Abstract
Collagen peptides have been used to identify binding sites for several important collagen receptors, including integrin α2β1, glycoprotein VI, and von Willebrand factor. In parallel, the structures of these collagen receptors have been reported, and their interactions with collagen peptides have been studied. Recently, the three-dimensional structure of the intact type I collagen fiber from rat tail tendon has been resolved by fiber diffraction. It is now possible to map the binding sites of platelet collagen receptors onto the intact collagen fiber in three dimensions. This minireview will discuss these recent findings and their implications for platelet activation by collagen.
Structural Architecture of Fibrillar Collagens
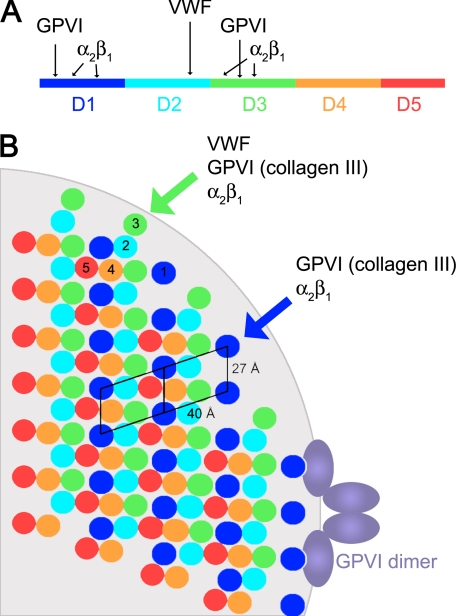
Collagen is the most abundant protein in the human body, where it is a vital component of the extracellular matrix and connective tissue, including blood vessels. There are 29 collagen types, of which seven (types I–III, V, XI, XXIV, and XXVII) are fibrillar, able to assemble as stable triple helices, which then form a complex higher order three-dimensional fibrous superstructure. At the simplest level, the collagen polypeptide sequence is composed of Gxx′ triplets containing glycine followed by variable residues that often include proline at position x and hydroxyproline at position x′. The three polypeptide chains that assemble to form a tropocollagen triple helix can be identical gene products, as for collagen III (three α1 chains), or may differ, as for collagen I (two α1 chains and one α2 chain). The tropocollagen triple helices assemble to form a supermolecular fiber. Studies over the past 3 decades (1–7) have established that the basic repeating unit within the collagen fiber contains five tropocollagens packed side-by-side in a pseudohexagonal arrangement, with a gap along the fiber axis between the end of one triple helix and the start of the next. This regularly repeated gap region gives rise to the characteristic banding pattern of the intact collagen fiber, with a spacing D of 67 nm between successive bands corresponding to the offset between adjacent tropocollagen molecules, the classical “quarter-stagger.” The densely packed (pseudohexagonal) regions extend over nearly half (0.46D) of a single D period, whereas the gap regions occupy the remaining length of the D period (0.54D). Each collagen molecule can be divided into five D periods (Fig. 1A). Collagen fibers have a circular cross-section, suggested by fiber diffraction data to be composed of concentric layers of fibrils with a thickness of ∼40 Å (8).
FIGURE 1.
Molecular packing of the type I collagen fiber. A, mapping the THPs specific for platelet receptors onto the collagen molecule. Each D segment is highlighted in a different color. B, schematic of the cross-section of a type I collagen fiber created by applying symmetry operators to the fiber diffraction structure (9). The cross-section was taken approximately at the beginning of segment D1. The light gray background shows the circumference of a 400-Å-diameter fiber at the same scale as the collagen triple helices. Because of interdigitation of the microfibrils, the relative position of the segments varies along the length of the fiber. The unit cell of Orgel et al. (9) was positioned such that the platelet receptor sites are accessible from the fiber surface; another possible orientation has been described (10).
In 2006, Orgel et al. (9) published the three-dimensional structure obtained using fiber diffraction of an intact collagen type I fiber from rat tail tendon. This structure revealed the complicated interdigitated arrangement of the triple helices within the collagen fiber. Each unit cell (corresponding to one D period in the mature collagen fiber) measured 678 Å long, 27 Å wide, and 40 Å deep and contained segments of five tropocollagen triple helices. As expected, a cross-section of the overlap region reveals approximate hexagonal packing of adjacent triple helices (Fig. 1). However, the tropocollagen triple helices do not behave as rigid rods, but rather, in the gap region, where there are just four triple helices, they adopt a right-handed twist around the microfibril to create a superstructure composed of five interdigitated triple helices. This twist dictates that there is variable exposure of the different segments of the collagen triple helix on the surface of the fiber.
The a unit cell axis in the structure measures 40 Å, consistent with the concentric ring spacing in the cross-section of a collagen fiber. The bc face of the unit cell (approximately perpendicular to the a unit cell axis; labeled 27 Å in Fig. 1B) would then represent the surface of the fiber typically exposed for interaction with collagen receptors. There are two possible ways to place the unit cell, resulting in different exposed collagen segments on the surface of the fiber. Orgel and colleagues (10) proposed an orientation that primarily exposes D periods 4 and 5 (Fig. 1); as described below, the opposite orientation, exposing segments D1 and D3, is more consistent with known platelet receptor interactions and is assumed in this minireview to represent the surface of the collagen fiber.
Fibrillar Collagens in the Blood Vessel Wall
The two most abundant collagens of the blood vessel wall are types I and III, both present in the subendothelial intima. Collagen I may be enriched in atherosclerotic plaque especially in its fibrous cap, but collagen III is also abundant. Several other collagens, including fibrillar type V, have been located within the vessel wall. Most information concerning receptor-collagen interactions is based on collagens I and III, given their availability from skin and placental material. Although the two collagens are highly conserved, collagen I α1 shows higher sequence identity to collagen II α1 than to either collagen I α2 or collagen III α1. Deductions concerning binding sites within heterotrimeric collagen I may be subject to some uncertainty because proof of binding has been obtained almost exclusively with homotrimeric THPs3 corresponding to collagens II and III that lack an α2 chain. Furthermore, the collagen α chains are staggered within the triple helix by one residue, yielding three potential isomeric positions for the α2 chain in heterotrimeric collagen I.
Use of Collagen Peptides to Identify Receptor-binding Sites
Given the complexity of the intact collagen fiber, most of our insights into the nature of platelet receptor-collagen interactions are based on model collagen-related peptides (11). Early studies identified a high-affinity site within collagen type I for the I-domain of integrin α2β1 (GFOGER, where O is hydroxyproline) (12) and a low-affinity site in collagen type III for GPVI composed of repeating GPO triplets (13). More recently, systematic analyses of human type III collagen have identified sequences that bind specific receptors. These studies used Toolkit III, which contains 57 overlapping peptides that cover the entire sequence of type III collagen; each peptide consists of 27 residues from the sequence, flanked by five GPP triplets and a terminal GPC triplet.
Integrin-binding Sites
The peptide-based approach has revealed a series of integrin-binding motifs within the collagens (12, 14). In collagen I, the highest affinity motif, GFOGER, lies in the third D period (D3) of each tropocollagen. Nearby lies a moderate affinity site composed of GMOGER (α1 chain) and GLOGER (α2 chain). Close to the N terminus of the molecule in D1 lie GROGER, another high-affinity site conserved across collagens I and III, and GLOGER. Within the C-terminal half of collagen I, only weak integrin-binding sites are found, GQRGER and GASGER, both of which bind only slightly to resting cells. Other low-affinity Gxx′GEx′ sites may also become relevant in activated platelets, enhancing the avidity of collagen for the integrin-bearing cell surface.
In collagen III, there is no α2 chain to consider, but homotrimeric Gxx′GER peptides do not reveal the whole story. The higher affinity GROGER and GMOGER motifs are conserved with collagen I α1; however, the ability of GLSGER in D4 to support binding is dependent on its context. GLSGER within a short THP acts as an intermediate-affinity site, but it is a very poor site at best when presented within 27 amino acids of native sequence as in Toolkit III (11), indicating that local sequence may interfere with integrin binding. Collagen III also contains a novel GLOGEN motif that binds both α2β1 and α1β1 (although the latter is not expressed on platelets) (14).
Glycoprotein VI
The recognition of GPVI by collagen is more complex. First, GPO-containing THPs bind GPVI with affinity that increases with the number of contiguous GPO triplets (15, 16). However, such tracts of GPO triplets are scarce. In collagen III, GPOGPOGPO occurs at positions −8 to 0, not present in collagen I, and this region binds GPVI quite well (17). The nearest equivalent in collagen I is at the C terminus of the molecule (D5), where GPOGPOGPOGPOGPO and GPOGPOGPOGPO may occur in the α1 and α2 chains, respectively. It is not certain that such long GPO tracts are fully hydroxylated though, which would reduce their potential for binding GPVI.
The best GPVI-binding motif in collagen III is a split GPO … GPOGPO motif in the middle of the molecule (D3) that may bind two or more GPVI molecules side-by-side (16, 17). The sequence of collagen III reveals no other such sites, although weak binding of GPVI occurs with a GPO … GPO site at the start of collagen III D4. It is possible that the corresponding GPO … GPOGPO occurring in D4 of collagen I α1 and α2 may serve, but the separation (two triplets) differs from that of the best site in collagen III (three triplets). How collagen I is recognized by GPVI remains to be established.
von Willebrand Factor
The recognition of collagen III by VWF appears to be straightforward: just a single, unambiguous motif is found in D2. The minimal motif is GPRGQOGVMGFO (18). Alanine-scanning substitution revealed those boldface residues to be crucial to binding. There are crucial differences in both the α1 and α2 chains of collagen I. It is hypothesized that the intact VWF-binding site requires assembly of the heterotrimer because neither the corresponding I α1 nor I α2 homotrimers will bind VWF.
Three-dimensional Mapping of Receptor-binding Sites on the Collagen Fiber
In parallel with the advances in understanding platelet receptor-binding collagen sequences, there have been impressive strides in determining the high-resolution crystal structures of the platelet receptors involved. The structure of the α2β1 I-domain has been determined alone and in complex with a GFOGER-containing collagen triple helix (19, 20). The GPIbα ectodomain has been solved alone (21) and in complex with VWF (22, 23) or thrombin (24). The structure of the collagen-binding A3-domain of VWF is known (25), and its binding site has been mapped by NMR (26). Recently, the structure of GPVI has been resolved (27), which identified a likely collagen-binding surface on GPVI consistent with mutational studies (28–31).
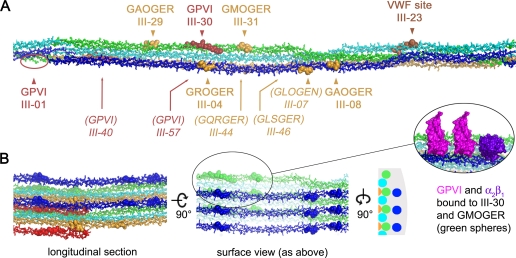
Despite this progress, it has been difficult to predict how the receptors recognize the surface of a large collagen fiber in three dimensions. The fiber diffraction structure of Orgel et al. (9) offers fresh insight. When mapping receptor-binding sites identified in THPs onto the intact collagen fiber, the first consideration is whether the corresponding motifs are actually accessible. As described earlier, it is assumed that the bc unit cell face in the structure of Orgel et al. represents the surface of the collagen fiber. The unit cell orientation consistent with accessibility of platelet receptor sites would expose most of the first D period segment (D1), the last third of D2, and the first half of D3. The right-handed twist of the microfibril dictates that the remaining portions of D2 and D3, as well as segments D4 and D5, are primarily buried within the collagen fiber (Fig. 2). The interpretations here assume also that homotrimeric collagen III adopts a similar interdigitated structure as that observed for heterotrimeric collagen I.
FIGURE 2.
Three-dimensional mapping of receptor-binding sites onto the collagen fiber. A, view of one D region of a collagen microfibril constructed using the structure of Orgel et al. (9). The tropocollagen triple helices are colored from blue to red according to D periods, as in Fig. 1. Receptor-binding sites determined using THPs are shown as spheres, with integrin α2β1 sites shown in yellow, GPVI sites in red, and the VWF site in brown. Surface-accessible sites are indicated in boldface with an arrowhead; sites typically buried within the microfibril are indicated in italics with an arrow. Only sites present in collagen III are marked. Corresponding sites from collagen I are listed in supplemental Table 1. B, views of the collagen fiber built up by neighboring microfibrils. The surface view corresponds to that shown in A and corresponds to a region of curvature, as shown by the schematic view of the transverse section. A close-up view of two GPVI molecules docked onto the III-30 site (16, 27) and the α2β1 I-domain docked onto the GMOGER site (20) is shown in the inset. In this panel, the receptor sites are shown as spheres colored according to the D period.
Integrin-binding Sites
According to this model, GROGER and GLOGER within D1 are fully accessible and would allow binding of the α2β1 I-domain without steric clashes (Fig. 2A). Sites GFOGER and GMOGER within D3 may be accessible depending on the degree of curvature in the collagen fiber surface (Fig. 1B, blue versus green circles). The GLOGEN site identified in collagen III as a ligand for α2β1 and α1β1 (peptide III-07) (14) is found on the outer surface of the microfibril, but on the side of the triple helix facing the neighboring tropocollagen. Thus, I-domains would be sterically excluded from this site in the intact fiber unless the triple helix was accessible from the side of the fiber or local fiber unfolding occurred. The final two sites, GQRGER (in collagen I) and GASGER, within D4 are buried within the microfibril and are unlikely to be accessible, except perhaps at the extreme ends of the collagen fiber (Fig. 2B, yellow spheres).
GPVI-binding Sites
Upon mapping the GPVI sites onto the fiber, it is striking that there are so few discrete loci where GPVI can bind within each D period. The type III collagen Toolkit peptides III-01 and III-30 fall within D1 and D3, respectively. Site III-01 would be exposed on the surface of the microfibril, and site III-30 may be accessible depending on the curvature; however, site III-57 is on the back of the microfibril and completely inaccessible. Interestingly, the GPO … GPO site at the beginning of D4, present in III-40, lies adjacent to III-01 on an underlying triple helix. This site would typically be buried, although it might become exposed at the ends of the collagen fiber, possibly creating a more extensive GPVI-binding site composed of neighboring III-01 and III-40 sites. In type I collagen, the III-01 site is completely absent, and the III-30 site is not conserved, so the partially exposed III-40 region would be the only known site available for GPVI binding.
The collagen fiber structure of Orgel et al. (9) is also consistent with the geometry of the GPVI dimer that occurs on the platelet surface (32, 33) and that was observed in the crystal structure (27). The spacing and orientation of the two putative collagen-binding grooves on a GPVI dimer match the location of GPVI-binding sites on every second unit cell in the collagen fiber structure. Thus, a GPVI dimer could simultaneously bind to a given site (i.e. III-01) on the ith and (i+2)th microfibrils, providing a structural basis for the avidity effect previously described for GPVI-Fc fusion binding to collagen (34).
VWF-binding Site
The single high-affinity site for VWF at the end of segment D2 (18) is exposed on the microfibril surface in the C-terminal half of the D period. Unlike the integrin and GPVI sites, the VWF site is mostly isolated from other receptor sites. Given that there is a single high-affinity VWF-binding site within the entire sequence of collagen type III, the exposed location of this site provides strong evidence that the available surface of the collagen fiber is composed of segments D1–D3 rather than segments D4 and D5. Furthermore, this orientation is consistent with exposure of all the high-affinity α2β1 sites (GROGER, GLOGER, GFOGER, and GMOGER), whereas the exposed D4/D5 model (10) for the collagen fiber would expose only the low-affinity GQRGER and GASGER sites adjacent to the collagenase cleavage site discussed by Perumal et al. (10).
Implications for Receptor Cross-talk and Receptor Activation
It is becoming clear that cooperative binding may occur when integrin- and GPVI-binding sites are close together. Thus, Toolkit peptide III-04, containing GROGERGxx′GPO, has affinity for both integrin and GPVI. In human platelets and wild-type mice, it binds integrin quite well, whereas in GPVI/FcRγ-deficient mice, it has poor affinity for integrin (17). This underscores the requirement for platelet activation in the recognition of even high-affinity integrin-binding motifs. Similarly, Toolkit peptide III-31, containing GPOGPOGxx′Gxx′Gxx′GMOGER, has higher affinity for platelets than peptide III-32, the sequence of which begins with GMOGER and lacks GPO triplets. Docking GPVI and the α2β1 I-domain onto the intact collagen fiber in the III-30/III-31 peptide region indicates that the receptors could potentially contact one another (Fig. 2B, inset). Because of its mucin-like stalk, the GPVI ectodomain would extend ∼220 Å from the platelet surface (27), which is comparable with the height of an activated integrin (35). There is evidence for both cross-talk and signal convergence between GPVI and α2β1 (36, 37); furthermore, recent work has shown that integrin signaling can occur via immunoreceptor tyrosine-based activation motifs in immunoreceptors such as FcγRIIa (38, 39). GPVI also colocalizes with the GPIb-V-IX complex, apparently via direct interaction between the ectodomains of GPVI and GPIbα (40), and both GPVI and GPIb have been reported in lipid raft fractions (41). However, the biological significance of this interaction is not clear because the VWF site in collagen is located ∼200–450 Å from the nearest GPVI sites.
There is also evidence for competition between receptors that bind to overlapping sites. Thus, SPARC (secreted protein acidic and rich in cysteine), discoidin domain receptors, and VWF all bind to a motif that includes the two triplets, GVMGFO, in segment D2 (42–44). VWF-dependent adhesion requires not just availability of that segment of collagen but also that competing species are at a low level or bind sufficiently weakly to allow VWF to bind vessel wall collagen. The observation of an inverse correlation between VWF levels and metastasis in mouse and human systems suggests that the consequences of such competition may be far-reaching (45).
Biological Implications of the Distribution of Receptor-binding Sites on the Collagen Fiber
The prevalence and spacing of specific receptor-binding sites on the surface of the collagen fiber play an important role in multiple stages of platelet activation and thrombus formation. The first stage occurs when GPIbα catches hold of VWF bound to exposed collagen, causing transient tethering of the platelet to the collagen surface. The VWF-binding site occurs only once in each D period; because of the repeating nature of the unit cells in the fiber, the VWF sites form a stripe running perpendicular to the main fiber axis. Thus, GPIbα ectodomains, with highly elongated stalk regions (46), allow a platelet under flow to come into contact with the collagen surface by tightly binding to widely spaced stripes of collagen-bound VWF in a manner reminiscent of hook-and-cable systems used to slow airplanes landing on the deck of an aircraft carrier. The next stage of thrombus formation involves binding to high-affinity integrin α2β1 sites such as GFOGER and platelet activation when GPVI becomes clustered upon binding to GPO repeats on the collagen fiber. The relative rarity of GPVI-binding sites and the low affinity of this interaction are consistent with a high threshold for platelet activation, which helps minimize thrombus formation under resting conditions. After GPVI signaling occurs, inside-out activation of integrin α2β1 allows tight binding to other Gxx′GER sites. The combination of frequent occurrence and high affinity of these sites ensures that the activated platelet will remain adhered firmly to the collagen surface as the thrombus begins to form.
The analysis presented here focuses on collagen fibers of large diameter with a uniform cross-section. Where fibers taper toward either end, some sites that were hidden in the context of a large-diameter fiber may become available. In the limit where a tapering fiber reduces to a single microfibril, then full accessibility of binding sites will result. Interestingly, it appears that maximum platelet reactivity occurs in small collagen structures corresponding to just one microfibril diameter.4 Some disorder is also needed to allow packing within the large fiber to adapt the quasihexagonal crystalline segments to a circular cross-section (8). The relative proportion of crystalline surfaces compared with disordered boundary regions will decrease as the diameter of the fiber is reduced, possibly exposing additional sites and thus allowing a fuller range of reactivity.
Finally, active remodeling of the collagen fiber organization may be critical for certain biological outcomes. For example, Perumal et al. (10) suggested that the collagenase cleavage site is revealed only after proteolytic clipping of the C-terminal telopeptide that folds back upon the microfibril. In the model presented here, the canonical collagenase site remains completely buried, so fiber reorganization would be needed for collagenase activity to proceed. Other proteins relevant to platelet activity can interact with this region, such as fibronectin, which binds a linear peptide near the collagenase site (47). Similarly, discoidin domain receptor 2 and SPARC bind nearby in segment D4. However, remote binding sites for the latter have also been identified (42, 44), so it may be that binding to an ancillary locus perturbs fiber organization and renders the primary site accessible. Although much work remains to be done to better understand the regulation of receptor interactions with fibrillar collagen, it is exciting to be able finally to begin to understand these processes in three dimensions.
Supplementary Material
This work was supported by the American Heart Association and the Ohio Eminent Scholar Program (to A. B. H.) and by the Wellcome Trust, the British Heart Foundation, and the Medical Research Council (to R. W. F.). This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
A. M. C. Simpson and R. W. Farndale, unpublished data.
- THP
- triple-helical peptide
- GP
- glycoprotein
- VWF
- von Willebrand factor.
REFERENCES
- 1.Hulmes D. J., Miller A. (1979) Nature 282, 878–880 [DOI] [PubMed] [Google Scholar]
- 2.Fraser R. D., MacRae T. P., Miller A., Suzuki E. (1983) J. Mol. Biol. 167, 497–521 [DOI] [PubMed] [Google Scholar]
- 3.Bella J., Eaton M., Brodsky B., Berman H. M. (1994) Science 266, 75–81 [DOI] [PubMed] [Google Scholar]
- 4.Kramer R. Z., Bella J., Brodsky B., Berman H. M. (2001) J. Mol. Biol. 311, 131–147 [DOI] [PubMed] [Google Scholar]
- 5.Fraser R. D., MacRae T. P., Miller A. (1987) J. Mol. Biol. 193, 115–125 [DOI] [PubMed] [Google Scholar]
- 6.Orgel J. P., Miller A., Irving T. C., Fischetti R. F., Hammersley A. P., Wess T. J. (2001) Structure 9, 1061–1069 [DOI] [PubMed] [Google Scholar]
- 7.Wess T. J., Hammersley A. P., Wess L., Miller A. (1998) J. Mol. Biol. 275, 255–267 [DOI] [PubMed] [Google Scholar]
- 8.Hulmes D. J., Wess T. J., Prockop D. J., Fratzl P. (1995) Biophys. J. 68, 1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orgel J. P., Irving T. C., Miller A., Wess T. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9001–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perumal S., Antipova O., Orgel J. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2824–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farndale R. W., Lisman T., Bihan D., Hamaia S., Smerling C. S., Pugh N., Konitsiotis A., Leitinger B., de Groot P. G., Jarvis G. E., Raynal N. (2008) Biochem. Soc. Trans. 36, 241–250 [DOI] [PubMed] [Google Scholar]
- 12.Knight C. G., Morton L. F., Onley D. J., Peachey A. R., Messent A. J., Smethurst P. A., Tuckwell D. S., Farndale R. W., Barnes M. J. (1998) J. Biol. Chem. 273, 33287–33294 [DOI] [PubMed] [Google Scholar]
- 13.Farndale R. W., Siljander P. R., Onley D. J., Sundaresan P., Knight C. G., Barnes M. J. (2003) Biochem. Soc. Symp. 70, 81–94 [DOI] [PubMed] [Google Scholar]
- 14.Raynal N., Hamaia S. W., Siljander P. R., Maddox B., Peachey A. R., Fernandez R., Foley L. J., Slatter D. A., Jarvis G. E., Farndale R. W. (2006) J. Biol. Chem. 281, 3821–3831 [DOI] [PubMed] [Google Scholar]
- 15.Knight C. G., Morton L. F., Onley D. J., Peachey A. R., Ichinohe T., Okuma M., Farndale R. W., Barnes M. J. (1999) Cardiovasc. Res. 41, 450–457 [DOI] [PubMed] [Google Scholar]
- 16.Smethurst P. A., Onley D. J., Jarvis G. E., O'Connor M. N., Knight C. G., Herr A. B., Ouwehand W. H., Farndale R. W. (2007) J. Biol. Chem. 282, 1296–1304 [DOI] [PubMed] [Google Scholar]
- 17.Jarvis G. E., Raynal N., Langford J. P., Onley D. J., Andrews A., Smethurst P. A., Farndale R. W. (2008) Blood 111, 4986–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lisman T., Raynal N., Groeneveld D., Maddox B., Peachey A. R., Huizinga E. G., de Groot P. G., Farndale R. W. (2006) Blood 108, 3753–3756 [DOI] [PubMed] [Google Scholar]
- 19.Emsley J., King S. L., Bergelson J. M., Liddington R. C. (1997) J. Biol. Chem. 272, 28512–28517 [DOI] [PubMed] [Google Scholar]
- 20.Emsley J., Knight C. G., Farndale R. W., Barnes M. J., Liddington R. C. (2000) Cell 101, 47–56 [DOI] [PubMed] [Google Scholar]
- 21.Emsley J., Cruz M., Handin R., Liddington R. (1998) J. Biol. Chem. 273, 10396–10401 [DOI] [PubMed] [Google Scholar]
- 22.Huizinga E. G., Tsuji S., Romijn R. A., Schiphorst M. E., de Groot P. G., Sixma J. J., Gros P. (2002) Science 297, 1176–1179 [DOI] [PubMed] [Google Scholar]
- 23.Dumas J. J., Kumar R., McDonagh T., Sullivan F., Stahl M. L., Somers W. S., Mosyak L. (2004) J. Biol. Chem. 279, 23327–23334 [DOI] [PubMed] [Google Scholar]
- 24.Dumas J. J., Kumar R., Seehra J., Somers W. S., Mosyak L. (2003) Science 301, 222–226 [DOI] [PubMed] [Google Scholar]
- 25.Huizinga E. G., Martijn van der Plas R., Kroon J., Sixma J. J., Gros P. (1997) Structure 5, 1147–1156 [DOI] [PubMed] [Google Scholar]
- 26.Nishida N., Sumikawa H., Sakakura M., Shimba N., Takahashi H., Terasawa H., Suzuki E., Shimada I. (2003) Nat. Struct. Biol. 10, 53–58 [DOI] [PubMed] [Google Scholar]
- 27.Horii K., Kahn M. L., Herr A. B. (2006) Blood 108, 936–942 [DOI] [PubMed] [Google Scholar]
- 28.O'Connor M. N., Smethurst P. A., Farndale R. W., Ouwehand W. H. (2006) J. Thromb. Haemost. 4, 869–873 [DOI] [PubMed] [Google Scholar]
- 29.Smethurst P. A., Joutsi-Korhonen L., O'Connor M. N., Wilson E., Jennings N. S., Garner S. F., Zhang Y., Knight C. G., Dafforn T. R., Buckle A., IJsseldijk M. J., de Groot P. G., Watkins N. A., Farndale R. W., Ouwehand W. H. (2004) Blood 103, 903–911 [DOI] [PubMed] [Google Scholar]
- 30.Lecut C., Arocas V., Ulrichts H., Elbaz A., Villeval J. L., Lacapère J. J., Deckmyn H., Jandrot-Perrus M. (2004) J. Biol. Chem. 279, 52293–52299 [DOI] [PubMed] [Google Scholar]
- 31.Kunicki T. J., Cheli Y., Moroi M., Furihata K. (2005) Blood 106, 2744–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arthur J. F., Shen Y., Kahn M. L., Berndt M. C., Andrews R. K., Gardiner E. E. (2007) J. Biol. Chem. 282, 30434–30441 [DOI] [PubMed] [Google Scholar]
- 33.Berlanga O., Bori-Sanz T., James J. R., Frampton J., Davis S. J., Tomlinson M. G., Watson S. P. (2007) J. Thromb. Haemost. 5, 1026–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miura Y., Takahashi T., Jung S. M., Moroi M. (2002) J. Biol. Chem. 277, 46197–46204 [DOI] [PubMed] [Google Scholar]
- 35.Takagi J., Petre B. M., Walz T., Springer T. A. (2002) Cell 110, 599–611 [DOI] [PubMed] [Google Scholar]
- 36.Inoue O., Suzuki-Inoue K., Dean W. L., Frampton J., Watson S. P. (2003) J. Cell Biol. 160, 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H., Kahn M. L. (2003) Mol. Cell. Biol. 23, 4764–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasirer-Friede A., Kahn M. L., Shattil S. J. (2007) Immunol. Rev. 218, 247–264 [DOI] [PubMed] [Google Scholar]
- 39.Boylan B., Gao C., Rathore V., Gill J. C., Newman D. K., Newman P. J. (2008) Blood 112, 2780–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arthur J. F., Gardiner E. E., Matzaris M., Taylor S. G., Wijeyewickrema L., Ozaki Y., Kahn M. L., Andrews R. K., Berndt M. C. (2005) Thromb. Haemost. 93, 716–723 [DOI] [PubMed] [Google Scholar]
- 41.Farndale R. W., Slatter D. A., Siljander P. R., Jarvis G. E. (2007) J. Thromb. Haemost. 5, (Suppl. 1) 220–229 [DOI] [PubMed] [Google Scholar]
- 42.Giudici C., Raynal N., Wiedemann H., Cabral W. A., Marini J. C., Timpl R., Bächinger H. P., Farndale R. W., Sasaki T., Tenni R. (2008) J. Biol. Chem. 283, 19551–19560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hohenester E., Sasaki T., Giudici C., Farndale R. W., Bächinger H. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18273–18277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konitsiotis A. D., Raynal N., Bihan D., Hohenester E., Farndale R. W., Leitinger B. (2008) J. Biol. Chem. 283, 6861–6868 [DOI] [PubMed] [Google Scholar]
- 45.Terraube V., Pendu R., Baruch D., Gebbink M. F., Meyer D., Lenting P. J., Denis C. V. (2006) J. Thromb. Haemost. 4, 519–526 [DOI] [PubMed] [Google Scholar]
- 46.Fox J. E., Aggerbeck L. P., Berndt M. C. (1988) J. Biol. Chem. 263, 4882–4890 [PubMed] [Google Scholar]
- 47.Erat M. C., Slatter D. A., Lowe E. D., Millard C. J., Farndale R. W., Campbell I. D., Vakonakis I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4195–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.