Abstract
Proteinase 3 (Pr3), the main target of anti-neutrophil cytoplasmic antibodies, is a neutrophil serine protease that may be constitutively expressed at the surface of quiescent circulating neutrophils. This raises the question of the simultaneous presence in the circulation of constitutive membrane-bound Pr3 (mPr3) and its plasma inhibitor α1-protease inhibitor (α1-Pi). We have looked at the fate of constitutive mPr3 at the surface of circulating blood neutrophils and of induced mPr3 on triggered neutrophils. We found significant Pr3 activity at the surface of activated neutrophils but not at the surface of quiescent neutrophils whatever the constitutive expression. This suggests that constitutive mPr3 is enzymatically inactive or its active site is not accessible to the substrate. Supporting this conclusion, we have not been able to demonstrate any interaction between constitutive mPr3 and α1-Pi, whereas induced mPr3 is cleared from the cell surface when activated cells are incubated with this inhibitor. But, unlike membrane-bound elastase that is also cleared from the surface of activated cells, mPr3 remained bound to the membrane when inhibited by elafin or by a low molecular weight chloromethyl ketone inhibitor, which shows that it binds more tightly to the neutrophil membrane. mPr3 may thus be present at the surface of circulating neutrophils in an environment replete with α1-Pi. The permanent presence of inactive Pr3 at the surface of quiescent neutrophils may explain why Pr3 is a major target of anti-neutrophil cytoplasmic antibodies, whose binding activates neutrophils and triggers inflammation, as in Wegener granulomatosis.
Proteinase 3 (Pr3)3 is a neutral serine protease (NSP) that is stored in the granules of circulating neutrophils (1, 2) and has been more recently located within secretory vesicles (3). Pr3, like its homologues neutrophil elastase (HNE) and cathepsin G (CG), participates in the intracellular degradation of phagocytized pathogens at inflammatory sites in combination with microbicidal peptides and the membrane-associated NADPH oxidase system (4). All three NSPs are also released from activated neutrophils and help destroy extracellular matrix components and regulate innate immunity, inflammation, and infection (5). Although NSPs are structurally and functionally related and are synthesized similarly (6), Pr3 differs from the other two by its bimodal, genetically determined, expression on the cell surface of quiescent neutrophils (7, 8). Thus, each individual has two subsets of neutrophils, mPr3high and mPr3low, whereas HNE and CG are not present in significant amount at the surface of resting neutrophils. Pr3 also differs from the other two NSPs by its storage within secretory vesicles that readily fuse with the plasma membrane (3). But it is not clear that this explains why Pr3 is constitutively expressed at the surface of a subpopulation of quiescent neutrophils. Supporting this hypothesis, it has been recently demonstrated that CD177 (also called NB1), which is also stored in secretory vesicles and has a bimodal membrane expression, is present on the plasma membrane of the same subset of neutrophils as Pr3 (9, 10).
The presence of Pr3 on the surface of quiescent neutrophils would favor neutrophil activation by anti-neutrophil cytoplasmic antibodies (ANCAs) during Wegener granulomatosis (WG) (11). This explains why this protease, unlike HNE and CG, is a risk factor for this autoimmune disease characterized by necrotizing inflammation particularly of the respiratory tract, kidneys, and by small vessel vasculitis (12). Binding of anti-Pr3 antibodies to tumor necrosis factor-α-primed neutrophils is impaired by α1-Pi (13), which suggests that mPr3 activity and the protease-antiprotease balance are involved in neutrophil activation during WG.
Measuring the Pr3 activity on the cell surface of quiescent and activated neutrophils requires specific substrates of Pr3 that were not available until recently (14, 15). Because of the storage of Pr3 in both secretory vesicles and primary granules and the presence of constitutive Pr3 at the surface of resting neutrophils, we have determined whether both constitutive and induced Pr3 are enzymatically active when bound to the cell surface, and how they are regulated by protease inhibitors. Pr3 activity is controlled by a variety of natural inhibitors, the most important of which are α1-Pi, elafin/trappin-2, and monocyte neutrophil elastase inhibitor. But none is specific for this protease, so it cannot be specifically targeted in vivo or ex vivo. We have previously shown that mHNE is rapidly cleared from the surface of activated neutrophils by α1-Pi and by EPI-hNE4, a low molecular weight recombinant inhibitor, with which it forms soluble, inactive complexes (16, 17). This raises the question of how mPr3 can be targeted by autoantibodies in the presence of α1-Pi, which efficiently inhibits its soluble form, although more slowly than it does HNE (18). We answered this question by investigating the enzymatic properties of mPr3 and its sensitivity to inhibitors. The behavior of mPr3 clearly differs from that of mHNE, which explains why it may be a preferential target for autoantibodies and so contributes to the pathogenicity of Wegener disease.
EXPERIMENTAL PROCEDURES
Materials
Human proteinase 3 (EC 3.4.21.76), human neutrophil elastase (EC 3.4.21.37), and human α1-Pi were obtained from Athens Research & Technology (Athens, GA). The calcium ionophore A23187, elafin, and EGTA were from Sigma. The specific elastase inhibitor EPI-hNE4 was a kind gift from F. Saudubray (Debiopharm, Lausanne, Switzerland). N,N-Dimethylformamide was from Merck. MeO-Suc-AAPA-CMK was from Enzyme System Products (Livermore, CA). PolymorphprepTM and LymphoprepTM were purchased from AbCys (Paris, France). Mouse IgG1 and goat F(ab′)2 fragment anti-mouse IgG (Fcγ)-FITC were from Beckman Coulter (Roissy, France). Anti-Pr3 mAbs (clone MCPR3-2) were purchased from Euromedex (Souffelweyersheim, France).
Isolation of Blood Neutrophils
Human neutrophils were purified from 16-ml samples of peripheral blood collected from healthy volunteers into EDTA-containing tubes essentially as reported previously (19). The neutrophil pellet recovered after lysing the erythrocytes was washed twice in PBS containing 4 mm EGTA. Cell viability was checked by trypan blue exclusion. Neutrophils were activated by suspending ∼3 × 106 cells/ml in PBS containing 1 mm CaCl2 and 1 mm MgCl2 and incubating them with A23187 (1 μm final) for 15 min at 37 °C (19). The resting and activated neutrophils were suspended in PBS/EGTA. We used activation with the calcium ionophore A23187 to optimize mPr3 exposure at the cell surface (20) and to avoid production of neutrophil extracellular traps, as occurs when interleukin-8 or phorbol esters are used, because they may interfere with flow cytometry analyses and enzyme assays (21).
Flow Cytometry Analysis
Flow cytometry was performed on a Beckman Coulter XL flow cytometer equipped with a 488 nm argon laser, and analyses were performed essentially as described in Ref. 19. To analyze specifically interaction of inhibitors with mPr3, mHNE was cleared from the cell surface by preincubating cells (2 × 105 to 5 × 105 quiescent or activated neutrophils) with a molar excess of EPI-hNE4 (5 × 10−8 to 5 × 10−7 m) (17). Any mPr3 antigen at the neutrophil surface was then detected prior to and after α1-Pi or elafin (5 × 10−7 to 10−6 m final) or CMK treatment (10 mm final), incubating the cells with monoclonal anti-Pr3 antibodies (clone MCPR3-2, Euromedex) diluted 1:50 for 30 min at 4 °C. After washing in PBS, cells were incubated with FITC-conjugated F(ab′)2 fragments from goat anti-mouse IgG (diluted 1:50) for 30 min at 4 °C. Mouse IgG1 isotype controls (diluted 1:50) were performed under the same experimental conditions. Data were recorded for at least 10,000 events and analyzed with the Expo32 software (Beckman Coulter, France).
Enzyme Assays
The activities of free and membrane-bound proteases were measured in PBS/EGTA. Free Pr3 and HNE were titrated with α1-Pi, the titer of which had been determined using bovine trypsin titrated with p-nitrophenyl-p′-guanidinobenzoate (22). mPr3 and mHNE activities were quantified by comparing the rates of hydrolysis of their specific FRET substrates (Abz)-VADnorVADRQ-(EDDnp) and (Abz)-APEEIMRRQ-(EDDnp) (15, 23) with that of titrated commercial proteases under the same experimental conditions. The concentration of Abz-peptidyl-EDDnp substrates was determined by measuring the absorbance at 365 nm, using ϵ365 nm = 17,300 m−1 cm−1 for EDDnp. Unactivated and activated (2 × 105 to 5 × 105 cells) neutrophils, or purified proteases used as controls, were incubated with 15 μm specific substrate in polypropylene microplate wells selected for their low binding properties (Hard-Shell Thin-Wall Microplates; MJ Research) at room temperature in activity buffer (10 mm PBS, 4 mm EGTA, pH 7.4). The fluorescence was recorded at λex = 320 nm and λem = 420 nm using a microplate fluorescence reader (Spectra Max Gemini; Molecular Devices) under continuous stirring.
Chromatographic Procedures and Analysis of Peptide Products
Once the enzyme reaction was complete, the reaction medium was incubated with 4 volumes of absolute ethanol for 15 min on ice and centrifuged at 13,000 × g for 10 min. The supernatant containing the hydrolysis products was recovered, air-dried under vacuum, and dissolved in 200 μl of 0.01% trifluoroacetic acid (v/v). Hydrolysis fragments were purified by rp-HPLC on a C18 column (2.1 × 30 mm or 2 × 33 mm, Uptisphere), using a P200 pump coupled to a Spectrasystem UV3000 detector (Thermo Separation Products), at a flow rate of 0.3 ml/min, with a linear (0–60%, v/v) gradient of acetonitrile in 0.01% trifluoroacetic acid over 20 min. Eluted peaks were monitored at three wavelengths (220, 320, and 360 nm) simultaneously, which allowed the direct identification of EDDnp-containing peptides prior to sequencing. Cleavage sites were identified by N-terminal sequencing in an Applied Biosystems Procise 494 protein sequencer attached to a model 140C micro-gradient system and a 610A data analysis system with the chemicals and program recommended by the manufacturer.
Release of Membrane-bound Proteases
The release of mPr3 and mHNE into PBS was checked by incubating cells (3 × 106 cells/ml) in buffer for up to 1 h and measuring the peptidase activities in supernatants cleared of cells by centrifugation for 5 min at 500 × g. Cell pellets were suspended in the same buffer, and membrane-bound activity was measured using the same procedure. The procedure was repeated using the same buffer supplemented with 1.5 m NaCl. Neutrophils were kept for 1 h at room temperature or 24 h at 37 °C under gentle stirring, collected by centrifugation at 500 × g for 15 min, and suspended in PBS containing 1.5 m NaCl (final concentration). The Pr3 and HNE activities in the supernatants of cells were measured and compared with the activities on unfractionated treated cells.
Inhibition of mPr3 by α1-Pi, Elafin, and MeO-Suc-AAPA- CMK
Inhibition of mPr3 by α1-Pi or elafin was analyzed under pseudo-first order conditions using a 1000-fold molar excess of inhibitor. Activated neutrophils (2 × 105 to 5 × 105 cells) corresponding to 5 × 10−10 m to 10−9 m active purified Pr3 were preincubated with EPI-hNE4 (10−8 m to 5 × 10−7 m) for 5 min at room temperature. The Pr3 substrate (15 μm) and α1-Pi or elafin (5 × 10−7 m to 10−6 m) were added at the same time to the reaction medium, and fluorescence was recorded continuously every 24 s for up to 4000 s. A control was made to check that mHNE remained fully inhibited by EPI-hNE4. The inhibition of mPr3 by α1-Pi or elafin was also checked under the same experimental conditions by preincubating the EPI-hNE4-treated cells with α1-Pi or elafin for 4000 s before adding the Pr3 substrate. mPr3 was also inhibited by incubating the cells with 10−4 m MeO-Suc-AAPA-CMK for 2 h. Controls using free Pr3 were performed under the same experimental conditions.
RESULTS
Enzymatic Activity and Stability of Proteinase 3 on the Cell Surface of Purified Neutrophils
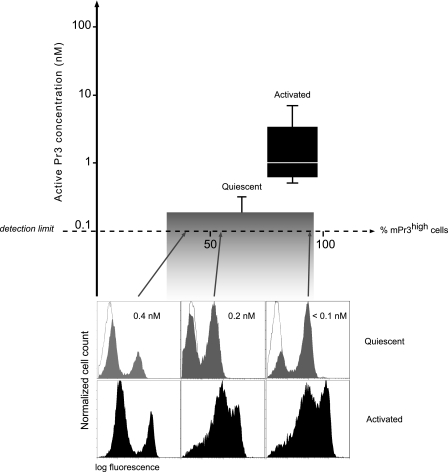
We first determined the Pr3 on the surfaces of resting and activated neutrophils from healthy donors by flow cytometry with anti-Pr3 antibodies. The resting neutrophils had a genetically determined distribution of the protease that resulted in a bimodal mPr3 expression (Fig. 1). The mPr3 activity remains low, generally below the detection limit (10−10 m) whatever the percentage of mPr3high quiescent cells, despite the high cell concentration (300,000 cells/150 μl). Any proteolytic activity detected was because of the partial activation of quiescent neutrophils that occurs during cell purification. This was confirmed by the parallel recording of a basal neutrophil elastase activity that should not be present at the surface of quiescent neutrophils (data not shown). These results strongly suggest that constitutive mPr3 is inactive. All the purified neutrophils had more Pr3 at their surface after they had been activated with the calcium ionophore A23187 (Fig. 1) or after incubation for 2 h in PBS at room temperature (data not shown). The Pr3 activity in the cell suspensions was increased by a factor of 5–20 (Fig. 1). We quantified the mPr3 concentration in the cell suspension by comparing the rate of hydrolysis of Abz-VADnorVADRQ-EDDnp by activated cell suspensions (300,000 cells/150 μl) with that of free, titrated Pr3. For this purpose, we assumed that mPr3 and soluble Pr3 hydrolyze their synthetic low molecular weight substrate similarly. The apparent concentration of mPr3 varied from <0.1 to 4 nm, and this concentration was independent of the percentage of mPr3high resting cells before activation (Fig. 1).
FIGURE 1.
Distribution and activity of membrane-bound proteinase 3 at the surface of quiescent and activated blood neutrophils. Top, box plot (n = 12) of Pr3 activity measured by incubating 300,000 cells/150 μl with the FRET substrate Abz-VADnorVADRQ-EDDnp. The lowest concentration of Pr3 activity that can be quantified is 0.1 nm. The width of the boxes reflects the percentage of mPr3high cells in each sample of quiescent and activated neutrophils, determined by flow cytometry. The selected blood samples are representative of the distribution of mPr3 in the population, which includes only 3% of monomodal mPr3low individuals (43). Bottom, flow cytometry analysis of representative neutrophil preparations from three healthy donors with different bimodal mPr3 distributions before (gray peaks) and after (black peaks) activation with the calcium ionophore A23187 and labeling with the anti-Pr3 mAbs (clone MCPR3–2). The thin lines represent nonspecific binding of isotype control IgG. Arrows indicate the percent of mPr3high quiescent neutrophils. The concentration of active mPr3 in each sample of quiescent neutrophils is indicated in nanomolar. It shows no correlation between the high/low mPr3 status and enzyme activity.
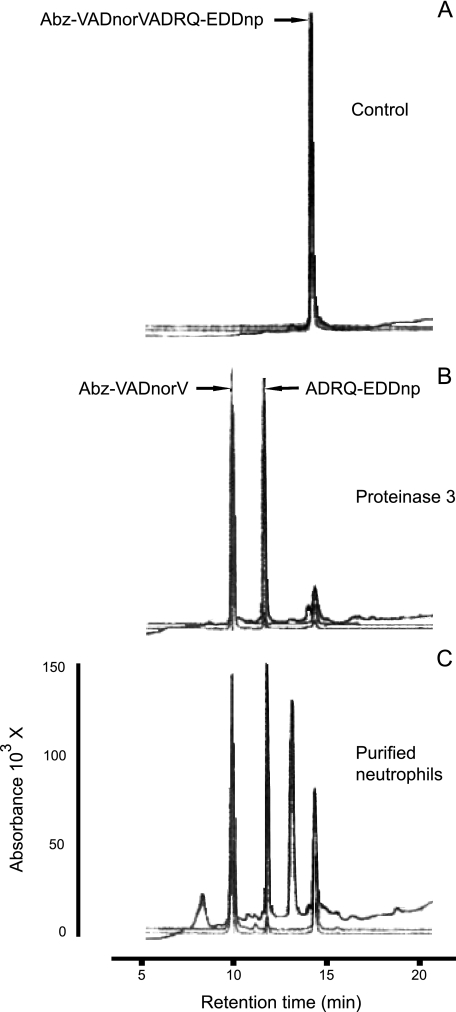
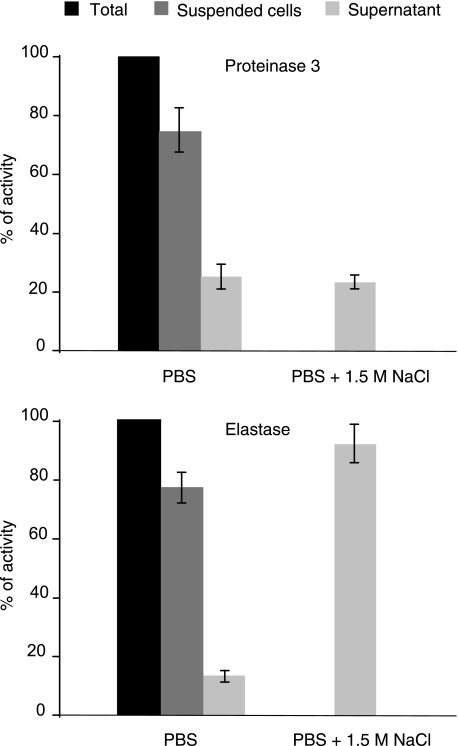
We ensured that no other neutrophil protease had cleaved the Pr3 substrate by checking that there was a single cleavage site at the expected norV-A bond of the FRET substrate. This was done by fractionation of the supernatant by rp-HPLC and N-terminal sequencing of the EDDnp-containing fragment (Fig. 2). We next measured the Pr3 activity in the supernatant after centrifuging the cell suspension at 500 × g for 5 min to ensure that the enzyme activity was mainly because of membrane-bound Pr3. About 25% of total activity was recovered in this supernatant (Fig. 3), and this percentage remained unchanged throughout a 2-h incubation before centrifugation. We checked by flow cytometry that the substrate did not release mPr3 from the membrane (data not shown). We also found that mPr3 was not released from the surface of activated cells placed in buffer containing 1.5 m NaCl, whereas almost all the mHNE was released (Fig. 3). Thus, mPr3 binds differently and more tightly to the cell surface than does mHNE. This raises the question of the inhibition of mPr3 at the cell surface. We investigated the interaction between constitutive and induced mPr3 with natural inhibitors, like α1-Pi and elafin, and synthetic inhibitors to better understand the function of these two mPr3 populations.
FIGURE 2.
Identification by rp-HPLC of the cleavage site within Abz-VADnorVADRQ-EDDnp FRET substrate before (A) and after hydrolysis by free proteinase 3 (B) and purified activated neutrophils (C). The cleavage sites were identified by N-terminal sequencing of the EDDnp-containing fragments having an absorbance peak at 360 nm (data not shown).
FIGURE 3.
Pr3 activity at the surface and in the supernatant of activated neutrophils. About 25% of the total Pr3 activity is released spontaneously when cells are suspended in PBS for at least 120 min, and no further release occurs following incubation with 1.5 m NaCl, whereas almost all the membrane-bound HNE is released into the supernatant after incubation with NaCl.
Inhibition of Membrane-bound Pr3 by Natural and Synthetic Inhibitors
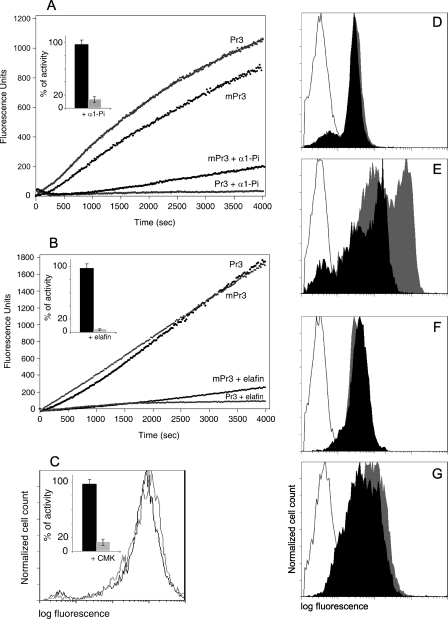
We first checked whether active Pr3 at the membrane surface of activated neutrophils was inhibited by α1-Pi, its main natural inhibitor. Because α1-Pi is not specific for Pr3 and preferentially inhibits HNE (18), we cleared mHNE from the cell surface using a molar excess of the low molecular weight HNE-specific inhibitor EPI-hNE4 (17). We ensured by kinetic analysis that no HNE activity remained in the reaction mixture, using an HNE-specific substrate, and that the Pr3 activity remained unchanged after EPI-hNE4 treatment (data not shown). We then added a large molar excess of α1-Pi (0.5–1 μm final concentration, close to its pathophysiological plasma concentration) to a suspension of triggered neutrophils (200–500 × 103 cells) whose mPr3 activity was 0.2 and 1 nm. We used soluble Pr3 as a control and adjusted it to obtain the same rate of hydrolysis. The rates of inhibition of soluble and mPr3 were then recorded under pseudo-first order conditions (I ≫ E) in the cell suspension and in the control. mPr3 was inhibited almost as rapidly as the soluble Pr3, but inhibition was not complete after 1 h of continuous recording (Fig. 4A). Because Pr3 is inhibited by α1-Pi more slowly than HNE, this was probably due to competition between the inhibitor and substrate for binding to newly exposed Pr3 molecules at the membrane surface of activated, unfixed cells. We confirmed this by incubating cells with the inhibitor for 1 h before adding the substrate. No activity was recorded under these conditions. The same result was obtained using a large molar excess of elafin (Fig. 4B) that fully inhibited mPr3 only when it was preincubated with the cell suspension, but not during an assay in the presence of the substrate. A molar excess of the low molecular weight chloromethyl ketone irreversible inhibitor MeO-Suc-AAPA-CMK also inhibited about 90% mPr3 activity after 2 h (Fig. 4C, inset). The next question was whether the newly formed mPr3-inhibitor complexes remain at the membrane surface.
FIGURE 4.
Fate of Pr3 at the neutrophil surface. Activities of soluble Pr3 and mPr3 prior to and after adding a molar excess of α1-Pi (A) or elafin (B). Insets show the percentages of inhibition after incubation for 1 h (mean of three analyses). Neutrophil samples in A and B are from two different healthy donors. Shown is the flow cytometry analysis of membrane-bound Pr3 on mPr3high quiescent blood neutrophils (D) and triggered neutrophils (E) before (gray peak) and after (black peak) incubation with α1-Pi, quiescent and triggered neutrophils before (gray peak) and after (black peak) incubation with elafin (F and G), and triggered neutrophils before (gray line) and after (black line) incubation with MeO-Suc-AAPA-CMK (C). The control isotype is depicted by a gray line (D–G). Neutrophils were labeled with MCPR3–2 mAb and revealed by FITC-conjugated anti-mouse IgG to visualize cell surface Pr3. The displacement of fluorescence in E shows that induced Pr3 is removed from the cell surface by α1-Pi but not by elafin (G) and by MeO-Suc-AAPA-CMK (C), whereas constitutive Pr3 is not (D).
Fate of Membrane-bound Pr3-Inhibitor Complexes
We studied the fate of mPr3-α1-Pi complexes by flow cytometry. Activated cells were first treated with EPI-hNE4 as before to remove mHNE, and the inactivity of HNE and activity of Pr3 were checked. A molar excess of α1-Pi was added to activated cells, which were then incubated with the MCPR3-2 anti-Pr3 mAb. This mAb was used because Western blotting showed that it still recognized Pr3 after binding to α1-Pi, despite the drastic structural rearrangements that occur during the formation of the complex between serpins and their target proteases. The fluorescent peak was not displaced by excess α1-Pi using mPr3high quiescent cells bearing constitutive mPr3 on their surface (Fig. 4D). This confirmed that constitutive mPr3 does not form an irreversible complex with the inhibitor. But whether this is because of an enzymatically inactive protease, or to impaired access to the active site as a result of a different exposure of constitutive mPr3, remains to be determined. The fluorescent peak was displaced when activated cells were incubated with α1-Pi, which strongly suggests that induced Pr3 is removed from the membrane when the complex is formed (Fig. 4E). But the fluorescent peak was not displaced when Pr3high quiescent cells and triggered cells were incubated with elafin (Fig. 4, F and G) or with the low molecular weight chloromethyl ketone inhibitor MeO-Suc-AAPA-CMK that also inhibits Pr3 at the surface of activated cells (Fig. 4C). We conclude that complexes formed between induced mPr3 and low molecular weight inhibitors remain at the cell surface. This differs from mHNE, which is cleared from the surface of triggered neutrophils by incubation with a low molecular weight recombinant inhibitor (17).
DISCUSSION
Pr3 is exposed at the surface of quiescent circulating neutrophils in a genetically determined fashion, unlike HNE or CG (7). We have previously developed Pr3 FRET substrates that can measure subnanomolar Pr3 concentrations (15, 23), one of them was used here to measure the Pr3 activity at the surface of quiescent and triggered neutrophils. When used at a concentration of 300,000 cells/150 μl, only activated neutrophils showed any significant activity against the Pr3-specific FRET substrate Abz-VADnorVADRQ-EDDnp. Some samples of quiescent cells retained detectable activity at their surface, but this was because of the propensity of neutrophils to become activated, especially during the erythrocyte removing step of their purification (24), and to the great sensitivity of the Pr3 FRET substrate (19). But this activity was independent of the mPr3 status of the cell population because some mPr3high cell suspensions had no detectable mPr3 activity. We conclude that constitutive mPr3 is inactive. This is why Pr3 may be present at the surface of circulating quiescent neutrophils without interacting with α1-Pi, despite its huge plasma concentration.
Assuming that membrane binding at the surface of triggered cells does not alter mPr3 kinetic properties, induced mPr3 would be about 10 times less concentrated than mHNE based on the rate of hydrolysis of its specific FRET substrate. But there is no specific pseudo irreversible Pr3 inhibitor presently available that would allow us to titrate mPr3 and to answer this question unambiguously, as we did for mHNE (16). Thus the enzymatic properties of mPr3 might be altered because of the way it is bound to the cell membrane. There is considerable evidence that mPr3 and mHNE bind differently to the membrane. For example, high salt concentrations led to the rapid release of mHNE from triggered neutrophil membranes but not to the release of mPr3. There is also a large cluster of positive charges at the HNE surface that is interrupted in Pr3, making it more electronegative than HNE (15). But mPr3 binding to neutrophil membranes is not only charge-dependent (25), because anchoring to the hydrophobic membrane leaflet (26–28) and covalent attachment to CD16/FcgRIIIb (29, 30) have been reported. Furthermore, mPr3 colocalizes with the adhesion molecule CD11b/CD18 (β2 integrin) (31). The glycosylphosphatidylinositol-anchored glycoprotein CD177 (NB1 antigen), which is bimodally expressed and is present on the same subset of neutrophils as Pr3 (9), has also been shown to mediate the exposure of mPr3 at the cell surface (32). This binding probably occurs via the single hydrophobic cluster on the surface of Pr3 but not on NE or CG (33). The fact that the NB1 antigen is stored in secretory vesicles and secondary granules, but not in primary granules (9), supports the idea that constitutive mPr3 originates from secretory granules, whereas induced Pr3, which is coexpressed with HNE and CG, is mainly stored in primary granules. This different storage within the cell could well result in different binding modes and/or changes in the protease structure at the cell surface that alter its enzymatic properties, but this needs further investigation. It is clear that the NB1 antigen is not the only binding site for mPr3 (34). von Vietinghoff et al. (10) found that there is an NB1-independent presentation early during differentiation in addition to membrane Pr3 presentation via the NB1 receptor. A proform that escapes granular targeting has been demonstrated during synthesis (35). Constitutive Pr3 could be an enzymatically inactive proform that still contains an N-terminal propeptide and is secreted during neutrophil maturation. Constitutive Pr3 could also be a proteolytically degraded and inactivated protease that still binds mAbs and ANCAs.
We confirmed that constitutive and induced mPr3 have different enzymatic properties by studying their interactions with α1-Pi. The simultaneous presence of active mPr3 and its inhibitors in the same physiological compartment has been explained by the resistance of mPr3 to naturally occurring inhibitors (20). But those experiments were done using fixed cells incubated with purified Pr3. Our present study shows that induced mPr3 is inhibited by α1-Pi and by low molecular weight inhibitors. Nevertheless, α1-Pi inhibits HNE faster than Pr3 (6), which means that transient extracellular Pr3 activity may remain when competition occurs between natural substrates and its main physiological inhibitor. Flow cytometry studies showed that mPr3 is cleared from the surface of triggered neutrophils by α1-PI, as is membrane-bound elastase (16). But mPr3 is not released from the cell surface by a canonical inhibitor like elafin or a low molecular weight chloromethyl ketone inhibitor, although these inhibitors fully inhibited its enzymatic activity. This again indicates that the active protease is more tightly bound to the neutrophil membrane than is mHNE, which is released from cell surfaces by its low molecular weight inhibitor EPI-hNE4 (17). Our flow cytometry studies also show that α1-Pi does not release constitutive mPr3 from quiescent neutrophils isolated from individuals having a significant population of mPr3high cells. This confirms that constitutive mPr3 is enzymatically inactive.
Constitutive mPr3 is reported to be a putative pathogenic factor in ANCA-associated vasculitis and chronic inflammatory diseases (36, 37), although this is still a matter of debate (38). But the binding of Pr3-ANCAs to cell surface Pr3, which results in neutrophil activation, remains the most attractive explanation for the contribution of these antibodies to the pathogenesis of Wegener granulomas (39). For mPr3 to interact with ANCAs, it should be present at the cell surface in an environment replete with α1-Pi that can bind to its active site and remove it from the cell surface. But we show here that constitutive mPr3 does not interact with α1-Pi, so it can remain at the surface of quiescent circulating neutrophils even in the presence of huge amounts of inhibitor. Constitutive mPr3 could still interact with circulating ANCAs under these conditions, which explains why mPr3high patients are more susceptible to a relapse during WG than are patients with low concentrations of constitutive mPr3 (11). A previous study reports that ANCAs do not bind to circulating neutrophils in WG patients (40), but this lack of binding could be due to the ANCA-induced activation of neutrophils and their subsequent adhesion to endothelium (38). Tumor necrosis factor-α-primed neutrophils incubated with excess α1-Pi are significantly less sensitive to activation by an anti-Pr3 mAb, suggesting that the inhibitor impairs mAb binding (13). Because induced mPr3 is present at the surface of primed neutrophils in addition to constitutive mPr3, this could be explained by the removal of induced mPr3 from the cell surface by α1-Pi, as was shown using cells that stably expressed the NB1 receptor (33). This also agrees with the recent observation that Pr3 is no longer present on neutrophil membranes when tumor necrosis factor-α activation is suppressed by α1-Pi (41). Other results show that Pr3-ANCAs interfere with the enzymatic activity of Pr3, but they were obtained using soluble Pr3 rather than membrane-bound Pr3 (42). Thus, there is no formal proof that the enzymatic activity of mPr3 modulates activation of neutrophils by anti-Pr3. α1-Pi preferentially inhibits HNE, so that mPr3 is not its main target (18). Hence, designing a serpin-like inhibitor that specifically removes induced mPr3 would help clarify the roles of constitutive and induced mPr3 in the pathogenicity of vasculitides, and serve as a putative therapeutic agent for controlling the neutrophil-induced inflammation that occurs in WG and related diseases.
Acknowledgment
We thank O. Parkes for editing the English text.
Footnotes
- Pr3
- proteinase 3
- mPr3
- membrane-bound proteinase 3
- HNE
- human neutrophil elastase
- mHNE
- membrane-bound human neutrophil elastase
- CG
- cathepsin G
- NSP
- neutral serine protease
- α1-Pi
- α1-protease inhibitor
- ANCAs
- antineutrophil cytoplasmic antibodies
- CMK
- chloromethyl ketone
- FITC
- fluorescein isothiocyanate
- MeO-Suc
- methoxysuccinyl
- FRET
- fluorescence resonance energy transfer
- PBS
- phosphate-buffered saline
- rp-HPLC
- reversed phase-high performance liquid chromatography
- WG
- Wegener granulomatosis
- mAb
- monoclonal antibody
- Abz
- ortho-aminobenzoic acid
- EDDnp
- N-(2,4-dinitrophenyl)ethylenediamine.
REFERENCES
- 1.Calafat J., Goldschmeding R., Ringeling P. L., Janssen H., van der Schoot C. E. (1990) Blood 75, 242–250 [PubMed] [Google Scholar]
- 2.Csernok E., Lüdemann J., Gross W. L., Bainton D. F. (1990) Am. J. Pathol. 137, 1113–1120 [PMC free article] [PubMed] [Google Scholar]
- 3.Witko-Sarsat V., Cramer E. M., Hieblot C., Guichard J., Nusbaum P., Lopez S., Lesavre P., Halbwachs-Mecarelli L. (1999) Blood 94, 2487–2496 [PubMed] [Google Scholar]
- 4.Pham C. T. (2006) Nat. Rev. Immunol. 6, 541–550 [DOI] [PubMed] [Google Scholar]
- 5.Taggart C. C., Greene C. M., Carroll T. P., O'Neill S. J., McElvaney N. G. (2005) Am. J. Respir. Crit. Care Med. 171, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 6.Korkmaz B., Moreau T., Gauthier F. (2008) Biochimie 90, 227–242 [DOI] [PubMed] [Google Scholar]
- 7.Halbwachs-Mecarelli L., Bessou G., Lesavre P., Lopez S., Witko-Sarsat V. (1995) FEBS Lett. 374, 29–33 [DOI] [PubMed] [Google Scholar]
- 8.Schreiber A., Busjahn A., Luft F. C., Kettritz R. (2003) J. Am. Soc. Nephrol. 14, 68–75 [DOI] [PubMed] [Google Scholar]
- 9.Bauer S., Abdgawad M., Gunnarsson L., Segelmark M., Tapper H., Hellmark T. (2007) J. Leukocyte Biol. 81, 458–464 [DOI] [PubMed] [Google Scholar]
- 10.von Vietinghoff S., Eulenberg C., Wellner M., Luft F. C., Kettritz R. (2008) Clin. Exp. Immunol. 152, 508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rossum A. P., Rarok A. A., Huitema M. G., Fassina G., Limburg P. C., Kallenberg C. G. (2004) J. Leukocyte Biol. 76, 1162–1170 [DOI] [PubMed] [Google Scholar]
- 12.Schreiber A., Luft F. C., Kettritz R. (2004) Kidney Int. 65, 2172–2183 [DOI] [PubMed] [Google Scholar]
- 13.Rooney C. P., Taggart C., Coakley R., McElvaney N. G., O'Neill S. J. (2001) Am. J. Respir. Cell Mol. Biol. 24, 747–754 [DOI] [PubMed] [Google Scholar]
- 14.Korkmaz B., Attucci S., Hazouard E., Ferrandiere M., Jourdan M. L., Brillard-Bourdet M., Juliano L., Gauthier F. (2002) J. Biol. Chem. 277, 39074–39081 [DOI] [PubMed] [Google Scholar]
- 15.Korkmaz B., Hajjar E., Kalupov T., Reuter N., Brillard-Bourdet M., Moreau T., Juliano L., Gauthier F. (2007) J. Biol. Chem. 282, 1989–1997 [DOI] [PubMed] [Google Scholar]
- 16.Korkmaz B., Attucci S., Jourdan M. L., Juliano L., Gauthier F. (2005) J. Immunol. 175, 3329–3338 [DOI] [PubMed] [Google Scholar]
- 17.Attucci S., Gauthier A., Korkmaz B., Delépine P., Martino M. F., Saudubray F., Diot P., Gauthier F. (2006) J. Pharmacol. Exp. Ther. 318, 803–809 [DOI] [PubMed] [Google Scholar]
- 18.Korkmaz B., Poutrain P., Hazouard E., de Monte M., Attucci S., Gauthier F. L. (2005) Am. J. Respir. Cell Mol. Biol. 32, 553–559 [DOI] [PubMed] [Google Scholar]
- 19.Korkmaz B., Attucci S., Juliano M. A., Kalupov T., Jourdan M. L., Juliano L., Gauthier F. (2008) Nat. Protoc. 3, 991–1000 [DOI] [PubMed] [Google Scholar]
- 20.Campbell E. J., Campbell M. A., Owen C. A. (2000) J. Immunol. 165, 3366–3374 [DOI] [PubMed] [Google Scholar]
- 21.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y., Zychlinsky A. (2004) Science 303, 1532–1535 [DOI] [PubMed] [Google Scholar]
- 22.Serveau C., Moreau T., Zhou G. X., ElMoujahed A., Chao J., Gauthier F. (1992) FEBS Lett. 309, 405–408 [DOI] [PubMed] [Google Scholar]
- 23.Korkmaz B., Attucci S., Moreau T., Godat E., Juliano L., Gauthier F. (2004) Am. J. Respir. Cell Mol. Biol. 30, 801–807 [DOI] [PubMed] [Google Scholar]
- 24.Fuchs T. A., Abed U., Goosmann C., Hurwitz R., Schulze I., Wahn V., Weinrauch Y., Brinkmann V., Zychlinsky A. (2007) J. Cell Biol. 176, 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owen C. A. (2008) Int. J. Biochem. Cell Biol. 40, 1246–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldmann W. H., Niles J. L., Arnaout M. A. (1999) Eur. J. Biochem. 261, 155–162 [DOI] [PubMed] [Google Scholar]
- 27.Hajjar E., Mihajlovic M., Witko-Sarsat V., Lazaridis T., Reuter N. (2008) Proteins 71, 1655–1669 [DOI] [PubMed] [Google Scholar]
- 28.Kantari C., Pederzoli-Ribeil M., Amir-Moazami O., Gausson-Dorey V., Moura I. C., Lecomte M. C., Benhamou M., Witko-Sarsat V. (2007) Blood 110, 4086–4095 [DOI] [PubMed] [Google Scholar]
- 29.David A., Fridlich R., Aviram I. (2005) Exp. Cell Res. 308, 156–165 [DOI] [PubMed] [Google Scholar]
- 30.Fridlich R., David A., Aviram I. (2006) J. Cell. Biochem. 99, 117–125 [DOI] [PubMed] [Google Scholar]
- 31.David A., Kacher Y., Specks U., Aviram I. (2003) J. Leukocyte Biol. 74, 551–557 [DOI] [PubMed] [Google Scholar]
- 32.von Vietinghoff S., Tunnemann G., Eulenberg C., Wellner M., Cristina Cardoso M., Luft F. C., Kettritz R. (2007) Blood 109, 4487–4493 [DOI] [PubMed] [Google Scholar]
- 33.Korkmaz B., Kuhl A., Bayat B., Santoso S., Jenne D. E. (2008) J. Biol. Chem. 283, 35976–35982 [DOI] [PubMed] [Google Scholar]
- 34.Hu N., Westra J., Kallenberg C. G. (2009) Autoimmun. Rev. 8, 510–514 [DOI] [PubMed] [Google Scholar]
- 35.Sköld S., Rosberg B., Gullberg U., Olofsson T. (1999) Blood 93, 849–856 [PubMed] [Google Scholar]
- 36.Witko-Sarsat V., Lesavre P., Lopez S., Bessou G., Hieblot C., Prum B., Noël L. H., Guillevin L., Ravaud P., Sermet-Gaudelus I., Timsit J., Grünfeld J. P., Halbwachs-Mecarelli L. (1999) J. Am Soc. Nephrol. 10, 1224–1233 [DOI] [PubMed] [Google Scholar]
- 37.van Rossum A. P., Limburg P. C., Kallenberg C. G. (2003) Clin. Exp. Rheumatol. 21, S64–68 [PubMed] [Google Scholar]
- 38.von Vietinghoff S., Schreiber A., Otto B., Choi M., Göbel U., Kettritz R. (2005) Clin. Nephrol. 64, 453–459 [DOI] [PubMed] [Google Scholar]
- 39.Van Rossum A. P., van der Geld Y. M., Limburg P. C., Kallenberg C. G. (2005) Kidney Int. 68, 537–541 [DOI] [PubMed] [Google Scholar]
- 40.Abdel-Salam B., Iking-Konert C., Schneider M., Andrassy K., Hänsch G. M. (2004) Kidney Int. 66, 1009–1017 [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto T., Kaneko T., Seto M., Wada H., Kobayashi T., Nakatani K., Tonomura H., Tono Y., Ohyabu M., Nobori T., Shiku H., Sudo A., Uchida A., Kurosawa D. J., Kurosawa S. (2008) Clin. Appl. Thromb. Hemost. 14, 186–192 [DOI] [PubMed] [Google Scholar]
- 42.van der Geld Y. M., Tool A. T., Videler J., de Haas M., Tervaert J. W., Stegeman C. A., Limburg P. C., Kallenberg C. G., Roos D. (2002) Clin. Exp. Immunol. 129, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rarok A. A., Stegeman C. A., Limburg P. C., Kallenberg C. G. (2002) J. Am. Soc. Nephrol. 13, 2232–2238 [DOI] [PubMed] [Google Scholar]