Abstract
Transcriptional repression through chromatin remodeling and histone deacetylation has been postulated as a driving force for tumorigenesis. We isolated and characterized a novel POZ domain Krüppel-like zinc finger transcription repressor, ZBTB5 (zinc finger and BTB domain-containing 5). Serial analysis of gene expression (SAGE) analysis showed that ZBTB5 expression is higher in retinoblastoma and muscle cancer tissues. Immunocytochemistry showed that ZBTB5 was localized to the nucleus, particularly nuclear speckles. ZBTB5 directly repressed transcription of cell cycle arrest gene p21 by binding to the proximal GC-box 5/6 elements and the two distal p53-responsive elements (bp −2323 ∼ −2299; bp −1416 ∼ −1392). Chromatin immunoprecipitation assays showed that ZBTB5 and p53 competed with each other in occupying the p53 binding elements. ZBTB5 interacted with co-repressor-histone deacetylase complexes such as BCoR (BCL-6-interacting corepressor), NCoR (nuclear receptor corepressor), and SMRT (silencing mediator for retinoid and thyroid receptors) via its POZ domain. These interactions resulted in deacetylation of histones Ac-H3 and Ac-H4 at the proximal promoter, which is important in the transcriptional repression of p21. MTT (3-(4,5-di meth yl thi azol-2-yl)-2,5-diphenyltetrazolium bromide) assays and fluorescent-activated cell sorter analysis revealed that ZBTB5 stimulated both cell proliferation and cell cycle progression, significantly increasing the number of cells in S-phase. Overall, our data suggest that ZBTB5 is a potent transcription repressor of cell cycle arrest gene p21 and a potential proto-oncogene stimulating cell proliferation.
The POZ domain,2 an evolutionarily conserved protein-protein interaction motif found in many regulatory proteins (1, 2), was originally identified in Drosophila melanogaster bric-à-brac, tramtrack, and broad complex transcription regulators and in many pox virus zinc finger proteins (3, 4). As many as 184 known human proteins, 96 Drosophila proteins, and 137 Caenorhabditis elegans proteins are estimated to contain the POZ domain (SMART data base). POZ domain proteins are involved in many critical cellular processes such as apoptosis (5), development (6, 7), ion channel activity (4), oncogenesis (8–10), and transcription (10–16). In particular, some of the POZ domain Krüppel-like zinc finger (POK) proteins are the major determinants of development, differentiation, and oncogenesis. For instance, promyelocytic leukemia zinc finger (PLZF)-null mice display severe defects in limb development and germ stem cell maintenance (7, 17). Th-POK (T-helper-inducing POZ/Krüppel-like factor, also known as cKrox) has been recently reported as a master regulator of T-cell lineage commitment (18). BCL-6 (B cell lymphoma transcription factor-6), PLZF, and HIC1 (Hypermethylated In Cancer) have been implicated in non-Hodgkin lymphoma, acute promyelocytic leukemia, and spontaneous malignant tumors, respectively (8, 9, 19). Recently, FBI-1 (also called Pokemon/LRF/ZBTB7A) was characterized as a proto-oncogenic transcription factor regulating ARF and Rb (retinoblastoma) genes (10, 20) and also as a critical determinant of B versus T lymphoid lineage fate (21).
The most striking and common property of POZ domain transcription factors is their ability to repress transcription via their POZ domains (12–16, 20), although a few actually activate transcription, such as FBI-1 and MIZ-1 in certain promoter contexts (22, 23). This characteristic probably underlies many biological processes controlled by these factors. The ability of the domain to interact with other key regulatory proteins such as corepressor proteins and other transcription factors appears to be important for repression. In particular, the POZ domains of human PLZF and BCL-6 have been shown to interact with SMRT/N-CoR, mSin3A, BCoR, and histone deacetylase (12–16, 20). Chromatin compaction by histone deacetylase complex recruited by the POZ domain was suggested to repress transcription in the case of PLZF-RARa fusion protein (13, 24, 26).
The cyclin-dependent kinase inhibitor p21 is a major player in cell cycle arrest in mammalian cells and the downstream cell-cycle regulator of the ARF-HDM2-p53-p21 pathway (27–29). The p21 gene, mainly regulated at the transcriptional level, is a transcriptional target of tumor suppressor p53 and plays a crucial role in mediating growth arrest when cells are exposed to DNA-damaging agents (Ref. 29 and references therein). Overexpression of p21 results in G1-, G2-, or S-phase arrest upon exposure to DNA-damaging agents (30–32). Whereas induction of p21 predominantly leads to cell cycle arrest, repression of p21 may have a variety of outcomes depending on the cellular context (Ref. 29 and references therein). Aside from p53, a variety of other factors including specificity proteins 1 and 3 (Sp1/Sp3), Smads, Ap2, STAT, BRCA1, E2F-1/E2F-3, and C/EBPα and -β activate the transcription of p21 (29 and references therein). In addition to its role responding to DNA damage, p21 has also been implicated in terminal differentiation, replicative senescence, and protection from p53-dependent and -independent apoptosis (Ref. 29 and references therein).
Sp1 family transcription factors that bind at the proximal promoter (bp −120 to −50) represent another group of major regulators that affect p21 gene expression (Ref. 29 and references therein). Sp1 is one of the best characterized transcription factors that bind to GC-rich DNA sequences in numerous cellular and viral genes (Refs. 33 and 34 and references therein). The six Sp1 binding GC boxes of the p21 proximal promoter have been shown to be important; mutation of the sites not only significantly affects transcription but also disrupts synergistic transcription activation by Sp1 and p53 and other signals that regulate p21 gene transcription (29, 35). Among the six GC boxes found in this region, GC-box 3 mediates p21 induction by various agents such as transforming growth factor-β, butyrate, the histone deacetylase inhibitor trichostatin A, lovastatin, and Ca2+. In contrast, GC-boxes 1 and 2 mediate transcriptional activation by phorbol esters and okadaic acid, the tumor suppressor protein BRCA1, and gut-enriched Krüppel-like factor (GKLF, KLF4). To date, no specific role has been attributed to the most proximal and overlapping GC boxes 5 and 6 (Ref. 29 and references therein). Together, these observations suggest that the specificity of utilizing different proximal GC-boxes under different p21 regulation conditions is important.
From the analysis of amino acid sequences of all available human POZ-domain proteins, we identified a novel ZBTB5 POK protein with a POZ-domain and two unique zinc finger domains. We investigated whether ZBTB5 could regulate any components of the ARF-HDM2-p53-p21 pathway and examined the mechanisms and physiological consequences of ZBTB5 action. ZBTB5 repressed transcription of the p21 gene and significantly increased cell proliferation. Our data suggest that ZBTB5 may be an important transcription regulator of p21 and may play a critical role in regulating important biological processes controlled by p21.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents
p21-Luc plasmid was kindly provided by Dr. Yoshihiro Sowa of the Kyoto Perpetual University of Medicine (Kyoto, Japan). The various pGL2-p21-Luc, pGL2-p53-Luc, pGL2-ARF-Luc, pGL2-HDM2-Luc, pcDNA3.1-p53, pcDNA3.1-Sp1, pG5-5x(GC-box)-Luc, corepressor expression vectors, and VP16-corepressors we used have been reported elsewhere or were prepared by us (20, 23, 25). The pcDNA3-ZBTB5 plasmid was prepared by cloning a cDNA fragment (KIAA0354) into pcDNA3.0 (Invitrogen). The GAL4-POZZBTB5 plasmid was prepared by cloning a cDNA fragment (encoding amino acids 1–135) into a pCMX-Gal4 plasmid. To prepare recombinant GST-POZZBTB5 and GST-ZFZBTB5 proteins, cDNA fragments encoding the POZ domain (amino acids 1–123) and zinc fingers (amino acids 612–766) were cloned into pGEX4T3 (Amersham Biosciences). All plasmid constructs were verified by DNA sequencing.
Antibodies against p21, p53, Sp1, glyceraldehyde-3-phosphate dehydrogenase, FLAG tag, Ac-H3, Ac-H4, HDAC3, and SMRT were purchased from Upstate (Charlottesville, VA), Chemicon (Temecula, CA), Calbiochem, and Santa Cruz Biotechnology (Santa Cruz, CA). Most of the chemical reagents were purchased from Sigma.
Cell Cultures
HEK293A, HCT116 p53+/+, and CV-1 cells were cultured in Dulbecco's modified eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). Saos-2 cells were cultured in McCoy's 5A medium (Invitrogen) supplemented with 15% fetal bovine serum.
Transcriptional Analysis of ARF-, HDM2-, p53-, p21-, and p53-responsive Promoters
pGL2-ARF-Luc, pGL2-HDM2-Luc, pGL2-p53-Luc, pG13-Luc, pG5-5x(GC-box)-Luc, and various pGL2-p21-Luc promoter reporter fusion plasmids as well as pcDNA3-ZBTB5, pcDNA3.1-p53, and pCMV-LacZ in various combinations were transiently transfected into various cell lines (HEK293A, HCT116, Saos-2, and CV-1) using Lipofectamine Plus reagent (Invitrogen). After 24–36 h of incubation, cells were harvested and analyzed for luciferase activity. Reporter activity was normalized with contransfected β-galactosidase activity for transfection efficiency.
Quantitative Real-time PCR of ZBTB5 mRNA Expression in Cells and RT-PCR of Total RNA Prepared from FVB Mouse Tissues
Total RNA was isolated from HEK293A cells using TRIzol reagent (Invitrogen). cDNAs were synthesized using 5 μg of total RNA, random hexamer (10 pmol), and Superscript reverse transcriptase II (200 units) in 20 μl using a reverse transcription kit (Invitrogen). qPCR was performed using SYBR Green Master Mix (Applied Biosystems). The following qPCR oligonucleotide primers sets were used: ZBTB5 forward, 5′-CCACTAGTGACTGCAGGCTG-3′, ZBTB5 reverse, 5′-CCTGCATAGGCCTGACGAA-3′; p21 forward, 5′-AGGGGACAGCAGAGGAAG-3′, p21 reverse, 5′-GCGTTTGGAGTGGTAGAAATCTG-3′; GAPDH forward, 5′-CCCCTTCATTGACCTCAACTAC-3′; GAPDH reverse, 5′-TCTCGCTCCTGGAAGATGG-3′.
To analyze ZBTB5 mRNA expression in FVB mouse, total RNA was isolated as described above from mouse brain, heart, liver, muscle, kidney, spleen, brown adipose tissues, and white adipose tissues, and RT-PCR was carried out using the following oligonucleotide primer sets: ZBTB5 mRNA forward, 5′-TTGCTGTTCACAGCTGCCAC-3′; reverse, 5′-TTAGCCTGCGGGCCTTCCAC-3′.
Western Blot Analysis
Cells were harvested and lysed in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8.0, 1% Nonidet P-40, 0.25% sodium deoxycholic acid, 150 mm NaCl, 1 mm EDTA, Complete Mini-Protease mixture). Cell extracts (40 μg) were separated using 12% SDS-PAGE gel electrophoresis, transferred onto Immun-BlotTM polyvinylidene difluoride membranes (Bio-Rad), and blocked with 5% skim milk (BD Biosciences). Blotted membranes were incubated with antibodies against FLAG tag (Sigma), glyceraldehyde-3-phosphate dehydrogenase (Chemicon), p21, p53, HDAC3, ZBTB5, and SMRT and then incubated with anti-mouse or rabbit secondary antibody conjugated with HRP (Vector Laboratories). Protein bands were visualized with ECL solution (PerkinElmer Life Sciences).
Knockdown of ZBTB5 mRNA by siRNA
Four siRNA against ZBTB5 mRNA were designed and purchased from Dharmacon (Lafayette, CO): siZBTB5-1, 5′-AACUUUACU-3′ and 5′-AGUAAAGUUAU-3′; siZBTB5-2, 5′-AGCUCGCAA-3′ and 5′-UUGCGAGCUCC-3′; siZBTB5-3, 5′-UCCUCAUUU-3′ and 5′-AAAUGAGGACG-3′; siZBTB5-4, 5′-UAAUGGAUG-3′ and 5′-CAUCCAUUACA-3′. siRNA (200 pmol) was transfected into HEK293A cells using Lipofectamine 2000 (Invitrogen). After transfection, the cells were harvested, total RNA was prepared, and RT-qPCR analysis of mRNA was performed as described above.
Quantitative Chromatin Immunoprecipitation (qChIP) Assays
HEK293A cells were transfected with increasing amounts of the ZBTB5 expression vector pcDNA3-FLAG-ZBTB5. The molecular interaction between ZBTB5 and p53 or Sp1 on the endogenous p21 promoter and histone modification at the p21 proximal promoter in the cells was analyzed by the standard qChIP assay protocol, as described elsewhere (20, 23, 25).
Levels of ZBTB5 binding at p53RE-1, -2, and Sp1 binding GC-box 5/6 sites were analyzed by anti-FLAG antibody (Sigma), and levels of binding of endogenous Sp1 and p53 proteins were analyzed by polyclonal antibodies against p53, and Sp1 (Santa Cruz Biotechnology). As a negative control, qChIP assays, IgG was used.
Quantitative PCR of chromatin immunoprecipitated DNA was carried out using the following oligonucleotide primer sets designed to amplify the upstream regulatory regions around p53 binding sites and the proximal promoter region of the p21 gene: p53RE-1 binding primers (bp, −2307 ∼ −1947), forward 5′-CTGTGGCTCTGATTGGCTTT-3′, reverse 5′-GGGTCTTTAGAGGTCTCCTGTCT-3′; p53RE-2 binding primers (bp, −1462 ∼ −1128), forward 5′-CCACAGCAGAGGAGAAAGAAG-3′, reverse 5′-GCTGCTCAGAGTCTGGAAATC-3′. p21 proximal promoter primers (bp −133 ∼ +30), forward 5′-GCGCTGGGCAGCCAGGAGCCT-3′, reverse 5′-CTGACTTCGGCAGCTGCTCAC-3′. To analyze histone H3 and H4 modification at the proximal promoter (bp, −131∼+100), forward 5′-GATCGGTACCGCGCTGGGCAGCCAGGAGCCT-3′, reverse 5′-TCGTCACCCGCGCACTTAGA-3′ primers were used.
Immunoprecipitation Assays
HEK293A cells (transfected with expression vector if necessary) were washed, pelleted, and resuspended in a lysis buffer supplemented with protease inhibitors (20 mm Tris-HCl, pH7.5, 150 mm NaCl, 10% glycerol, 1% Triton X-100). Cell lysate was precleared, and the supernatant was incubated overnight with anti-ZBTB5 (or anti-p53, anti-Sp1 anti-Myc, anti-FLAG, anti-corepressor) antibody on a rotating platform at 4 °C followed by incubation with protein A-Sepharose Fast Flow beads. Beads were collected, washed, and resuspended in equal volumes of 5× SDS loading buffer. Immunoprecipitated proteins were separated with 12% SDS-PAGE. The Western blot assay was performed as described above using appropriate antibodies.
Mammalian Two-hybrid Assays
CV-1 cells were co-transfected with pG5-Luc, pGal4-POZZBTB5, pVP16-corepressors, and pCMV-LacZ using Lipofectamine Plus (Invitrogen). After 36 h of transfection, cells were harvested and assayed for luciferase activity. Luciferase activity was then normalized with co-transfected β-galactosidase activity.
GST Fusion Protein Purification, in Vitro Transcription, and Translation of Corepressors, p53, or Sp1, and Pulldown Assays
Recombinant GST, GST-POZZBTB5, and GST-ZFZBTB5 fusion proteins were prepared from Escherichia coli BL21 (DE3) grown for 4 h at 37 °C in medium containing 1 mm isopropyl 1-thio-β-d-galactopyranoside. The E. coli were lysed and purified using glutathione-agarose 4 bead affinity chromatography (Peptron, Taejeon, Korea). The purified proteins were then resolved with 12% SDS-PAGE to quantitate and assess purity. Corepressor, p53, and Sp1 polypeptides were prepared by incubating 1 μg of pcDNA3-corepressor, pcDNA3.1-p53, or pcDNA3.1-Sp1 expression plasmid with TNT Quick-coupled Transcription/Translation Extract (Promega) containing 40 μl of TNT Quick Master Mix and 2 μl of [35S]methionine (1175.0 Ci/mol) (PerkinElmer Life Sciences) at 30 °C for 90 min. Polypeptide expression levels were then analyzed by running 1 μl of the total mixture through 12% SDS-PAGE and autoradiography.
For GST fusion protein pulldown assays, GST fusion protein-agarose bead complexes were incubated with in vitro translated [35S]methionine-labeled corepressors, p53, and Sp1 polypeptides at 4 °C for 4 h in HEMG buffer (40 mm HEPES, 100 mm KCl, 0.2 mm EDTA, 5 mm MgCl2, 0.1% Nonidet P-40, 10% glycerol, 1.5 mm DTT, protease inhibitors). The reaction mixtures were centrifuged, pellets were rinsed, and the bound proteins were separated using 12% SDS-PAGE. Gels were then exposed to x-ray film using an image-intensifying screen (Eastman Kodak Co.).
Preparation of Recombinant Adenovirus Overexpressing ZBTB5
ZBTB5 cDNA was cloned into the adenovirus E1 shuttle vector pCA14 (Microbix; Ontario, Canada) to generate pCA14-ZBTB5. The pCA14-ZBTB5 shuttle vector was linearized by XmnI digestion, and the adenovirus vector vmdl324Bst (from Dr. Verca at the University of Fribourgh, Switzerland), containing the Ad5 genome deleted in the E1 and E3 region, was also linearized with BstBI digestion. The linearized pCA14-ZBTB5 and the vmdl324Bst digested with BstBI were co-transformed into E. coli BJ518 for homologous recombination. Proper homologous recombinant adenoviral plasmid was digested with PacI and transfected into HEK293 cells to generate adenovirus expressing ZBTB5 (dl324-ZBTB5). Propagation and titration of the recombinant virus was carried out by standard methods. PCR amplification and DNA sequencing using primers specific to ZBTB5 confirmed the adenovirus genotype.
Electromobility Shift Assay (EMSA)
EMSAs were carried out as described previously (20, 23, 25). The probe sequences of Sp1 response elements on the p21 proximal promoter or the sequences of p53 response elements on the p21 distal promoter used in EMSA were as follows (only top strands are shown): GC-box 1, 5′-GATCGGGAGGGCGGTCCCG-3′; GC-box 2, 5′-GATCTCCCGGGCGGCGCG-3′; GC-box 3, 5′-GATCCGAGCGCGGGTCCCGCCTC-3′; GC-box 4, 5′-GATCCTTGAGGCGGGCCCG-3′; GC-box 5/6, 5′-GATCGGGCGGGGCGGTTGTATATCA-3′; p53RE-1, 5′-GATCCGTTAGAGGAAGAAGACTGGGCATGTCTG-3′; p53RE-2, 5′-GATCCATCAGGAACATGTCCCAACATGTTGAGCTC-3′.
Immunocytochemistry
HEK293A cells were transfected with pcDNA3-FLAG-ZBTB5 plasmid, washed, and fixed with cold methanol/formaldehyde. Cells were permeabilized, washed, blocked with horse serum, and incubated with mouse anti-FLAG primary antibody. After thorough washing, cells were further incubated with fluorescein isothiocyanate-conjugated anti-mouse IgG secondary antibody and, finally, soaked with solution containing 4, 6-diamidino-2-phenylindole (1 mg/ml). The cells were mounted and examined with a Carl Zeiss LSM 510 confocal laser scanning microscope.
Oligonucleotide Pulldown Assays
HEK293A cells were lysed in HKMG buffer (10 mm HEPES, pH 7.9, 100 mm KCl, 5 mm MgCl2, 10% glycerol, 1 mm DTT, and 0.5% Nonidet P-40). Cellular extracts were incubated with 1 μg of biotinylated double-stranded oligonucleotides (p53RE-1, p53RE-2, and Sp1–5/6) for 16 h. The sequences of the oligonucleotides are as follows (only top strands are shown): Sp1–5/6, 5′-CCTTGAGGCGGGCCCGGGCGGGGCGGTTGTATATCAGGGC-3′; p53RE-1, 5′-GTCAGGAACATGTCCCAACATGTTGAGCTC-3′; p53RE-2, 5′-TAGAGGAAGAAGACTGGGCATGTCTGGGCA-3′. To collect DNA-bound proteins, the mixtures were incubated with streptavidin-agarose beads for 2 h, washed with HKMG buffer, and precipitated by centrifugation. The precipitate was analyzed by Western blot assay using the antibody against ZBTB5 as described above.
FACS Analysis
HEK293A cells were transfected with ZBTB5 expression vector or siZBTB5 RNA. The cells were washed, fixed with methanol, and stained with solution containing propidium iodide (50 μg/ml) and ribonuclease A (100 μg/ml) for 30 min at 37 °C in the dark. DNA content, cell cycle profiles, and forward scatter were analyzed with a FACSCalibur (BD Biosciences) flow cytometer with emission detection at 488 nm (excitation) and 575 nm (peak emission). Data were analyzed using ModFit LT 2.0 (Verity Software House, Inc.) and WindMDI 2.8 (Joseph Trotter, Scripps Research Institute).
MTT Assay
Confluent HEK293A cells grown on 10-cm culture dishes were transfected with ZBTB5 expression vector or siZBTB5 RNA, then transferred to 6-well culture dishes and grown for 0–4 days. At 0, 1, 2, 3, and 4 days, cells were incubated for 1 h at 37 °C with 20 μl/well MTT (2 mg/ml). Precipitates were dissolved with 1 ml of dimethyl sulfoxide. Cellular proliferation was determined from the conversion of MTT to formazan using a SpectraMAX 250 (Molecular Device Co., Sunnyvale, CA) at 570 nm.
Preparation of Anti-ZBTB5 Antibody
To obtain a rabbit polyclonal antibody against ZBTB5 protein, a white rabbit was immunized subcutaneously with a mixture of recombinant polypeptides, GST-POZ (amino acids 1–123), and GST-ZFDBD (amino acids 612–766) 7 times at 2-week intervals. Blood was collected, incubated at 37 °C for 90 min, and centrifuged. The supernatant was incubated with the protein A/G-agarose beads (Santa Cruz Biotechnology). The beads were collected and washed, and the antibody was eluted.
RESULTS
ZBTB5 Is a New BTB/POK Protein That Represses Transcription of the p21 and HDM2 Genes of the p53 Pathway
We isolated and characterized a novel member of the human POZ protein family, ZBTB5 (zinc finger and BTB domain-containing 5 or KIAA0354), which encodes a protein of 677 amino acid residues. ZBTB5 has a POZ domain at its N terminus (amino acids 1–123) and two zinc finger domains at its C terminus (amino acids 613–664) (Fig. 1A; supplemental Fig. 1). Serial analysis of gene expression (SAGE) analysis shows that ZBTB5 is expressed in most human tissues, and our RT-PCR analysis of mouse total RNA showed that mouse Zbtb5 is also expressed ubiquitously, with particularly high expression in spleen and white adipose tissues (supplemental Fig. 3B). Immunocytochemistry revealed nuclear localization of ZBTB5 (supplemental Fig. 3C). Interestingly, ZBTB5 is highly expressed in retina and muscle cancer tissues (cgap.nci.nih.gov).
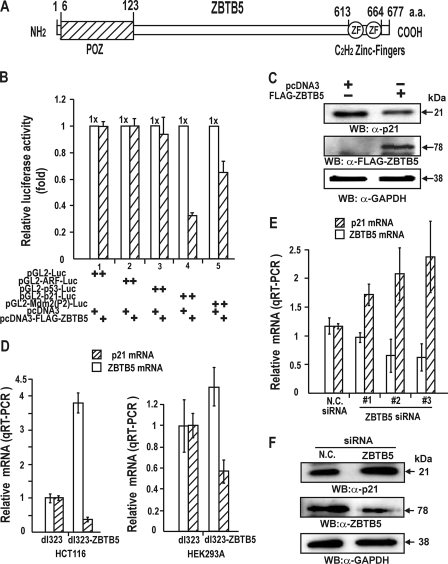
FIGURE 1.
ZBTB5 represses transcription of the p21 and HDM2 genes in HEK293A cells. A, structure of the ZBTB5 protein. Open box, POZ domain; numbered open circles, zinc fingers. a.a., amino acids. B, transcription assays of p53 pathway genes by ZBTB5. ZBTB5 expression vector and promoter-luciferase fusion reporter plasmid were transiently co-transfected into HEK293A cells, and luciferase activity was measured. C, Western blot (WB) analysis of HEK293A cell lysates transiently transfected with ZBTB5 expression vector. GAPDH, control. D, RT-qPCR analysis of the total RNA isolated from HCT116 and HEK293A cells transfected with either control adenovirus (dl324) or recombinant adenovirus (dl324-ZBTB5) overexpressing ZBTB5. E, RT-qPCR analysis of the endogenous p21 and ZBTB5 mRNA after HEK293A cells were transfected with three different siRNAs targeting ZBTB5 or nonsilencing siRNA. Knockdown of ZBTB5 mRNA derepressed p21 gene expression. F, Western blot analysis of HEK293A cell lysates transiently transfected with ZBTB5 siRNA. N.C., scrambled siRNA negative control.
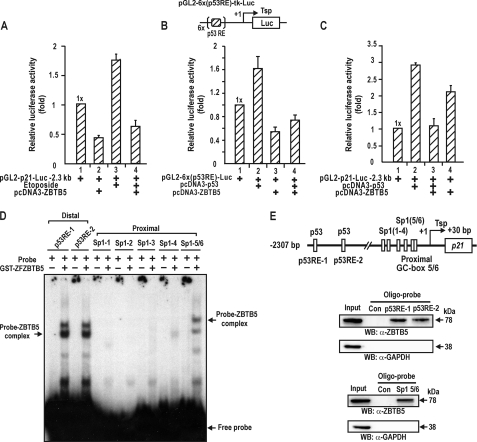
FIGURE 3.
ZBTB5 represses transcription activation by p53. ZBTB5 binds to the distal p53 binding elements and proximal GC-box 5/6 of the p21 gene. A, transcription analysis in HEK293A cells. Etoposide treatment of the cells increased p21 gene expression, which was repressed by ZBTB5. kb, kilobases. B, transcriptional activation of pGL2-6x(p53RE)-Luc by ectopic p53 was repressed by ZBTB5 in Saos-2 cells lacking p53. p53RE, distal p53 binding element of p21. C, transcription analysis. HCT116 p53−/− cells lacking p53 were transiently co-transfected with a mixture of an expression vector of p53 and/or ZBTB5 and pGL2-p21-Luc wild type (−2.3 kilobases), and luciferase activity was measured. D, EMSA. Two [α-32P]dATP-labeled p53RE-1 and -2 probes and Sp1 binding GC-box 5/6 probes were incubated with GST-ZFZBTB5 (0.5 μg) and separated by 4% nondenaturing PAGE. ZBTB5 bound to the distal p53 binding elements and GC-box 5/6 of p21. ZFZBTB5, zinc finger DNA binding domain of ZBTB5. E, oligonucleotide pulldown assay of ZBTB5 binding to the p53 binding elements and proximal GC-box 5/6. HEK293A cells extracts were incubated with biotinylated double-stranded oligonucleotides. The mixtures were further incubated with streptavidin-agarose beads and precipitated by centrifugation. The precipitate was analyzed by Western blot (WB) assay using antibodies against ZBTB5 and Sp1.
Recently, several reports have implicated POZ domain proteins such as FBI-1, BCL-6, and Miz-1 in cell cycle regulation, differentiation, development, and oncogenesis (10, 20, 22). We investigated whether ZBTB5 influenced expression of genes of the p53 pathway, which are important in the regulation of cell cycle. ZBTB5 expression vector and various promoter-Luc fusion reporter constructs were transiently co-transfected and analyzed for reporter luciferase gene expression in HEK293A cells. ZBTB5 repressed transcription of p21 and HDM2 gene expression by 70 and 35%, respectively (Fig. 1B). The ectopic ZBTB5 expressed by plasmid or recombinant ZBTB5 adenovirus repressed endogenous p21 gene transcription (Fig. 1, C and D). Alternatively, knock-down of ZBTB5 mRNA by siRNA derepressed endogenous p21 transcription in HEK293A cells (Fig. 1, E and F). Overall, our data suggest that ZBTB5 is a transcription repressor of p21 gene.
ZBTB5 Represses Transcription of Cell Cycle Arrest p21 Gene and Repression by ZBTB5 Is Dependent on p53 Binding Elements
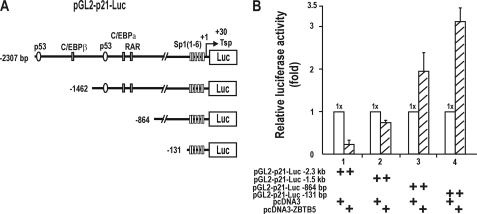
We examined which regulatory elements of the p21 promoter were important for transcriptional repression of p21 by ZBTB5 in HEK293A cells. As ZBTB5 repressed transcription of the endogenous p21 gene (Fig. 1B). ZBZTB5 repressed transcription of two different promoter constructs (−1462 and −2307 bp) by 30–80%, and repression was particularly potent with the −2.3-kilobase promoter. Interestingly, ZBTB5 was not able to repress transcription of shorter promoter constructs (−864 and −131 bp) and instead significantly activated transcription of p21 (190–310%). ZBTB5 transcription activation was particularly potent with the shortest promoter construct (−131 bp), which is highly concentrated in Sp1 binding GC-boxes (Fig. 2, A and B).
FIGURE 2.
ZBTB5 represses transcription of the p21 gene by acting on the distal regulatory element containing p53 binding sites. A, structure of various p21 promoter constructs tested. B, transcription assays. HEK293A cells were transiently co-transfected with ZBTB5 expression vector and pGL2-p21-Luc reporter plasmids with variable upstream sequences and analyzed for luciferase activity.
It appears that ZTB5 significantly repressed transcription of the p21 promoter bearing a −2.3-kilobase upstream sequence containing the two distal p53-binding sites but that repression was somewhat weak with the −1.5-kilobase construct with one p53 binding element. These data suggest that transcriptional repression by ZBTB5 may involve p53 and distal p53 binding elements.
ZBTB5 Represses Transcriptional Activation of p21 by p53 and Binds to the p53 Binding Elements and Proximal Sp1 Binding GC-box 5/6
Because ZBTB5 only significantly repressed transcription of the promoter with distal p53 binding sites, we suspected that the repression mechanisms involved p53 and distal p53 binding elements. We investigated whether ZBTB5 could block the transcriptional activation of p21 by etoposide-activated p53 or by ectopic p53 in HEK293A cells and HCT116 p53−/− cells lacking p53. In HEK293A cells, treatment with the DNA-damaging agent etoposide increased p21 gene expression by inducing p53, which was again repressed by ZBTB5 (Fig. 3A). Additional transcriptional analysis of pG5–6x(p53RE)-Luc with five copies of p53 binding elements of the p21 gene in the proximal promoter showed that ZBTB5 blocked transcription activation by p53 in Saos-2 cells (Fig. 3B). Ectopic p53 expression in HCT116 p53−/− cells increased p21 gene expression, which was repressed by ZBTB5 (Fig. 3C). Interestingly, ZBTB5 increased p21 gene expression in HCT116 p53−/− cells, which is relevant with the transcription activation of the short p21 promoter lacking p53 binding element in HEK293A cells (Fig. 2B).
It is also important to note that ZBTB5 repressed transcription without the induced or ectopic p53 in HEK293A cells on the two promoter constructs (Fig. 3, A and B). These data suggest that ZBTB5 may inhibit transcription of the p21 gene by directly acting on the distal p53 binding elements. Indeed, EMSA showed that the zinc finger DNA binding domain of ZBTB5 can bind to p53RE-1 and -2 (Fig. 3, D and E). Moreover, oligonucleotide pulldown assays showed that endogenous ZBTB5 binds the elements (Fig. 3E). These results imply a potential binding competition between p53 and ZBTB5 on the two p53 binding elements and that the binding competition may be important in transcription repression. In addition, ZBZTB5 also bound to the proximal Sp1 binding GC-box 5/6, although relatively weakly compared with the p53 binding elements in EMSA (Fig. 3, D and E).
ZBTB5 and p53 Compete with Each Other in Binding to the p53 Binding Elements, but ZBTB5 Dramatically Increases Sp1 Binding to the Proximal GC-box 5/6 of Endogenous p21 Gene
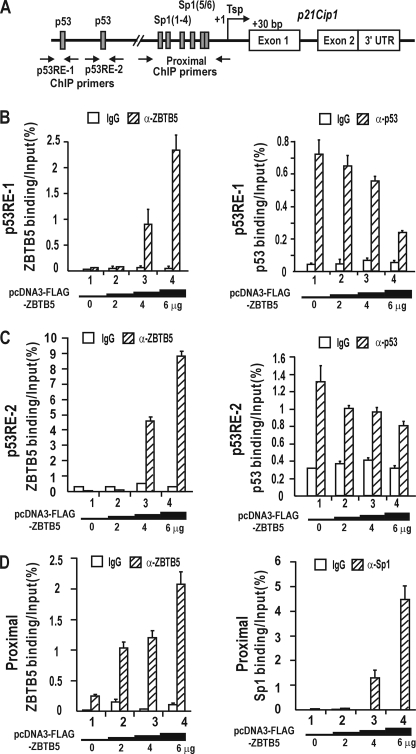
We tested whether the ZBTB5 and p53 proteins compete for sites on the endogenous p21 gene using quantitative ChIP assays. A FLAG-ZBTB5 expression vector was transiently transfected into HEK293A cells, and binding interactions were analyzed on the p53RE-1 and -2 and the proximal GC boxes of endogenous p21. ChIP assays using antibodies against FLAG tag and p53 revealed that ZBTB5 bound to p53RE-1 and -2 by competing with p53 in a dose-dependent manner (Fig. 4, B and C). In contrast, qChIP assays of binding of ZBTB5 and Sp1 on the proximal promoter of endogenous p21 revealed that ZBTB5 not only binds to the region but also dramatically increases Sp1 binding to the region (Fig. 4D), which may explain the synergistic transcription activation on the short p21 promoter construct.
FIGURE 4.
ZBTB5 competes with p53 in binding to the distal p53 binding elements and increases Sp1 binding to the GC-box 5/6 in vivo. A, structure of the endogenous p21 gene. Distal p53 binding elements and proximal GC-rich elements are indicated. Arrows indicate binding positions of the qChIP oligonucleotide primers. Tsp +1, transcription start site. UTR, untranslated region. B and C, qChIP assay of binding competition between p53 and FLAG-ZBTB5 on the distal p53 binding regions of endogenous p21 in HEK293A cells. ZBTB5 competed with p53 in binding to the elements. The cells were transfected with increasing amounts (0–6 μg) of FLAG-ZBTB5 expression vector. D, qChIP assay of FLAG-ZBTB5 and Sp1 binding on the proximal promoter region of endogenous p21 in HEK293A cells. ZBTB5 increases Sp1 binding to the elements. The cells were transfected with increasing amounts of FLAG-ZBTB5 expression vector (0–6 μg). Antibodies against FLAG tag, p53, Sp1, and IgG were used in ChIP assays.
Although the functional significance of the binding of FLAG-ZBTB5 to the proximal promoter GC boxes in the short promoter context remains unclear, our data suggest that ZBTB5 may repress transcription of the endogenous p21 gene by binding competition with p53, thus interfering with p53 binding onto the distal p53 binding elements and communication between p53 and proximal promoter bound Sp1 on the endogenous p21 gene.
ZBTB5 Interacts with p53 or Sp1, and the Interaction May Be Important in the Transcriptional Regulation of p53 or Sp1 Target Genes
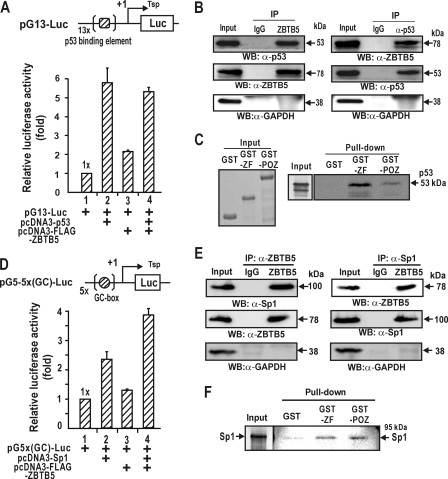
Because transcription repression or activation can be achieved by protein-protein interaction between transcription factors, we investigated whether ZBTB5 interacts with either p53 or Sp1. Co-immunoprecipitation and Western blot assays of HEK293A cells transfected with FLAG-ZBTB5 expression vector revealed that ZBTB5 and p53 or Sp1 interact with each other in vivo (Fig. 5, B and E). The GST fusion protein pulldown assay also showed that the GST-ZFZBTB5 domain and GST-POZZBTB5 interacted with p53 or Sp1 in vitro, suggesting that ZBTB5 interact directly with p53 or Sp1 (Fig. 5, C and F).
FIGURE 5.
ZBTB5 interacts directly with p53 and inhibits transcription activation of pG13-Luc by p53. A, transcriptional activation of pG13-Luc by ectopic p53 can be repressed by ZBTB5 in Saos-2 cells. pG13 contains 13 copies of the p53-binding element. B, co-immunoprecipitation of ZBTB5 and p53. HEK293A cell lysates were immunoprecipitated (IP) using anti-ZBTB5 antibody and analyzed by Western blotting (WB) using anti-p53 antibody. The lysates were also immunoprecipitated by anti-p53 antibody and analyzed by Western blotting using anti-ZBTB5 antibody. C, in vitro GST fusion protein pulldown assays. Left, SDS-PAGE gel of recombinant GST, GST-ZFZBTB5, and GST-POZZBTB5 proteins. Right, recombinant GST, GST-POZZBTB5, or GST-ZFZBTB5 was incubated with [35S]methionine-labeled p53, pulled down, and resolved by 10% SDS-PAGE. The gel was then exposed to x-ray film. Input, 10% of the p53 added in the binding reactions. D, transcriptional activation of pG5-5x(GC-box)-Luc by Sp1 can be synergistically activated by ZBTB5 in HEK293A. pG5-5x(GC-box)-Luc contains five copies of the putative Sp1 binding sites. E, co-immunoprecipitation of ZBTB5 and Sp1. HEK293A cell lysates were immunoprecipitated using anti-ZBTB5 antibody and analyzed by Western blotting using anti-Sp1 antibody. The lysates were also immunoprecipitated by anti-Sp1 antibody and analyzed by Western blotting using anti-ZBTB5 antibody. F, in vitro GST fusion protein pulldown assays. Recombinant GST, GST-POZZBTB5, and GST-ZFZBTB5 was incubated with [35S]methionine-labeled Sp1, pulled down, and resolved by 10% SDS-PAGE. The gels were then exposed to x-ray film. Input, 10% of the labeled Sp1 added in the binding reactions.
To address the functional significance of such protein-protein interactions, we examined whether ZBTB5 affected transcription activation by either p53 or Sp1 on artificial test promoter constructs designed to analyze transcription activation by the two factors. In the case of p53, as shown in the above, transcriptional activation of p21 by p53 was repressed by ZBTB5 (Fig. 3 A and B). Furthermore, transcription of the p53-responsive gene, pGL2-6x(p53RE)-Luc, which contains the p53 binding elements of p21, was repressed by ZBTB5 in Saos-2 cells (Fig. 3C). We observed similar results with pG13-Luc with 13 putative p53 binding sites (Fig. 5A). The data potentially indicate that the protein interactions may contribute to inhibiting transcriptional activation by p53, probably by decreasing p53 binding on the distal p53 binding elements (Fig. 4, B and C).
In contrast to p53, ZBTB5 synergistically activated transcription on the test promoter pG5-5x(GC-box)-Luc, indicating that the Sp1-ZBTB5 interaction might be an important in synergistic transcription activation (Fig. 5D). This discovery is in line with transcription activation of the short proximal promoter of p21 (−131 bp) by ZBTB5 (Fig. 2B) and increased Sp1 binding by ZBTB5 (Fig. 4D) on the proximal promoter of endogenous p21 gene, which is loaded with six Sp1 binding GC boxes.
The POZ Domain of ZBTB5 Interacts with the Corepressor-HDAC Complex to Deacetylate Histones Ac-H3 and Ac-H4 at the Proximal Promoter of Endogenous p21
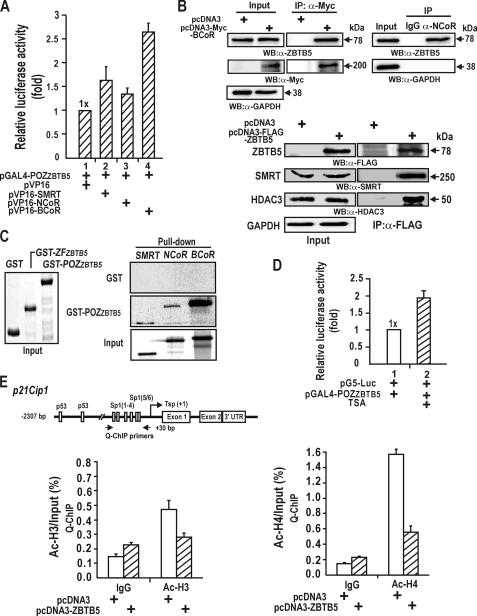
ZBTB5 repressed transcription by direct binding competition with p53 at the distal p53 binding elements (Figs. 1B, 2B, 3, A and B, and Fig. 4, B and C). Transcriptional repressors, including some POZ-domain proteins like PLZF and BCL-6, often repress transcription through interaction with corepressors such as SMRT, NCoR, BCoR, and mSin3A. Mammalian two-hybrid assays in HEK293A cells using pG5-Luc, pGal4-POZZBTB5, and pVP16-corepressor fusion protein expression vectors demonstrated that the POZ domain interacted with SMRT, BCoR, and NCoR (Fig. 6A). Co-immunoprecipitation and Western blot analysis of HEK293A cell extracts or HEK293A cell extracts transfected with FLAG-ZBTB5 expression vector using anti-SMRT co-repressor and anti-HDAC3 antibodies revealed that ZBTB5 interacted with BCoR, NCoR, SMRT, and HDAC3 in vivo (Fig. 6B), indicating that ZBTB5 may inhibit transcription on the p21 proximal promoter by interacting with the co-repressor-HDAC complex via its POZ domain. In addition, GST fusion protein pulldown assays using the recombinant GST-POZZBTB5 protein and in vitro translated [S35]methionine labeled co-repressor polypeptides showed that the POZ domain of ZBTB5 can interact directly with SMRT, BCoR, and NCoR (Fig. 6C).
FIGURE 6.
The POZ domain of ZBTB5 interacts directly with corepressors SMRT, NCoR, and BCoR, and ZBTB5-corepressor-HDAC complexes deacetylate histones Ac-H3 and Ac-H4. A, mammalian two-hybrid assays of protein-protein interactions between the POZ-domain and corepressor proteins. HEK293A cells were transfected with pG5-Luc, pGal4-POZZBTB5, and pVP16-corepressor expression plasmids, and luciferase activity was measured. B, co-immunoprecipitation of ZBTB5, BCoR, NCoR, SMRT, and HDAC3. Cell lysates prepared from HEK293A cells or HEK293A transfected with FLAG-ZBTB5 expression vector were immunoprecipitated (IP) using anti-FLAG or ZBTB5 antibody and analyzed by Western blotting (WB) using anti-Myc-BCoR, NCoR, SMRT, and HDAC3 antibodies. C, in vitro GST fusion protein pulldown assays. Recombinant GST or GST-POZZBTB5 was incubated with [35S]methionine-labeled corepressors, pulled down, and resolved by 10% SDS-PAGE. The gels were then exposed to x-ray film. Input, 10% of the corepressors added in the binding reactions. D, trichostatin A (TSA) treatment derepressed transcriptional repression of pG5-Luc by the Gal4-POZ ZBTB5 domain. Plasmid mixtures of pG5-Luc and the pGal4-POZZBTB5expression plasmid were transiently co-transfected into HEK293A cells. Trichostatin A treatment derepressed transcription of the reporter gene, implicating the involvement of HDACs in transcriptional repression by ZBTB5. E, qChIP assays of histone modification at the proximal promoter of the endogenous p21 gene using antibodies against Ac-H3 and Ac-H4. Cells were transfected with FLAG-ZBTB5 and immunoprecipitated with the indicated antibodies, IgG, Ac-H3, or Ac-H4. ZBTB5 deacetylated histones Ac-H3 and Ac-H4 at the proximal promoter. UTR, untranslated region.
Corepressor complexes recruited by transcriptional repressors often contain HDAC proteins, and ZBTB5 fits this pattern. These HDACs deacetylate the histones of nearby nucleosomes to repress transcription. Treatment of HEK293A cells with the HDAC inhibitor trichostatin A after co-transfection with pG5-Luc, and pGal4-POZZBTB5 significantly affected transcriptional repression by the POZ domain and resulted in a significant increase in transcription (Fig. 6D). These data implicate the involvement of HDACs in transcriptional repression by ZBTB5.
Corepressor-HDACs recruited by ZBTB5 may deacetylate the histones of nearby nucleosomes around the proximal promoter. Accordingly, we used ChIP to examine whether the acetylation status of histones H3 and H4 at the proximal promoter of the endogenous p21 gene was altered by ZBTB5-corepressor-HDACs complexes in HEK293A cells transfected with FLAG-ZBTB5 expression vector. The complex significantly decreased acetylated histones H3 and H4 at the proximal promoter of p21 by 40–65% (Fig. 6E).
ZBTB5 Stimulates Cell Proliferation and Increases the Number of Cells in S-phase
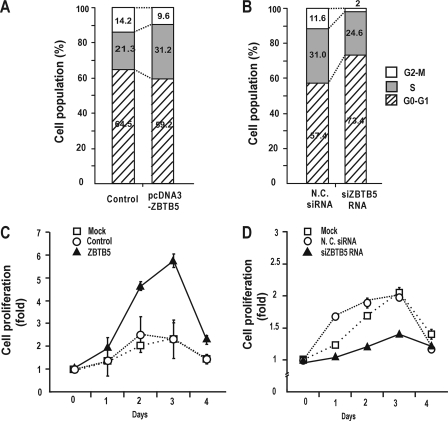
ZBTB5 potently repressed transcription of p21, which is a major regulator of cell cycle arrest. HEK293A cells transfected with ZBTB5 expression vector were analyzed for cell cycle progression by FACS. ZBTB5 stimulated cell cycle progression and increased the number of HEK293A cells in S phase (21.3% in control versus 31.2% in HEK293A-ZBTB5) (Fig. 7A). Knockdown of endogenous ZBTB5 mRNA by siZBTB5 RNA resulted in a decrease in the number of cells in S-phase (31.0% in control versus 24.6% in HEK293A-ZBTB5) and a concomitant increase in the number of cells in the G0-G1 phases (Fig. 7B). MTT assays showed that overexpression of ZBTB5 significantly increased cell proliferation, and knockdown of ZBTB5 mRNA by siRNA decreased cell proliferation in HEK293A cells (Fig. 7, C and D). Overall, our data suggest that ZBTB5 potently stimulates cell growth and proliferation and may be one of the major regulators of cell proliferation by regulating p21 gene expression.
FIGURE 7.
ZBTB5 stimulates cell proliferation and increases the number of cells in S-phase. A, B, FACS analysis of cell cycle progression. HEK293A cells were transfected with ZBTB5 expression vector or control vector, cultured, and stained with propidium iodide. Cell proliferation was measured by FACS. Alternatively, the cells were transfected with siRNA against ZBTB5 mRNA, and cell cycle progression was analyzed. N.C., scrambled siRNA negative control. C, D, MTT assay of HEK293A cells grown for 1, 2, 3, and 4 days. The cells were transfected with either control pcDNA3 vector or pcDNA3-FLAG-ZBTB5 expression vector and analyzed for cell growth. Alternatively, the cells were treated with either negative control siRNA or siZBTB5 RNA. All assays were performed in triplicate. Error bars are included but are too tight to see.
DISCUSSION
We found that ZBTB5 repressed transcription of the p21 and HDM2 genes. Our investigation on transcription regulation of the cell cycle arrest gene p21 by ZBTB5 revealed that p21 is the direct target of ZBTB5. ZBTB5 regulates transcription of the p21 gene through a molecular mechanism that involves p53 and the two upstream p53-responsive elements. ZBTB5 bound to distal p53 binding elements by competing with p53 and repressed the contribution of p53 to transcription. The site has been shown to mediate the induction of p21 gene expression by genotoxic stresses. Accordingly, DNA-damaging signals that result in p53-medicated induction of p21 gene can be blocked by ZBTB5. Overall, these molecular features of ZBTB5 may explain how ZBTB5 acts as a regulatory protein of cell growth and proliferation and, potentially in oncogenesis, by inhibiting p21 transcription.
ZBTB5 binds to the proximal Sp1 binding GC box 5/6, which is a direct target of regulation by Sp1 and Sp family members. Intriguingly, ZBTB5 binding to this particular site increased transcription activation of short promoter by Sp1, although in the much longer promoter context, ZBTB5 repressed transcription of p21. In line with this finding, ChIP assays showed that ZBTB5 significantly increases Sp1 binding to the proximal promoter region, which may explain transcription activation on the short proximal promoter.
Protein interaction between proximal promoter-bound Sp1 and distal p53 is important in the transcriptional activation of the p21 gene, even in the basal level of p53. It appears that the presence of p53 and p53 binding element affects the role of ZBTB5 either as a transcription repressor or activator of the p21 gene. Although ZBTB5 represses transcription of p21 in the p53+/+ HEK293A and HCT116 p53+/+ cells, ZBTB5 stimulates transcription significantly, even on the longer p21 promoter construct or endogenous p21 gene in HCT116 p53−/− cells, probably by the molecular interaction between ZBTB5 and Sp1 on the proximal promoter.3 ZBTB5 may disrupt protein-protein interaction by Sp1 and p53 on the p21 gene to repress transcription. This observation raised the possibility that ZBTB5 is a unique POK family transcription regulator that can act either as a positive or negative regulator of p21 transcription depending on the cellular p53 status, i.e. mutation, absence, or presence.
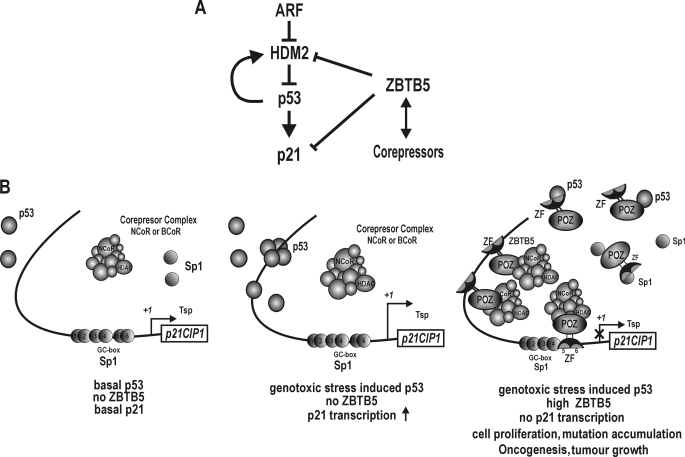
Based on our data, we propose a hypothetical model of transcriptional regulation of p21 by ZBTB5 in p53-positive cells (Fig. 8). Under normal cellular conditions where p53 expression is low and ZBTB5 expression is absent or lower than Sp1, the p21 gene is expressed in low basal levels driven by Sp1, and cells proliferate normally. Challenge with a genotoxic stress induces production of tumor suppressor p53, which binds to the distal p53 responsive elements and activates transcription of p21 by interacting with the Sp1 bound at the proximal GC boxes. The induced p21 arrests cell cycle progression, allowing cells to repair DNA damage. In cells without DNA damage where ZBTB5 expression is high, ZBTB5 represses transcription directly by binding to both the distal p53 binding elements and proximal GC-box 5/6. ZBTB5 bound to the regulatory elements recruits the corepressor-HDAC complex, causing deacetylation of histone Ac-H3 and Ac-H4 around the proximal promoter and repressing transcription.
FIGURE 8.
Hypothetical model of transcriptional regulation of cell cycle arrest gene p21 by ZBTB5. A, p53 pathway and ZBTB5 targets. ZBTB5 represses transcription of the HDM2 and p21 genes. Solid line with arrowhead (→), transcriptional activation; solid line with ⊥, transcriptional repression. Solid line with double arrowhead (↔), molecular interaction. B, hypothetical model of transcriptional repression of p21 by ZBTB5 under three different cellular conditions. ZBTB5 represses transcription of cell cycle arrest gene p21 by binding to the two distal p53 binding elements by competition with p53. ZBTB5 also binds to the proximal Sp1 binding GC-box 5/6, with an unclear function in the transcriptional repression of the endogenous p21 gene by ZBTB5. ZBTB5 recruits co-repressor-HDAC complexes, which deacetylate histones Ac-H3 and Ac-H4 at the proximal promoter to repress transcription. Tsp +1, transcription start site. ZF, zinc finger DNA binding domain. ×, transcription repression.
When cells are under genotoxic stress and ZBTB5 expression is high or in cancerous tissues that have high levels of both p53 and ZBTB5, ZBTB5 represses transcription directly by binding to both the distal p53 binding elements and proximal GC-box 5/6. Although p53 expression is also highly induced under these conditions, p53 has to compete with ZBTB5 to bind to the distal p53 binding elements and is also affected by molecular interactions between p53 and ZBTB5 that further impede binding. Although p53 is present, transcription of p21 is potently repressed by ZBTB5. Cells proliferate without cell cycle arrest, mutations accumulate, and cells are likely to undergo oncogenic transformation (Fig. 8). These series of molecular events may be important in the oncogenesis of retinoblastoma and muscle cancer, where expression of ZBTB5 is high.
Although it can bind to the distal p53 binding elements to repress transcription of p21, ZBTB5 also has characteristics of Sp1 family Krüppel-like zinc finger proteins and binds to some of the GC boxes that are similar to the GC boxes recognized by Sp1. Our findings suggest that some GC boxes recognized by Sp1 may be transcriptional activation targets of ZBTB5 and that Sp1-ZBTB5 binding competition or enhancement may be a general mechanism of transcriptional regulation of some ZBTB5 target genes.
Molecular interactions occurring both in the proximal and distal promoter of the p21 gene are unique and may also be applicable to the transcription regulation of other genes such as HDM2. Indeed, the HDM2 gene has one binding site for p53 in the P2 promoter region and is transcriptionally activated by p53. In contrast, p21 has two p53 binding elements. The difference in the binding site number may explain why ZBTB5 repressed transcription of p21 more potently.
In our laboratory we have observed that other POZ-domain transcription factors such as FBI-1 (Pokemon), ZBTB2, and PLZF repress transcription of p21 by acting on the distal p53 binding elements and proximal Sp1 binding GC-box.4 The common theme of transcriptional regulation of cell cycle regulator gene p21 by POZ-domain class transcription factors is that distal p53 binding elements are the primary target sites of transcription repression, which is eventually conveyed into the histone deacetylation of the proximal promoter.
Although the molecular events involving ZBTB5 on the p53 binding elements are relatively straightforward, their action in the short proximal promoter seems more complex, probably because Sp1 family members, MIZ-1, c-Myc, BCL-6, FBI-1, and ZBTB5 are integrated into the region to either activate or repress transcription. ZBTB5 activates transcription by acting on the short proximal promoter. ZBTB5 may act as transcription activator in the region by interacting with transcription regulators such as Sp family members and MIZ-1 that can bind to the juxtaposed regulatory elements. The physiological importance of the transcription activation of the short p21 promoter (−131 bp) by ZBTB5 is unclear at present because ZBTB5 apparently represses endogenous p21 gene transcription. Taken together, our findings indicate that ZBTB5 may play a critical role in regulating important biological processes such as DNA repair and cell growth, differentiation, and apoptosis by regulating the transcription of p21 and HDM2 of the p53 pathway.
Supplementary Material
This work was supported by Korean Science and Engineering Foundation of the Korean Ministry of Science and Technology Medical Research Center Grant R13-2002-054-05002-0 (to M.-W. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
D.-I. Koh and M.-W. Hur, unpublished data.
W.-I. Choi, and H.-W. Hur, unpublished data.
- POZ
- poxvirus and zinc finger
- POK
- poxviruses and zinc finger (POZ) and Krüppel
- Pokemon
- POK erythroid myeloid ontogenic factor
- ARF
- alternative reading frame gene
- BCL-6
- B-cell lymphoma-6
- BCoR
- BCL-6-interacting corepressor
- BTB
- bric-a brac tramtrack broad complex
- ChIP
- chromatin immunoprecipitation
- qChIP
- quantitative ChIP
- CV-1
- African green monkey kidney cells
- EMSA
- electromobility shift assay
- FACS
- fluorescent activated cell sorter
- FBI-1
- factor that binds to the inducer of short transcripts of human immunodeficiency virus-1
- GST
- glutathione S-transferase
- HDM2
- human analogue of mouse double minute oncogene
- LacZ
- β-galactosidase gene
- Luc
- luciferase gene
- NCoR
- nuclear receptor corepressor
- PLZF
- promyelocytic leukemia zinc finger protein
- SMRT
- silencing mediator for retinoid and thyroid receptors
- Sp1
- specificity protein 1
- RT
- reverse transcription
- RT-qPCR
- reverse transcription-quantitative PCR
- qPCR
- quantitative PCR
- ZBTB5
- zinc finger bric-a brac tramtrack broad complex protein 5
- ZFDBD
- zinc finger DNA binding domain
- HDAC
- histone deacetylase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- siRNA
- small interfering RNA.
REFERENCES
- 1.Bardwell V. J., Treisman R. (1994) Genes Dev. 8, 1664–1677 [DOI] [PubMed] [Google Scholar]
- 2.Albagli O., Dhordain P., Deweindt C., Lecocq G., Leprince D. (1995) Cell Growth Differ. 6, 1193–1198 [PubMed] [Google Scholar]
- 3.Koonin E. V., Senkevich T. G., Chernos V. I. (1992) Trends Biochem. Sci. 17, 213–214 [DOI] [PubMed] [Google Scholar]
- 4.Aravind L., Koonin E. V. (1999) J. Mol. Biol. 285, 1353–1361 [DOI] [PubMed] [Google Scholar]
- 5.Yamochi T., Kaneita Y., Akiyama T., Mori S., Moriyama M. (1999) Oncogene 18, 487–494 [DOI] [PubMed] [Google Scholar]
- 6.Farkas G., Gausz J., Galloni M., Reuter G., Gyurkovics H., Karch F. (1994) Nature 371, 806–808 [DOI] [PubMed] [Google Scholar]
- 7.Barna M., Hawe N., Niswander L., Pandolfi P. P. (2000) Nat. Genet. 25, 166–172 [DOI] [PubMed] [Google Scholar]
- 8.Chen Z., Brand N. J., Chen A., Chen S. J., Tong J. H., Wang Z. Y., Waxman S., Zelent A. (1993) EMBO J. 12, 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerckaert J. P., Deweindt C., Tilly H., Quief S., Lecocq G., Bastard C. (1993) Nat. Genet. 5, 66–70 [DOI] [PubMed] [Google Scholar]
- 10.Maeda T., Hobbs R. M., Merghoub T., Guernah I., Zelent A., Cordon-Cardo C., Teruya-Feldstein J., Pandolfi P. P. (2005) Nature 433, 278–285 [DOI] [PubMed] [Google Scholar]
- 11.Deltour S., Guerardel C., Leprince D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14831–14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhordain P., Albagli O., Lin R. J., Ansieau S., Quief S., Leutz A., Kerckaert J. P., Evans R. M., Leprince D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin R. J., Nagy L., Inoue S., Shao W., Miller W. H., Jr., Evans R. M. (1998) Nature 391, 811–814 [DOI] [PubMed] [Google Scholar]
- 14.Dong S., Zhu J., Reid A., Strutt P., Guidez F., Zhong H. J., Wang Z. Y., Licht J., Waxman S., Chomienne C., Chen Z., Zelent A., Chen S. J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3624–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang C. C., Ye B. H., Chaganti R. S., Dalla-Favera R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6947–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynh K. D., Fischle W., Verdin E., Bardwell V. J. (2000) Genes Dev. 14, 1810–1823 [PMC free article] [PubMed] [Google Scholar]
- 17.Costoya J. A., Hobbs R. M., Barna M., Cattoretti G., Manova K., Sukhwani M., Orwig K. E., Wolgemuth D. J., Pandolfi P. P. (2004) Nat. Genet. 36, 653–659 [DOI] [PubMed] [Google Scholar]
- 18.Sun G., Liu X., Mercado P., Jenkinson S. R., Kypriotou M., Feigenbaum L., Galéra P., Bosselut R. (2005) Nat. Immunol. 6, 373–381 [DOI] [PubMed] [Google Scholar]
- 19.Chen W., Cooper T. K., Zahnow C. A., Overholtzer M., Zhao Z., Ladanyi M., Karp J. E., Gokgoz N., Wunder J. S., Andrulis I. L., Levine A. J., Mankowski J. L., Baylin S. B. (2004) Cancer Cell 6, 387–398 [DOI] [PubMed] [Google Scholar]
- 20.Jeon B. N., Yoo J. Y., Choi W. I., Lee C. E., Yoon H. G., Hur M. W. (2008) J. Biol. Chem. 283, 33199–33210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maeda T., Merghoub T., Hobbs R. M., Dong L., Maeda M., Zakrzewski J., van den Brink M. R., Zelent A., Shigematsu H., Akashi K., Teruya-Feldstein J., Cattoretti G., Pandolfi P. P. (2007) Science 316, 860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan R. T., Saito M., Basso K., Niu H., Dalla-Favera R. (2005) Nat. Immunol. 6, 1054–1060 [DOI] [PubMed] [Google Scholar]
- 23.Choi W. I., Jeon B. N., Park H., Yoo J. Y., Kim Y. S., Koh D. I., Kim M. H., Kim Y. R., Lee C. E., Kim K. S., Osborne T. F., Hur M. W. (2008) J. Biol. Chem. 283, 29341–29354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy L., Kao H. Y., Chakravarti D., Lin R. J., Hassig C. A., Ayer D. E., Schreiber S. L., Evans R. M. (1997) Cell 89, 373–380 [DOI] [PubMed] [Google Scholar]
- 25.Lee J. A., Suh D. C., Kang J. E., Kim M. H., Park H., Lee M. N., Kim J. M., Jeon B. N., Roh H. E., Yu M. Y., Choi K. Y., Kim K. Y., Hur M. W. (2005) J. Biol. Chem. 280, 28061–28071 [DOI] [PubMed] [Google Scholar]
- 26.Grignani F., De Matteis S., Nervi C., Tomassoni L., Gelmetti V., Cioce M., Fanelli M., Ruthardt M., Ferrara F. F., Zamir I., Seiser C., Grignani F., Lazar M. A., Minucci S., Pelicci P. G. (1998) Nature 391, 815–818 [DOI] [PubMed] [Google Scholar]
- 27.el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 28.Toledo F., Wahl G. M. (2006) Nat. Rev. Cancer 6, 909–923 [DOI] [PubMed] [Google Scholar]
- 29.Gartel A. L., Radhakrishnan S. K. (2005) Cancer Res. 65, 3980–3985 [DOI] [PubMed] [Google Scholar]
- 30.el-Deiry W. S., Harper J. W., O'Connor P. M., Velculescu V. E., Canman C. E., Jackman J., Pietenpol J. A., Burrell M., Hill D. E., Wang Y., Wiman K.G., Mercer W. E., Kastan M. B., Kohn K. W., Elledge S. J., Kinzler K. W., Vogelstein B. (1994) Cancer Res. 54, 1169–1174 [PubMed] [Google Scholar]
- 31.Niculescu A. B., 3rd, Chen X., Smeets M., Hengst L., Prives C., Reed S. I. (1998) Mol. Cell. Biol. 18, 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogryzko V. V., Wong P., Howard B. H. (1997) Mol. Cell. Biol. 17, 4877–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. (1987) Cell 51, 1079–1090 [DOI] [PubMed] [Google Scholar]
- 34.Kaczynski J., Cook T., Urrutia R. (2003) Genome Biology 4, 206.1–206.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koutsodontis G., Tentes I., Papakosta P., Moustakas A., Kardassis D. (2001) J. Biol. Chem. 276, 29116–29125 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.