Abstract
Through the action of its membrane-bound type I receptor, transforming growth factor-β (TGF-β) elicits a wide range of cellular responses that regulate cell proliferation, differentiation, and apo pto sis. Many of these signaling responses are mediated by Smad proteins. As such, controlling Smad activity is crucial for proper signaling by TGF-β and its related factors. Here, we show that TGF-β induces phos pho ryl a tion at three sites in the Smad3 linker region in addition to the two C-terminal residues, and glycogen synthase kinase 3 is responsible for phos pho ryl a tion at one of these sites, namely Ser-204. Alanine substitution at Ser-204 and/or the neighboring Ser-208, the priming site for glycogen synthase kinase 3 in vivo activity, strengthened the affinity of Smad3 to CREB-binding protein, suggesting that linker phos pho ryl a tion may be part of a negative feedback loop that modulates Smad3 transcriptional activity. Thus, our findings reveal a novel aspect of the Smad3 signaling mechanism that controls the final amplitude of cellular responses to TGF-β.
Transforming growth factor-β (TGF-β)2 is the prototype of a large family of secreted polypeptide growth factors that regulate a multitude of cellular processes affecting proliferation, differentiation, and apoptosis (1, 2). It is now generally accepted that the plethora of biological activities of TGF-β is initiated by the binding of the ligand to a heteromeric complex of two types of transmembrane receptors: TβRI and TβRII, each equipped with an intrinsic serine/threonine kinase (3). Ligand occupancy causes an association between TβRII and TβRI, which results in phosphorylation of TβRI by the constitutively active TβRII. The phosphorylated TβRI then triggers activation of Smad2 and/or Smad3 by phosphorylation at the C-terminal serine residues, forcing Smad2 and Smad3 to dissociate from the membrane-bound receptors and form a heteromeric complex with Smad4 (4, 5). Phosphorylation of Smad2 and Smad3 also enables them to accumulate in the nucleus (6), where Smad3 but not full-length Smad2 directly binds to DNA. However, the affinity of Smad3 to DNA does not support a one-on-one stoichiometry binding model in vivo (7); instead, Smad3 relies on cooperative binding with other transcription factors to elicit respective Smad-mediated transcriptional responses (3, 8). The Smad transcriptional complexes have the ability to either activate or repress transcription of a selected set of target genes depending on the nature of associated cofactors and the status of local chromatin structure in the context of signal receiving cells. It is now clear that this Smad-mediated signaling pathway is controlled by or functions in conjunction with Smad-independent mechanisms, such as those governed by MAPKs (9, 10). These non-Smad signaling conduits can modulate Smad activity to custom fit signaling outputs to a particular need, generating a myriad of cellular responses to TGF-β.
The Smad proteins consist of an N-terminal (MH1) domain that binds DNA and a C-terminal (MH2) domain that interacts with type I receptors, other Smad proteins, and various transcriptional coactivators/corepressors (11). These two highly conserved domains are separated by a less conserved linker region. There are four SP/TP sites for proline-directed kinases in both Smad2 and Smad3 linker regions (12). However, except for the first TP site, flanking sequences around the other three SP sites of Smad2 are quite different from those of Smad3, suggesting potentially different modes of regulation between these two proteins. Previously, epidermal growth factor, hepatocyte growth factor, the Ras oncogene, and other activators of the MAPK pathway have been shown to induce phosphorylation of Smad2 and/or Smad3 at these linker sites (12–14). In addition, during cell cycle progression, the Smad3 linker can also be phosphorylated by activated cyclin-dependent kinases (CDKs) during the G1/S phase (15). Many of these phosphorylation events have been reported to have an antagonistic role on Smad3 activity (12, 14, 15), which may be a mechanism for overriding TGF-β-induced growth arrest by cancer cells expressing high levels of CDKs or oncogenic Ras. Conversely, a synergistic activation effect by linker phosphorylation on Smad3 activity has also been reported (13). It is possible that each phosphorylation event produces a different impact on Smad3 activity through a different underlying molecular mechanism. Further detailed studies of individual phosphorylation sites are needed to clarify the contribution of these linker phosphorylation events to Smad3 function.
Here, we report that TGF-β can induce phosphorylation of Smad3 at Thr-179, Ser-204, and Ser-208. We show that glycogen synthase kinase 3 (GSK3) directly phosphorylates Smad3 at Ser-204, whereas a different kinase may be responsible for phosphorylation of Ser-208, which is a prerequisite priming site for GSK3 activity in vivo. We further demonstrate that mutations at Ser-204 and Ser-208 strengthened the transcriptional activity of Smad3 by enhancing its affinity to CBP. Thus, our findings reveal a novel aspect of the Smad3 signaling mechanism, entailing a negative feedback control by phosphorylation of the Smad3 linker region.
EXPERIMENTAL PROCEDURES
Expression Plasmids
Full-length Smad3 cDNA (16) without the start codon was subcloned into pCMV5-FLAG (17) to generate N-terminal FLAG-tagged Smad3. Smad3 mutations were generated using a PCR-based strategy and subcloned into pCMV5-FLAG. All PCR-amplified regions were verified by sequencing. Plasmids for GST-Smad3 (16), GST-Smad3NL, GST-Smad3C (18), and Myc-Smurf2 (19) have been described previously. GST-Smad3L was constructed by inserting the Pro-147–Gln-222 fragment of Smad3 into the BamHI/XhoI sites of pGEX-4T-2 (Amersham Biosciences). An expression plasmid for Myc-tagged CBP was a gift from Dr. S. H. Snyder's lab.
Antibodies and Reagents
The rabbit polyclonal antibody against pSer-423/pSer-425 of Smad3 was generated using a synthetic peptide PSIRCS(pS)V(pS) in collaboration with Rockland Immunochemicals, Inc. (Gilbertsville, PA). Rabbit antiserums against Smad3 pThr-179, pSer-204, or pSer-208 were gifts from Dr. F. Liu (15). The monoclonal antibody to GSK3 was obtained from Cell Signaling Technology (Beverly, MA). GSK3 inhibitor SB216763 and ALK5 inhibitor SB431542 were obtained from Tocris Bioscience (Ellisville, MO), whereas MEK1/2 inhibitor U0126, p38 inhibitor SB203580, and JNK inhibitor SP600125 were purchased from Calbiochem (La Jolla, CA). All other antibodies and reagents used in this study have been described previously (20, 21, 22).
Cell Culture and Transfection Conditions
Cultural conditions for C2C12 cells have been described previously (23). HEK293 (CRL-1573, ATCC), wild type, and Smad3−/− mouse embryonic fibroblasts (MEFs) (24) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). Hep3B (HB8064, ATCC), mink lung epithelial cells including wild type (Mv1Lu) (CCL64, ATCC), TβRI-defective RIBL17 (25), and RIBL17 cells that stably express TβRI (26) were maintained in minimal essential Eagle's medium with Earle's balanced salt solution supplemented with nonessential amino acids and 10% FBS. AML12 cells (CRL2254, ATCC) were maintained in Dulbecco's modified Eagle's medium/F12 medium containing 10% FBS, 40 ng/ml dexamethasone and ITS+ premix (VWR Scientific). Transfections of Hep3B and HEK293 cells were done using FuGENE 6 (Roche Applied Science), while transfection of MEFs was carried out with Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. Oligofectamine (Invitrogen) was used for small interfering RNA transfection.
Western Blotting, Immunoprecipitation, and Cytoplasmic and Nuclear Fractionation
The methods for Western blotting and immunoprecipitation have been described previously (19). Separation of cytoplasmic and nuclear fractions were performed by harvesting cells in buffer A containing 10 mm Tris-HCl, pH 8.0, 10 mm NaCl, 2 mm MgCl2, and 5 mm dithiothreitol with protease and phosphatase inhibitors. Nonidet P-40 was added at a final concentration of 0.5%. After an incubation of 10–15 min on ice, the cytoplasmic fraction (supernatant) was isolated by centrifugation at 3,000 rpm for 5 min. The pellets were washed twice with buffer A and incubated with buffer B containing 20 mm Hepes, pH 7.9, 420 mm NaCl, 1.5 mm MgCl2, 5 mm dithiothreitol, and 10% glycerol with protease and phosphatase inhibitors for 30 min on ice. After centrifugation at 13,000 rpm for 10 min, the supernatant was collected as nuclear fraction.
In Vitro Kinase Assay
Recombinant GST or GST-Smad3 fusion proteins (1 μg) purified from bacteria were incubated with a Myc-tagged TβRI cytoplasmic domain immunoprecipitated from transfected HEK293 cells or commercially available activated recombinant JNK2, p38α, Erk2, and GSK3β (Calbiochem) in 1× kinase buffer (10 mm Hepes-KOH, pH 7.5, 5 mm MgCl2, 5 mm MnCl2, and 5 mm CaCl2) at 30 °C. Either 5 μCi of [γ-32P]ATP (5,000 μCi/mmol) or 1 mm ATP was included in the reaction. After a 30-min incubation, the reaction was stopped by adding an equal volume of 2× SDS sample buffer and subjected to autoradiography or anti-phospho-Smad3 blotting after electrophoresis using 10% SDS-PAGE.
Transcription Reporter Assay
MEFs seeded into 12-well plates were transfected with (CAGA)12-Luc (27) (0.25 μg), pRK-βgal (28) (0.05 μg), vector control, or Smad3 plasmid (0.01 μg). Twenty-four h after transfection, cells were treated with 4 ng/ml TGF-β in Dulbecco's modified Eagle's medium containing 0.2% FBS for overnight. Cells were then harvested to measure luciferase and β-galactosidase activities. All assays were performed in duplicate and repeated more than three times, and all values were normalized for transfection efficiency against β-galactosidase activity.
Phosphorylation Site Mapping by Mass Spectrometry
To map the GSK3-dependent phosphorylation site within the Smad3 linker, GST-Smad3L phosphorylated by GSK3β in vitro was excised from SDS-PAGE after Coomassie Blue staining. The gel slice was subjected to digestion with trypsin and/or Glu-C. The peptides were extracted and analyzed by liquid chromatography coupled directly on-line with a linear ion trap mass spectrometer (Thermo Scientific, San Jose, CA) as described previously (29). The raw tandem mass spectrometry data were searched using SEQUEST (Thermo Electron, San Jose, CA) against a data base consisting of the human Smad3 protein sequence. The identified phosphopeptides were further examined by manual inspection of the tandem mass spectrometry spectra to confirm identification of the correct peptide sequence and sites of phosphorylation.
Adenovirus Infection and Proliferation Assay
Smad3 and Smad3 S204A/S208A double mutant (2SPAP) cDNA were subcloned into a pDUALCCM-CMVEGFP shuttle vector (Vector Biolabs, Philadelphia, PA) and transferred into an adenovirus genome and produced adenovirus constitutively expressing Smad3 and Smad3 (2SPAP). GFP control adenovirus was also obtained from Vector Biolabs. Infection of AML12 cells was performed in a 60-mm plate at a multiplicity of infection of 100 with a single virus for 2 h in plain Dulbecco's modified Eagle's medium/F12 medium. Infected cells were recovered in regular medium for 24 h and split into 24-well plates. For proliferation assay, cells were incubated with TGF-β (4 ng/ml) for 40 h, and 3H-thymidine was added at the last 4 h.
RESULTS
TGF-β Induces Phosphorylation of Smad3 at Thr-179, Ser-204, and Ser-208 in Vivo
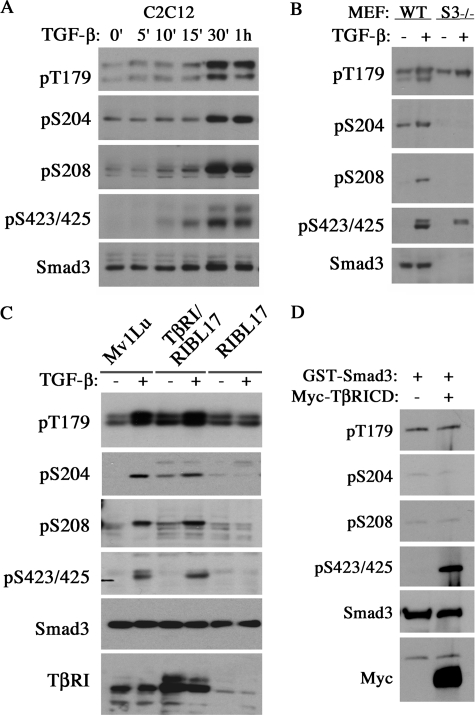
A major event leading to Smad3 activation is the TGF-β-induced, TβRI-mediated phosphorylation at two C-terminal serine residues, Ser-423 and Ser-425, which triggers dissociation of Smad3 from its receptors to form a complex with Smad4 and accumulate in the nucleus (3, 8). Phosphorylation in the Smad3 linker region connecting two conserved MH1 and MH2 domains has been considered as part of the mechanisms responsible for cross-talks between TGF-β and other kinase pathways such as ERK, JNK, and p38, as well as cyclin-dependent kinase CDK2/4 (12–15). When tested initially in mouse C2C12 myoblasts, however, we were surprised to find that TGF-β not only induced C-terminal phosphorylation but also phosphorylation of Thr-179, Ser-204, and Ser-208 in the linker region (Fig. 1A), as evident by Western blot analysis using phospho-specific antibodies (15). The onset of these phosphorylation events had similar kinetics as the C-terminal phosphorylation (Fig. 1A). The specificity of the phospho-antibodies was confirmed in Smad3-deficient MEFs (Fig. 1B). To determine whether the TGF-β-induced linker phosphorylation on Smad3 requires the TβRI, we examined the phosphorylation status of Smad3 in RIBL17 cells, a line of the TβRI-defective mink lung epithelial cells (25). Whereas in wild type mink lung cells (Mv1Lu) TGF-β was able to induce phosphorylation at Thr-179, Ser-204, and Ser-208 residues, it failed to do so in the RIBL17 cells (Fig. 1C). Stable expression of TβRI restored TGF-β-induced linker phosphorylation in RIBL17 cells (Fig. 1C), indicating that the type I receptor is essential. However, it appeared that TβRI is not the kinase that directly phosphorylates Smad3 at these linker sites because purified cytoplasmic domain of TβRI with an intact kinase domain failed to phosphorylate the Smad3 linker sites in vitro, albeit it was capable to phosphorylate the Ser-423 and Ser-425 at the C terminus (Fig. 1D).
FIGURE 1.
TGF-β induces Thr-179, Ser-204, and Ser-208 phosphorylation in the linker region. A, time courses of phosphorylation at Thr-179, Ser-204, and Ser-208 of Smad3 induced by TGF-β in C2C12 cells. Total cell lysates were probed for endogenous Smad3 with phospho-specific and Smad3 antibodies. B, phosphorylation of linker residues in wild type (WT) or Smad3−/− MEFs. Cells were serum-starved for overnight and then treated with TGF-β for 1 h. C, phosphorylation of linker residues in wild type (Mv1Lu), TβRI stably transfected RIBL17, and TβRI-defective (RIBL17) mink lung epithelial cells as determined in B. D, in vitro kinase assay using purified GST-Smad3 and Myc-tagged cytoplasmic domain of TβRI.
GSK3 Is Required for TGF-β-induced Phosphorylation of Ser-204 on Smad3
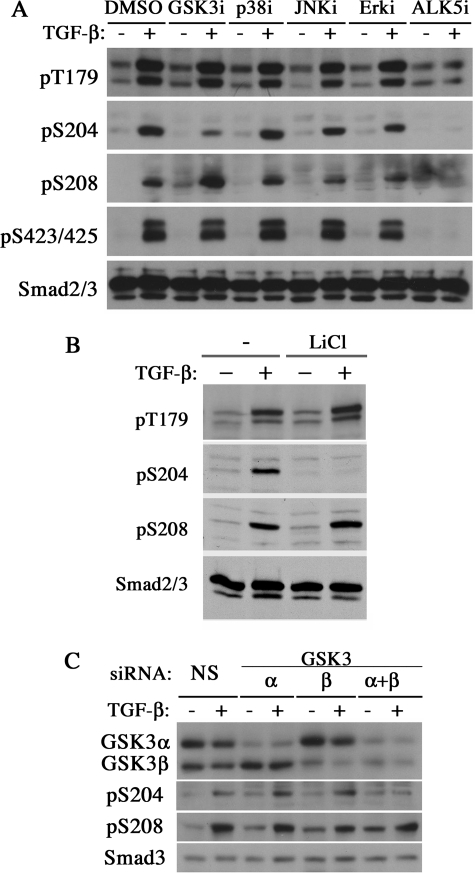
Thr-179, Ser-204, and Ser-208 of Smad3 are situated immediately N-terminal to a proline residue in the linker region, making them plausible sites for phosphorylation by proline-directed protein kinases. Previously, Thr-179 was shown as a phosphorylation site for CDKs, whereas all three residues were shown as sites for ERK induced by epidermal growth factor (14, 15). A separate study also reported that Ser-208 could be phosphorylated by JNK in response to hepatocyte growth factor stimulation (13). Because these MAP kinases can be activated by TGF-β as the consequence of non-Smad signaling, we examined whether they had a role in the TGF-β-induced linker phosphorylation of Smad3 in AML12 cells. By treating the cells in the presence of specific kinase inhibitors, we found that neither ERK, p38, nor JNK kinases had any effect on the status of TGF-β-induced linker phosphorylation (Fig. 2A). Similar results were obtained in C2C12 cells (data not shown). These results suggest that one or several different kinases are likely responsible for phosphorylation at the above three sites. Indeed, we found that phosphorylation of Ser-204 was significantly inhibited by a selective GSK3 inhibitor, SB216763 (Fig. 2A). Treating C2C12 cells with LiCl, a potent GSK3 inhibitor, also diminished TGF-β-induced Ser-204 phosphorylation (Fig. 2B), whereas phosphorylation at all three sites was blocked by SB431542, a TβRI kinase-specific inhibitor (Fig. 2A). These data reiterated the requirement of TβRI in linker phosphorylation and suggest that GSK3 can phosphorylate Smad3 at least at one site within the linker region.
FIGURE 2.
GSK3 is required for the TGF-β-induced phosphorylation at Ser-204. A, phosphorylation of linker residues in AML12 cells treated with dimethyl sulfoxide (DMSO), SB208530 (p38 inhibitor (i); 10 μm), SP600125 (JNK inhibitor; 10 μm), U0126 (ERK inhibitor; 10 μm), SB216763 (GSK3 inhibitor; 10 μm), and SB431542 (TβRI inhibitor; 10 μm). The cells were serum-starved overnight before receiving inhibitor treatment for 30 min and then TGF-β for an additional 1 h. B, phosphorylation of Ser-204 and Ser-208 in C2C12 cells with or without LiCl (20 mm) treatment for 30 min and an additional 1 h of TGF-β treatment. C, phosphorylation of Ser-204 and Ser-208 in Hep3B cells transfected with control, small interfering RNAs against GSK3α, GSK3β, or both. Two days after transfection, cells were treated with TGF-β for 1 h and analyzed by Western blot for GSK3, pSer-204, pSer-208, and total Smad3. siRNA, small interfering RNA; NS, non-silencing control.
To corroborate the above findings, we probed the role of GSK3 in Ser-204 phosphorylation with two previously verified human GSK3 small interfering RNAs (30) in Hep3B cells. The separate knocking down of either GSK3α or GSK3β had little effect on Ser-204 phosphorylation, whereas knockdown of both GSK3 genes simultaneously diminished the phosphorylation at this site (Fig. 2C). As expected, phosphorylation of Ser-208 or the two C-terminal residues Ser-423/Ser-425 was not affected by GSK3-specific small interfering RNA (Fig. 2C). These results confirmed that GSK3 activity is required for TGF-β-induced Ser-204 phosphorylation.
Ser-208 Serves as Priming Site for Phosphorylation of Ser-204 by GSK3 in Vivo
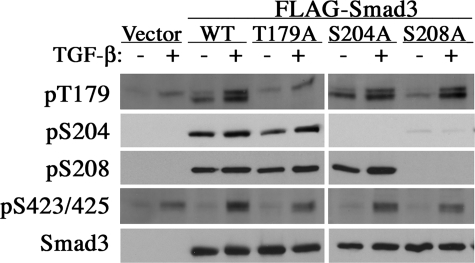
GSK3 recognizes a (S/T)XXX(S/T) consensus motif on its substrates, in which the first serine or threonine residue of this motif is the actual GSK3 phosphorylation site. Phosphorylation by GSK3 in vivo requires a priming event at the second serine or threonine residue, usually catalyzed by a different kinase (31). Ser-204 is the only potential GSK3 site in the entire linker region of Smad3. Because phosphorylation of Ser-208 slightly precedes that of Ser-204 (Fig. 1A), this phosphorylation could potentially be the priming event for the GSK3 action at Ser-204. To test this hypothesis, we created point mutations at Thr-179, Ser-204, and Ser-208, respectively, and introduced these Smad3 mutants into the Smad3-deficient cells. Our results showed that alanine substitution at Ser-208 but not Thr-179 abolished phosphorylation at Ser-204, whereas phosphorylation at Ser-208 or Thr-179 was not affected by the mutation of other residues (Fig. 3). These results suggest that prior phosphorylation at Ser-208 is likely required for the subsequent phosphorylation at Ser-204 in vivo.
FIGURE 3.
Ser-208 as a priming site for phosphorylation at Ser-204. Smad3−/− MEFs were transfected with FLAG-tagged Smad3 and various linker phosphorylation site mutants. Two days later, cells were treated with TGF-β for 1 h and analyzed as in Fig. 2C. WT, wild type.
GSK3 Directly Phosphorylates Smad3 at Ser-204
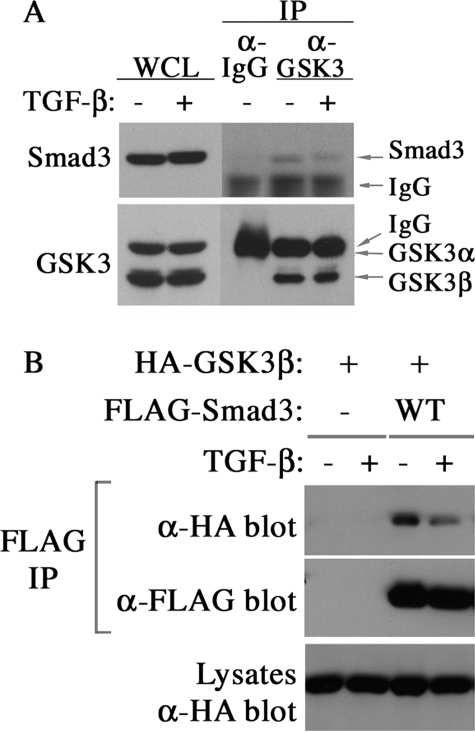
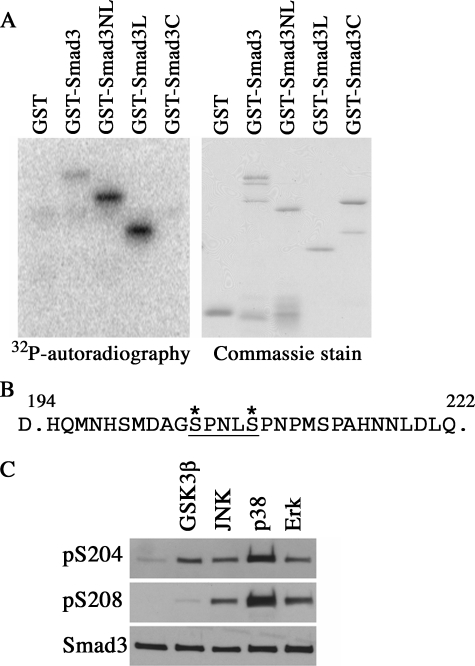
A recent study identified Smad3 in a complex with GSK3 (32), and we also observed the physical interaction between endogenous GSK3 and Smad3 in C2C12 cells (Fig. 4A) or between the transfected HA-GSK3β and FLAG-Smad3 in Hep3B cells (Fig. 4B) as shown by co-immunoprecipitation assays. The affinity of Smad3 to GSK3 was decreased after TGF-β stimulation, possibly due to a rapid dissociation after phosphorylation. This argues that Ser-204 may be phosphorylated directly by GSK3. To investigate this possibility, we performed in vitro kinase assays on various full-length or truncated Smad3 fragments using purified recombinant GSK3β and found that full-length Smad3, the N-terminal domain plus the linker region (Smad3NL), and the linker fragment (Smad3L) can be phosphorylated by GSK3β (Fig. 5A). In contrast, the C-terminal domain alone (Smad3C) or the GST control was not phosphorylated (Fig. 5A). Analysis of the phosphorylated GST-Smad3L fusion proteins by tandem mass spectrometry identified a phospho-peptide generated by Glu-C digestion corresponding to residues from 194 to 222 of Smad3 (Fig. 5B). A phosphoserine residue, Ser-204, is present in this peptide, but surprisingly, Ser-208 was also found to be phosphorylated (Fig. 5B). This result was likely caused by the diminished requirement of priming phosphorylation by GSK3 in vitro. To further confirm the direct phosphorylation by GSK3 in the linker region, the in vitro kinase assays were repeated on GST-Smad3 fusion protein, and Western blot analysis was conducted using Ser-204 and Ser-208 phospho-specific antibodies. As shown in Fig. 5C, Ser-204 was phosphorylated by GSK3β, ERK, JNK, and p38 in vitro, as was Ser-208, but the efficiency of GSK3β toward Ser-208 was lower in comparison with other kinases. Taken together, the above results demonstrated that Smad3 could be directly phosphorylated by GSK3 at Ser-204.
FIGURE 4.
Physical interaction between Smad3 and GSK3. A, co-immunoprecipitation assays of endogenous GSK3 and Smad3 in C2C12 cells. Cells were stimulated ± TGF-β for 1 h, lysed, and subjected to immunoprecipitation using either anti-mouse IgG or monoclonal antibody against GSK3. GSK3-bound Smad3 was detected by anti-Smad3 blot. B, co-IP assays of hemagglutinin (HA)-tagged GSK3β and FLAG-tagged Smad3 in transfected Hep3B cells. Two days after transfection, cells were treated with TGF-β for 1 h and subjected to anti-FLAG immunoprecipitation. GSK3β that was brought down along with Smad3 was detected by anti-hemagglutinin blot. The expression level of transfected hemagglutinin-GSK3β in whole cell lysates (WCL) were shown at the bottom.
FIGURE 5.
GSK3 directly phosphorylates Smad3 at Ser-204. A, autoradiography of various GST-Smad3 fusion proteins phosphorylated in vitro for 30 min by recombinant GSK3 in the presence of 32P-labeled ATP. Total inputs of purified GST, GST-Smad3, Smad3NL, Smad3L, and Smad3C fusion proteins were shown by Coomassie staining in the right panel. B, mass spectrum detection of a phosphopeptide corresponding to residues His-194 to Gln-222 of Smad3. Phosphorylated Ser-204 and Ser-208 were denoted with an asterisk. C, in vitro kinase assay of purified GST-Smad3 by recombinant GSK3β, JNK, p38, or ERK. Phosphorylation of Ser-204 or Ser-208 was detected by phospho-specific antibodies in Western analysis. Equal loading of substrates was confirmed by anti-Smad3 blot.
Phosphorylation of Smad3 at Ser-204 and Ser-208 Inhibits Smad3 Activity
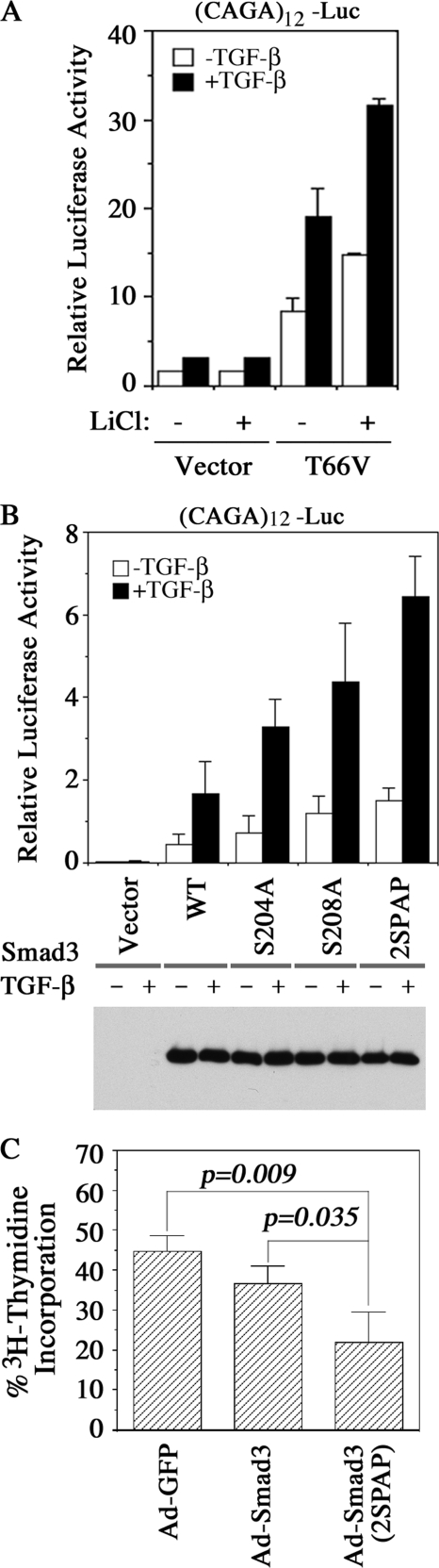
To determine the role of GSK3-induced phosphorylation of Ser-204, we first measured the effect of the GSK3 inhibitor, LiCl, on the transcriptional activity of Smad3. GSK3 was recently shown to phosphorylate Smad3 at Thr-66 in the N terminus, thereby causing Smad3 to be unstable (32). To exclude the effect of Thr-66 phosphorylation, we performed this experiment with a T66V mutant Smad3 in MEFs isolated from Smad3-deficient embryos. In the absence of Smad3, TGF-β cannot activate transcription from the (CAGA)12-Luc reporter (27), but this activity can be restored upon transfection of Smad3 T66V, and suppressing GSK3 activity with LiCl treatment further augmented this activity (Fig. 6A). To specifically assess the effect of Ser-204 and Ser-208 phosphorylation on Smad3, we measured the transcriptional activity of Smad3 S204A and S208A mutants and found that these mutants were more potent in restoring TGF-β activation of the (CAGA)12-Luc reporter than their wild type counterparts (Fig. 6B). The S204A/S208A double mutant (2SPAP) exhibited even higher activity in this rescuing experiment (Fig. 6B). Because the expression levels of these mutants were comparable with that of the wild type Smad3, their elevated transcriptional activity was likely due to changes in the phosphorylation status.
FIGURE 6.
Ser-204 and/or Ser-208 mutants are hyperactive. A, Smad3-dependent (CAGA)12-Luc reporter assay. Smad3−/− MEFs were transfected with (CAGA)12-Luc, pRK-βgal, and FLAG-tagged Smad3 T66V plasmids as indicated. Twenty-four h after transfection, cells were treated ± TGF-β for overnight in the absence or presence of 10 mm LiCl. B, effect of Smad3 mutants S204A, S208A, and the Ser-204/Ser-208 double mutant (2SPAP) on the (CAGA)12-Luc reporter assay in Smad3−/− MEFs. Experiments were performed as in A. Twenty-four h after transfection, cells were treated ± TGF-β for overnight. Expression of Smad3 and its mutant derivatives in Smad3−/− MEFs after transfection shown in the bottom panel. C, effect of the Smad3 Ser-204/Ser-208 double mutant (2SPAP) on TGF-β-induced cell growth arrest. AML12 cells were infected with adenoviruses expressing GFP, Smad3, or Smad3 (2SPAP), respectively. Cells were then treated ± TGF-β for 40 h and assessed for proliferation. The data are presented as the percentage of [3H]thymidine incorporation in the presence of TGF-β compared with untreated cells. WT, wild type; Ad, adenovirus.
Next, we examined the effects of S204A and S208A mutants on the growth inhibitory response of TGF-β by measuring [3H]thymidine incorporation in AML12 cells that were infected with adenoviruses expressing wild type Smad3 or the S204A/S208A double mutant, 2SPAP. Control GFP adenovirus-infected cells exhibited a normal TGF-β-induced inhibitory response on DNA synthesis in the presence of 10% FBS, as their proliferation was reduced to 44% in the presence of TGF-β (Fig. 6C). Overexpression of wild type Smad3 further reduced the proliferation in the presence of TGF-β to 37% as expected. Expression of the Smad3 2SPAP mutant rendered cells even more sensitive to TGF-β-induced growth inhibition, as proliferation was reduced to 21% in comparison with that of untreated cells (Fig. 6C). Taken together, these results indicate that phosphorylation of Smad3 at the linker region by GSK3 inhibits Smad3 activity.
Phosphorylation of Smad3 at Ser-204 Affects Neither Stability nor Nuclear Accumulation of Smad3
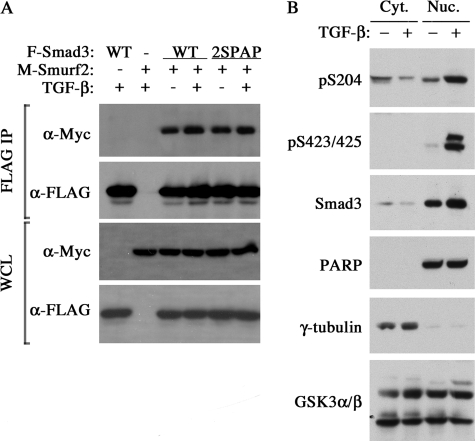
A recent report showed that phosphorylation of Smad1 linker by ERK and GSK3 enhances interaction between Smad1 and Smurf1, thereby promoting Smad1 degradation (33). Because Smad3 binds to the ubiquitin ligase Smurf2 through a PY motif, upstream of Ser-204/Ser-208 in the linker region (19), it is possible that phosphorylation of Ser-204 affects Smad3 stability by strengthening the interaction between Smad3 and Smurf2. However, this result was not observed in our experiments as the Ser-204 and/or Ser-208 mutants bound Smurf2 with the same affinity as wild type Smad3 (Fig. 7A), and no change in stability of these mutants was detected compared with wild type Smad3 (data not shown), which was consistent with reports by other laboratories (32, 33).
FIGURE 7.
Lack of effect of Ser-204 phosphorylation on Smurf2 binding and nuclear accumulation. A, co-IP assays of Myc-tagged (M) Smurf2 and FLAG-tagged (F) Smad3 proteins in transfected Hep3B cells after stimulated with or without TGF-β for 1 h. Expression levels of transfected proteins in whole cell lysates (WCL) were shown at the bottom. B, nuclear and cytoplasmic distribution of Smad3 proteins in AML12 cells treated with TGF-β for 1 h. The content of each form of Smad3 in the nuclear and cytoplasmic fractions was determined by Western analysis. PARP and γ-tubulin were used as nuclear (Nuc.) and cytoplasmic (Cyt.) specific protein markers, respectively. WT, wild type; PARP, poly (ADP-ribose) polymerase.
Phosphorylation within Smad3 linker regions has been shown to prevent nuclear accumulation (12). To elucidate whether phosphorylation of Ser-204 has this effect, we examined the intracellular distribution of Smad3 in AML12 cells that had been deprived of serum for overnight and then treated with TGF-β for 1 h. After subfractionating cells into cytoplasmic and nuclear fractions, we analyzed the content of various forms of Smad3 in these two fractions by Western blotting. As expected, TGF-β treatment increased nuclear but decreased cytoplasmic content of total Smad3 (Fig. 6B). Under this condition, the C-terminally phosphorylated form of Smad3 (pSer-423/pSer-425) was detected exclusively in the nucleus, whereas the majority of Ser-204 phosphorylated form of Smad3 was also found in the nuclear fractions (Fig. 7B). These results indicate that phosphorylation of Ser-204 does not prevent nuclear accumulation of Smad3.
Phosphorylation of Ser-204 Reduces Ability of Smad3 to Bind CBP
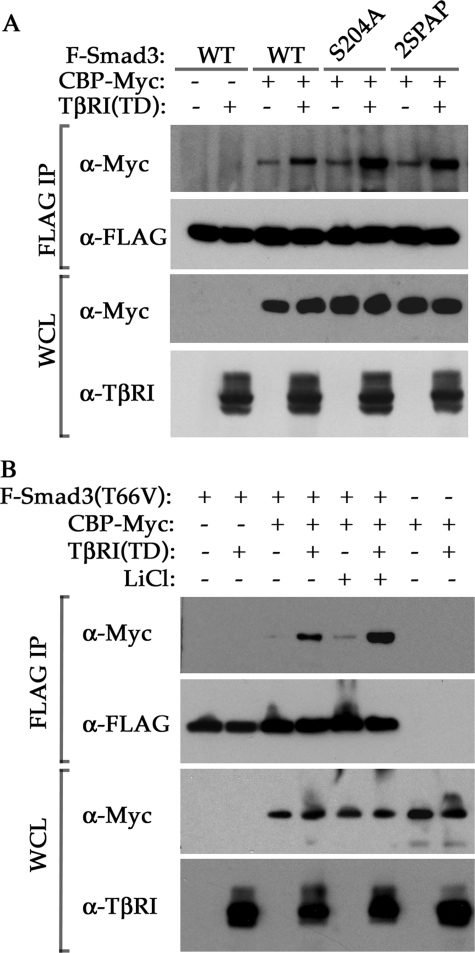
In light of the increased transcriptional activity of the S204A and 2SPAP mutants (Fig. 7B) and a previous report showing that the linker region of Smad3 can interact physically with transcription coactivator p300/CBP (34, 35), we tested whether phosphorylation of Ser-204 affects the binding between Smad3 and CBP. We cotransfected HEK293 cells with Myc-tagged CBP and FLAG-tagged Smad3 along with the constitutively active TβRI(TD) and then determined binding affinity using immunoprecipitation assays. As shown in Fig. 8A, activation of the TGF-β pathway by TβRI(TD) induced the interaction between Smad3 and CBP, but the S204A and 2SPAP mutants exhibited even stronger affinity toward CBP after TβRI activation. We also examined the effect of endogenous GSK3 on the interaction between Smad3 and CBP using Smad3-T66V in the absence or presence of LiCl. As shown in Fig. 8B, suppressing GSK3 activity with LiCl greatly enhanced the physical interaction between Smad3 and CBP, suggesting that phosphorylation of Ser-204 by GSK3 likely inhibits the Smad3 transcriptional activity by reducing its interaction with CBP.
FIGURE 8.
Mutation in Ser-204 or Ser-208 enhances TGF-β-induced interaction between Smad3 and CBP. A, co-IP assays of Myc-tagged CBP and various FLAG-tagged (F) Smad3 proteins in the presence or absence of constitutively active TβRI(TD) in transfected HEK293 cells. Cells were lysed and subjected to anti-FLAG immunoprecipitation. Smad3-bound CBP was determined using anti-Myc Western blot. The expression levels of transfected CBP and TβRI(TD) proteins in whole cell lysates (WCL) were shown at the bottom. B, effect of GSK3 activity on the physical interaction between Smad3 and CBP. A co-IP assay of Myc-tagged CBP and FLAG-tagged Smad3 T66V was performed as in A. Cells were treated ± 10 mm LiCl for 1 h before lysis. WT, wild type.
DISCUSSION
In this report, we showed that upon TGF-β stimulation, Smad3 undergoes a relatively rapid phosphorylation at three residues in the linker region in addition to C-terminal phosphorylation and identified GSK3 as the kinase responsible for one of the sites, Ser-204. We further showed that an alanine substitution at Ser-204 strengthened the ability of Smad3 to bind CBP, implying that phosphorylation at this site may decrease the transcriptional activity of Smad3. Thus, our findings suggest that by eliciting these two phosphorylation events simultaneously, TGF-β likely calls on a negative feedback loop to modulate the amplitude of final signaling output.
The structurally diverse linker region of Smad proteins harbors a number of kinase recognition motifs and a Smurf binding site. Numerous studies have shown that this region is the convergence point by multiple signaling pathways to attenuate or otherwise modulate downstream cellular responses to the TGF-β family of ligands. For instance, there are four ERK consensus sites present in Smad1, Smad5, and Smad8 of the bone morphogenetic protein pathway; phosphorylation at these sites in response to fibroblast growth factors is associated with Smurf1 binding and cytoplasmic retention (33, 36). Balancing bone morphogenetic protein signaling through integrating fibroblast growth factor input at the Smad1 linker region is important for maintaining tissue homeostasis during development (37, 38). There are also four MAPK sites in Smad2 and Smad3 linkers; phosphorylation at these sites was attributed as one of the mechanisms that the Ras oncogene uses to counteract the tumor suppressive function of TGF-β (12). Phosphorylation within the Smad3 linker was also shown to be catalyzed by CDK2 and CDK4 during the entry into mitosis, leading to decreased expression of CDK inhibitor p15 and thus a subdued TGF-β-mediated growth inhibition (15). The PY motif in the linker region recognized by the HECT domain E3 ligase Smurf is critical in curtailing Smad functions by allowing Smurf-mediated ubiquitination and proteasomal degradation (19, 39, 40). Thus, the notion that TGF-β also taps into the linker region to moderate Smad3 transcriptional activity is consistent with the regulatory function of this domain.
Together with a previous study of Smad1, the current finding on Smad3 linker phosphorylation revealed a general aspect of Smad signaling mechanism. However, not all kinases or the consequential effects of the linker phosphorylation are known. Although ample experimental data have implicated activation of MAPK pathways as part of the non-Smad TGF-β signaling mechanism, our results nevertheless pointed to GSK3, not any MAPK, as the kinase that directly phosphorylates Ser-204 upon TGF-β stimulation. This is reminiscent of the BMP-induced Smad1 linker phosphorylation, which is also independent of MAPK (33). Recently, GSK3 was shown to phosphorylate Thr-66 outside the linker region, which has a role in controlling the basal level of Smad3 through degradation (32). In our experiments, alanine substitution of Ser-204 not only had no effect on Smad3 stability, but it also failed to influence Smurf2 binding, suggesting that the negative feedback control exerted by GSK3 over Smad3 differs from its activity at Thr-66 in both location and effect. However, because in most experiments TGF-β treatment was carried out in serum-starved cells, a condition in which GSK3 is constitutively active, phosphorylation of Ser-204 by GSK3 might require input from an unknown priming kinase induced by TGF-β that phosphorylates Ser-208. The different priming kinase activities likely represent two faces of the same GSK3 function as a repressive kinase of TGF-β signaling. Earlier studies also showed cytoplasmic retention of Smad3 phosphorylated by MAPK/Ras and mutated Smad3 lacking all linker phosphorylation sites, suggesting linker phosphorylation could lead to nuclear exclusion of Smad3 precluding it from participating in target gene transcription (12). A recent report showed that phosphorylation of the Smad1 linker by ERK and GSK3 enables Smad1 to be recognized by Smurf1, which in addition to inducing ubiquitination and proteasomal degradation of Smad1, can dislodge Smad1 binding to nuclear translocation factor Nup214 due to steric hindrance, thus leading to cytoplasmic retention of Smad1 (33). However, we found a majority of the Ser-204 phosphorylated form of Smad3 in the nucleus upon TGF-β stimulation, and alanine substitution at either Ser-204 or Ser-208 did not alter the physical interaction between Smurf2 and Smad3. Instead, the S204A and S208A mutant of Smad3 exhibited heightened affinity toward CBP, suggesting that phosphorylation at these two sites may have direct impact on Smad3 transcriptional activity. The discrepancy between these observations is likely a reflection of the nature of linker mutations used in different experiments and awaits further resolution by more detailed studies.
Acknowledgments
We thank Drs. F. Liu for Smad3 linker phospho-specific antibodies, J. Massagué for RIBL17 cells, X. H. Feng and R. Derynck for RI14 cells, C. X. Deng for Smad3 knockout mice, X. He for HA-GSK3β plasmid, S. H. Snyder for Myc-CBP plasmid, P. ten Dijke for (CAGA)12-Luc reporter construct, and L. Y. Tang and K. Sixt for assistance.
This work was supported, in whole or in part, by the National Institutes of Health, NCI, Center for Cancer Research, Intramural Research Program and is funded in part by National Institutes of Health Contract N01-CO-12400 from NCI.
- TGF-β
- transforming growth factor-β
- CREB
- cAMP-responsive element-binding protein
- CBP
- CREB-binding protein
- JNK
- c-Jun NH2-terminal kinase
- ERK
- extracellular signal-regulated kinase
- pSer
- phosphoserine
- pThr
- phosphothreonine
- MAPK
- mitogen-activated protein kinase
- CDK
- cyclin-dependent kinase
- GSK3
- glycogen synthase kinase 3
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- MEF
- mouse embryonic fibroblast
- luc
- luciferase
- IP
- immunoprecipitation
- TβRI(TD)
- constitutively active TβRI.
REFERENCES
- 1.Massagué J. (1998) Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 2.Derynck R., Miyazono K. (2008) in The TGF-beta Family (Derynck R., Miyazono K. eds) pp. 29–43, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 3.Feng X. H., Derynck R. (2005) Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 4.Wu J. W., Hu M., Chai J., Seoane J., Huse M., Li C., Rigotti D. J., Kyin S., Muir T. W., Fairman R., Massagué J., Shi Y. (2001) Mol. Cell 8, 1277–1289 [DOI] [PubMed] [Google Scholar]
- 5.Chacko B. M., Qin B. Y., Tiwari A., Shi G., Lam S., Hayward L. J., De Caestecker M., Lin K. (2004) Mol. Cell 15, 813–823 [DOI] [PubMed] [Google Scholar]
- 6.Schmierer B., Hill C. S. (2005) Mol. Cell. Biol. 25, 9845–9858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y., Wang Y. F., Jayaraman L., Yang H., Massagué J., Pavletich N. P. (1998) Cell 94, 585–594 [DOI] [PubMed] [Google Scholar]
- 8.Schmierer B., Hill C. S. (2007) Nat. Rev. Mol. Cell Biol. 8, 970–982 [DOI] [PubMed] [Google Scholar]
- 9.Derynck R., Zhang Y. E. (2003) Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 10.Moustakas A., Heldin C. H. (2005) J. Cell Sci. 118, 3573–3584 [DOI] [PubMed] [Google Scholar]
- 11.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 12.Kretzschmar M., Doody J., Timokhina I., Massagué J. (1999) Genes Dev. 13, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori S., Matsuzaki K., Yoshida K., Furukawa F., Tahashi Y., Yamagata H., Sekimoto G., Seki T., Matsui H., Nishizawa M., Fujisawa J., Okazaki K. (2004) Oncogene 23, 7416–7429 [DOI] [PubMed] [Google Scholar]
- 14.Matsuura I., Wang G., He D., Liu F. (2005) Biochemistry 44, 12546–12553 [DOI] [PubMed] [Google Scholar]
- 15.Matsuura I., Denissova N. G., Wang G., He D., Long J., Liu F. (2004) Nature 430, 226–231 [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Feng X., We R., Derynck R. (1996) Nature 383, 168–172 [DOI] [PubMed] [Google Scholar]
- 17.Hoodless P. A., Haerry T., Abdollah S., Stapleton M., O'Connor M. B., Attisano L., Wrana J. L. (1996) Cell 85, 489–500 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y., Feng X. H., Derynck R. (1998) Nature 394, 909–913 [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Chang C., Gehling D. J., Hemmati-Brivanlou A., Derynck R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita M., Fatyol K., Jin C., Wang X., Liu Z., Zhang Y. E. (2008) Mol. Cell 31, 918–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita M., Ying S. X., Zhang G. M., Li C., Cheng S. Y., Deng C. X., Zhang Y. E. (2005) Cell 121, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu L., Hébert M. C., Zhang Y. E. (2002) EMBO J. 21, 3749–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying S. X., Hussain Z. J., Zhang Y. E. (2003) J. Biol. Chem. 278, 39029–39036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X., Letterio J. J., Lechleider R. J., Chen L., Hayman R., Gu H., Roberts A. B., Deng C. (1999) EMBO J. 18, 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cárcamo J., Weis F. M., Ventura F., Wieser R., Wrana J. L., Attisano L., Massagué J. (1994) Mol. Cell. Biol. 14, 3810–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng X. H., Zhang Y., Wu R. Y., Derynck R. (1998) Genes Dev. 12, 2153–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng X. H., Filvaroff E. H., Derynck R. (1995) J. Biol. Chem. 270, 24237–24245 [DOI] [PubMed] [Google Scholar]
- 29.Yu L. R., Conrads T. P., Uo T., Kinoshita Y., Morrison R. S., Lucas D. A., Chan K. C., Blonder J., Issaq H. J., Veenstra T. D. (2004) Mol. Cell. Proteomics 3, 896–907 [DOI] [PubMed] [Google Scholar]
- 30.Aza-Blanc P., Cooper C. L., Wagner K., Batalov S., Deveraux Q. L., Cooke M. P. (2003) Mol. Cell 12, 627–637 [DOI] [PubMed] [Google Scholar]
- 31.Cohen P., Frame S. (2001) Nat. Rev. Mol. Cell Biol. 2, 769–776 [DOI] [PubMed] [Google Scholar]
- 32.Guo X., Ramirez A., Waddell D. S., Li Z., Liu X., Wang X. F. (2008) Genes Dev. 22, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sapkota G., Alarcón C., Spagnoli F. M., Brivanlou A. H., Massagué J. (2007) Mol. Cell 25, 441–454 [DOI] [PubMed] [Google Scholar]
- 34.Wang G., Long J., Matsuura I., He D., Liu F. (2005) Biochem. J. 386, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prokova V., Mavridou S., Papakosta P., Kardassis D. (2005) Nucleic Acids Res. 33, 3708–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kretzschmar M., Doody J., Massagué J. (1997) Nature 389, 618–622 [DOI] [PubMed] [Google Scholar]
- 37.Pera E. M., Ikeda A., Eivers E., De Robertis E. M. (2003) Genes Dev. 17, 3023–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aubin J., Davy A., Soriano P. (2004) Genes Dev. 18, 1482–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu H., Kavsak P., Abdollah S., Wrana J. L., Thomsen G. H. (1999) Nature 400, 687–693 [DOI] [PubMed] [Google Scholar]
- 40.Lin X., Liang M., Feng X. H. (2000) J. Biol. Chem. 275, 36818–36822 [DOI] [PubMed] [Google Scholar]