured for 12 h, were stimulated with IL-8 (100 ng/ml) for 24 h. Culture supernatants were assayed for SEAP activity. Results are expressed as fold activity over the nontransfected control (A). The cell pellets were extracted and assayed for β-galactosidase activity. Cells, transfected with following constructs were cultured for 12 h and then incubated with IL-8 (100 ng/ml) for 2 h and NF-κB assayed from nuclear extracts (B).
To further prove IL-8-mediated activation of NF-κB is TRAF2-independent but TRAF6-dependent, 12-h cultured cells (50% of the cells from the above experiment) after transfection with different dominant-negative constructs were stimulated with IL-8 (100 ng/ml) for 2 h, and nuclear extracts were analyzed to detect NF-κB. The data as shown in Fig. 6B indicate that the IL-8-induced NF-κB activation was 4.4-fold, and this induction was almost unaffected in the cells transfected with dominant-negative constructs of TRADD, FADD, or TRAF2. Cells transfected with dominant-negative constructs of TRAF6, NIK, IKK, or IκBα did not induce NF-κB activation mediated by IL-8. From this data it is clear that TRAF6 is involved in IL-8-mediated activation of NF-κB.
IL-8 Recruits IRAK and TRAF6 for Induction of NF-κB
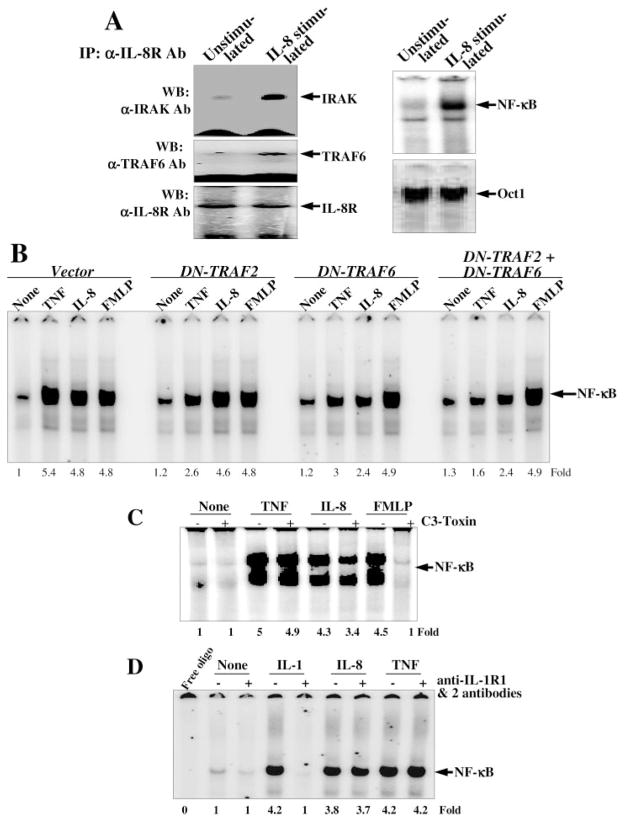
As we have shown that dominant-negative TRAF6 abrogates IL-8-mediated NF-κB activation, and this signaling is mediated through IL-8 receptors, we were interested in identifying the molecules involved in this pathway. U-937 cells were stimulated with 100 ng/ml IL-8 for 2 h, and cell extracts were immunoprecipitated by anti-IL-8R antibody. The IRAK, TRAF6, and IL-8R were detected as immune complexes in Western blots. The levels of IRAK and TRAF6 were increased significantly in IL-8-stimulated cells without changing the level of IL-8R. The IL-8-mediated activation of NF-κB was observed in the nuclear fraction of IL-8-stimulated cells without changing the level of Oct1. The data suggest that IL-8 stimulation leads to recruitment of IRAK and TRAF6, which stimulates NF-κB activation, but not the housekeeping transcription factor, Oct1.
IL-8 Activates NF-κB Mostly through TRAF6 and Partially through Rho-GTPase
As TNF induces cells to activate NF-κB through recruitment of TRAF2 and TRAF6, and formyl peptide (FMLP) induces cells through activation of Rho-GTPase, we compared the cell activation induced by TNF, IL-8, or FMLP in TRAF2 and/or TRAF6 dominant-negative cells. The GFP-positive cell numbers were 23, 25, or 24% in dominant-negative TRAF2-, TRAF6-, or TRAF2- and TRAF6-transfected cells, respectively. TNF induced NF-κB in vector-, DN-TRAF2-, DN-TRAF6-, or DN-TRAF2-, and DN-TRAF6-transfected cells by 5.4-, 2.6-, 3-, or 1.6-fold, respectively. Under similar conditions IL-8 induced 4.8-, 4.6-, 2.4-, or 2.4-fold, respectively and FMLP induced 4.8-, 4.8-, 4.9-, or 4.9-fold respectively (Fig. 7B). The results suggest that TNF-induced NF-κB activation is abrogated either by DN-TRAF6 and/or DN-TRAF2, whereas IL-8-induced NF-κB activation is inhibited by DN-TRAF6, but not by DN-TRAF2-transfected cells. FMLP-induced NF-κB activation is not affected either by DN-TRAF2- and/or DN-TRAF6-transfected cells.
Fig. 7.
A, effect of IL-8 on recruitment of TRAF6 and IRAK. U-937 cells were stimulated with 100 ng/ml IL-8 for 2 h and then 0.5-mg extract proteins were immunoprecipitated with anti-IL-8R antibody. IRAK, TRAF6, and IL-8R were detected by Western blot. The NF-κB and Oct1 were assayed from nuclear extracts by EMSA. B, effect of TNF, IL-8, and FMLP on activation of NF-κB on TRAF2 and/or TRAF6 dominant-negative-transfected cells. U-937 cells were transfected with dominant-negative constructs of TRAF2 and/or TRAF6 as described. Cells were then stimulated with TNF (100 pM), IL-8 (100 ng/ml), or FMLP (100 nM) for 2 h. NF-κB was assayed from nuclear extracts. C, effect of C3 toxin on TNF-, IL-8-, or FMLP-induced NF-κB activation. U-937 cells, treated with C3 toxin (10 μg/ml) for 12 h were stimulated with TNF (100 pM), IL-8 (100 ng/ml), or FMLP (100 nM) for 2 h. NF-κB was assayed from nuclear extracts. D, effect of anti-IL-1R1 and -IL-1R2 antibodies on IL-8-induced NF-κB activation. U-937 cells were preincubated with anti-IL-1R1 and -IL-1R2 antibodies (1 μg each) for 2 h and then stimulated with IL-1 (1 nM), IL-8 (100 ng/ml), or TNF (0.1 nM) for 2 h. Nuclear extracts were prepared and assayed for NF-κB.
As IL-8-induced NF-κB activation was not completely inhibited in dominant-negative TRAF6 or TRAF2 and TRAF6-transfected cells a possible role for Rho-GTPase was determined. U-937 cells were pretreated with 10 μg/ml C3 toxin (Clostridium botulinum C3 transferase; an inhibitor of Rho-GTPase) for 12 h, then stimulated with TNF, IL-8, or FMLP for 2 h, and NF-κB was assayed from nuclear extracts. As shown in Fig. 7C, TNF-induced NF-κB activation was unaffected, IL-8-induced NF-κB activation was partially inhibited, and FMLP-induced NF-κB activation was completely inhibited in C3 toxin-pre-treated cells. The results suggest that IL-8-induced NF-κB activation is partially mediated by Rho-GTPase.
IL-8 Does Not Share IL-1 Receptors to Induce NF-κB
IL-1 interacts with its receptors and recruits IRAK for cell signaling. As IL-8 recruited IRAK for NF-κB activation, we were interested in determining the role of IL-8 in sharing IL-1 receptors. U-937 cells were incubated with anti-IL-1R1 and -IL-1R2 antibodies and then induced with IL-1, IL-8, or TNF for 2 h. The nuclear extracts were then prepared and assayed for NF-κB. The IL-1, IL-8, or TNF induced NF-κB by 4.2-, 3.8-, or 4.2-fold, respectively. Anti-IL-1R1 and -IL-1R2 antibody-preincubated cells showed complete inhibition of IL-1-, but not IL-8-or TNF-induced NF-κB activation (Fig. 7D). The result suggests that IL-8-induced NF-κB activation is preceded through interaction of its own receptors, but not IL-1 receptors.
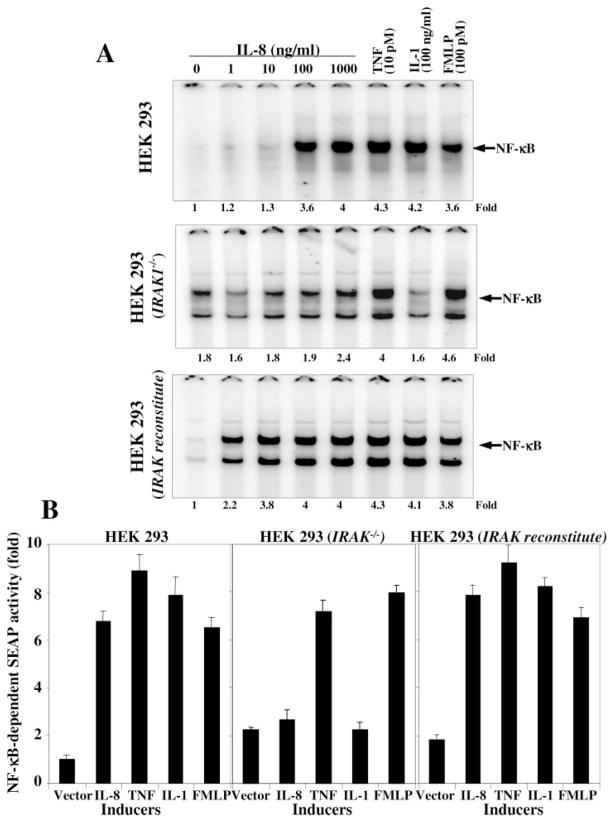
IL-8-induced NF-κB Activation Is Not Observed in IRAK Knock-out Cells
To detect the role of IRAK on IL-8-mediated NF-κB activation further IRAK knock-out or reconstituted HEK293 cells were stimulated with different concentrations of IL-8, 10 pM TNF, 100 ng/ml IL-1, or 100 pM FMLP for 2 h, and nuclear extracts were assayed for NF-κB. As shown in Fig. 8A, the IL-8 induced NF-κB in a dose-dependent manner on HEK293 IRAK-reconstituted cells, but not in knock-out cells. TNF-induced NF-κB activation was partially inhibited in IRAK knock-out cells. IL-1- but not FMLP-induced NF-κB activation was completely blocked in IRAK knock-out cells. The results suggest the involvement of IRAK in IL-8-induced NF-κB activation. Both IRAK-reconstituted and knock-out cells were transiently transfected with NF-κB-SEAP reporter gene and stimulated with different concentrations of IL-8, 10 pM TNF, 100 ng/ml IL-1, or 100 pM FMLP for 24 h. The SEAP activity was assayed from culture supernatant. As shown in Fig. 8B, the SEAP activity was increased with IL-8 in a dose-dependent manner in IRAK-reconstituted cells, but not in knock-out cells. The SEAP activity followed the same pattern as shown for NF-κB activation.
Fig. 8. Effect of IL-8, TNF, IL-1, or FMLP on NF-κB activation in HEK293, HEK293 (IRAK−/−), and HEK293 (IRAK-reconstituted) cells.
Cells were stimulated with different concentrations of IL-8, 10 pM TNF, 100 ng/ml IL-1, or 100 pM FMLP for 2 h. The nuclear extracts were prepared and assayed for NF-κB (A). Cells were transfected with NF-κB-SEAP construct and then culture for 12 h. Transfected cells were stimulated with IL-8 (100 ng/ml), TNF (10 pM), IL-1 (100 ng/ml), or FMLP (100 pM) for 24 h, and culture supernatant was assayed for SEAP (B) and indicated as fold of activation against vector control.
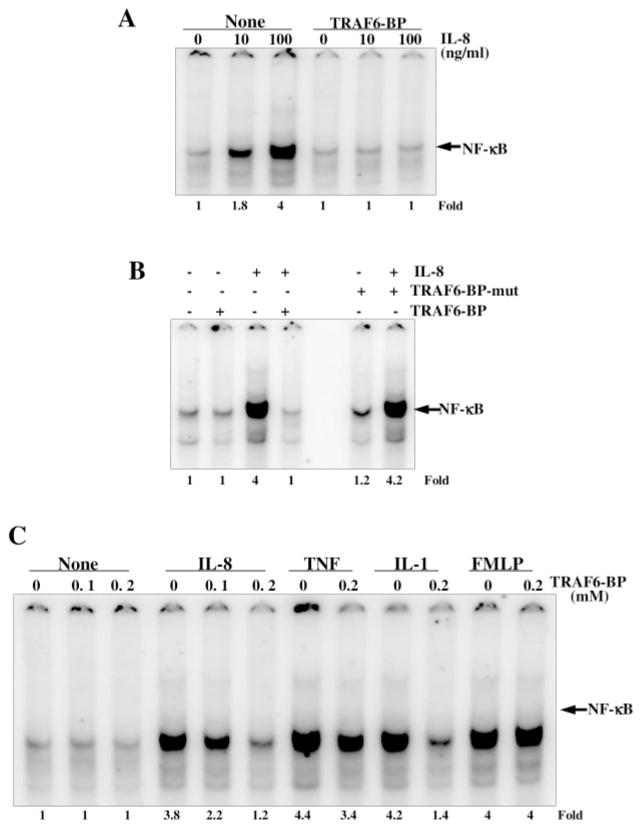
TRAF6-binding Peptide (TRAF6-BP) Blocks IL-8-induced NF-κB Activation
To detect the role of TRAF6 on IL-8-mediated NF-κB activation, mouse macrophages (RAW264.7) were incubated with 200 μM TRAF6-BP and then stimulated with 10 and 100 ng/ml IL-8 for 2 h. Nuclear extracts were then assayed for NF-κB. As shown in Fig. 9A, the IL-8 induced NF-κB in RAW264.7 cells in a dose-dependent manner and TRAF6-BP completely inhibited IL-8-induced NF-κB activation. Cells were pretreated with 200 μM of each synthetic TRAF6-BP and TRAF6-BP mutant (TRAF6-BP-mut) for 2 h and then stimulated with 100 ng/ml IL-8 for 2 h. IL-8 induced NF-κB in RAW264.7 cells and TRAF6-BP, but not TRAF6-BP-mut inhibited IL-8-induced NF-κB activation (Fig. 9B). The TRAF6-BP inhibitory peptide-pretreated cells showed partial inhibition of TNF-induced, almost complete inhibition of IL-1-induced, but no inhibition of FMLP-induced NF-κB activation (Fig. 9C). These results suggest the involvement of TRAF6 on IL-1- or IL-8-mediated NF-κB activation.
Fig. 9. Effect of TRAF6-binding peptide (TRAF6-BP) on IL-8-, TNF-, IL-1-, or FMLP-induced NF-κB activation.
RAW264.7 cells were preincubated with 200 μM TRAF6-BP for 2 h and then stimulated with 10 and 100 ng/ml IL-8 for 2 h. Nuclear extracts were assayed for NF-κB (A). Cells were incubated with 200 μM of TRAF6-BP or TRAF6-BP mutant (TRAF6-BP-mut) peptides for 2 h and then stimulated with IL-8 (100 ng/ml) for 2 h. Nuclear extracts were assayed for NF-κB (B). Cells were incubated with indicated concentrations of TRAF6-BP for 2 h and then stimulated with IL-8 (100 ng/ml), TNF (10 pM), IL-1 (100 ng/ml), or FMLP (100 pM) for 2 h. Nuclear extracts were prepared and assayed for NF-κB (C).
DISCUSSION
IL-8 is a neutrophil and lymphocyte chemoattractant and activator that may play an important role in mediating events at sites of inflammation (52). The molecular mechanism of inflammation induced by TNFα and IL-1 has been extensively studied. Several reports suggest the importance of IL-8 in inflammation, but its mechanism of action is not well understood. Several inducers of inflammation (like IL-1 and TNFα) activate NF-κB, which then activates several genes, including its own inducers involved in inflammation and tumorigenesis. IL-8 is one of the genes activated by NF-κB (28–30). As NF-κB is involved in cell proliferation and angiogenesis, we were interested in deciphering IL-8-mediated signaling pathways in tumor cells. IL-8 induces NF-κB activation as detected by EMSA (Fig. 1A), NF-κB-dependent reporter gene activation (SEAP) (Fig. 1, D and E), and the downstream ICAM-1, VCAM-1 and Cox-2 as well as Cox-2-dependent luciferase gene expression (Fig. 3). This activation is specific for IL-8 as anti-IL-8 antibody blocks IL-8-induced NF-κB activation (Fig. 1B). IL-8-induced NF-κB activation is not caused by endotoxin, because anti-TLR4 antibody did not block this activation (Fig. 1C). IL-8 caused activation of NF-κB by activation of IKK (Fig. 2B) followed by phosphorylation and degradation of IκBα (Fig. 2A). Activation of NF-κB is shown by disappearance of p65 from the cytosol and the subsequent translocation in the nucleus (Fig. 2, C and D). Most cell types showed a dose-dependent activation of NF-κB in response to IL-8 (Fig. 4). Unlike TNF, IL-8 did not induce NF-κB in glioma (H4) cells. We detected 125I-IL-8 binding for different cells (45) and found more low affinity IL-8 receptors in H4 cells (high/low affinity IL-8 receptors for HeLa, U-937, THP-1, RAW264.7, H4, PBMC, or neutrophils were 5848/4650, 5452/4218, 5104/4028, 7126/4572, 1848/8124, 6184/3892, or 18418/6841 per cell, respectively).
Apart from NF-κB, IL-8 activated JNK and MAPK in a concentration- and time-dependent manner (Fig. 5). TNF activates NF-κB through sequential interaction of the TNF receptor with TRADD and TRAF2 (53, 54, 55). Further TRAF2 interacts with NIK and IKK and thereby activates NF-κB (53, 54). IL-1 activates NF-κB through recruitment of IRAK and TRAF6, which leads to activation of NF-κB and different MAP kinases including JNK and p38 MAPK (32). The chemokines interact with G-protein coupled receptors and activate NF-κB through activation of GTPase (40–43, 56). Upon expression of DN-TRAF6, -NIK, -IKK, or - IκBα, the IL-8 mediated NF-κB activation was severely inhibited (Fig. 6). Recruitment of TRAF6 through IRAK leads to activation of different MAP kinases including p38 MAPK and JNK (32–39). Our data suggest that IL-8-mediated cell activation proceeds through recruitment of TRAF6 and IRAK, which possibly activate MAPK and JNK. Several lines of evidence suggest that MAPKs can participate in the regulation of NF-κB transcriptional activity (57, 58). The involvement of TRAF6 in inflammation is well established. Inflammatory agents such as IL-1 and bacterial LPS recruit IRAK4, which phosphorylates IRAK1. IRAK1 then activates TRAF6 and transduces the downstream signal to activate NIK followed by IKK, finally activating NF-κB and thereby leading to the genesis of inflammation (33–37). It is clear that TRAF6 is required for IL-8-induced activation of NF-κB as TRAF6-binding peptide, which inhibits TRAF6-mediated response, inhibited IL-8-induced NF-κB activation (Fig. 9). Though TNF-induced NF-κB activation is TRAF2-dependent and TRAF6-independent, partial inhibition of NF-κB in TRAF6 dominant-negative-transfected cells might reflect the transfection efficiency. In IRAK1 knock-out cells IL-8-mediated NF-κB is almost not observed (Fig. 8). We have shown that IL-8-mediated activation of NF-κB occurs through sequential events: recruitment of IRAK and TRAF6, activation of NIK and IKK, phosphorylation and subsequent degradation of IκBα, and translocation of p65 to nucleus. The nuclear translocation of p65 leads to binding to NF-κB responsive promoter and activates its dependent genes, which are involved in inflammation. So far this is the first report where IL-8 has been shown to activate NF-κB, and this activation mostly occurs through a TRAF6-dependent pathway.
FMLP interacts with its G-protein-coupled receptors and induces NF-κB through activation of Rho-GTPase, because C3 toxin completely abrogates FMLP-induced NF-κB activation (Fig. 7C). Though IL-8 receptors are also G-protein-coupled receptors, the signaling pathway followed by IL-8 is still similar to TNF or IL-1. Partial activation of NF-κB was observed at higher concentrations of IL-8 in IRAK1 knock-out cells suggesting involvement of Rho-GTPase. IL-1 interacts with its receptors and activates NF-κB through activation of IRAK. Our data suggest that IL-8-mediated activation of NF-κB is also preceded through activation of IRAK without sharing IL-1Rs (Fig. 7D). However a recent report showed that the Kaposi’s sarcoma-associated virus G protein-coupled receptor, homologous to the IL-8R activates AP-1 and CREB, but not NF-κB via Erk1/2 and PI3K/Akt pathways (44). Another report suggests that the CXC chemokine (LIX) activates NF-κB and dependent proinflammatory cytokines via PI3K, NIK, IKK, and IκBα (59). These reports further strengthen our observations.
IL-8 is involved in cancer progression through the stimulation of angiogenesis. How IL-8 enhances metastasis is still not properly understood. In this report we have provided data on IL-8-mediated induction of NF-κB and NF-κB-dependent genes, which may play a role in new blood vessel formation during tumor progression. To conclude we can say that NF-κB activation is one of the important pathways used by IL-8 during inflammation and angiogenesis. This also makes IL-8 an important drug target in diseases like rheumatoid arthritis, osteoporosis, cardiovascular disorders, and neoplasms, which involve aberrant regulation of IL-8 or NF-κB.
Acknowledgments
We thank Dr. David McMillan for carefully reading the manuscript.
Footnotes
This work was supported by the core grant of Centre for DNA Fingerprinting and Diagnostics (CDFD) and Department of Biotechnology (DBT), Government of India, and National Institutes of Health/RCMI Grant RR03045-16.
The abbreviations used are: IL-8, interleukin-8; C3 toxin, C. botulinum C3 transferase; CE, cytoplasmic extract; Cox, cyclooxygenase; DN, dominant-negative; EMSA, electrophoretic mobility shift assay; FMLP, formyl methionyl leucyl phenylalanine; ICAM, intracellular adhesion molecule; IκBα, inhibitory subunit of κB; IKK, IκBα kinase; IRAK, interleukin-1 receptor-associated kinase; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; NE, nuclear extract; NF-κB, nuclear transcription factor κB; PBMC, peripheral blood mono-nuclear cells; SEAP, secretory alkaline phosphatase; TRADD, TNF receptor-associated death domain; TRAF, TNF receptor-associated factor; TRAF6-BP, TRAF6-binding peptide; VCAM, vascular cell adhesion molecule; GFP, green fluorescent protein; GST, glutathione S-transferase; Ab, antibody; MEK, MAP kinase kinase; PI3K, phosphatidylinositol 3-kinase; ERK, extracellular signal-regulated kinase; DAPI, 4,6-diamidino-2-phenylindole.
References
- 1.Koch AE, Polverini PJ, Kunkel SL, Harlow LA, DiPietro LA, Elner VM, Elner SG, Strieter RM. Science. 1992;258:1798–1801. doi: 10.1126/science.1281554. [DOI] [PubMed] [Google Scholar]
- 2.Strieter RM, Polverini PJ, Arenberg DA, Walz A, Opdenakker G, Van Damme J, Kunkel SL. J Leukocyte Biol. 1995;57:752–762. doi: 10.1002/jlb.57.5.752. [DOI] [PubMed] [Google Scholar]
- 3.Murdoch C, Monk PN, Finn A. Cytokine. 1999;11:704–712. doi: 10.1006/cyto.1998.0465. [DOI] [PubMed] [Google Scholar]
- 4.Salcedo R, Ponce ML, Young HA, Wasserman K, Ward JM, Kleinman HK, Oppenheim JJ, Murphy WJ. Blood. 2000;96:34–40. [PubMed] [Google Scholar]
- 5.Schnitzel W, Monschein U, Besemer J. J Leukoc Biol. 1994;55:763–770. doi: 10.1002/jlb.55.6.763. [DOI] [PubMed] [Google Scholar]
- 6.Burrows SD, Doyle ML, Murphy KP, Franklin SG, White JR, Brooks I, McNulty DE, Scott MO, Knutson JR, Porter D, Young PR, Hensley P. Biochemistry. 1994;33:12741–12745. doi: 10.1021/bi00209a002. [DOI] [PubMed] [Google Scholar]
- 7.Rampart M, Van Damme J, Zonnekeyn L, Herman AG. Am J Pathol. 1989;135:21–25. [PMC free article] [PubMed] [Google Scholar]
- 8.Huber AR, Kunkel SL, Todd RF, 3rd, Weiss SJ. Science. 1991;254:99–102. doi: 10.1126/science.1718038. [DOI] [PubMed] [Google Scholar]
- 9.Smith WB, Gamble JR, Clark-Lewis I, Vadas MA. Immunol. 1993;78:491–497. [PMC free article] [PubMed] [Google Scholar]
- 10.Derevianko A, D’Amico R, Graeber T, Keeping H, Simms HH. J Leukoc Biol. 1997;62:268–276. doi: 10.1002/jlb.62.2.268. [DOI] [PubMed] [Google Scholar]
- 11.Dewald B, Thelen M, Baggiolini M. J Biol Chem. 1988;263:16179–161884. [PubMed] [Google Scholar]
- 12.Fidler IJ, Singh RK, Yoneda J, Kumar R, Xu L, Dong Z, Bielenberg DR, McCarty M, Ellis LM. Cancer J. 2000;6:S225–S226. [PubMed] [Google Scholar]
- 13.Folkman J, Camphausen K. Science. 2001;293:227–228. doi: 10.1126/science.1062892. [DOI] [PubMed] [Google Scholar]
- 14.Nor JE, Christensen J, Mooney DJ, Polverini PJ. Am J Pathol. 1999;154:375–384. doi: 10.1016/S0002-9440(10)65284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fidler IJ, Ellis LM. Cell. 1994;79:185–188. doi: 10.1016/0092-8674(94)90187-2. [DOI] [PubMed] [Google Scholar]
- 16.Folkman J. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 17.Yang SK, Eckmann L, Panja A, Kagnoff MF. Gastroenterology. 1997;113:1214–1223. doi: 10.1053/gast.1997.v113.pm9322516. [DOI] [PubMed] [Google Scholar]
- 18.Smith DR, Polverini PJ, Kunkel SL, Orringer MB, Whyte RI, Burdick MD, Wilke CA, Strieter RM. J Exp Med. 1994;179:1409–1415. doi: 10.1084/jem.179.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh RK, Varney ML, Ino K, Vose JM, Bierman PJ, Talmadge JE. Exp Hematol. 2000;28:499–507. doi: 10.1016/s0301-472x(00)00145-4. [DOI] [PubMed] [Google Scholar]
- 20.Inoue K, Slaton JW, Eve BY, Kim SJ, Perrotte P, Balbay MD, Yano S, Bar-Eli M, Radinsky R, Pettaway CA, Dinney CP. Clin Cancer Res. 2000;6:2104–2119. [PubMed] [Google Scholar]
- 21.Sen R, Baltimore D. Cell. 19986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 22.Baeuerle PA, Baichwal VR. Adv Immunol. 1997;65:111–137. [PubMed] [Google Scholar]
- 23.Baichwal VR, Baeuerle PA. Curr Biol. 1997;7:R94–R96. doi: 10.1016/s0960-9822(06)00046-7. [DOI] [PubMed] [Google Scholar]
- 24.Surh YJ, Chun KS, Cha HH, Han SS, Keum YS, Park KK, Lee SS. Mutat Res. 2001:480–481. 243–268. doi: 10.1016/s0027-5107(01)00183-x. [DOI] [PubMed] [Google Scholar]
- 25.Aggarwal BB, Natarajan K. Eur Cytokine Netw. 1996;7:93–124. [PubMed] [Google Scholar]
- 26.Karashima T, Sweeney P, Kamat A, Huang S, Kim SJ, Bar-Eli M, McConkey DJ, Dinney CP. Clin Cancer Res. 2003;9:2786–2797. [PubMed] [Google Scholar]
- 27.Christman JW, Sadikot RT, Blackwell TS. Chest. 2000;117:1482–1487. doi: 10.1378/chest.117.5.1482. [DOI] [PubMed] [Google Scholar]
- 28.Message SD, Johnston SL. J Leukoc Biol. 2004;75:5–17. doi: 10.1189/jlb.0703315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brasier AR, Jamaluddin M, Casola A, Duan W, Shen Q, Garofalo RP. J Biol Chem. 1998;273:3551–3661. doi: 10.1074/jbc.273.6.3551. [DOI] [PubMed] [Google Scholar]
- 30.Yang D, Chertov O, Oppenheim JJ. J Leukoc Biol. 2001;69:691–697. [PubMed] [Google Scholar]
- 31.Driessler F, Venstrom K, Sabat R, Asadullah K, Schottelius AJ. Clin Exp Immunol. 2004;135:64–73. doi: 10.1111/j.1365-2249.2004.02342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye H, Arron JR, Lamothek B, Cirilli M, Kobayashi T, Shevdeq NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M, Duk K, Singh S, Pikeq JW, Darnay BG, Choi Y, Wu H. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 33.Huang Q, Yang J, Lin Y, Walker C, Cheng J, Liu ZG, Su B. Nat Immunol. 2003;5:98–103. doi: 10.1038/ni1014. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Z, Johnson HJ, Nie H, Qin J, Bird TA, Li X. J Biol Chem. 2003;278:10952–10956. doi: 10.1074/jbc.M212112200. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida Y, Kumar A, Koyama Y, Peng H, Arman A, Boch JA, Auron PE. J Biol Chem. 2004;279:1768–1776. doi: 10.1074/jbc.M311498200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Jimi E, Bothwell AL. J Immunol. 2003;171:3620–3626. doi: 10.4049/jimmunol.171.7.3620. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Tupper JC, Bannerman DD, Winn RK, Rhodes CJ, Harlan JM. Infect Immun. 2003;71:4414–4420. doi: 10.1128/IAI.71.8.4414-4420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Neill LA, Greene C. J Leukocyte Biol. 1998;63:650–657. [PubMed] [Google Scholar]
- 39.Baud V, Liu ZG, Bennett B, Suzuki N, Xia Y, Karin M. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Browning DD, Pan ZK, Prossnitz ER, Ye RD. J Biol Chem. 1997;272:7995–8001. doi: 10.1074/jbc.272.12.7995. [DOI] [PubMed] [Google Scholar]
- 41.Pan ZK, Kravchenko VV, Ye RD. J Biol Chem. 1995;270:7787–7790. doi: 10.1074/jbc.270.14.7787. [DOI] [PubMed] [Google Scholar]
- 42.Pan ZK. Biochim Biophys Acta. 1998;1443:90–98. doi: 10.1016/s0167-4781(98)00198-5. [DOI] [PubMed] [Google Scholar]
- 43.Chen LY, Zuraw BL, Ye RD, Pan ZK, Rho A. J Biol Chem. 2004;279:7208–7212. doi: 10.1074/jbc.M309542200. [DOI] [PubMed] [Google Scholar]
- 44.Cannon ML, Cesarman E. Oncogene. 2004;23:514–523. doi: 10.1038/sj.onc.1207021. [DOI] [PubMed] [Google Scholar]
- 45.Takada Y, Aggarwal BB. Blood. 2004;104:4113–4121. doi: 10.1182/blood-2004-04-1607. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Commane M, Burns C, Vithalani K, Cao Z, Stark GR. Mol Cell Biol. 1999;19:464–4652. doi: 10.1128/mcb.19.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manna SK, Samanta S, Samanta AK. J Immunol. 1997;159:5042–5052. [PubMed] [Google Scholar]
- 48.Sreenivasan Y, Sarkar A, Manna SK. Oncogene. 2003;22:4356–4369. doi: 10.1038/sj.onc.1206486. [DOI] [PubMed] [Google Scholar]
- 49.Manna SK, Zhang HJ, Yan T, Oberley LW, Aggarwal BB. J Biol Chem. 1998;273:13245–13254. doi: 10.1074/jbc.273.21.13245. [DOI] [PubMed] [Google Scholar]
- 50.Sarkar A, Sreenivasan Y, Ramesh GT, Manna SK. J Biol Chem. 2004;279:33768–33781. doi: 10.1074/jbc.M403424200. [DOI] [PubMed] [Google Scholar]
- 51.Coliins AR. Bioessays. 1999;21:238–244. [Google Scholar]
- 52.Mukaida M. Int J Hematol. 2000;72:391–398. [PubMed] [Google Scholar]
- 53.Karin M, Delhase M. Proc Natl Acad Sci U S A. 1998;95:9067–9069. doi: 10.1073/pnas.95.16.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu H, Shu HB, Pan MG, Goeddel DV. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 55.Yoshizumi M, Fujita Y, Izawa Y, Suzaki Y, Kyaw M, Ali N, Tsuchiya K, Kagami S, Yano S, Sone S, Tamaki T. Exp Cell Res. 2004;292:1–10. doi: 10.1016/j.yexcr.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Huang S, Chen LU, Zuraw BL, Ye RD, Pan ZK. J Biol Chem. 2001;276:40977–40981. doi: 10.1074/jbc.M105242200. [DOI] [PubMed] [Google Scholar]
- 57.Schulze-Osthoff K, Ferrari D, Riehemann K, Wesselborg S. Immunobiol. 1997;198:35–49. doi: 10.1016/s0171-2985(97)80025-3. [DOI] [PubMed] [Google Scholar]
- 58.Stambe C, Atkins RC, Hill PA, Paterson DJN. Kidney Int. 2003;64:2121–2132. doi: 10.1046/j.1523-1755.2003.00324.x. [DOI] [PubMed] [Google Scholar]
- 59.Chandrasekar B, Melby PC, Sarau HM, Raveendran M, Perla RP, Marelli-Berg FM, Dulin NO, Singh IS. J Biol Chem. 2003;278:4675–4686. doi: 10.1074/jbc.M207006200. [DOI] [PubMed] [Google Scholar]