Abstract
Increasing evidence suggests that the cytoplasmic tail of membrane type 1 matrix metalloproteinase (MT1-MMP) is subject to phos pho ryl a tion and that this modification may influence its enzymatic activity at the cell surface. In this study, phos pho ryl a ted MT1-MMP is detected using a phospho-specific antibody recognizing a protein kinase C consensus sequence (phospho-TXR), and a MT1-MMP tail peptide is phos pho ryl a ted by exogenous protein kinase C. To characterize the potential role of cytoplasmic residue Thr567 in these processes, mutants that mimic a state of either constitutive (T567E) or defective phos pho ryl a tion (T567A) were expressed and analyzed for their functional effects on MT1-MMP activity and cellular behavior. Phospho-mimetic mutants of Thr567 exhibit enhanced matrix invasion as well as more extensive growth within a three-dimensional type I collagen matrix. Together, these findings suggest that MT1-MMP surface action is regulated by phos pho ryl a tion at cytoplasmic tail residue Thr567 and that this modification plays a critical role in processes that are linked to tumor progression.
Largely composed of a mixture of collagens, laminins, and vitronectin, the extracellular matrix (ECM)2 serves as both a physical scaffold and a barrier against cell invasion. It has become increasingly evident that the structural condition of the ECM plays a unique role in regulating cell behavior. Proteolysis of integral components of the basement membrane disturbs the barrier provided by the ECM. Without physical restriction, cells invade the surrounding environment in an unregulated manner. The ability of matrix metalloproteinases (MMPs) to collectively degrade nearly all ECM constituents allows this class of enzymes to function in a diverse range of physiological processes (1, 2). Of the anchored MMPs, membrane type 1 matrix metalloproteinase (MT1-MMP) was the first to be discovered and has been most thoroughly characterized. Unlike soluble MMPs, MT1-MMP has a stretch of hydrophobic amino acids that traverse the cell membrane, followed by a short cytoplasmic tail composed of 20 amino acids (3). The advantage of cell surface localization is 2-fold. Surface restriction allows MT1-MMP to modify the immediate pericellular environment, overcoming physical constraints imposed by the ECM (2). Localization at the cell surface also places tethered MMPs in an optimal position to function at invadapodia, highly specialized areas of the cell membrane that form during focalized cell invasion (4). Although information regarding the role of the cytoplasmic tail is relatively limited (5, 6), this domain may function as a bridge to the intracellular machinery.
MT1-MMP has an essential role in matrix remodeling during physiological processes (7, 8). Conversely, its enzymatic activity is key to acquiring a metastatic phenotype in a variety of tumor cells, including lung, colon, breast, and cervical carcinomas (2, 9–11). The ability to alter the physical structure of the pericellular environment, while triggering the activation and modification of several cell surface proteins, identifies a central role for MT1-MMP in influencing cellular behavior (12). In return, stringent cellular regulation of MT1-MMP enzymatic activity is necessary to prevent aberrant proteolysis.
Increasing evidence suggests that the cytoplasmic tail of MT1-MMP may regulate its activity at the cell surface. It has been demonstrated that MT1-MMP is internalized from the cell surface and that this process requires the presence of the cytoplasmic domain (5, 6). Tail truncation restricts MT1-MMP to the cell surface, suggesting that this domain contains sequence(s) that either mediate internalization or are required for physical interaction with another protein that facilitates its internalization (5, 6). The mechanism regulating this process has yet to be determined. Interestingly, both invasion and migration are down-regulated in cells where MT1-MMP is restricted to the cell surface (5, 6). These data suggest a correlation between internalization and matrix turnover, where MT1-MMP activity is either abrogated or enhanced under appropriate stimuli.
Reversible phosphorylation is widely recognized as a key post-translational modification that regulates protein function. The cytoplasmic domain of MT1-MMP has three potential phosphorylation sites: Thr567, Tyr573, and Ser577. Recent work by Nyalendo et al. (13) indicates that MT1-MMP is phosphorylated at tyrosine residue Tyr573, and that this modification influences cell migration. Several surface proteins are regulated by phosphorylation at multiple residues. In the MT1-MMP cytoplasmic tail, Thr567 has homology with the consensus sequence for both protein kinase C (TXR) and ERK1/2 (XTP) (14), suggesting the possibility that active MT1-MMP might also be regulated through phosphorylation of this cytoplasmic tail residue. In the present study, we report that MT1-MMP bears a threonine phosphorylation site in its cytoplasmic tail and that this modification plays an important role in regulating several aspects of carcinoma cell behavior, including invasion and three-dimensional growth.
EXPERIMENTAL PROCEDURES
Materials
Anti-FLAG monoclonal M2 antibody and peroxidase-conjugated secondary antibodies were purchased from Sigma. Anti-MT1-MMP (hinge or catalytic domain) antibodies were purchased from Chemicon or Abcam. Antibodies directed against phosphothreonine were obtained from Cell Signaling Technologies (Danvers, MA) or Abcam. Calyculin A (serine-threonine phosphatase inhibitor) was purchased from Cell Signaling Technologies. Super Signal-enhanced chemiluminescence reagents were purchased from Pierce. Rat tail collagen type I and Matrigel Basement Membrane Matrix were purchased from BD Biosciences. Gold chloride was purchased from Fisher. TIMP-2 (tissue inhibitor of metalloproteinase-2) was graciously provided by Dr. Rafael Fridman (Wayne State University, Detroit, MI).
Cell Culture
The breast cancer cell line MDA MB 231 was kindly provided by Dr. V. Craig Jordan (Fox Chase Cancer Center, Philadelphia, PA) and was maintained in minimal essential medium containing 10% fetal bovine serum. Cell culture media and reagents were purchased from Mediatech (Herndon, VA).
DNA Constructs and Generation of Stable Cell Lines
The human MT1-MMP cDNA with C-terminal FLAG tag (DYKDDDDK) was kindly provided by Dr. Duanqing Pei (University of Minnesota, Minneapolis, MN). The open reading frame sequences of MT1-MMP cDNA were TA-subcloned into eukaryotic expression vector pCR3.1-Uni (Invitrogen) to obtain the wild type construct. To facilitate different studies, the wild type construct was further FLAG-tagged in the stalk region, as described (15). Subsequently, the T567A and T567E point mutations were generated on the wild type cDNA and on the tagged constructs using QuikChange (Stratagene). Transfection of cells was done using FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions, and stable cell lines were generated using G418 selection. These cells were selected because they express endogenous MT1-MMP, insuring proper activation of the proMT1-MMP zymogen, as well as key components of the pro-MMP-2 activation system (i.e. pro-MMP-2 and TIMP-2), thereby facilitating the assay of potential functional changes brought about by phosphorylation. Stable cell lines were generated using G418 selection, and cells were sorted by fluorescence-activated cell sorting using an antibody against the FLAG epitope tag (M2). To ensure equivalent expression levels, cells were sorted using anti-FLAG M2 every 3–5 passages. All experiments were performed with freshly sorted cell populations.
Kinase Assays
Analysis of PKCδ-dependent phosphorylation of a purified synthetic MT1-MMP tail peptide was performed as described previously (16). Briefly, substrate (MT1-MMP tail peptide or histone H1 (45 μg/ml)) was incubated with recombinant PKCδ (3.5 nm) in 45 μm α-glycerol phosphate buffer, pH 7.0, containing 0.9 mm dithiothreitol, 9.0 mm MgCl2, 0.45 mm CaCl2, and 4.5 μm ATP in the presence or absence of the activators phosphatidylserine (45 μg/ml) and diacylglycerol (1.6 μg/ml) (as indicated). Reactions were initiated by the addition of 5 μCi of [γ-32P]ATP and were terminated by the addition of 50 μl of 3× Laemmli SDS stop solution and thermal denaturation at 100 °C for 5 min (16) prior to separation on a 12% SDS-polyacrylamide gel and autoradiography.
Gelatin Zymography
Gelatinase activities in conditioned media were determined using SDS-polyacrylamide gel electrophoresis zymography. Conditioned media (20 μl) from an equivalent number of cells were electrophoresed without reduction on SDS-polyacrylamide gels prepared with 9% acrylamide containing 0.1% gelatin. SDS was removed through a 1-h incubation in 2.5% Triton X-100, and gels were incubated in 20 mm glycine, 10 mm CaCl2, 1 μm ZnCl2 (pH 8.3) at 37 °C for 24 h prior to staining for gelatin with Coomassie Blue. Enzyme activity was visualized as zones of gelatin clearance within the gels.
MT1-MMP Immunoprecipitation and Immunoblotting
For Western blotting of whole cell lysates, cells were lysed using 50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, and the protein concentration of lysates was analyzed using the Bio-Rad DC detection kit and bovine albumin standards. Cell lysates (50 μg) were electrophoresed on 9% SDS-polyacrylamide gels, transferred to polyvinylidene difluoride membrane, and blocked with 3% bovine serum albumin in 50 mm Trizma (Tris base) (pH 7.5), 300 mm NaCl, 0.2% Tween 20 (TBST). Membranes were incubated for 1 h at room temperature with a 1:1000 dilution of FLAG M2 monoclonal antibody in 3% bovine serum albumin/TBST. Immunoreactive bands were visualized with a peroxidase-conjugated anti-rabbit IgG (1:4000 in 3% bovine serum albumin/TBST) and enhanced chemiluminescence. For immunoprecipitation analyses, cells were serum-starved in the presence of the broad spectrum MMP inhibitor GM6001 (Chemicon, Temecula, CA), switched to serum containing medium for 3 h, collected with lysis buffer (above), and subjected to immunoprecipitation using anti-MT1-MMP (hinge antibody, 1:4000 dilution) and protein G beads. Immunoprecipitates were electrophoresed on 9% polyacrylamide gels and subjected to Western blotting using anti-MT1-MMP catalytic domain (1:4000) or anti-phospho-TXR antibodies (1:1000) followed by ECL detection, as described above. Alternatively, cells were treated with the serine-threonine phosphatase inhibitor Calyculin A (20 nm, 10 min), collected as above, and subjected to immunoprecipitation using anti-phospho-Thr antibody (2 μg). Immunoprecipitates were then subjected to Western blotting with anti-FLAG M2.
Biotin Surface Labeling
To label cell surface proteins, cells were incubated with the non-cell-permeable biotin analogue Sulfo-NHS-LCBiotin (1 mg/ml) for 25 min on ice, washed three times in cold 100 mmol/liter glycine in phosphate-buffered saline and once in cold phosphate-buffered saline, and lysed in modified radioimmunoprecipitation assay buffer (50 mmol/liter Tris (pH 7.5), 150 mmol/liter NaCl, 1% Triton X-100, 5 mmol/liter EDTA). Equivalent amounts (50–70 μg) of lysate, as determined by the Bio-Rad protein assay, were incubated with 25–30 μl NeutrAvidin protein overnight at 4 °C with gentle agitation. The NeutrAvidin biotin-labeled lysate complex was then washed five times in modified radioimmunoprecipitation assay lysis buffer, resuspended in modified radioimmunoprecipitation assay lysis buffer plus sample dilution buffer with 2-mercaptoethanol, and prepared for Western blot analysis.
Cellular Migration
To assess cell motility, migration through transwell membranes coated with a thin layer of collagen (100 μg/ml, 37 °C, 1 h) on both the upper and lower surfaces was evaluated as described below for invasion using an incubation time of 18 h. To conduct in vitro scratch wound assays, cells were plated in 8-well plates, cultured to confluence, and serum-starved overnight. Two scratch wounds were made in each well using a micropipette tip. Two points were randomly selected, marked for each scratch, and photographed using a digital camera at 0, 24, and 48 h. Five relative measurements were taken for each of the four points for each experimental condition using the MetaMorph Imaging System (Universal Imaging Corp., Downington, PA). These resulting five measurements for each point were averaged and then normalized based on the initial measurement for that point at 0 h. The four normalized values were then averaged for each experimental condition. The data include results from three separate assays.
To measure haptotactic migration, a colloidal gold migration assay was utilized. The collagen-colloidal gold coating was prepared as previously described (17, 18) with minor modification. Briefly, glass coverslips (22 mm2) in a 6-well plate were coated with rat tail type I collagen (BD Biosciences) at a concentration of 5 μg/cm2 by slowly evaporating 2 ml of a 12 μg/ml collagen solution in 0.02 n acetic acid at room temperature. The colloidal gold particles were prepared as follows. Distilled water (11.3 ml) was combined with 6 ml of 36.5 mm Na2CO3 and 1.5 ml of 17.4 mm AuCl4H (Fisher). The solution was heated with a burner just to boiling, and 1.8 ml of 0.1% formaldehyde (diluted from 36.5–38% formalin) was added. In order to prevent denaturation of collagen, the solution was then cooled to below 70 °C. The cooled solution was layered over collagen-coated coverslips (2 ml each) and incubated at room temperature for 1 h to allow absorption of gold particles to the coated collagen. The unabsorbed gold salts were carefully rinsed off with culture medium, preventing drying of the coverslip. 1 × 103 cells were plated onto the coated coverslip and allowed to migrate for 24–48 h. The coverslips were then rinsed carefully with cold phosphate-buffered saline on ice and fix with 3.7% formaldehyde on ice for 10 min. The coverslips were briefly rinsed with phosphate-buffered saline following distilled water and mounted on slides with gelvatol. The images of phagokinetic track were acquired by dark field with a Nikon D100 camera mounted on a Nikon Microphot-FXA microscope. The images were then analyzed by Adobe Photoshop CS.
Cellular Invasion
Type I collagen was dissolved in 0.5 m acetic acid at a concentration of 2 mg/ml. For invasion experiments, the collagen stock was neutralized with 100 mm Na2CO3 (pH 9.6) to a final concentration of 0.4 mg/ml. Transwell inserts (0.8 μm; BD Biosciences) were coated on the underside with 500 μl of collagen diluted to a concentration of 100 μg/ml at 37 °C for 1 h. Collagen gels were prepared in the inner well by adding 50 μl of collagen (20 μg) at room temperature and allowing gels to air-dry overnight. Collagen-coated inserts were then washed with minimum essential medium three times to remove salts and used immediately. Cells were trypsinized and washed with serum-free medium, and 1 × 105 cells were added to the inner invasion chamber in a volume of 200 μl. The outer wells contained 400 μl of culture medium. Cells were allowed to invade for 24–48 h as indicated; non-invading cells were removed from inner wells using a cotton swab, and invading cells adherent to the bottom of the membrane were fixed and stained using a Diff-Quick staining kit (DADE AG, Miami, FL). Invading cells were enumerated by dividing membranes into four quadrants and counting the number of cells in three distinct areas for each quadrant under a ×10 objective using an ocular micrometer. Assays were performed in triplicate.
For Matrigel invasion assays, the inner well of transwell inserts (0.8 μm; BD Biosciences) were coated with 100 μl of Matrigel Basement Membrane Matrix at a concentration of 1 mg/ml and allowed to permeabilize at room temperature for 1 h. Unpermeabilized liquid was removed, and filters were washed with serum-free medium and allowed to air dry. Cells were trypsinized and washed with serum-free medium, and 3.75 × 105 cells were added to the inner invasion chamber in a volume of 500 μl. The outer well contained 750 μl of cell culture medium. Cells were allowed to invade for 18 h, as indicated; non-invading cells were removed from inner wells using a cotton swab, and invading cells adherent to the bottom of membrane were fixed and stained using the Diff-Quick staining kit (DADE AG). Invading cells were enumerated by dividing membranes into four quadrants and counting the number of cells in three distinct areas for each quadrant under a ×10 objective using an ocular micrometer.
Three-dimensional Collagen Culture
Three-dimensional cultures were prepared by diluting type I rat tail collagen (BD Biosciences) with 10× minimum Eagle's medium (Invitrogen) to a final concentration of 1.5 mg/ml. Cells (5 × 104) were added to the collagen mixture prior to solidification. In parallel control experiments, the MMP inhibitor TIMP-2 was included at a final concentration of 5 μg/ml (19). Following growth for 5–8 days in three-dimensional collagen, gels were dissolved using bacterial collagenase (2 mg/ml; Worthington), and cell number was evaluated by hemocytometry, as described (19). Experiments were repeated in triplicate. In control experiments, solidified collagen gels were prepared as described above, with 5 × 104 cells being added to the gel surface following collagen solidification.
RESULTS
MT1-MMP Is Phosphorylated on Cytoplasmic Thr567
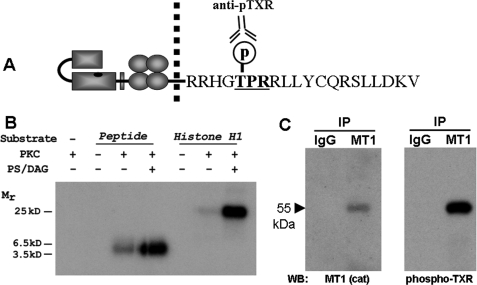
To investigate the hypothesis that MT1-MMP undergoes phosphorylation at residue Thr567 (Fig. 1A), in vitro kinase assays were conducted using a synthetic peptide containing the MT1-MMP tail sequence as substrate and recombinant PKCδ. Tail peptide was incubated in the presence or absence of purified PKC, and reactions were initiated in the presence or absence of the PKC-activating cofactors diacylglycerol and phosphatidylserine (16). PKC-stimulated phosphorylation of the MT1-MMP tail peptide was readily noted on autoradiograms (Fig. 1B), whereas 32P incorporation was not observed in the absence of PKC and was greatly diminished in the absence of PKC activators diacylglycerol and phosphatidylserine. Histone H1, a recognized substrate for PKC, was used in these experiments as a positive control for 32P incorporation. To further investigate the hypothesis that MT1-MMP undergoes phosphorylation at residue Thr567, MT1-MMP was immunoprecipitated from whole cell lysates using an antibody recognizing the hinge domain of MT1-MMP, electrophoresed, electroblotted, and probed using an antibody specific to the phosphorylated PKC recognition sequence, phospho-TXR (Fig. 1A). As shown in Fig. 1C, the phosphothreonine antibody recognized a 55-kDa species co-migrating with active MT1-MMP. Taken together, these studies suggest that MT1-MMP can be phosphorylated at residue Thr567 and that this event can occur in a PKC-dependent manner.
FIGURE 1.
Analysis of MT1-MMP cytoplasmic tail residue Thr567. A, schematic of the MT1-MMP cytoplasmic tail sequence showing the location of Thr567 within the PKC consensus recognition sequence TXR. B, in vitro kinase assays were conducted using a synthetic peptide containing the MT1-MMP tail sequence as substrate and recombinant PKCδ. Reactions were carried out in the presence or absence of purified PKC and were initiated in the presence or absence of diacylglycerol (DAG) and phosphatidylserine (PS), as indicated. Polypeptides were resolved by electrophoresis and analyzed by autoradiography. C, MT1-MMP was immunoprecipitated from whole cell lysates using an antibody recognizing the hinge domain of MT1-MMP. Following electrophoresis and electroblotting, immunoprecipitate (IP) blots were probed with antibody directed against the MT1-MMP catalytic domain (left) or anti-phospho-TXR (right).
Expression of MT1-MMP Thr567 Mutants in Human Breast Carcinoma Cells
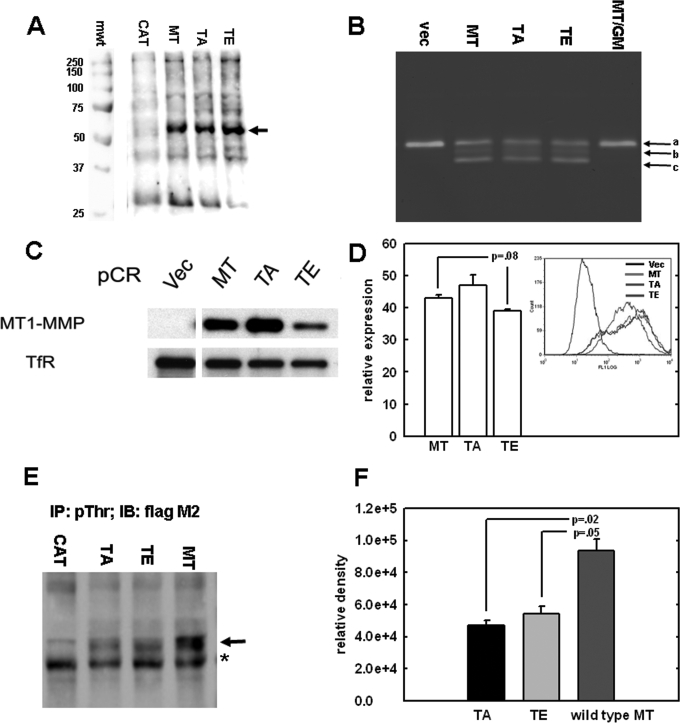
Based on the observation that MT1-MMP can be phosphorylated at Thr567, studies were undertaken to characterize the effect of this modification on cell behavior. To date, studies investigating the functional role of the cytoplasmic tail have primarily involved deletion of the entire domain. This approach makes it difficult to determine whether the observed effects are due to the absence of specific sequences in the cytoplasmic domain or whether functional phenotypes are a pleiotropic effect of truncation. In order to investigate the potential functional consequence(s) of Thr567 modification, a more specific approach was developed. Using site directed mutagenesis, Thr567 was mutated to glutamic acid (T567E) to mimic constitutive phosphorylation or alanine (T567A) to represent a phospho-defective mutation. Wild type MT1-MMP (MT in Fig. 2) and cells transfected with vector alone (CAT) were used as controls. Constructs were transfected into MDA-MB231 breast carcinoma cells, and stable clones were obtained by G418 selection, fluorescence-activated cell sorting, and immunoblotting for MT1-MMP expression. Analysis of whole cell lysates from wild type, T567A, and T567E demonstrated equivalent MT1-MMP expression (Fig. 2A). Direct activation of pro-MMP-2 (progelatinase-A) is a classical function of MT1-MMP (3, 20–23). MMP-2 is responsible for degrading type IV collagen, a major component of the basement membrane (3). Gelatin zymography was used to determine the effect of Thr567 mutation on the ability of MT1-MMP to catalyze pro-MMP-2 activation. Relative to wild type cells, no reproducible change in pro-MMP-2 activation was demonstrated by T567A or T567E mutant cell lines (Fig. 2B), indicating that Thr567 mutation does not alter the ability of MT1-MMP to act as an upstream effector of MMP-2 activity. Since previous studies indicate that the cytoplasmic tail alters the surface presentation of active MT1-MMP by regulating its internalization from the cell surface (5, 6), studies were conducted to determine whether alteration of residue Thr567 modulates basal MT1-MMP surface presentation. Surface labeling (Fig. 2C) and fluorescence-activated cell sorting analysis (Fig. 2D) indicate surface presentation of wild type and mutant MT1-MMP, with the T567E mutant exhibiting relatively lower surface expression compared with wild type MT1-MMP. In control experiments, cells expressing wild type MT1-MMP or the T567E and T567A mutants were immunoprecipitated with a phospho-Thr antibody, followed by Western blotting with FLAG M2 antibody to detect only exogenous epitope-tagged MT1-MMP (Fig. 2, E and F). These data show a significant decrease in detection of phospho-Thr-modified MT1-MMP in cells expressing mutant proteins.
FIGURE 2.
Analysis of MT1-MMP Thr567 mutants. A, expression of MT1-MMP constructs. MDA-MB-231 cells were transfected with FLAG epitope-tagged constructs containing either wild type MT1-MMP (MT), empty vector (CAT), or mutations at cytoplasmic residue Thr567 that either mimic (TE) or prevent (TA) phosphorylation. Cells were lysed, and lysates were subjected to Western blotting using anti-FLAG M2 antibodies as described under “Experimental Procedures.” mwt, molecular weight standards in kDa. B, gelatin zymography was performed on serum-free conditioned medium from cells transfected with empty vector (vec), wild type MT1-MMP (MT), or the T567A and T567E mutants (TA and TE, respectively). Controls included cells transfected with wild type MT1-MMP cultured in the presence of the broad spectrum MMP inhibitor GM6001 (MT/GM). Arrows a–c, the migration positions of pro-MMP-2, intermediate MMP-2, and active MMP-2, respectively. C, surface expression of MT1-MMP constructs. Stable cell lines were generated that display comparable levels of MT1-MMP expression in whole cell lysates. To evaluate surface presentation, surface proteins were first biotinylated with a non-cell-permeable biotin analog and lysed, and lysates were precipitated with NeutrAvidin to isolate surface-associated proteins, followed by analysis for MT1-MMP by Western blotting using anti-FLAG M2, as described under “Experimental Procedures.” D, fluorescence-activated cell sorting analysis was conducted using monoclonal M2 FLAG antibody and AlexaTM 488-conjugated anti-mouse IgG secondary antibody to quantify the amount of cell surface MT1-MMP as described under “Experimental Procedures.” E, cells transfected with wild type or mutant MT1-MMP were treated with the serine-threonine phosphatase inhibitor Calyculin A (20 nm, 10 min), and lysates were subjected to immunoprecipitation using anti-phospho-Thr antibody (2 μg). Immunoprecipitates were then subjected to Western blotting with anti-FLAG M2, as described under “Experimental Procedures.” The arrow denotes the migration position of MT1-MMP. *, nonspecific binding. F, densitometric quantitation of triplicate repeat experiments as in E.
Thr567 Modification and Cell Migration
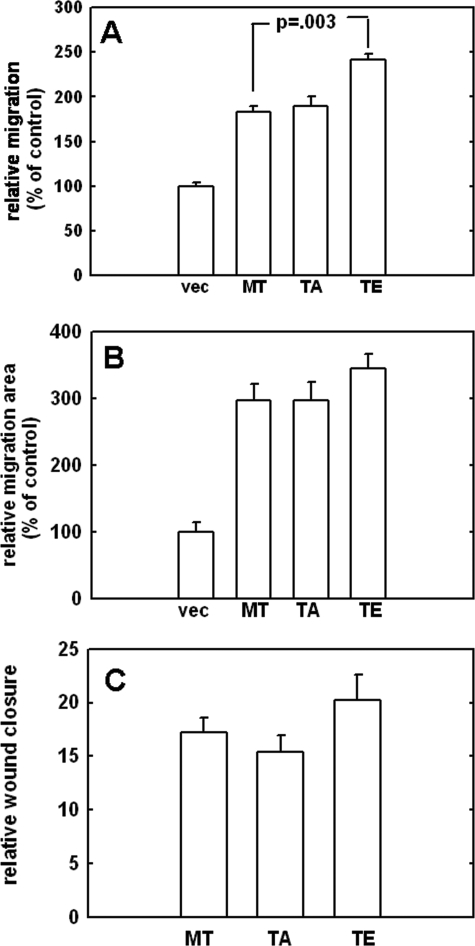
Acquisition of MT1-MMP expression stimulates migration in a variety of cell types and requires focalized degradation of ECM components. In addition to modifying the ECM, MT1-MMP promotes migration through interaction with and modification of various cell adhesion molecules (24, 25). A variety of assays were used to determine whether Thr567 mutation affects the ability of MT1-MMP to promote cell migration. A Boyden chamber migration assay was first used to investigate possible phenotypic differences. Relative to vector controls, all MT1-MMP-expressing cells exhibited enhanced migration (Fig. 3A). Further, the T567E mutant exhibited a small but statistically significant increase in chemotactic migration relative to wild type MT1-MMP (p = 0.003) (Fig. 3A). Collagen-driven single cell motility was then analyzed by a colloidal gold assay (17, 26), wherein glass coverslips were coated with colloidal gold, overlaid with type I collagen, and seeded as single cells at low density, followed by quantitation of the phagokinetic tracks made by migrating cells. Although expression of MT1-MMP increased cell migration relative to vector controls (vec), no significant difference in motility was observed between wild type, T567A, and T567E cells (Fig. 3B). Previous studies have shown that the cytoplasmic tail is required to promote motility in colloidal gold assays (6). Our findings indicate that although this domain is required to promote motility in this particular assay, this function is not influenced by Thr567 status. Another important physiological function of MT1-MMP is its ability to stimulate migration during wound closure. Assays were conducted to investigate the potential role of Thr567 mutation in cell migration within this context. As demonstrated in Fig. 3C, there was no significant difference in the rate of wound closure between wild type, T567A, and T567E mutants.
FIGURE 3.
Analysis of cell migration. A, the effect of Thr567 mutation on chemotactic migration was assessed using a Boyden chamber migration assay. The underside of the chamber was coated with type I collagen (10 μg/ml). Cells were serum-starved overnight, incubated in Boyden chambers, and allowed to migrate for 3.5 h at 37 °C prior to enumeration of migrating cells. Results are the averages of three independent experiments and are shown as percentages relative to vector controls (designated 100%). B, the colloidal gold migration assay was used to evaluate collagen-driven single cell motility. Cells were serum-starved and allowed to migrate for 12 h prior to quantitation of phagokinetic tracks by computer-assisted image analysis. Results are the averages of three independent experiments and are shown as percentages relative to vector controls (designated 100%). C, the effect of Thr567 mutation on experimental wound closure was assessed using a scratch wound assay. Cells were plated to confluence and serum-starved overnight. Scratches were mechanically introduced to the monolayer using a micropipette tip. Cells were incubated at 37 °C for 18 h. Results are expressed as relative wound closure compared with wound width at time 0.
Thr567 Modification Enhances Cell Invasion
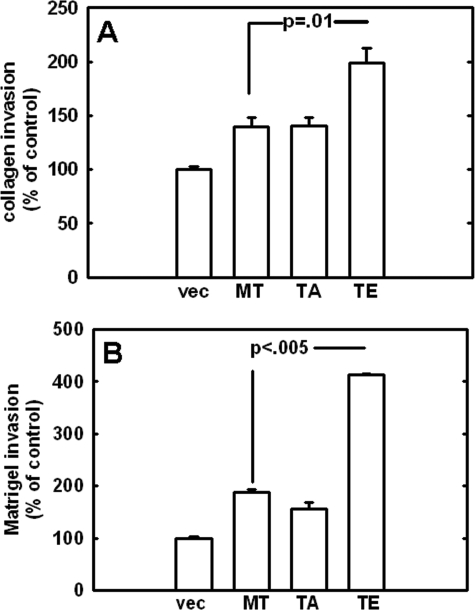
MT1-MMP enzymatic activity during cellular invasion is critical to establishing the metastatic potential of a carcinoma (19). Degradation of key matrix components is a prerequisite for invasion of many tissues and allows cells to invade neighboring tissues unchecked. To assess the potential effect of Thr567 mutation on invasion efficiency, the ability of MT1-MMP mutants to penetrate type I collagen gels was first assessed, since previous studies have demonstrated MT1-MMP to be a potent interstitial collagenase (7, 19). Relative to vector controls, all MT1-MMP-expressing cells exhibited enhanced ability to invade type I collagen gels, with T567E cells as the most invasive (Fig. 4A). Because the ability to traverse the basement membrane is often critical to metastasis, the ability of MT1-MMP mutants to invade Matrigel, a basement membrane extract, was investigated. As with type I collagen, T567E exhibited statistically significant increased invasion through a Matrigel barrier (Fig. 4B). Collectively, the observed phenotypic differences suggest that the phospho-mimetic modification at Thr567 may elicit an increased invasive response.
FIGURE 4.
Analysis of cell invasion. The effect of Thr567 mutation on cell invasion was analyzed using Boyden chambers overlaid with gels composed of type I collagen (A) or Matrigel (B), as described under “Experimental Procedures.” Cells were serum-starved overnight prior to incubation in coated Boyden chambers for 18 h at 37 °C and enumeration of invading cells. Results are the averages of three independent experiments and are shown as percentages relative to vector controls (designated 100%).
Thr567 Modification Enhances Three-dimensional Growth
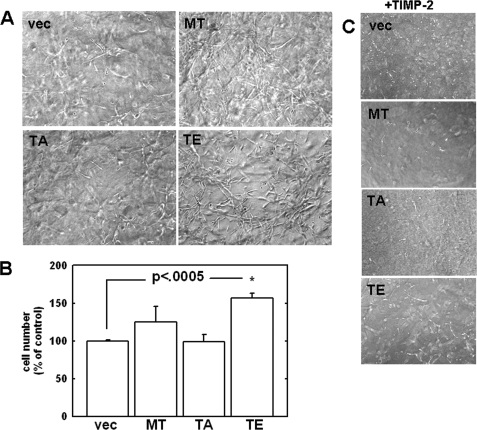
Expression of MT1-MMP confers a distinct growth advantage to tumor cells within a three-dimensional matrix both in vitro and in vivo relative to control cells (2, 19). Although these studies highlight MT1-MMP-dependent type I collagenolysis as a requirement for growth in this environment, the specific domain(s) responsible for this effect were not reported. To determine the potential role of cytoplasmic tail modification on MT1-MMP-mediated three-dimensional growth, cells were seeded within three-dimensional type I collagen gels and cultured for 8 days prior to dissolution of the gel and quantitation of cell proliferation (19). Because TIMP-2 acts as an inhibitor of MT1-MMP activity (19), control experiments were conducted in the presence of TIMP-2. Because MDA-MB-231 cells have a mesenchymal-like phenotype, they do not grow as multicellular aggregates. However, cells expressing wild type MT1-MMP exhibit reproducibly enhanced proliferation within a three-dimensional matrix relative to untransfected control cells (Fig. 5A), growing as loose, elongated clusters. Furthermore, the T567E mutant exhibits a more aggressive proliferation within three-dimensional collagen gels (Fig. 5, A and B). The enhanced three-dimensional growth is suppressed in control matrices containing TIMP-2, confirming the requirement of MT1-MMP enzymatic activity (Fig. 5C). In additional control experiments, analysis of cell growth atop planar two-dimensional collagen surfaces illustrates similar growth rates for wild type and T567E and T567A mutants (data not shown). These data suggest that the phosphorylation state of cytoplasmic Thr567 (as represented by T567E) may regulate growth within a three-dimensional environment and thereby confer a distinct pathological advantage to more invasive cells that have penetrated into the collagen-rich extracellular microenvironment.
FIGURE 5.
Analysis of growth in three-dimensional collagen gels. A, three-dimensional cultures were prepared by diluting type I rat tail collagen with complete medium to a final concentration of 1.5 mg/ml. A, cells (5 × 104) were seeded into the collagen mixture prior to solidification. Gels were allowed to incubate at 37 °C for 8 days and were photographed using phase-contrast microscopy. B, quantitation of proliferation within three-dimensional collagen gels was performed by dissolution of the gels using bacterial collagenase, followed by hemocytometry, as described under “Experimental Procedures” (19). C, in control experiments, the MMP inhibitor TIMP-2 was also included at a final concentration of 5 μg/ml (right), and the experiment was performed as in A.
DISCUSSION
Studies involving the epidermal growth factor receptor indicate that the cytoplasmic domain contains a PKC consensus sequence at Thr654 (28–30). Phosphorylation at this site inhibits mitogenic signal transduction from epidermal growth factor receptor (29) and prevents receptor internalization by restricting the receptor to cell surface caveolae (31). Similar studies show that additional membrane proteins, including the human insulin receptor (32) and IL-2 receptor (33, 34), have cytoplasmic domains that are substrates for PKC. In a similar fashion, threonine phosphorylation could potentially modulate MT1-MMP trafficking and cell function.
Although the enzymatic activity of MT1-MMP has a major role in both physiological and pathological processes, events regulating its activity are not yet fully understood. A growing body of work demonstrates that acquisition of MT1-MMP expression alone promotes a metastatic phenotype, resulting in enhanced cell migration (24, 34, 35), invasion of the basement membrane (3, 36), and three-dimensional growth (27). Increasing evidence suggests that the cytoplasmic tail may function as a regulatory domain controlling these processes. This region modulates endocytosis of the enzyme, thereby regulating the level of active MT1-MMP at the cell surface (5, 6). As a functional consequence, tail-truncated mutants were found to be less efficient at cell migration and invasion (6). More recent work indicates that tyrosine phosphorylation at the cytoplasmic tail plays a functional role in cell migration, since mutants that cannot undergo phosphorylation at this residue are less efficient at promoting cell migration in response to chemoattractants (13). The mechanism(s) underlying the ability of the tail to function in this capacity is unclear.
Results yielded in the present studies underscore a pivotal role for the cytoplasmic tail in regulating MT1-MMP function. Although regulatory factors that control this event are currently unknown, our findings support the occurrence of phosphorylation at Thr567 in the MT1-MMP cytoplasmic tail. Characterization of Thr567 mutants that mimic constitutive phosphorylation (T567E) demonstrate that this specific modification functions in the regulation of several cellular events tied to MT1-MMP activity. In particular, the current data suggest that, although the surface presentation is somewhat diminished, the T567E mutant displays enhanced collagenolytic activity relative to wild type MT1-MMP. Although elucidation of the precise mechanism for increased collagenolysis requires further experimentation, it is possible that T567E mutation (and, by extension, Thr567 phosphorylation) may enhance catalytic efficiency of collagen cleavage through a conformational alteration of the proteinase. Alternative mechanisms are also feasible. For example, T567E mutation may result in altered surface retention or retention in a specific membrane microdomain in which collagenolytic activity is more efficient. Other MT1-MMP-trafficking mechanisms may also be affected by cytoplasmic tail alteration.
The T567E mutant exhibits enhanced invasion through a basement membrane extract and type I collagen relative to control, wild type, and T567A cell lines. This phenotype is of pathological significance, because basement membrane penetration, followed by invasion of the underlying stroma, facilitates metastasis and disease progression.
Interestingly, although there was no significant difference between cell lines in wound closure or single cell random motility, as detected by migration on colloidal gold, the presence of a phospho-mimetic at Thr567 does confer an advantage in chemotactic migration. Successful motility in this assay requires cytoskeletal polarization at the leading edge of the cell and three-dimensional invasion through the filter in response to chemical stimuli (collagen on the underside of the filter in this example). In contrast, colloidal gold and wound closure assays assess two-dimensional migration. In vivo, the physical localization of MT1-MMP is critical during cell migration and invasion through the surrounding matrix. Although the mechanism underlying the superior function of T567E in chemotactic migration and ECM invasion is unclear, it is possible that phosphorylation at this residue may alter the ability of MT1-MMP to localize at the invading front of the cell. This may occur through a direct mechanism or by indirectly interfering with the ability of MT1-MMP to spatially interact with other cell molecules that function in lamellipodia formation.
The cell adhesion molecule CD44 plays a direct role in directing MT1-MMP localization to lamellipodia (37). During motility, CD44 localizes to the leading edge of the cell by binding the cytoskeletal component F actin with its cytoplasmic domain (38). CD44 binds the hemopexin domain of MT1-MMP through its stem, indirectly linking MT1-MMP to the cell cytoskeleton. Disruption of this association inhibits the interaction between the two molecules and diminishes the ability of MT1-MMP to localize to lamellipodia (37). Although improperly localized MT1-MMP retains the ability to activate MMP-2, it is unable to promote matrix turnover or cell invasion (39). The implication of this is that regulated localization at invadapodia is tightly linked to MT1-MMP function. It is plausible that Thr567 phosphorylation may alter the ability of the protein to spatially interact with the invasion machinery during cytoskeletal reorganization of the leading edge. Alternately, the phenotypic differences demonstrated by T567E and T567A mutants may be due to changes in MT1-MMP trafficking and membrane microdomain localization, similar to data showing that PKC-induced phosphorylation of epidermal growth factor receptor Thr654 modulates mitogenic signaling by regulating both internalization and localization to caveolae (29, 31). A corresponding alteration in trafficking and cellular locale would influence activities that require proper localization of MT1-MMP, including migration, invasion, and protein interactions.
Perhaps the most interesting phenotypic finding is the ability of the T567E phospho-mimetic mutation to enhance three-dimensional growth within a collagen matrix. Cancer progression is characterized by increased growth atop the basement membrane, followed by invasion and proliferation within the three-dimensional environment of the stromal matrix (27, 40, 41). Composed largely of type I collagen or fibrin (40, 41), invasive carcinomas must demonstrate a distinct growth advantage within the constrained environment provided by the stroma. It has been established that MT1-MMP expression is required for three-dimensional growth (27), and we have previously demonstrated that collagen invasion by MDA-MB-231 cells is an MT1-MMP-dependent process (42). Although these studies identify MT1-MMP-dependent type I collagenolysis as a requirement for this observed growth advantage, the specific domain(s) responsible for this action have yet to be delineated. Although T567E cells demonstrate enhanced proliferation in these assays, analysis of cell growth atop collagen surfaces illustrates similar growth rates for wild type, T567E, and T567A mutants. These data suggest that the phosphorylation state at cytoplasmic tail residue Thr567 may function as a molecular switch to either enhance or inhibit growth within a matrix. Lack of a phenotypic difference between mutants atop collagen surfaces suggests that the advantage afforded by this modification is specific to more invasive cells that are functioning within a three-dimensional environment, similar to that encountered in the stroma.
In addition to modulating internalization (5, 6), the results presented herein indicate that the cytoplasmic tail may regulate several aspects of MT1-MMP enzymatic function through phosphorylation of the unique Thr567 residue. These findings suggest that strategies to block Thr567 phosphorylation may subdue cellular characteristics associated with more aggressive tumors, including enhanced invasion of and proliferation within dense collagen gels, and may represent an alternative strategy to peptide hydroxamate-based MMP inhibitors.
Acknowledgment
We thank Dr. Evelyn Maizels (Northwestern University) for assistance with phosphopeptide analysis.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grant RO1 CA86984 (to M. S. S.), RO1 CA109545 (to M. S. S.), and CA86984-07S1 (to M. S. S. and N. M. M.).
- ECM
- extracellular matrix
- MMP
- matrix metalloproteinase
- MT1-MMP
- membrane type 1 matrix metalloproteinase
- PKC
- protein kinase C.
REFERENCES
- 1.Sternlicht M. D., Werb Z. (2001) Annu. Rev. Cell Dev. Biol. 17, 463–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabeh F., Ota I., Holmbeck K., Birkedal-Hansen H., Soloway P., Balbin M., Lopez-Otin C., Shapiro S., Inada M., Krane S., Allen E., Chung D., Weiss S. J. (2004) J. Cell Biol. 167, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato H., Takino T., Okada Y., Cao J., Shinagawa A., Yamamoto E., Seiki M. (1994) Nature 370, 61–65 [DOI] [PubMed] [Google Scholar]
- 4.Buccione R., Orth J. D., McNiven M. A. (2004) Nat. Rev. Mol. Cell Biol. 5, 647–657 [DOI] [PubMed] [Google Scholar]
- 5.Jiang A., Lehti K., Wang X., Weiss S. J., Keski-Oja J., Pei D. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uekita T., Itoh Y., Yana I., Ohno H., Seiki M. (2001) J. Cell Biol. 155, 1345–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S. A., Mankani M., Robey P. G., Poole A. R., Pidoux I., Ward J. M., Birkedal-Hansen H. (1999) Cell 99, 81–92 [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z., Apte S. S., Soininen R., Cao R., Baaklini G. Y., Rauser R. W., Wang J., Cao Y., Tryggvason K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4052–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yana I., Seiki M. (2002) Clin. Exp. Metastasis 19, 209–215 [DOI] [PubMed] [Google Scholar]
- 10.Zhai Y., Hotary K. B., Nan B., Bosch F. X., Muñoz N., Weiss S. J., Cho K. R. (2005) Cancer Res. 65, 6543–6550 [DOI] [PubMed] [Google Scholar]
- 11.Itoh Y., Seiki M. (2006) J. Cell Physiol. 206, 1–8 [DOI] [PubMed] [Google Scholar]
- 12.Barbolina M. V., Stack M. S. (2008) Semin. Cell. Dev. Biol. 19, 24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyalendo C., Michaud M., Beaulieu E., Roghi C., Murphy G., Gingras D., Béliveau R. (2007) J. Biol. Chem. 282, 15690–15699 [DOI] [PubMed] [Google Scholar]
- 14.Marshall C. J. (1994) Curr. Opin. Genet. Dev. 4, 82–89 [DOI] [PubMed] [Google Scholar]
- 15.Wu Y. I., Munshi H. G., Sen R., Snipas S. J., Salvesen G. S., Fridman R., Stack M. S. (2004) J. Biol. Chem. 279, 8278–8289 [DOI] [PubMed] [Google Scholar]
- 16.Maizels E. T., Peters C. A., Kline M., Cutler R. E., Jr., Shanmugam M., Hunzicker-Dunn M. (1998) Biochem. J. 332, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albrecht-Buehler G. (1977) Cell 11, 395–404 [DOI] [PubMed] [Google Scholar]
- 18.Tjia J. S., Moghe P. V. (2002) Ann. Biomed. Eng. 30, 851–866 [DOI] [PubMed] [Google Scholar]
- 19.Hotary K. B., Allen E. D., Brooks P. C., Datta N. S., Long M. W., Weiss S. J. (2003) Cell 114, 33–45 [DOI] [PubMed] [Google Scholar]
- 20.Strongin A. Y., Collier I., Bannikov G., Marmer B. L., Grant G. A., Goldberg G. I. (1995) J. Biol. Chem. 270, 5331–5338 [DOI] [PubMed] [Google Scholar]
- 21.Butler G. S., Butler M. J., Atkinson S. J., Will H., Tamura T., Schade van Westrum S., Crabbe T., Clements J., d'Ortho M. P., Murphy G. (1998) J. Biol. Chem. 273, 871–880 [DOI] [PubMed] [Google Scholar]
- 22.Will H., Atkinson S. J., Butler G. S., Smith B., Murphy G. (1996) J. Biol. Chem. 271, 17119–17123 [DOI] [PubMed] [Google Scholar]
- 23.Atkinson S. J., Crabbe T., Cowell S., Ward R. V., Butler M. J., Sato H, Seiki M., Reynolds J. J., Murphy G. (1995) J. Biol. Chem. 270, 30479–30485 [DOI] [PubMed] [Google Scholar]
- 24.Kajita M., Itoh Y., Chiba T., Mori H., Okada A., Kinoh H., Seiki M. (2001) J. Cell Biol. 153, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endo K., Takino T., Miyamori H., Kinsen H., Yoshizaki T., Furukawa M., Sato H. (2003) J. Biol. Chem. 278, 40764–40770 [DOI] [PubMed] [Google Scholar]
- 26.Chen J. D., Helmold M., Kim J. P., Wynn K. C., Woodley D. T. (1994) Dermatology 188, 6–12 [DOI] [PubMed] [Google Scholar]
- 27.Hotary K., Allen E., Punturieri A., Yana I., Weiss S. J. (2000) J. Cell Biol. 149, 1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter T., Ling N., Cooper J. A. (1984) Nature 311, 480–483 [DOI] [PubMed] [Google Scholar]
- 29.Davis R. J., Czech M. P. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4080–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowen S., Stanley K., Selva E., Davis R. J. (1991) J. Biol. Chem. 266, 1162–1169 [PubMed] [Google Scholar]
- 31.Mineo C., Gill G. N., Anderson R. G. (1999) J. Biol. Chem. 274, 30636–30643 [DOI] [PubMed] [Google Scholar]
- 32.Lewis R. E., Cao L., Perregaux D., Czech M. P. (1990) Biochemistry 29, 1807–1813 [DOI] [PubMed] [Google Scholar]
- 33.Gallis B., Lewis A., Wignall J., Alpert A., Mochizuki D. Y., Cosman D., Hopp T., Urdal D. (1986) J. Biol. Chem. 261, 5075–5080 [PubMed] [Google Scholar]
- 34.Shackelford D. A., Trowbridge I. S. (1986) J. Biol. Chem. 261, 8334–8341 [PubMed] [Google Scholar]
- 35.Gingras D., Bousquet-Gagnon N., Langlois S., Lachambre M. P., Annabi B., Béliveau R. (2001) FEBS Lett. 507, 231–236 [DOI] [PubMed] [Google Scholar]
- 36.Tsunezuka Y., Kinoh H., Takino T., Watanabe Y., Okada Y., Shinagawa A., Sato H., Seiki M. (1996) Cancer Res. 56, 5678–5683 [PubMed] [Google Scholar]
- 37.Mori H., Tomari T., Koshikawa N., Kajita M., Itoh Y., Sato H., Tojo H., Yana I., Seiki M. (2002) EMBO J. 21, 3949–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naor D., Sionov R. V., Ish-Shalom D. (1997) Adv. Cancer Res. 71, 241–319 [DOI] [PubMed] [Google Scholar]
- 39.Nakahara H., Howard L., Thompson E. W., Sato H., Seiki M., Yeh Y., Chen W. T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7959–7964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 41.Liotta L. A., Kohn E. C. (2001) Nature 411, 375–379 [DOI] [PubMed] [Google Scholar]
- 42.Tam E. M., Wu Y. I., Butler G. S., Stack M. S., Overall C. M. (2002) J. Biol. Chem. 277, 39005–39014 [DOI] [PubMed] [Google Scholar]