Abstract
Recent studies indicate that FoxO transcription factors play an important role in promoting muscle atrophy. To study mechanisms mediating effects of FoxO proteins on muscle wasting, FoxO1-estrogen receptor fusion proteins that are activated by treatment with 4-hydroxytamoxifen (4-OH-T) were stably transfected in C2C12 skeletal myoblasts using the pBABE retroviral system and grown into multinucleated skeletal myotubes. Activation of FoxO1 resulted in significant muscle atrophy, which was accompanied by DNA fragmentation, evidenced by terminal deoxynucleotidyl transferase dUTP-mediated nick end labeling. Cells expressing a DNA-binding-deficient form of FoxO1 also exhibited significant atrophy on FoxO1 activation but no hallmark signs of apoptosis. FoxO1 activation resulted in a significant increase in muscle atrophy F-box (MAFbx)/atrogin-1, muscle-specific RING finger protein 1 (MuRF-1), and Bcl-2-interacting mediator of cell death (Bim) gene expression, with no significant increase in Bcl-2/adenovirus E1B 19-kDa-interacting protein 3 (BNip3) gene expression. Although the ability of FoxO1 to induce MuRF-1 gene expression appeared to be independent of DNA binding, expression of MAFbx/atrogin-1 and Bim was significantly blunted in cells expressing DNA-binding-deficient FoxO1. BNip3 gene expression was significantly elevated in DNA-binding-deficient mutant cells. These findings indicate that FoxO1 promotes skeletal muscle atrophy through induction of proteolytic and apoptotic machinery via DNA-binding-dependent and -independent mechanisms.
Keywords: atrophy, MAFbx/atrogin-1, Bim, BNip3, terminal deoxynucleotidyl transferase dUTP-mediated nick end labeling
skeletal muscle atrophy attributed to prolonged bed rest, inactivity, chronic disease, and/or aging is a culmination of cellular events involving increased protein degradation, mitochondrial dysfunction, and loss of myonuclei (21). A variety of molecular mechanisms appear to operate in a coordinated fashion to regulate skeletal muscle atrophy, and recent data suggest that the winged-helix transcription factor FoxO1 plays a crucial role in regulating this process (22, 32). Expression of FoxO1 in rodent and cell culture models has been shown to decrease skeletal muscle size, in part through its ability to upregulate genes associated with proteasome-mediated protein breakdown, including muscle atrophy F-box (MAFbx)/atrogin-1 and muscle-specific RING finger protein 1 (MuRF-1) (22, 32).
In addition to proteasome-mediated protein degradation, the remodeling of atrophying skeletal muscle appears to be associated with upregulation of programmed cell death (apoptosis) machinery, involving disruption of mitochondrial integrity and nuclear damage (2, 10, 11). For instance, induction of mitochondria-associated proapoptotic genes and DNA fragmentation occurs in muscle atrophy resulting from mechanical unloading and denervation (1, 2, 30). Although the role of FoxO1 in apoptosis of atrophying skeletal muscle has yet to be discerned, FoxO1 and related protein family members have been linked to promotion of apoptosis in various cells through upregulation of cell death receptors and various proapoptotic signaling genes (14). The Bcl-2 family of proteins encompasses an array of proteins capable of promoting or repressing apoptosis on exposure to a variety of stimuli (for review see Ref. 13). Bcl-2-interacting mediator of cell death (Bim) and the Bcl-2/adenovirus E1B 19-kDa-interacting protein 3 (BNip3), known gene targets of FoxO proteins (29, 33), are powerful inducers of apoptosis through their ability to trigger mitochondrial disruption (6, 35). Although the ability of FoxO1 to modulate gene expression can occur through direct binding to specific response elements with the promoters of target genes, including Bim (29), FoxO1 can also affect gene expression through expression of other FoxO1-responsive genes that can be altered through mechanisms that do not require direct DNA binding (20, 28, 37). Thus it is possible that FoxO1 may promote muscle atrophy and proteolytic and/or apoptotic gene expression through mechanisms other than direct promoter binding.
Therefore, the purpose of this study was to examine the link between FoxO1 expression, protein degradation, and apoptosis during skeletal muscle atrophy via the use of an inducible cell culture model to overexpress the FoxO1 protein in differentiated skeletal muscle cells. Furthermore, to elucidate the contribution of promoter binding to FoxO1-mediated skeletal muscle atrophy, a DNA-binding-deficient FoxO1 mutant muscle cell line was also utilized. It was hypothesized that 1) FoxO1 expression within differentiated skeletal muscle cells would result in skeletal muscle atrophy, which would be associated with loss of muscle protein, DNA damage, and increased expression of genes associated with protein ubiquitination and apoptosis, and 2) FoxO-1-induced muscle atrophy and associated changes in gene expression would be mediated through DNA-binding-dependent and -independent mechanisms.
METHODS
Retroviral transfections.
FoxO1-estrogen receptor (FoxO1-ER) fusion proteins that are activated by treatment with 4-hydroxytamoxifen (4-OH-T) were stably expressed in C2C12 skeletal myoblasts using a pBABE retroviral system, as previously reported (4). Briefly, cells were seeded in flat-bottom culture plates and grown in DMEM supplemented with 10% FBS (culture medium). At ∼50% confluence, cells were placed in culture medium supplemented with Polybrene (10 μg/ml) and transfected overnight with one of four retroviral constructs. Positively transfected cells were selected by incubation of myoblasts in culture medium supplemented with puromycin (Specialty Media, Temecula, CA; 3.5 μg/ml) for 24 h. The three retroviral constructs included FoxO1AAA-ER, FoxO1AAA/Arg215-ER, and FoxO1EV-ER. AAA corresponds to the alanine mutations of the three putative FoxO1 Akt phosphorylation sites, Thr24, Ser256, and Ser319 (16). AAA/Arg215 contains an additional mutation, H215R, which is in the DNA-binding domain and disrupts promoter binding (27). Empty vector (EV) constructs [empty cassette (no FoxO1-ER expression)] and nontransfected C2C12 cells served as controls.
Skeletal myotube culture.
Mouse C2C12 cells (28) (American Type Culture Collection) were maintained in low-glucose DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (culture medium). At 80–90% confluence, cells were switched to DMEM supplemented with 2% horse serum (differentiation medium) for 4 days. On day 4, myotubes were treated with differentiation medium supplemented with vehicle (ethanol, 0.1% final concentration) or 500 nM 4-OH-T (Sigma-Aldrich, St. Louis, MO) for up to 48 h.
Protein and DNA quantification.
For protein quantification, C2C12 myotubes were collected in a buffer containing 10 mM MgCl2, 10 mM KH2PO4, 1 mM EDTA, 5 mM EGTA, 50 mM β-glycerolphosphate, 10 mM PMSF, 10 mM leupeptin, 10 mM aprotinin, and 10 mM Na3VO4. Total protein concentration was assayed with a detergent-compatible protein assay kit (DC Protein Assay, Bio-Rad, Hercules, CA) and quantified by spectrophotometry using a microtiter plate reader (SpectraMax 190, Molecular Devices, Sunnyvale, CA). DNA was isolated from C2C12 myotubes by phenol-chloroform extraction according to a standard Tripure procedure (Roche Applied Science, Indianapolis, IN) and quantified by UV spectrophotometry.
Histology.
FoxO1AAA-ER, FoxO1AAA/Arg215-ER, FoxO1EV-ER, and nontransfected C2C12 myotubes were grown on 18-mm glass coverslips (Fisher Scientific, Pittsburgh, PA) and coated with cell attachment matrix (ECL, Upstate, Charlottesville, VA), and immunohistochemistry was performed 24 h after treatment with vehicle or 4-OH-T. Terminal deoxynucleotidyltransferase dUTP-mediated nick end labeling (TUNEL) was performed using a rhodamine-based In Situ Cell Death Detection Kit (Roche Applied Science). Briefly, each slide was rinsed twice for 5 min with sterile 1× PBS. TUNEL reagent was prepared as directed in the protocol, and 25 μl were loaded onto each slide. Slides were covered with a square glass coverslip and incubated for 1 h at 37°C inside a water-jacketed CO2 incubator (Nuaire, Plymouth, MN). Slides were washed three additional times with 1× PBS. For assessment of myotube development and integrity, skeletal myotubes were counterstained with a mouse monoclonal sarcomeric α-actinin antibody (Sigma-Aldrich; 1:2,000 dilution) and then incubated with a fluorescein-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch, West Grove, PA; 1:200 dilution). 4′,6-Diamidino-2-phenylindole (Vector Labs, Burlingame, CA) was used to visualize myonuclei. Cells were observed using a fluorescence microscope (model IX70, Olympus, Melville, NY) equipped with a digital camera and image-processing software (Spot-RT, Diagnostic Instruments, Sterling Heights, MI). The percentage of TUNEL-positive myonuclei (nuclei present in myotubes containing ≥2 myonuclei) was quantified in five areas of each slide, and the mean was calculated for comparison across the respective conditions.
Gene expression analysis.
Total RNA was isolated using an RNeasy Mini column (Qiagen, Valencia, CA), purified by DNase digestion (Turbo DNase, Ambion, Foster City, CA), and stored at −80°C until further processing. Total RNA was separated by agarose gel electrophoresis, and ribosomal bands were examined to ensure sample quality. cDNA was synthesized using 10 ng of total RNA, and PCR amplification was performed using a one-step quantitative real-time PCR kit (Superscript III, Invitrogen) and gene-specific Taqman primers and probes (Applied Biosystems, Foster City, CA) designed against MAFbx (Mm00499518_m1), MuRF-1 (Mm01188690_m1), Bim (Mm00437795_m1), BNip3 (Mm00833810_g1), and GAPDH (Mm99999915_g1). Quantitative real-time PCR were carried out in triplicate using a Real-Time Detection System (model 7500, Applied Biosystems). Analyses were performed to verify the dynamic range and confirm consistency among the amplification efficiencies of the various target genes. Data were expressed via the comparative cycle threshold (Ct) method, in which ΔCt values were calculated for all samples as follows: Ct(target gene) − Ct(housekeeping gene), where the target genes were MAFbx, MuRF-1, Bim, and BNip3 and the housekeeping gene was GAPDH. Relative changes in gene expression were then calculated for each target gene via the 2−ΔΔCt method, in which ΔCt values determined for each of the experimental (4-OH-T-treated) samples were subtracted from the ΔCt value from the calibrator (vehicle-treated) sample. The stability of the GAPDH housekeeping gene was confirmed through Ct analysis, which revealed that the range for average Ct values for the various conditions was within 0.5 Ct.
Protein ubiquitination assay.
Total cell homogenates (500 μg of protein) from vehicle- and 4-OH-T-treated samples were subjected to immunoprecipitation via magnetic bead separation (Invitrogen). Briefly, homogenates were precleared in a slurry of protein A/G-coated magnetic beads and then incubated overnight with 5 μg of total ubiquitin antibody (catalog no. SC-8017, Santa Cruz Biotechnology, Santa Cruz, CA). Subsequently, samples were incubated for 1 h with protein A/G-coated magnetic beads, resuspened in Laemmeli sample buffer, and boiled for 5 min. For determination of total ubiquitination, samples were resolved by SDS-PAGE on 4–15% polyacrylamide gradient gels (Jule, Milford, CT), which were incubated overnight in krypton protein stain (Thermo Scientific, Rockford, IL) and then developed via infrared detection using the Odyssey system.
Statistical analyses.
A two-way factorial ANOVA (P ≤ 0.05) was used to determine differences between conditions for all dependent variables. Newman-Keuls post hoc analyses (P ≤ 0.05) were used to locate differences between means if a significant main effect and/or interaction was found.
RESULTS
Activation of DNA binding is not required for FoxO1-induced atrophy of skeletal myotubes.
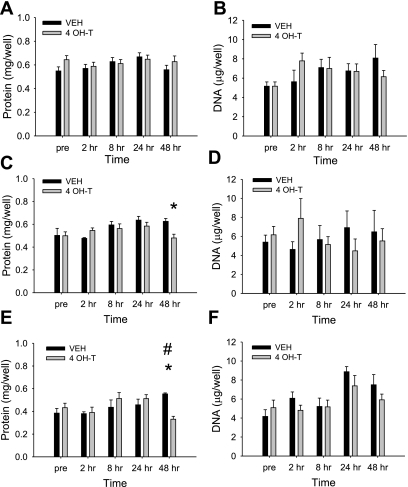
As shown in Fig. 1A, no significant difference in total protein was observed in nontransfected C2C12 myotubes (control) after treatment with vehicle or 4-OH-T across the time course. A similar response in total protein was observed in C2C12 cells stably transfected with the empty pBABE vector (FoxO1EV-ER; data not shown). In contrast, total protein was significantly decreased in the myotubes expressing FoxO1AAA-ER after 48 h of treatment with 4-OH-T compared with vehicle (Fig. 1C). This reduction in protein did not appear to be tightly coupled with DNA binding, since a similar effect of 4-OH-T on protein content was seen in cells expressing a FoxO1-ER fusion protein where DNA-binding activity was impaired (Fig. 1E). As shown in Fig. 1, B, D, and F, total DNA content was not changed in any of the myotubes (P > 0.05), and the total number of nuclei was not different in any of the conditions after treatment with vehicle or 4-OH-T (data not shown).
Fig. 1.
Total protein and DNA data for nontransfected C2C12 cells (A and B) and FoxO1AAA-ER (C and D) and FoxO1AAA/Arg215-ER (E and F) cells before and 2, 8, 24, and 48 h after treatment with vehicle (VEH) or 4-hydroxytamoxifen (4-OH-T). *Significant difference between VEH and 4-OH-T. #Significantly different from Pre for 4-OH-T treatment. n = 6 for each condition.
DNA binding is required for FoxO1-induced apoptosis in skeletal myotubes.
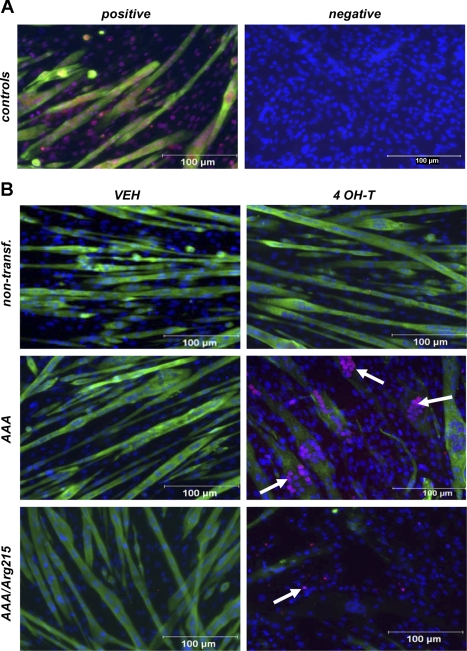
As shown in Fig. 2B, no loss of sarcomeric α-actinin or TUNEL staining was observed in nontransfected myotubes after 24 h of treatment with 4-OH-T. A similar response in staining was observed in the FoxO1EV-ER cells (images not shown). In contrast, FoxO1AAA-ER and FoxO1AAA/Arg215-ER mutants exhibited a profound loss of sarcomeric α-actinin staining after treatment with 4-OH-T (Fig. 2B). Activation of FoxO1 through the administration of 4-OH-T resulted in a near-complete loss of sarcomeric α-actinin by 24 h, which only manifested in ∼20% loss of total protein content by 48 h after treatment (Fig. 1, C and E). Inasmuch as gene array data did not show a significant loss of expression of muscle-specific genes (see supplemental data in the online version of this article), it is possible that FoxO1 promotes early degradation of differentiated muscle proteins, perhaps through upregulation of ubiquitin ligase gene expression (see below). FoxO1AAA-ER myotubes exhibited significant increases in TUNEL-positive nuclei after treatment with 4-OH-T (Fig. 2B, arrows). In contrast, FoxO1AAA/Arg215-ER myotubes exhibited scant signs of TUNEL staining after treatment with 4-OH-T (Fig. 3).
Fig. 2.
Sarcomeric α-actinin and terminal deoxynucleotidyl transferase dUTP-mediated nick end labeling (TUNEL) of C2C12 skeletal myotubes. Magnification ×200. A: DNase-treated positive control (3 U/ml) and negative control (no primary sarcomeric α-actinin antibody or TUNEL reagent). B: nontransfected, FoxO1AAA-ER (AAA), and FoxO1AAA/Arg215-ER (AAA/Arg215) cells treated with vehicle or 4 OH-T for 24 h. Arrows indicate TUNEL-positive myonuclei.
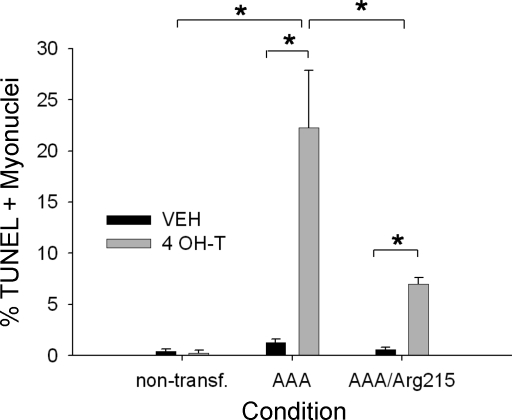
Fig. 3.
Quantification of TUNEL-positive myonuclei for conditions represented in Fig. 2. *Significantly different (P < 0.05) from all other conditions.
A significant number of TUNEL-positive nuclei were observed in FoxO1AAA-ER (∼22%, n = 3) and FoxO1AAA/Arg215-ER (∼8%, n = 3) myotubes after 24 h of treatment with 4-OH-T (Fig. 3). However, the increase TUNEL labeling was significantly greater in the FoxO1AAA-ER cells than in the FoxO1AAA/Arg215-ER cells (P < 0.05). Significant TUNEL labeling was not present in myotube nuclei in any of the control conditions after 4-OH-T treatment.
Impaired DNA binding results in differential gene expression conferred through FoxO1 activation.
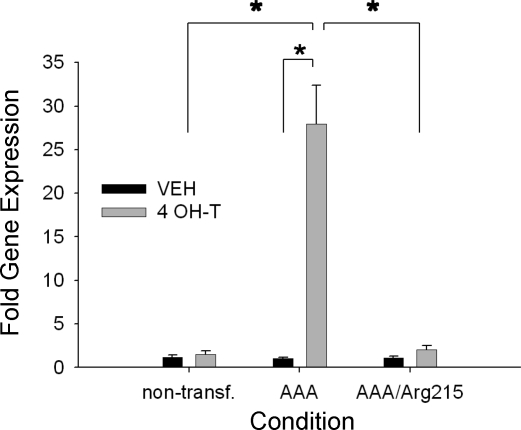
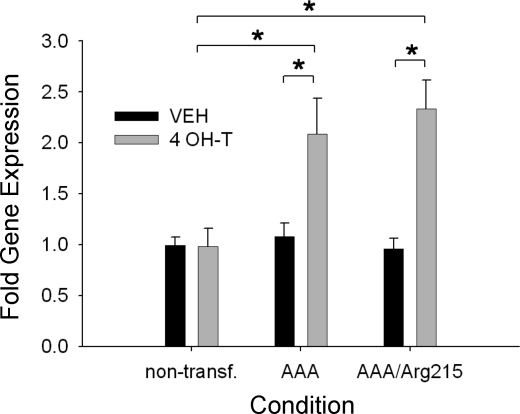
Treatment with 4-OH-T did not alter the expression of target genes (below) in nontransfected C2C12 cells or FoxO1EV-ER cells, so expression data for all gene targets were compared with data for the nontransfected C2C12 cells. As shown in Fig. 4, MAFbx/atrogin-1 gene expression was increased ∼27-fold after 24 h of treatment with 4-OH-T in the FoxO1AAA-ER cells. In contrast, MAFbx gene expression was unchanged in the nontransfected cells and FoxO1AAA/Arg215-ER cells after exposure to 4-OH-T (Fig. 4).
Fig. 4.
MAFbx relative gene expression for nontransfected, FoxO1AAA-ER, and FoxO1AAA/Arg215-ER myotubes after 24 h of treatment with vehicle or 4 OH-T. Brackets denote statistical significance (*P < 0.05). n = 4–6 per condition.
MuRF-1 gene expression was elevated in a similar manner in the FoxO1AAA-ER (∼2-fold) and FoxO1AAA/Arg215-ER (∼2.2-fold) cells after 4-OH-T treatment and was greater in both of these transfected cells than in the nontransfected cells (Fig. 5), indicating that FoxO1 can regulate the expression of MuRF-1 through a mechanism that is independent of DNA binding.
Fig. 5.
MuRF-1 relative gene expression for nontransfected, FoxO1AAA-ER (AAA), and FoxO1AAA/Arg215-ER (AAA/Arg215) myotubes after 24 h of treatment with vehicle or 4 OH-T. Brackets denote statistical significance (*P < 0.05). n = 4–6 per condition.
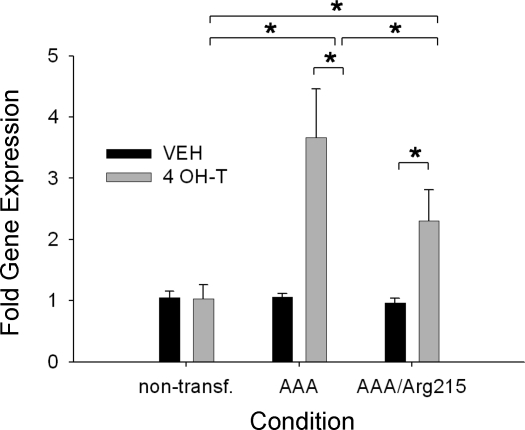
Gene expression for Bim was significantly elevated in the FoxO1AAA-ER (∼3.5-fold) and FoxO1AAA/Arg215-ER (∼2-fold) cells and was greater in both of these transfected cells than in the nontransfected cells (Fig. 6). The increase in Bim gene expression was significantly greater in the FoxO1AAA-ER than FoxO1AAA/Arg215-ER cells, indicating that FoxO1 regulates the expression of Bim through DNA-binding-dependent and -independent mechanisms.
Fig. 6.
Bim relative gene expression for nontransfected, FoxO1AAA-ER, and FoxO1AAA/Arg215-ER myotubes after 24 h of treatment with vehicle or 4 OH-T. Brackets denote statistical significance (*P < 0.05). n = 4–6 per condition.
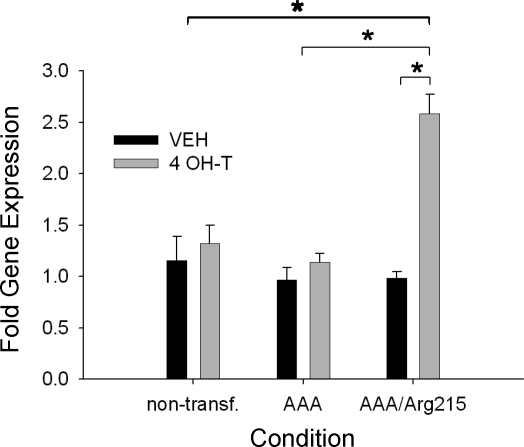
BNip3 gene expression was unchanged in the FoxO1AAA-ER cells after administration of 4-OH-T (Fig. 7). FoxO1AAA/Arg215-ER cells exhibited a significant increase in BNip3 gene expression after 4-OH-T treatment (∼2.2-fold), indicating that FoxO1 activation in the absence of DNA binding results in an alternative apoptotic gene expression profile.
Fig. 7.
BNip3 relative gene expression for nontransfected, FoxO1AAA-ER, and FoxO1AAA/Arg215-ER myotubes after 24 h of treatment with vehicle or 4 OH-T. Brackets denote statistical significance (*P < 0.05). n = 4–6 per condition.
DNA binding is required for FoxO1-induced protein ubiquitination of skeletal myotube proteins.
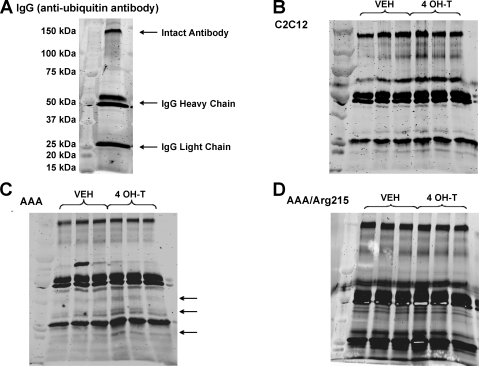
For examination of the effects of FoxO1 on the ubiquitination of muscle proteins, ubiquitinated proteins were immunoprecipitated from muscle cells after 48 h of treatment with vehicle or 4-OH-T. To better distinguish ubiquitinated protein bands from antibody proteins present in the immunoprecipitates, we resolved the SC-8019 ubiquitin antibody via SDS-PAGE and then stained the gel to determine antibody protein banding. Staining revealed prominent bands of ∼150, 50, and 25 kDa, representing the intact antibody, IgG heavy chain, and IgG light chain fragments, respectively (Fig. 8A). As expected, treatment of nontransfected C2C12 skeletal myotubes with 4-OH-T did not result in a detectable increase in protein ubiquitination compared with vehicle treatment (Fig. 8B). However, after treatment with 4-OH-T, FoxO1AAA-ER myotubes elicited signs of increased ubiquitination, evidenced by darker protein bands, particularly at 15–40 kDa (Fig. 8C). Despite a significant loss of total protein, protein ubiquitination in FoxO1AAA/Arg215-ER myotubes treated with 4-OH-T did not appear to increase significantly compared with those treated with vehicle (Fig. 8D). These results suggest that DNA-binding-independent effects of FoxO1 on protein content are not mediated by increasing ubiquitination of muscle proteins.
Fig. 8.
Total protein ubiquitination of vehicle- and 4 OH-T-treated skeletal myotubes. A: krypton staining of IgG anti-ubiquitin antibody. B–D: krypton staining of C2C12, FoxO1AAA-ER, and FoxO1AAA/Arg215-ER myotubes. Arrows denote areas of protein banding displaying increased ubiquitination after 48 h of tamoxifen treatment. n = 3 per condition.
DISCUSSION
This study demonstrates that muscle atrophy resulting from FoxO1 protein activation is the culmination of multiple mechanisms, including increased activity of protein ubiquitination and cellular apoptotic machinery. Although the ability of FoxO1 to induce muscle atrophy does not appear to be dependent on DNA binding, its effects on protein ubiquitination and apoptosis, which may be necessary for the removal of select proteins and myonuclei during the atrophy process, respectively, appear to be contingent on direct interaction between FoxO1 and promoter binding sites.
Strong evidence linking FoxO1 expression to skeletal muscle atrophy through the upregulation of genes associated with muscle protein degradation, including MAFbx/atrogin-1 and MuRF-1, has been provided through the use of in vitro and in vivo models (15, 19, 22, 25, 26, 32). As reported previously (22, 36), our findings show that the responsiveness of MuRF-1 to FoxO1 activation is relatively blunted compared with the robust increase in MAFbx/atrogin-1 gene expression. Although it cannot be discerned from our data, the differential response in expression of these proteins induced through FoxO1 activation may be reflective of different roles of MAFbx/atrogin-1 and MuRF-1 in regulating muscle homeostasis. For instance, overexpression of MAFbx/atrogin-1 in C2C12 skeletal myotubes has been shown to be a powerful inducer of muscle atrophy (5), whereas Hirner et al. (19) failed to show muscle atrophy in the presence of genetic overexpression of MuRF-1 in the rodent model. Rather, they observed interaction of MuRF-1 with several genes associated with carbohydrate metabolism, including pyruvate dehydrogenase, phosphoinositide-dependent kinase-2, and phosphoinositide-dependent kinase-4. Thus it is conceivable that MuRF-1 plays a more significant role in regulating the metabolic milieu within atrophying skeletal muscle as opposed to actively promoting protein degradation. Our findings also indicate that the ability of FoxO1 to regulate MAFbx/atrogin-1 gene expression is strongly dependent on DNA binding, whereas gene expression for MuRF-1 did not appear to be tightly coupled to direct FoxO1-DNA binding. Although FoxO1 has been shown to directly interact with the MuRF-1 promoter region (36), our data indicate that this ability may not be necessary to augment MuRF-1 gene expression in skeletal muscle. This contention is supported by Ramaswamy et al. (24), who demonstrated that overexpression of the related FoxO factor FoxO3a augmented gene expression independent of DNA binding. Further investigation into potential protein-protein interactions and/or promoter assembly mechanisms involved in mediating FoxO1-induced muscle atrophy is warranted.
Coupled with these changes in proteasome-associated genes, we observed disparate findings in terms of protein ubiquitination following the activation of FoxO1 in the various cell lines. Specifically, total protein ubiquitination appeared to be elevated in the FoxO1AAA-ER cell line after treatment with 4-OH-T, but not in FoxO1AAA/Arg215-ER cells. His215, which is located in the DNA-binding domain of all FoxO and other forkhead proteins, is thought to participate directly in sequence-specific interactions involved in DNA (7), and mutation of this residue greatly impairs the binding of FoxO1 to DNA target sites in in vitro assays. The finding that mutation of this residue prevents the effects of FoxO1-ER fusion proteins in C2C12 cells suggests that some of these effects may be mediated through mechanisms that do not require a direct interaction between FoxO1 and specific DNA-binding sequences. Given the potent ability of MAFbx/atrogin-1 to induce protein degradation (5), it is likely that the suppressed induction of MAFbx/atrogin-1 observed in the FoxO1-DNA-binding-deficient mutants may account for limited protein ubiquitination after FoxO1 activation. Inasmuch as muscle atrophy and total protein loss were still observed in the DNA-binding-deficient condition, despite a blunting of MAFbx/atrogin-1 gene expression and total protein ubiquitination, it is likely that alternative mechanisms beyond proteasome-mediated degradation contribute to muscle atrophy following FoxO1 activation.
One of the more noted cellular events associated with the remodeling of atrophied skeletal muscle fibers is a loss of myonuclei (3), likely triggered through the activation of proapoptotic pathways (10–12). Although studies of the role of FoxO1 in apoptosis within differentiated muscle are lacking, the activation of FoxO1 and related FoxO proteins (e.g., FoxO3a and FoxO4) has been widely implicated in initiating a cascade of cellular events resulting in apoptosis and/or cell cycle arrest in proliferatively active cells. Gene array data collected in our laboratory demonstrating increased expression of DNA damage-responsive and proteolytic genes [i.e., GADD45β and ubiquitin specific protease 2, respectively] following FoxO1 activation in differentiated myotubes corroborate the proapoptotic and protein degradatory role of FoxO1 in differentiated skeletal muscle (see supplemental data).
Apoptotic signaling induced via FoxO1 expression appeared contingent on DNA binding, inasmuch as DNA fragmentation and proapoptotic Bim gene expression were less prevalent in FoxO1AAA/Arg215-ER than FoxO1AAA-ER cells. The significant increase in TUNEL-positive myonuclei and Bim gene expression observed in the FoxO1AAA/Arg215-ER cell line likely was the result of trace DNA binding, inasmuch as this mutant cell line results in a suppression, but not an elimination, of FoxO1-DNA binding (X. Zhang and T. Unterman, unpublished observations), as previously discussed. It is conceivable that the abrogated FoxO1-DNA binding resulted in a delay, as opposed to a loss, of apoptotic signaling transduction. However, because of technical difficulties associated with more prolonged exposures of cells to 4-OH-T (i.e., significant cell detachment and frailty of remaining cells), we were unable to obtain reliable data for the histochemical measurements of apoptosis at later time points. However, pilot gene expression analysis of cells exposed to a broader time course (2, 8, 24, and 48 h) of treatment (data not shown) indicated that there was no significant change in the pattern of gene expression. Thus it appears that the differences in apoptotic signaling between the FoxO1AAA-ER and FoxO1AAA/Arg215-ER mutant cell lines were due to differences in DNA-binding capability, rather than temporal differences in apoptotic signaling.
Surprisingly, BNip3 demonstrated a disparate response, in that gene expression was unchanged in FoxO1AAA-ER cells but was significantly elevated in the DNA-binding-deficient FoxO1 cell line. During apoptosis, BNip3 appears to function as an allosteric protein, requiring Bax or Bak to facilitate cell death (8). However, when apoptosis is not activated, which appears to be the case in the FoxO1AAA/Arg215-ER cells, BNip3 can function to disrupt mitochondrial integrity, possibly triggering alternative forms of cell death (e.g., necrosis) and/or autophagy (9, 17, 18, 35). Given the ability of the FoxO isoforms to interact directly with a diverse array of nonrelated transcription factors (34), it is possible that, in the absence of direct promoter binding, FoxO1 may be acting to modulate subsets of genes through protein-protein interactions and, subsequently, activate cell destruction machinery that is distinct from the pathways observed in the DNA-binding-capable mutants. We did not observe a significant disruption of sarcolemma integrity, evidenced by insignificant lactate dehydrogenase release, on exposure to 4-OH-T (T. McLoughlin, unpublished observations), indicating that it is unlikely that the muscle atrophy observed after FoxO1 activation (DNA-binding capable or deficient) was due to necrosis. Although it cannot be definitively concluded in the present study, a link between FoxO expression and autophagy in skeletal muscle has been provided by Mammucari et al. (23), who demonstrated that activation of Akt blocked FoxO3a activation and the induction of autophagy in muscle. Thus autophagic processes, primed through upregulation of BNip3 signaling, may be acting to facilitate muscle atrophy, particularly in cells expressing the DNA-binding-deficient form of FoxO1. Given that we did not observe any significant increase in caspase-3 activity following FoxO1 activation in any of the cell lines studied (see supplemental data), it is possible that nonlethal forms of cellular disruption (e.g., autophagy) involving other effector molecules may be at play, acting to remove harmful and/or damaged mitochondria and unnecessary nuclei, in an attempt to remodel, rather than eradicate, skeletal muscle cells (9).
Findings from this study provide further insight into the complex role of FoxO1 in biological regulation of skeletal muscle. Specifically, the remodeling of atrophying skeletal muscle induced through FoxO1 activation may rely on DNA-binding-dependent and -independent mechanisms. Although it is always difficult to extend in vitro work to complex, intact physiological systems, it is possible that the elevations of FoxO1 expression may be a potent driving force for the breakdown and removal of muscle proteins and elimination of nuclei through activation of apoptotic machinery associated with various muscle atrophy conditions, including aging, immobilization, and various degenerative muscle diseases. Given the multipronged strategy of FoxO1 in regulating cellular function, careful consideration should be given to the development of future pharmaceutical, gene therapy, and/or physical rehabilitation approaches in attempting to thwart FoxO1-induced muscle atrophy.
GRANTS
This project was partially funded through National Institutes of Health Grants T32 HL-07692-12 (T. J. McLoughlin), AR-45617 (K. A. Esser), and DK-41430 (T. G. Unterman) and the Department of Veterans Affairs Merit Review Program (T. G. Unterman).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Nissam Hay and Nathan Majewski for packaging of the retroviral constructs and guidance in retroviral transfections, Wenwei Zhang for preparation of RNA, and Rebecca R. Suffolk for assistance with growing cells and performing histochemistry experiments.
REFERENCES
- 1.Adhihetty PJ, O'Leary MF, Chabi B, Wicks KL, Hood DA. Effect of denervation on mitochondrially mediated apoptosis in skeletal muscle. J Appl Physiol 102: 1143–1151, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Allen DL, Linderman JK, Roy RR, Bigbee AJ, Grindeland RE, Mukku V, Edgerton VR. Apoptosis: a mechanism contributing to remodeling of skeletal muscle in response to hindlimb unweighting. Am J Physiol Cell Physiol 273: C579–C587, 1997 [DOI] [PubMed] [Google Scholar]
- 3.Alway SE, Degens H, Krishnamurthy G, Smith CA. Potential role for Id myogenic repressors in apoptosis and attenuation of hypertrophy in muscles of aged rats. Am J Physiol Cell Physiol 283: C66–C76, 2002 [DOI] [PubMed] [Google Scholar]
- 4.Bastie CC, Nahle Z, McLoughlin T, Esser K, Zhang W, Unterman T, Abumrad NA. FoxO1 stimulates fatty acid uptake and oxidation in muscle cells through CD36-dependent and -independent mechanisms. J Biol Chem 280: 14222–14229, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708, 2001 [DOI] [PubMed] [Google Scholar]
- 6.Chipuk JE, Fisher JC, Dillon CP, Kriwacki RW, Kuwana T, Green DR. Mechanism of apoptosis induction by inhibition of the anti-apoptotic BCL-2 proteins. Proc Natl Acad Sci USA 105: 20327–20332, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364: 412–420, 1993 [DOI] [PubMed] [Google Scholar]
- 8.Crow MT. Hypoxia, BNip3 proteins, and the mitochondrial death pathway in cardiomyocytes. Circ Res 91: 183–185, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res 64: 4286–4293, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Dirks A, Leeuwenburgh C. Apoptosis in skeletal muscle with aging. Am J Physiol Regul Integr Comp Physiol 282: R519–R527, 2002 [DOI] [PubMed] [Google Scholar]
- 11.Dirks AJ, Leeuwenburgh C. The role of apoptosis in age-related skeletal muscle atrophy. Sports Med 35: 473–483, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Dupont-Versteegden EE, Murphy RJ, Houle JD, Gurley CM, Peterson CA. Activated satellite cells fail to restore myonuclear number in spinal cord transected and exercised rats. Am J Physiol Cell Physiol 277: C589–C597, 1999 [DOI] [PubMed] [Google Scholar]
- 13.Frenzel A, Grespi F, Chmelewskij W, Villunger A. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis 14: 584–596, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene 27: 2312–2319, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol 37: 1974–1984, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem 274: 17184–17192, 1999 [DOI] [PubMed] [Google Scholar]
- 17.Hamacher-Brady A, Brady NR, Gottlieb RA, Gustafsson AB. Autophagy as a protective response to Bnip3-mediated apoptotic signaling in the heart. Autophagy 2: 307–309, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, Gustafsson AB. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ 14: 146–157, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Hirner S, Krohne C, Schuster A, Hoffmann S, Witt S, Erber R, Sticht C, Gasch A, Labeit S, Labeit D. MuRF1-dependent regulation of systemic carbohydrate metabolism as revealed from transgenic mouse studies. J Mol Biol 379: 666–677, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Hirota K, Daitoku H, Matsuzaki H, Araya N, Yamagata K, Asada S, Sugaya T, Fukamizu A. Hepatocyte nuclear factor-4 is a novel downstream target of insulin via FKHR as a signal-regulated transcriptional inhibitor. J Biol Chem 278: 13056–13060, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Hoffman EP, Nader GA. Balancing muscle hypertrophy and atrophy. Nat Med 10: 584–585, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem 279: 41114–41123, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 6: 458–471, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2: 81–91, 2002 [DOI] [PubMed] [Google Scholar]
- 25.Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J 21: 140–155, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. FoxO transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117: 399–412, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG. Regulation of glucose-6-phosphatase gene expression by protein kinase Bαand the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem 275: 36324–36333, 2000 [DOI] [PubMed] [Google Scholar]
- 28.Schuur ER, Loktev AV, Sharma M, Sun Z, Roth RA, Weigel RJ. Ligand-dependent interaction of estrogen receptor-α with members of the forkhead transcription factor family. J Biol Chem 276: 33554–33560, 2001 [DOI] [PubMed] [Google Scholar]
- 29.Shore AM, White PC, Hui RC, Essafi A, Lam EW, Rowe M, Brennan P. Epstein-Barr virus represses the FoxO1 transcription factor through latent membrane protein 1 and latent membrane protein 2A. J Virol 80: 11191–11199, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siu PM, Pistilli EE, Alway SE. Apoptotic responses to hindlimb suspension in gastrocnemius muscles from young adult and aged rats. Am J Physiol Regul Integr Comp Physiol 289: R1015–R1026, 2005 [DOI] [PubMed] [Google Scholar]
- 32.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 14: 395–403, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, Jr, DiStefano PS, Chiang LW, Greenberg ME. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 296: 530–534, 2002 [DOI] [PubMed] [Google Scholar]
- 34.van der Vos KE, Coffer PJ. FOXO-binding partners: it takes two to tango. Oncogene 27: 2289–2299, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Vande Velde C, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, Hakem R, Greenberg AH. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol Cell Biol 20: 5454–5468, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, Bodine SC. The glucocorticoid receptor and FoxO1 synergistically activate the skeletal muscle atrophy associated MuRF-1 gene. Am J Physiol Endocrinol Metab 295: E785–E797, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao HH, Herrera RE, Coronado-Heinsohn E, Yang MC, Ludes-Meyers JH, Seybold-Tilson KJ, Nawaz Z, Yee D, Barr FG, Diab SG, Brown PH, Fuqua SA, Osborne CK. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J Biol Chem 276: 27907–27912, 2001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.