Abstract
INI1/hSNF5/BAF47/SMARCB1 is an HIV-1 integrase (IN)-binding protein that modulates viral replication in multiple ways. A minimal IN-binding domain of INI1, S6 (amino acids 183–294), transdominantly inhibits late events, and down-modulation of INI1 stimulates early events of HIV-1 replication. INI1 both stimulates and inhibits in vitro integration depending on IN concentration. To gain further insight into its role in HIV-1 replication, we purified and biochemically characterized INI1. We found that INI1 forms multimeric structures. Deletion analysis indicated that the Rpt1 and Rpt2 motifs form the minimal multimerization domain. We isolated mutants of INI1 that are defective for multimerization using a reverse yeast two-hybrid system. Our results revealed that INI1 residues involved in multimerization overlap with IN-binding and nuclear export domains and are required for nuclear retention and co-localization with IN. Multimerization-defective mutants are also defective for mediating the transdominant effect of INI1-S6-(183–294). Furthermore, we found that INI1 is a minor groove DNA-binding protein. Although IN binding and multimerization are required for INI1-mediated inhibition, the acceptor DNA binding property of INI1 may be required for stimulation of in vitro strand transfer activities of IN. Binding of INI1 to IN results in the formation of presumably inactive high molecular weight IN-INI1 complexes, and the multimerization-defective mutant was unable to form these complexes. These results indicate that the multimerization and IN binding properties of INI1 are necessary for its ability to both inhibit integration and influence assembly and particle production, providing insights into the mechanism of INI1-mediated effects in HIV-1 replication.
HIV-13 replication is a dynamic process that is modulated by the interaction of several host cellular proteins (1). A genome-wide siRNA-mediated knockdown indicated that hundreds of host factors are involved in the stimulation or inhibition of HIV-1 replication (2). Understanding the interplay between the host proteins and the HIV-1 viral proteins is essential to fully comprehend the dynamic relationship between the virus and the host.
INI1/hSNF5/BAF47/SMARCB1 is a core component of the SWI/SNF chromatin-remodeling complex. It interacts directly with the HIV-1-encoded integrase (IN) required for the integration of the viral DNA into the host chromosome (3, 4). IN mediates the insertion of viral cDNA into host chromosomal DNA by sequential steps of 3′ processing and strand transfer (or joining) (4, 5). INI1 binds directly to HIV-1 IN in vitro and in vivo and modulates several steps of HIV-1 replication (3, 6–8). The ectopically expressed minimal IN-binding domain of INI1 transdominantly and potently inhibits HIV-1 assembly and particle production (8). The inhibitory effect is dependent on IN-INI1 interaction and is abrogated when an IN mutant defective for interaction with INI1 is used (8). Furthermore, particle production is minimal in cells lacking INI1, and reintroduction of INI1 into these cells can partially correct the defect (6). These results indicate that INI1 is required for HIV-1 late events. Additional studies have indicated that INI1 is selectively incorporated into HIV-1 but not other retroviral and lentiviral particles (9). Virally encapsidated INI1 is required for post-entry early events of HIV-1 replication prior to integration (6). These studies indicate that producer cell-associated as well as virion-associated INI1 is required for HIV-1 replication. Contrary to these proviral functions of INI1, siRNA-mediated knockdown studies indicate that INI1 in the target cells inhibits early events of HIV-1 replication (7). These studies indicate that whereas INI1 in the target cells may act as an antiviral host protein, HIV-1 may subvert the INI1 antiviral effect, and HIV-1 may utilize this host factor for late events in the producer cells and for early preintegration events in the target cells. Interestingly, in an earlier study, we demonstrated that partially purified INI1 both inhibits and stimulates in vitro integration in a manner dependent on IN concentration (3). Although INI1 stimulates in vitro strand transfer reactions at low IN concentrations, it inhibits the reaction at high concentrations (3). Further structure-function analysis of INI1 is required to understand this complex and dual role of INI1 during HIV-1 replication.
INI1 gene is also a tumor suppressor that is biallelically deleted in aggressive pediatric cancers known as rhabdoid tumors (10). INI1 mutations have been found in other soft tissue cancers (11–13). The mechanism of INI1-mediated tumor suppression is not fully understood. INI1 protein has two highly conserved domains that are imperfect direct repeats (termed Rpt1 and Rpt2) of each other and a third conserved coiled coil domain (termed homology region 3 or HR3) at the C terminus. The Rpt1 and Rpt2 domains appear to be involved in protein-protein interaction with various cellular and viral proteins (3, 14–18). Additionally, the Rpt2 domain harbors a masked nuclear export signal, and the C-terminal domain is involved in inhibiting the nuclear export of the protein in the steady state. INI1 exhibits nonspecific DNA binding activity (18). The cancer-associated mutations occur throughout the open reading frame of the INI1 gene, suggesting that mutation in any one of the INI1 domains may inactivate the protein and that multiple domains are required for its function (19–21).
To gain further insight into the mechanism of its action, we purified INI1 protein to homogeneity and characterized it biochemically. Here we report, for the first time, that INI1 forms dimeric and higher order multimeric structures. We have characterized the multimerization domain of INI1 and found that multimerization and IN binding activities of INI1 are required for inhibition of in vitro integration. Furthermore, we found that the multimerization, IN binding, and nuclear export properties of INI1 are important for transdominant effects. In addition, we found that INI1 possesses a minor groove DNA binding activity and that the nonspecific acceptor DNA binding activity of INI1 may be required for stimulation of in vitro integration. Finally, we found that multimerization of the full-length protein is necessary for its ability to be retained in the nucleus and to co-localize with HIV-1 IN in the nucleus. Thus, our studies provide novel insights into the mechanism by which INI1 regulates HIV-1 replication.
MATERIALS AND METHODS
Plasmids
The plasmids pGEX-INI1, pGEX-IN, pSH2-INI1, and pGADNotINI1 and deletion fragments pGADNotINI1-(183–294), pSH2IN, pBABEpuro-INI1, pCGNINI1, and pQE32-INI1 have been described previously (18). Details of the construction of pQE32-INI1-(141–304) and generation of the random mutagenesis library of pGADNotINI1-(183–294) are as described in the supplemental material. CFP-INI1, DD5, and DD6 constructs were generated by PCR amplification and subcloning of the subsequent fragments at the EcoRI-BamHI sites of pECFP vector (Clontech).
Purification of Proteins
His6-INI1 and His6-INI1-(183–294) were purified to homogeneity as described in the supplemental material. His6-LEDGF was purified as reported (27) except that His6-LEDGF was not digested with PreScission protease, and two purification steps, viz. Ni-NTA and Mono-S Sepharose, were used. The partially purified protein was dialyzed against IN dialysis buffer (3). C-terminal His-tagged IN was purified as described (3).
Yeast Two-hybrid Analysis and GST Pulldown Assays
These tests were performed as described, and details are provided in the supplemental “Materials and Methods” (3).
Glycerol Gradient Centrifugation and Gel Filtration Chromatography
The details of the methods used are provided in the supplemental “Materials and Methods.” Briefly, Mono-Q Sepharose eluate of His-INI1 was separated on a 15–35% glycerol gradient at 40,000 rpm (SW41 Ti rotor) for 24 h. Fractions (400–450 μl) were collected and subjected to trichloroacetic acid precipitation. The precipitated proteins were subjected to Western blot analysis using an anti-His monoclonal antibody as probe. To compare the oligomeric status of wild type and mutant INI1 proteins, the protein was incubated in 500 μl of the buffer for 3 h on ice and then subjected to glycerol gradient centrifugation as described above.
Gel filtration chromatography was performed in 20 mm Tris-HCl (pH 7.5), 100 mm NaCl, 1 mm dithiothreitol, with 200 μg of His-INI1-(183–294) eluate from the Mono-Q Sepharose column. Fractions (500 μl) were collected and subjected to Western blot analysis using anti-His antibody as probe.
Joining Assay, DNA Binding Assay, and EMSA
The joining assays were performed with 32P-labeled 3′ preprocessed duplex DNA (U5.5 and U5.4) as donor DNA and pcDNA as target DNA as described previously (3) using the indicated amounts of proteins except that ∼7 ng of radiolabeled substrate and 100 ng of target DNA were used. Salt concentrations were varied between 10 and 150 mm NaCl.
For agarose-based gel retardation assay about 8 and 16 pmol of hydroxylapatite eluate of the wild type INI1 or mutant INI1 were incubated with 200 ng of pcDNA in buffer containing 20 mm HEPES-KOH (pH 6.8), 100 mm NaCl, 0.1 mm EDTA, 1 mm dithiothreitol, 10% glycerol, 3 mm MnCl2, and protease inhibitors at 30 °C for 1 h. Protein-DNA complexes were resolved by 1% agarose gel electrophoresis and stained with ethidium bromide.
For EMSA ∼7 ng of radiolabeled 3′ preprocessed duplex DNA (U5.5 and U5.4) was incubated with 5 pmol of hydroxylapatite eluate of wild type INI1 in strand transfer reaction buffer and protease inhibitors at 37 °C for 10 min. 10× cold viral LTR DNA was added to the reaction mixture. Minor groove inhibitors were added to the reaction mixture as indicated. About 10 μl of reaction mixture was run on a 6% native polyacrylamide gel made in Tris borate-EDTA, and the gel was run at 10 mA for 2 h. The gel was dried and subjected to autoradiography.
p24 Enzyme-linked Immunosorbent Assay and Confocal Microscopy
These were carried out as described previously, and the details are provided in the supplemental material (8, 23).
RESULTS
Purified Recombinant INI1 Is a Multimer
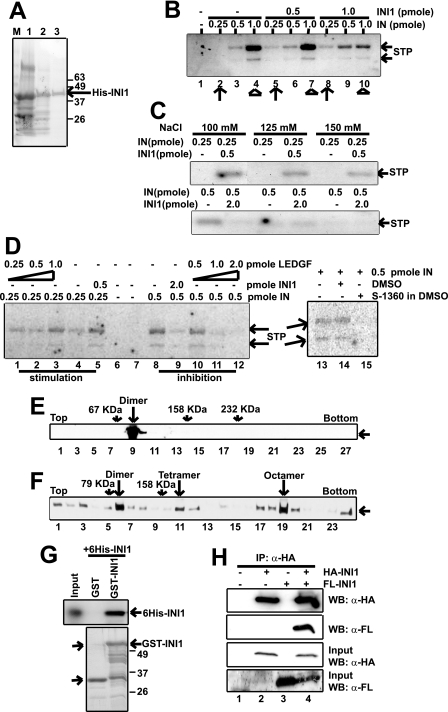
To understand the structure-function relationships of INI1, we biochemically purified bacterially expressed His-tagged INI1 (His-INI1) through three consecutive chromatography steps, viz. Ni-NTA-agarose, hydroxylapatite, and Mono-Q Sepharose columns (see supplemental “Materials and Methods”). After elution through the Mono-Q Sepharose column, INI1 was purified to near homogeneity (>95%; Fig. 1A). To test whether recombinant INI1 is an active protein, we carried out in vitro strand transfer reactions using different concentrations of purified INI1 and different IN:INI1 molar ratios. Previous reports had indicated that INI1 is able to activate and inhibit in vitro strand transfer activity of HIV-1 IN depending on the IN concentration and IN:INI1 molar ratios (3). We employed the hydroxylapatite eluate for carrying out IN reactions because of the poor yield after the Mono-Q column purification step. INI1 stimulated joining of the substrate to the target DNA in a dose-dependent manner at low concentrations (0.25 pmol) of IN (Fig. 1B, compare lane 2 with lanes 5 and 8, indicated by arrows) but inhibited strand transfer reaction at higher concentration of IN (1.0 pmol) (Fig. 1B, compare lanes 4 and 7 with lane 10, indicated by arrowheads). At or near the physiological salt concentration (100, 125, and 150 mm) of NaCl, we found that 0.5 pmol of INI1 stimulated integrase activity at lower integrase input (0.25 pmol) and a lower INI1:IN ratio (2:1) but inhibited integrase activity at higher integrase input (0.5 pmol) and a higher INI1:IN ratio (4:1) (Fig. 1C). At physiological salt concentrations, a higher amount of INI1 was required to obtain robust inhibition of integrase activity. Similar results were obtained with another IN-binding protein, LEDGF. We found that purified full-length LEDGF stimulated IN strand transfer activity at a low IN concentration and an IN:LEDGF ratio of 1:4 and inhibited the reaction at a high IN concentration and an IN:LEDGF ratio of 1:2 (Fig. 1D, lanes 1–12). Furthermore, the addition of 5.3 μm IN inhibitor (S-1360) resulted in inhibition of integrase strand transfer activities under these conditions (Fig. 1D, lanes 13–15).
FIGURE 1.
Active INI1 is a multimer. A, Coomassie Blue staining of protein preparations from different stages of His-INI1 purification, viz. Ni-NTA, hydroxylapatite, and Mono-Q Sepharose. B, in vitro integration assay with increasing amounts of IN (0.25, 0.5, and 1 pmol) and INI1 (0.5 and 1 pmol) as indicated. C, in vitro integration assay at physiological salt concentration as indicated. For stimulation, 0.25 pmol of IN and 0.5 pmol of INI1 were used. For inhibition, 0.5 pmol of IN and 2 pmol of INI1 were used. D, in vitro integration assay with increasing concentrations of IN (0.25 and 0.5 pmol) and LEDGF (0.25, 0.5, 1, and 2 pmol) as indicated and with 5.3 μm integrase inhibitor S-1360. Lane 14 is a control for the addition of inhibitor, where vehicle (DMSO) has been added. E, 15–35% glycerol gradient centrifugation of <5 nm Mono-Q His-INI1. Fractions were analyzed by Western blot using anti-His antibody as probe. BSA, aldolase, and catalase were run in parallel gradients. F, 15–35% glycerol gradient centrifugation of >100 nm Mono-Q His-INI1. Analysis was carried out as described in E. Aldolase was used as a molecular weight marker. G, GST pulldown assay using GST, GST-tagged INI1 immobilized on glutathione-Sepharose beads, and purified His-INI1. Bound proteins were analyzed by Western blot using anti-His antibody as probe. H, coimmunoprecipitation (IP) of FLAG-INI1 using HA-INI1 as bait. 293T cells were transfected with either HA-INI1 or FLAG-INI1 or both, and cell lysates were immunoprecipitated using anti-HA agarose. Immunoprecipitated proteins were analyzed by Western blot (WB) using anti-FLAG and anti-HA antibodies as probes. STP, strand transfer product.
During the course of these studies, we observed that INI1 forms a higher order structure and has a tendency to aggregate at high concentration. This and additional observations using in vitro and in vivo binding studies suggested that INI1 is a multimer, as detailed below. To determine whether INI1 exists as a multimer in solution, purified INI1 eluted from the Mono-Q Sepharose column was subjected to glycerol gradient centrifugation. Purified marker proteins were run as a control to estimate the apparent molecular weight relative to the fractions. At low concentrations (<5 nm), purified INI1 fractionated in a single peak after BSA (67 kDa) in glycerol gradients (Fig. 1E). However, at higher protein concentrations (>100 nm), INI1 migrated in multiple peaks (Fig. 1F). The first peak of INI1 was observed just after the 79-kDa marker, indicating that it is a dimer. Furthermore, other peaks of INI1 migrated with approximate molecular masses indicative of tetramers and octamers and higher order forms (Fig. 1F). Taken together, these studies demonstrated that recombinant INI1 is a dimer in solution at low concentrations but forms higher order structures at high concentrations.
To further confirm that INI1 self-associates, we carried out other in vitro and in vivo interaction assays. A GST pulldown assay was carried out using GST-INI1 and His6-INI1 expressed in bacteria. The bacterial lysates expressing His6-INI1 were treated with DNase I, and the binding reaction was carried out in the presence of ethidium bromide to avoid DNA-mediated association (22). In this assay, although the control GST bound to beads failed to pull down His-INI1, GST-INI1 was able to interact robustly with His-INI1, confirming that INI1 interacts with itself in vitro (Fig. 1G). To investigate whether INI1 self-associates in vivo, 293T cells were transfected with HA-tagged INI1 (HA-INI1) or FLAG-tagged INI1 (FL-INI1) alone or together. The lysates were pretreated with micrococcal nuclease to avoid scoring for a DNA-mediated association. FLAG-INI1 was able to coimmunoprecipitate HA-INI1 (Fig. 1H, lane 4) but not when expressed alone (Fig. 1H, lane 2). Taken together these results demonstrate that INI1 self-associates both in vitro and in vivo.
Conserved Rpt1 and Rpt2 Domains of INI1 Are Involved in Self-association
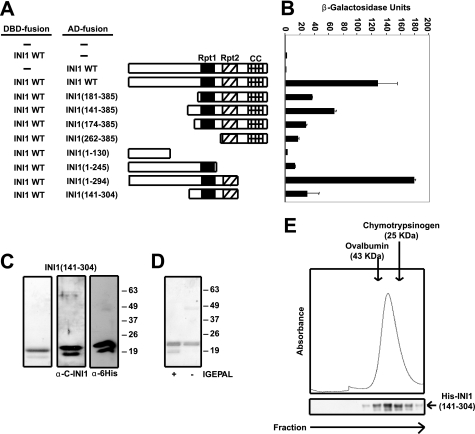
To determine which domain of INI1 is involved in self-association, a deletion analysis was carried out using the yeast two-hybrid assay. A series of truncation mutants of INI1 as a fusion to GAL4AD (Fig. 2A) were tested for their ability to interact with full-length INI1 fused to the LexADBD (Fig. 2B). GAL4AD-INI1 efficiently interacted with LexADBD-INI1 in yeast (Fig. 2B). Mutants with deletion in the N terminus of INI1 that contained the Rpt1, Rpt2, and HR3 domains retained their ability to interact with full-length INI1, whereas the N-terminal fragment INI1-(1–130) failed to interact with LexADBD-INI1 (Fig. 2B). Deletion fragments INI1-(1–245) and INI1-(262–385), containing either the Rpt1 or Rpt2 motif, respectively, retained their ability to bind LexADBD-INI1 (Fig. 2B) although to a lesser extent (10 and 14%, respectively). Truncation of the C-terminal coiled coil domain of INI1 did not affect the ability of INI1-(1–294) to bind LexADBD-INI1 robustly (Fig. 2B). Interestingly, the mutant INI1-(1–294), which retained both the Rpt and the N-terminal domains, exhibited robust activity. These results suggested that although the N-terminal fragment itself is not sufficient for self-association, it may be required for proper folding of INI1 that allows efficient self-association. The fact that INI1-(141–304) retained its ability to bind LexADBD-INI1 (Fig. 2B) demonstrates that the Rpt1 and Rpt2 motifs form the minimal multimerization domain.
FIGURE 2.
A region containing the Rpt1 and Rpt2 motifs comprises the minimal multimerization domain of INI1. A, schematics of wild type (WT) INI1 and various INI1 deletion mutants fused to GAL4AD. Rpt1, repeat 1 motif; Rpt2, repeat 2 motif; CC, coiled coil domain. B, quantitative liquid yeast two-hybrid assay (β-galactosidase/ONPG assay) of LexADBD-fused wild type INI1 and different deletion fragments and wild type INI1 fused to GAL4AD. C, Coomassie Blue staining and Western blot analysis using the indicated antibodies of Mono-Q Sepharose-purified INI1-(141–304). D, Coomassie Blue staining of INI1-(141–304) purified in the presence and absence of detergent (IGEPAL) showing the faster migrating band to be an artifact of detergent solubilization. E, Superdex 200 HR 10/30 gel filtration of Mono-Q Sepharose-purified INI1-(141–304). Fractions collected were analyzed by Western blot using anti-His antibody as probe. Chymotrypsinogen A (25 kDa) and ovalbumin (43 kDa) were used as molecular mass standards.
Minimal Multimerization Domain of INI1 Is a dimer in Solution
To determine whether the minimal multimerization domain of INI1 can self-associate in solution, deletion fragment S7 (amino acids 141–304) was expressed in bacteria and purified through three chromatographic columns, viz. Ni-NTA, hydroxylapatite, and Mono-Q Sepharose, respectively. The purified INI1-(141–304) polypeptide migrated in SDS-PAGE as two bands (Fig. 2C, left panel). Our analysis indicated that the faster migrating polypeptide is neither a degradation product of INI1-(141–304) nor a bacterial contaminant because: (i) antibodies against both the N-terminal His epitope tag and the C terminus of INI1 recognized the two polypeptides in the purified INI1-(141–304) preparation (Fig. 2C, middle and right panels), and (ii) mass spectrometric analysis revealed a single polypeptide species in purified preparations (data not shown). We found that the faster migrating polypeptide was an artifact of detergent solubilization, as it was absent when the protein was purified in the absence of IGEPAL, and the single band stained positively with INI1 antibody (Fig. 2D and data not shown). These results indicated that we were able to purify the deletion fragment to apparent homogeneity.
To investigate the multimeric form of INI1-(141–304) we subjected the purified protein to gel filtration chromatography using a Superdex 200 HR 10/30 column. INI1-(141–304) eluted from the column soon after ovalbumin (43 kDa) and before chymotrypsinogen A (25 kDa), demonstrating that it is a dimer (∼42 kDa) (Fig. 2E). Western blot analysis of fractions eluted from the gel filtration column using an antibody against the His epitope tag confirmed that the eluted protein was INI1-(141–304) (Fig. 2E).
Isolation and Characterization of Multimerization-defective Mutants of INI1
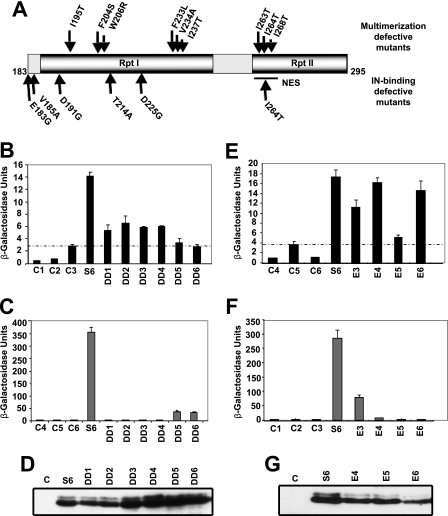
To investigate the functional significance of multimerization of INI1 in HIV-1 replication, we isolated a panel of mutants of the minimal multimerization domain of INI1, S6 (amino acids 183–294), using a reverse yeast two-hybrid system. A random mutation library of INI1-S6-(183–294) fused to GAL4AD (8) was screened against LexADB-INI1 to isolate a panel of multimerization-defective mutants with one or two amino acid substitutions (Table 1). These mutations were in either the Rpt1 or the Rpt2 motif and were mostly hydrophobic (Fig. 3A). These results suggested that hydrophobic interactions are likely to be important for INI1 self-association. Furthermore, among the residues mutated, amino acids Phe-204, Trp-206, Phe-233, Ile-237, Ile-263, Ile-264, and Ile-268 were partially or fully conserved phylogenetically (data not shown). In addition, several clusters of mutations were observed in Rpt1, and mutations in the Rpt2 motif overlapped with the nuclear export signal (NES) of INI1 (Fig. 3A). Previously we had identified mutants of INI1-(183–294) that are defective for interaction with IN (8). Interestingly, we found that several residues of INI1-(183–294) necessary for IN binding were distinct from those required for INI1 multimerization (Fig. 3A and Table 2). Although IN binding-defective mutants were more frequent in the Rpt1 domain, multimerization-defective mutants were found in both the Rpt1 and Rpt2 regions.
TABLE 1.
Interaction of dimerization-defective S6 with INI1 and IN
| S6clone | Mutation in Rpt1 | Mutation in Rpt2 | Interaction with INI1a | Interaction with INa |
|---|---|---|---|---|
| % | % | |||
| S6 | 100 | 100 | ||
| DD1 | F233L | 33 | <1 | |
| DD2 | F204S | 33 | <1 | |
| DD3 | V234A,I237T | 27 | <1 | |
| DD4 | I195T,W206R | 28 | <1 | |
| DD5 | I263T | <5 | 10 | |
| DD6 | I264T,I268T | <1 | 10 |
a Percentage of β-galactosidase activity representing the interaction of wild type S6 protein with wild type INI1 or IN.
FIGURE 3.
Isolation and characterization of INI1-(183–294) mutants defective for multimerization. Amino acid residues involved in multimerization of INI1 are overlapping but distinct from those involved in IN binding. A, identity and position of multimerization-defective mutants of INI1-(183–294). B, quantitative yeast two-hybrid assay (β-galactosidase/ONPG assay) of GAL4AD-fused wild type and mutant INI1-(183–294) with full-length INI1 fused to LexADBD. C, quantitative yeast two-hybrid assay (β-galactosidase/ONPG assay) of GAL4AD-fused wild type and mutant INI1-(183–294) with LexADBD-fused wild type IN. D, Western blot analysis of wild type and mutant INI1-(183–294) containing yeast extracts using anti-GAL4AD antibody as probe. E, quantitative yeast two-hybrid assay (β-galactosidase/ONPG assay) of GAL4AD-fused wild type and mutant INI1-(183–294) and LexADBD-fused wild type INI1. F, quantitative yeast two-hybrid assay (β-galactosidase/ONPG assay) of GAL4AD-fused wild type and mutant INI1-(183–294) and LexADBD-fused wild type IN. G, Western blot analysis of wild type and mutant INI1-(183–294) containing yeast extracts using anti-GAL4AD antibody as probe.
TABLE 2.
Interaction of IN-binding mutants of S6 with INI1 and IN
| S6clone | Mutationin Rpt1 | Mutationin Rpt2 | Interactionwith INa | Interactionwith INI1a |
|---|---|---|---|---|
| % | % | |||
| S6 | 100 | 100 | ||
| E3 | D225G | 26 | 55 | |
| E4 | T214A | <2 | 93 | |
| E5 | V185A | I264T | <1 | 10 |
| E6 | E183G,D191G | <1 | 81 |
a Percentage of β-galactosidase activity representing the interaction of wild type S6 protein with wild type INI1 or IN.
To further investigate the relationship between IN binding and multimerization, we analyzed two panels of mutants of S6 INI-(183–294) for their ability to bind to both INI1 and IN, using a yeast two-hybrid system scoring for β-galactosidase activity (Fig. 3, Table 1). The first panel consisted of mutants isolated as defective for binding to INI1 using the reverse two-hybrid system. The second panel included mutants of S6 INI1-(183–294) that were previously isolated as defective for IN binding (8). We found that the IN binding activity of S6 was stronger than its ability to bind to INI1 (Fig. 3, B–F). In general, the mutants that were defective for multimerization (DD1–DD6) were also defective for IN binding (Fig. 3, B and C). However, there was an inverse correlation between the residual multimerization and IN binding activities of these mutants. The mutants DD1(F233L), DD2(F204S), DD3(V234A,I237T), and DD4(I195T,W206R), which retained partial multimerization (∼30% when compared with that of control INI1-(183–294), were completely defective for binding to IN (compare B and C in Fig. 3). Conversely, the mutants DD5 (I263T) and DD6 (I264T,I268T), which harbored mutations in Rpt2 that were completely defective for INI1 binding, retained a partial ability to bind to IN (∼10%) (Table 1; Fig. 3, B and C).
Similarly, in the reciprocal experiment in which IN binding-defective mutants were tested for INI1 binding, we found that many of the S6 mutants retained their ability to bind to INI1 (Table 2; Fig. 3, E and F). Although the mutants E4 (T214A) and E6 (E183G,D191G) were able to bind INI1 efficiently, the mutant E3 (D225G) bound to INI1 partially, and the mutant E5 (V185A,I264T) was defective (10% activity compared with wild type) in binding to S6 (Fig. 3, E and F; Table 2). These results indicated that although the multimerization domains overlapped with IN-binding domain, there were clear differences between the two interactions. Amino acid residues Glu-183, Asp-191, and Thr-214 in the Rpt1 motif, which are not involved in multimerization, were critical for IN binding. These results indicated that specific residues in the Rpt1 region are necessary for IN binding and are distinct from those required for multimerization. Western blot analysis of yeast extracts containing wild type or mutant INI1-(183–294) proteins using an anti-GAL4AD antibody showed that the proteins accumulated stably in yeast cells (Fig. 3, D and G).
Effect of Multimerization on Nuclear Export of S6 INI1-(183–294) and on Inhibition of HIV-1 Particle Production
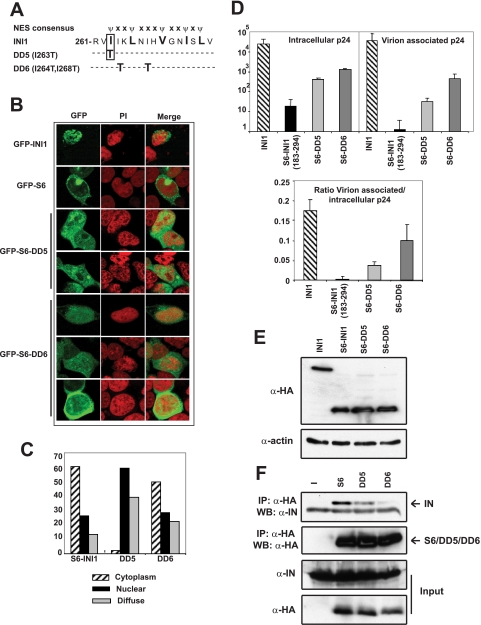
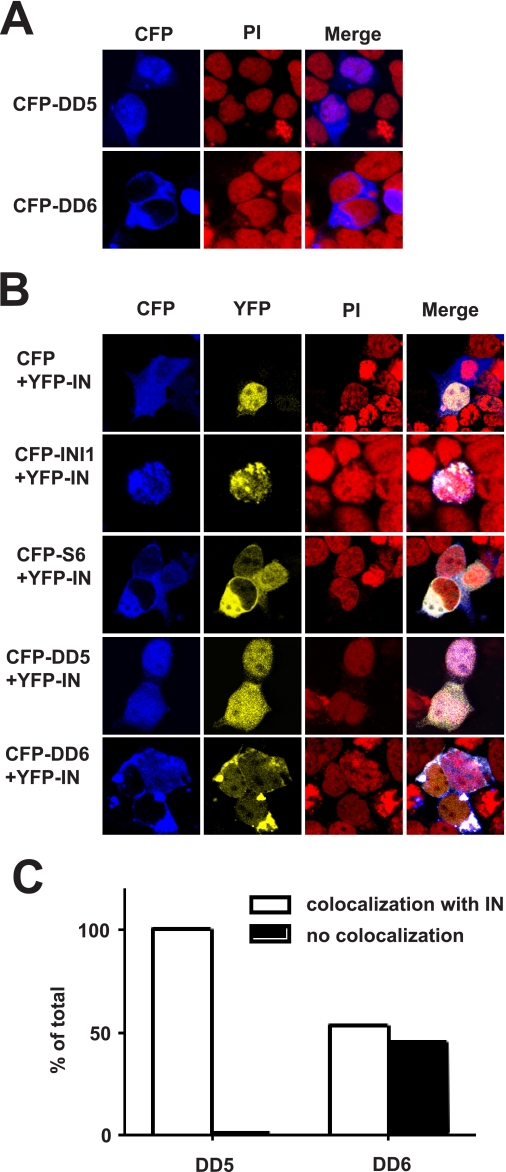
Previously, we demonstrated that INI1-S6-(183–294) harboring the minimal IN-binding domain potently inhibits HIV-1 particle production (8). This fragment is ectopically expressed due to the unmasking of an NES in INI1 (23). In many transcription factors, the nuclear export domains often overlap with multimerization domains (24). Similarly, we observed that the multimerization-defective mutations found in the Rpt2 region of INI1-(183–294)/S6 overlap with the NES region. The NES of INI1 resides within amino acids 263–276, and the hydrophobic residues within this region that match the NES consensus are essential for nuclear export (Ref. 23 and Fig. 4A). We noted that the residue mutated in DD5 (I263T), was one of the five hydrophobic residues essential for nuclear export within this region (Fig. 4A and Ref. 23). On the other hand, the residues mutated in DD6 (I264T,I268T) fell outside the NES consensus sequence (Fig. 4A and Ref. 23). Therefore, we hypothesized that mutation in DD5, but not in DD6, may disrupt nuclear export. The two mutants DD5 (I263T) and DD6 (I264T,I268T), were expressed as GFP fusion proteins in HeLa cells, and the subcellular localization was determined via confocal microscopy. Although GFP-INI1-(183–294)/S6 localized primarily to the cytoplasm (Fig. 4B), the DD5 mutant exhibited reduced cytoplasmic localization and was nuclear in the majority of cells (59%) (Fig. 4, B and C); the DD6 mutant exhibited cytoplasmic localization in the majority of the cells similar to that of S6 (Fig. 4, B and C). These results confirmed our previous observations of nuclear export of INI1 and demonstrated that some residues required for multimerization and nuclear export overlap.
FIGURE 4.
Requirement of multimerization and nuclear export properties for the inhibition of particle production by INI1-S6-(183–294). A, location of DD5 and DD6 mutations with respect to NES. B, confocal microscopy of GFP-fused S6 and DD5 and DD6 mutants. C, subcellular localization of S6 and DD5 and DD6 mutants. Values are expressed as percentages. D, intracellular, virion-associated, and ratio of virion-associated to intracellular p24 in the presence of wild type INI1 or S6, DD5, and DD6 mutants. E, expression of INI1, S6, DD5 and DD6 proteins in 293T cells. F, coimmunoprecipitation (IP) of YFP-IN with HA-tagged S6, DD5, and DD6. Inputs of YFP-IN and HA-tagged proteins are shown. WB, Western blot; PI, propidium iodide.
To determine the contribution of the multimerization and/or nuclear export properties of INI1-S6-(183–294) for its ability to inhibit HIV-1 particle production, the mutants were co-expressed along with HIV-1 viral vectors in 293T cells, and particle production was assessed by measuring the intracellular and virion-associated p24 levels by enzyme-linked immunosorbent assay (Fig. 4D). These results indicated that INI1-(183–294) robustly inhibited virion-associated p24 levels, consistent with previous reports (Fig. 4D, top right panel, and Ref. 8). This decrease was accompanied with a decrease in intracellular p24 levels (Fig. 4D, top left panel). In addition, we found that there was a decrease in the ratio of virion associated to intracellular p24 levels (Fig. 4D, bottom panel), suggesting that S6 inhibits viral particle production by both decreasing the intracellular p24 and by further affecting particle release. Interestingly, the mutations in DD5 (I263T) and DD6 (I264T,I268T) partially rescued the inhibition of particle production (Fig. 4D, top and bottom panels). Wild type and mutant proteins were expressed at similar levels, indicating that rescue in particle production is not due to a defect in expression (Fig. 4E). These results demonstrate that a defect in the multimerization of DD5 and DD6 and a defect in the nuclear export properties of DD5 significantly abrogated their ability to mediate the dominant negative effect.
Because multimerization and IN-binding residues were overlapping, we investigated whether DD5 and DD6 were defective for binding to IN in vivo. We tested the ability of DD5 and DD6 to bind to YFP-IN in 293T cells by coimmunoprecipitation assay. Although HA-tagged INI1-(183–294)/S6 robustly associated with YFP-IN (Fig. 4F), DD5 (I263T) showed an ∼2-fold reduction and DD6 (I264T,I268T) demonstrated a 10-fold reduction in YFP-IN binding activity as compared with the wild type fragment (Fig. 4F). These results suggested that multimerization, IN binding, and cytoplasmic localization were necessary for INI1-(183–294)/S6 to inhibit viral particle production. These studies, taken together, implicate a role for INI1 in maintaining viral protein levels and in the assembly/release of viral particles in the cytoplasm of the producer cells.
Co-localization of IN with INI1
To determine whether the mutations that affect properties of the truncated version of INI1/S6 also affect the full-length protein, the mutations DD5 (I263T) and DD6 (I264T,I268T) were introduced into full-length INI1 by site-directed mutagenesis. To test the ability of IN to co-localize with INI1 and its mutants, INI1, S6, and full-length DD5 and DD6 mutants were expressed as fusion to cyan fluorescent protein, and IN was expressed as fusion to yellow fluorescent protein. We had demonstrated previously that INI1 is primarily nuclear and that the truncated version of the protein (S6) is cytoplasmic (Ref. 8 and Figs. 4 and 5). Here, we first tested the localization properties of DD5 and DD6. As expected, DD5 mutants, within the context of full-length protein, localized primarily to the nuclear compartment with some diffuse localization (Fig. 5A). Interestingly, the DD6 mutant within the context of the full-length protein, with an intact NES, was able to localize to the cytoplasm (Fig. 5A). These results confirm the presence of NES in INI1 and further suggest that multimerization may be important for keeping the protein in the nucleus. We then tested the co-localization of IN with various proteins. IN is primarily nuclear. We found that when IN was co-expressed with INI1, it tightly co-localized with INI1 (Fig. 5B). Furthermore, as long as INI1 mutant proteins retained the ability to bind to IN, they appeared to dictate the subcellular localization of IN (Fig. 5B). For example, expression of S6 (a cytoplasmic mutant of INI1) resulted in cytoplasmic localization of IN (Fig. 5B). IN also co-localized with the DD5 mutant, as it retained the ability to bind to IN (Fig. 5B). Interestingly, IN only partially co-localized with the DD6 mutants (Fig. 5, B and C). These results are consistent with our observation that the DD6 mutant of INI1 is partially defective for binding to IN (Fig. 4F). Together, these results are consistent with our findings that IN and INI1 co-localize in the nucleus in vivo and that this co-localization is dependent on INI1 multimerization and IN binding.
FIGURE 5.
Co-localization of IN with INI1 and mutants. A, confocal microscopy of cells expressing full-length INI1 harboring DD5 and DD6 mutations. B, confocal microscopy to determine the co-localization of yellow fluorescent protein (YFP)-IN with cyan fluorescent protein (CFP)-INI1 and its mutants. C, graphic representation of percentage of cells exhibiting co-localization of IN and INI1 harboring either DD5 or DD6 mutants. PI, propidium iodide.
Modulation of in Vitro Integration by INI1 Is Mediated by Its Ability to Multimerize and Bind to IN
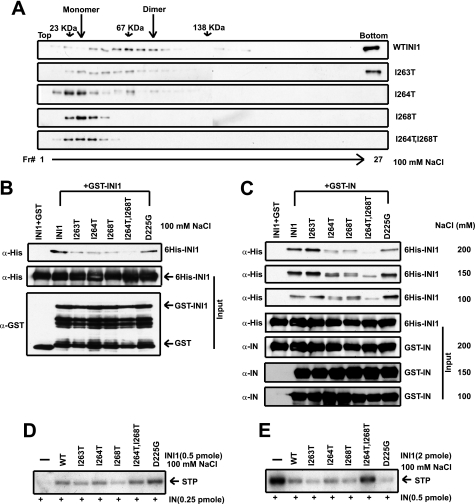
To determine the effect of the multimerization and IN binding properties of INI1, wild type and mutant proteins (DD5, DD6, as well as proteins with single mutations from DD6 (I264T and I268T) were purified through two chromatographic steps, viz. Ni-NTA and hydroxylapatite columns. These proteins were tested for their ability to multimerize, bind to IN, and modulate in vitro strand transfer reactions.
To determine the multimerization status, the purified proteins were subjected to glycerol gradient centrifugation at a low concentration (∼5 nm) to separate various oligomeric forms of INI1. Although wild type INI1 migrated as a dimer and a higher molecular weight species, the mutant DD5 (I263T) was partially defective for multimerization forming both monomeric and dimeric and a higher molecular weight species (Fig. 6A). The DD6A and DD6C mutants with I264T and I268T substitutions, respectively, were defective in multimerization. Although I264T formed less than 10% dimers, I268T formed only monomers (Fig. 6A). Furthermore, the mutant DD6 (I264T,I268T) formed only monomers at this concentration. These results indicate that these mutants indeed are defective for multimerization.
FIGURE 6.
Effect of multimerization-defective mutants of full-length INI1 on in vitro integration. A, 15–35% glycerol gradient centrifugation of wild type INI1 and DD mutants. Fractions were analyzed by Western blot using anti-His antibody as probe. B, GST pulldown experiment with GST or GST-fused INI1 and HAP eluate of wild type and mutant His-INI1 as shown. Bound proteins were analyzed by Western blot using anti-His antibody as probe. The same blots were stripped and probed with anti-GST antibodies. C, GST pulldown of wild type INI1 and DD mutants and E3 with glutathione-Sepharose 4B beads coupled to GST or GST-IN at different salt concentrations. Bound proteins were analyzed by Western blot using anti-His antibody as probe. The same blots were stripped and probed with anti-IN (integrase) antibodies. D, in vitro integration (stimulation) assay with wild type (WT), DD mutants, and E3 INI1 HAP eluate at 100 mm salt. STP, strand transfer product. E, in vitro integration (inhibition) assay with wild type, DD mutants and E3 INI1 HAP eluate at 100 mm salt.
We next tested the ability of these mutants to bind to INI1 and IN in a GST pulldown assay (Fig. 6, B and C). As a control, we also tested D225G (a mutant originally isolated as IN binding-defective S6) within the context of full-length INI1 (8). The GST pulldown assay indicated that INI1 mutants were defective for interaction with IN to a variable degree. The DD6 (I264T,I268T) mutant bound to IN the least, and the highest defect was observed at 200 mm salt (Fig. 6C). The mutants with single substitutions (DD6A and DD6C) also exhibited partial IN binding defects. DD5 mutant was able to bind to IN at all salt concentrations (Fig. 6C). Taken together these studies demonstrate that whereas Ile-263 is specifically required for multimerization of INI1, Ile-264 and Ile-268 are involved in both multimerization and IN binding.
Effect of Multimerization and IN Binding Activities of INI1 in Modulating Strand Transfer Activities
To determine whether multimerization and IN binding activities are required for INI1 to function, we next tested the ability of INI1 mutants to modulate in vitro strand transfer activities. The hydroxylapatite eluates of wild type and mutant INI1 proteins were tested for their ability to stimulate and inhibit strand transfer reaction in a manner dependent on IN concentration and the IN:INI1 molar ratio. At 100 mm NaCl concentration, wild type INI1 and the mutant proteins were able to stimulate HIV-1 IN activity (Fig. 6D), although the mutant I268T exhibited reduced (∼50%) activity (Fig. 6D). Similar results were obtained at 150 mm salt, although the overall efficiency of integration activity was reduced (data not shown). We then tested the mutants for their ability to inhibit IN joining activities at high IN concentrations. The results indicated that although wild type INI1 showed robust inhibition, the DD6 (I264T,I268T) mutant, which is most defective for both multimerization and IN binding, was unable to inhibit integrase activity (Fig. 6E). These results were in contrast to the lack of effect of DD6 mutant on the stimulation of IN activity. These studies taken together demonstrate that the ability of INI1 to stimulate IN joining activity is independent of multimerization and that IN binding and the ability to inhibit strand transfer activity are dependent on these protein-protein interactions.
Mechanism of INI1-mediated Stimulation and Inhibition of IN Joining Activities
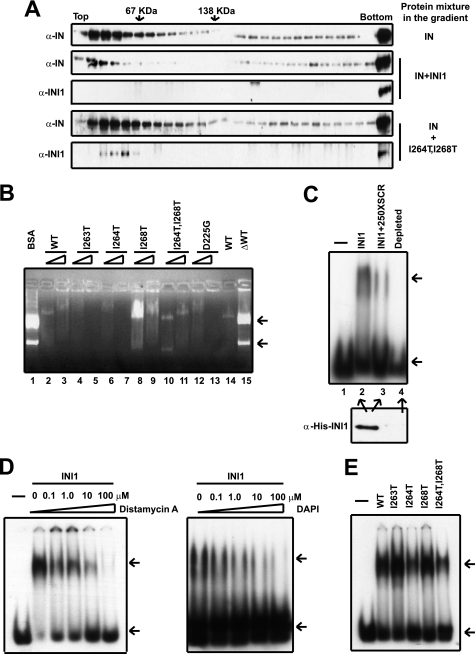
Our results, for the first time, indicated that inhibition of strand transfer activity in vitro is dependent on the ability of INI1 to bind to IN. We hypothesized that INI1 could inhibit IN joining activity by binding to IN and then by either disrupting the homomeric interactions of IN or by creating nonfunctional multimeric forms of IN and INI1. To test either of these possibilities, we used glycerol gradient centrifugation to study the nature of IN-INI1 complexes in solution. Purified IN and INI1 proteins were mixed together at a molar ratio of 1:4 and then subjected to glycerol gradient centrifugation. As a control, IN alone was also subjected to glycerol gradient centrifugation. The results of these experiments indicated that IN separated as monomeric, dimeric, and higher molecular weight forms upon glycerol gradient centrifugation (Fig. 7A, top panel). As indicated previously, at these concentrations, INI1 also forms dimers (refer to Fig. 1E). However, when IN was mixed with INI1, lower order forms of these proteins were either reduced (as for IN, Fig. 7A, second panel) or completely eliminated (as for INI1, third panel). These results suggested that the interaction of IN and INI1 may result in a presumably inactive, large, multimeric, high molecular weight complex that sediments at the bottom of the gradient (Fig. 7A). If this were the case, then the mutant of INI1 DD6 (I264T,I268T) that is defective for multimerization and IN binding would be defective for reducing the dimeric structures of IN. Consistent with this idea, contrary to wild type INI1, the presence of mutant DD6 (I264T,I268T) did not affect the sedimentation of IN. Furthermore, DD6 (I264T,I268T) separated as a monomer in the same gradient (Fig. 7A, bottom two panels). The lack of formation of high molecular weight complexes correlates with the inability of DD6 (I264T,I268T) mutant to inhibit IN joining activity in vitro and indicates, for the first time, that one mechanism by which INI1 may inhibit integration reaction is by forming inactive higher order IN-INI1 complexes.
FIGURE 7.
Correlation of multimerization and DNA binding properties of INI1 to modulate in vitro integration. A, 15–35% glycerol gradient centrifugation of integrase and integrase incubated with wild type INI1 or I264T,I268T. Fractions were analyzed by Western blot using anti-INI1 and anti-integrase antibodies as probes. B, gel retardation assay with HAP eluate (8 and 16 pmol) of wild type INI1 (WT) and DD mutants (including E3) and pCDNA plasmid as explained under “Materials and Methods.” C, EMSA of HAP eluate of INI1 (5 pmol) with 23-mer viral LTR double-stranded DNA radiolabeled with γ-32P ATP in the presence of 10× cold oligos. 250× scrambled LTR DNA was used for the competition experiment. His-INI1 was immunodepleted with anti-His antibody. D, EMSA of HAP INI1 (5 pmol) with radiolabeled viral LTR DNA in the presence of increasing amounts of distamycin A and DAPI as indicated. E, EMSA of HAP wild type and mutant INI1 (5 pmol) with radiolabeled viral LTR DNA.
INI1 Binds to Minor Groove of LTR DNA
The multimerization and IN binding-deficient mutant of INI1 DD6 (I264T, I268T), which was most defective for the inhibition of joining activities, was not defective for stimulation of IN joining activities in vitro (Fig. 6, D and E). These results suggest either that the residual IN binding activity of these mutants is sufficient to stimulate integration reaction or that the ability of INI1 to stimulate IN joining activity is due to some other function of INI1. Previously, we had demonstrated that INI1 has the ability to bind to DNA nonspecifically (18). We surmised that the nonspecific DNA binding activities of INI1 could modify the DNA such that it would be more accessible for IN-mediated strand transfer activities and that the mutants defective for protein-protein interactions may not be defective for DNA binding activities. To investigate this further, we tested the DNA binding properties of wild type and mutant INI1 proteins at two different protein concentrations (8 and 16 pmol). The wild type protein bound to plasmid DNA, as indicated by the lack of migration of DNA into the gel from the wells (Fig. 7B, lanes 2 and 3). As a control, the addition of BSA had no effect on the DNA migration (Fig. 7B, lane 1). When INI1 was depleted from the extract (Fig. 7B, lane 15), there was a lack of DNA binding activity in the extract, as indicated by the appearance of DNA migration into the gel, similar to that of the control BSA (Fig. 7B, compare lanes 1 and 15), suggesting that DNA binding activity in the extract was due to INI1. Interestingly, we found that all of the mutants except mutant I268T were able to retard the DNA similar to findings in the wild type protein (Fig. 7B). The mutant I268T exhibited a partial defect in DNA binding, in that although the plasmid DNA entered the gel in the presence of this mutant, it was retarded to some extent (Fig. 7B, lane 8). These results are consistent with our hypothesis that the DNA binding activities of INI1 may be required to stimulate IN strand transfer reactions.
IN has the ability to bind to both the target and LTR DNAs. Therefore, we tested whether INI1 would bind to the radiolabeled LTR donor in an EMSA (Fig. 7C). The binding of INI1 to radiolabeled LTR DNA was evident in the EMSA, which was competed by an excess of nonspecific cold scrambled DNA, indicating that the binding was not sequence-specific (Fig. 7C, lanes 2 and 3). When the protein preparation was depleted of INI1, the gel shift was not observed, indicating that the mobility shift observed was due to the activity of INI1 protein (Fig. 7C, lane 4). To further characterize the DNA binding property of INI1, we tested to see whether it binds to the minor groove of DNA using DNA intercalating agents such as distamycin A and DAPI. These drugs specifically disrupt the binding of proteins to the minor groove of DNA (25, 26). For example, tau, a microtubule-associated protein that is a well known DNA minor groove binder, binds a 13-mer DNA with a protein:DNA molar ratio of 1:1 (26), whereas in our experiments INI1 bound a 23-mer DNA with a protein:DNA molar ratio of ∼30:1. One explanation for this effect is that INI1 is a higher order multimer. Distamycin A and DAPI inhibit a known DNA minor groove-binding transcription factor, hTBP, with an IC50 of ∼1 μm but do not inhibit major groove-binding proteins like EGR1 and WT1 (25). When distamycin A and DAPI were added to the reaction containing radiolabeled LTR and INI1 protein, we found that they strongly inhibited the DNA binding activity of INI1, with the drugs at 0.1 μm inhibiting more than half of the INI1 DNA binding activity (Fig. 7D). These results established, for the first time, that INI1 binds to the minor groove of DNA. The binding of tau to the minor groove is inhibited by distamycin A at an inhibitor:protein ratio of 10:1 (26), whereas distamycin A and DAPI maximally inhibited INI1 binding to DNA at an inhibitor:protein ratio of 7:1 and 5:1, respectively.
We then tested to determine whether the multimerization and IN-binding mutants of INI1 were defective for LTR binding activity. We found that all of the mutants retained the ability to bind to LTR under the current conditions (Fig. 7E). Although there was some variability in the degree of LTR DNA binding, this variability was not significant in repeated experiments. Overall these results indicated that wild type and mutant INI1 proteins are able to bind to the minor groove of LTR. This result is slightly different from the result that we observed for binding of INI1 and the I268T mutant to plasmid DNA. We found that whereas the mutant I268T was somewhat defective for binding to plasmid DNA, it was not defective for binding to LTR DNA. This difference can be explained by a difference in the binding affinity of the protein to the topologically complex plasmid DNA as compared with the 23-mer LTR DNA. These results taken together indicate that differential ability of the wild type and mutant I268T to bind to plasmid DNA correlate with the ability of INI1 to stimulate IN joining activity.
DISCUSSION
INI1 was the first host protein to be identified as a binding partner for HIV-1 IN. However, the role of this protein in HIV-1 replication is not completely understood, due in part to a lack of biochemical and structural information relating to this protein. In this report, we have attempted to purify and characterize INI1 biochemically. We report here, for the first time, that INI1 forms dimeric, tetrameric, octameric, and higher order structures depending on the concentrations of the protein. We have shown that the Rpt1 and Rpt2 regions of INI1 are necessary for multimerization, and the N-terminal domain of INI1, although not necessary, is required for efficient self-association. It is interesting to note that the purified fragment S7-(141–304), which harbors homomeric interaction domains, is a dimer in solution even at higher concentrations. Therefore, it is possible that although the minimal domain needed for dimerization includes the Rpt1 and Rpt2 regions, the N-terminal domain may contribute to higher order interactions leading to multimerization. Future structural information will likely shed light on these properties of INI1.
Knowledge of the biochemical properties of INI1 is essential to comprehend the role that this protein has in HIV-1 replication. As the multimerization property may be necessary to exhibit its effect, we have isolated mutants of INI1 that are defective for this function. Our effort yielded mutants in the Rpt1 and Rpt2 motifs. Most of the mutated residues were partially or fully conserved and hydrophobic in nature, suggesting that hydrophobic interaction may be critical for self-association. Interestingly, a majority of the mutations in the Rpt2 motif are concentrated near the N terminus of the NES or overlap with it. Comparative analysis of the self-association of multimerization-defective mutants indicated that whereas Rpt1 mutants are ∼30% as active as wild type INI1-(183–294), Rpt2 mutants are only <5% as active as the wild type protein, suggesting that amino acid residues in the Rpt2 motif are critical for multimerization. This is contrary to findings in IN binding-defective mutations of INI1-(183–294), where the majority of the mutations were found in Rpt1. Although the mutants that were found to be defective in self-association were also defective in binding to IN, the degree of the defect was inversely correlated. For example, among the multimerization-defective mutants analyzed, DD5 (I263T) and DD6 (I264T,I268T) mutants were most defective for multimerization and were least defective for IN binding. Furthermore, many of the Rpt1 mutants (T214A,D225G and E183G,D191G) that were defective for binding IN were not defective for multimerization. These results suggest that the two functions of INI1, i.e. multimerization and IN binding, are mediated by overlapping regions but require distinct residues. We suggest that the amino acid residues in the Rpt1 motif, in addition to playing a role in multimerization, may make direct contact with IN. This is consistent with our previous report that there is direct association between the Rpt1 motif of INI1 and integrase (18). Finally, the observation that multimerization of INI1 is required for its association with IN was reproduced with the full-length INI1 protein.
Because the residues involved in multimerization overlapped with the NES sequence, we also investigated the nuclear export ability of these mutants. Our previous studies had identified a NES consensus sequence in INI1 between residues 263 and 275. We reported that in addition to the residues that exactly match the known consensus for NES (265 ΨXXXΨXXΨXΨ), an additional upstream residue, Ile-263, is required for nuclear export (23). Our current results strengthen these previous results in that the mutant DD5 (I263T) with a mutation in a single residue in the NES consensus was defective for nuclear export, and the mutant DD6 (I264T,I268T), which was mutated in hydrophobic residues outside the NES consensus sequence, was not defective for nuclear export. These results suggest that multimerization and nuclear export are mediated by overlapping residues in INI1. We utilized a pair of mutants that are differentially defective for nuclear export, but commonly defective for multimerization, to assess the contribution of these properties to mediate transdominant inhibition of HIV-1 replication. Our results indicate that both multimerization and nuclear export are necessary for mediating the transdominant effect. Interestingly, we found that expression of the transdominant negative mutant not only inhibited particle production, but it also significantly decreases intracellular p24 protein levels. This latter effect could either be due to a decrease in protein expression itself or to a decrease in the overall stability of Gag and Gag-Pol proteins. Because the two mutants that were defective for either multimerization alone or for both multimerization and IN binding partially rescued the defect in p24 levels, we believe that these functions are important for both p24 protein expression/stability as well as particle production. More experiments are needed to address the effect of these mutants on intracellular viral protein levels. Nevertheless, our results demonstrate the intricate relationship among nuclear export, multimerization, and IN binding, due to the overlapping of the domains responsible for these functions.
To determine the influence of multimerization and/or IN binding on in vitro integrase activity, we tested the ability of the mutants I263T, I264T, I268T, D225G, and I264T,I268T to modulate integrase activity. Overall, we found that multimerization and the ability to bind integrase were critical for INI1-mediated inhibition of integrase activity, whereas the ability of INI1 to stimulate integrase activity was independent of these functions. At this point, we cannot rule out the possibility that the residual IN binding properties of the INI1 mutants are sufficient for stimulation function. Interestingly, the mutant I268T was partially defective in its ability to stimulate integrase activity, although it retained IN binding. We surmised that the other possible function of INI1, such as nonspecific DNA binding activity, could be important in the strand transfer reaction. Therefore, we tested the mutants for their ability to bind to both the donor oligo duplex derived from LTR and the acceptor plasmid DNA. Consistent with previous results, we found that INI1 robustly bound to the acceptor plasmid. Among the mutants, only I268T demonstrated a partial defect in DNA binding in an agarose gel retardation assay, consistent with the idea that acceptor DNA binding by INI1 is critical for its ability to stimulate integrase function in the strand transfer reaction. However, it should be noted here that even the most defective IN mutant exhibited residual IN binding activity, and INI1-integrase interaction may still be necessary for INI1 stimulatory activity. Thus, the residual binding of the DD6 (I264T,I268T) to integrase may be sufficient for its ability to stimulate but not to inhibit strand transfer activities.
In this report we have demonstrated that INI1 has the ability to bind to the minor groove of double-stranded DNA. The addition of purified INI1 protein resulted in gel shift of LTR oligonucleotides, which was abrogated when the INI1 protein was depleted from the preparation. Furthermore, distamycin A and DAPI, the two DNA intercalating agents that prevent the interaction of proteins to the minor groove, disrupted the interaction of INI1 with LTR DNA. This property of INI1 could also explain the ability of INI1 to stimulate the joining activity. However, this property alone is not a sufficient explanation, as all of the mutants, including the one that was slightly defective for IN joining activities, bound to LTR DNA. We propose that the ability of INI1 to bind acceptor DNA is required for its ability to stimulate IN joining activities. At this time, we cannot rule out the possibility that INI1 binding to LTR DNA could also be important for its stimulatory activity in vitro. These results may help define the mechanism of INI1-dependent modulation of HIV-1 integrase activity. These studies allow us to assess the role of INI1 in HIV-1 replication, which may ultimately lead to the development of novel therapeutic strategies.
Supplementary Material
Acknowledgments
We thank Dr. V. Prasad at Albert Einstein College for critically reading the manuscript, Drs. U. Maitra and S. Almo at Albert Einstein College for useful discussions, and Dr. Neamati at the University of Southern California for the generous gift of integrase inhibitor (S-1360).
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI039951-11. This work was also supported by an Irma T. Hirchl/Monique Weill-Caulier scholar award (to G. V. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods.”
- HIV-1
- human immunodeficiency virus, type 1
- IN
- HIV-1-encoded integrase
- Ni-NTA
- nickel-nitrilotriacetic acid
- EMSA
- electrophoretic mobility shift assay
- LTR
- long terminal repeat
- LEDGF
- lens epithelium-derived growth factor
- BSA
- bovine serum albumin
- HA
- hemagglutinin
- NES
- nuclear export signal
- DAPI
- 4′,6-diamidino-2-phenylindole
- ONPG
- o-nitrophenyl-β-d-galactopyranoside
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- HAP
- hydroxylapatite.
REFERENCES
- 1.Sorin M., Kalpana G. V. (2006) Curr. HIV Res. 4, 117–130 [DOI] [PubMed] [Google Scholar]
- 2.Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008) Science 319, 921–926 [DOI] [PubMed] [Google Scholar]
- 3.Kalpana G. V., Marmon S., Wang W., Crabtree G. R., Goff S. P. (1994) Science 266, 2002–2006 [DOI] [PubMed] [Google Scholar]
- 4.Van Maele B., Busschots K., Vandekerckhove L., Christ F., Debyser Z. (2006) Trends Biochem. Sci. 31, 98–105 [DOI] [PubMed] [Google Scholar]
- 5.Engelman A., Cherepanov P. (2008) PLoS Pathog. 4, e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorin M., Yung E., Wu X., Kalpana G. V. (2006) Retrovirology 3, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maroun M., Delelis O., Coadou G., Bader T., Ségéral E., Mbemba G., Petit C., Sonigo P., Rain J. C., Mouscadet J. F., Benarous R., Emiliani S. (2006) J. Biol. Chem. 281, 22736–22743 [DOI] [PubMed] [Google Scholar]
- 8.Yung E., Sorin M., Pal A., Craig E., Morozov A., Delattre O., Kappes J., Ott D., Kalpana G. V. (2001) Nat. Med. 7, 920–926 [DOI] [PubMed] [Google Scholar]
- 9.Yung E., Sorin M., Wang E. J., Perumal S., Ott D., Kalpana G. V. (2004) J. Virol. 78, 2222–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Versteege I., Sévenet N., Lange J., Rousseau-Merck M. F., Ambros P., Handgretinger R., Aurias A., Delattre O. (1998) Nature 394, 203–206 [DOI] [PubMed] [Google Scholar]
- 11.Hulsebos T. J., Plomp A. S., Wolterman R. A., Robanus-Maandag E. C., Baas F., Wesseling P. (2007) Am. J. Hum. Genet. 80, 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modena P., Lualdi E., Facchinetti F., Galli L., Teixeira M. R., Pilotti S., Sozzi G. (2005) Cancer Res. 65, 4012–4019 [DOI] [PubMed] [Google Scholar]
- 13.Sévenet N., Sheridan E., Amram D., Schneider P., Handgretinger R., Delattre O. (1999) Am. J. Hum. Genet. 65, 1342–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozenblatt-Rosen O., Rozovskaia T., Burakov D., Sedkov Y., Tillib S., Blechman J., Nakamura T., Croce C. M., Mazo A., Canaani E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4152–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu D. Y., Kalpana G. V., Goff S. P., Schubach W. H. (1996) J. Virol. 70, 6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee D., Sohn H., Kalpana G. V., Choe J. (1999) Nature 399, 487–491 [DOI] [PubMed] [Google Scholar]
- 17.Cheng S. W., Davies K. P., Yung E., Beltran R. J., Yu J., Kalpana G. V. (1999) Nat. Genet. 22, 102–105 [DOI] [PubMed] [Google Scholar]
- 18.Morozov A., Yung E., Kalpana G. V. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1120–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitz U., Mueller W., Weber M., Sévenet N., Delattre O., von Deimling A. (2001) Br. J. Cancer 84, 199–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sévenet N., Lellouch-Tubiana A., Schofield D., Hoang-Xuan K., Gessler M., Birnbaum D., Jeanpierre C., Jouvet A., Delattre O. (1999) Hum. Mol. Genet. 8, 2359–2368 [DOI] [PubMed] [Google Scholar]
- 21.Yuge M., Nagai H., Uchida T., Murate T., Hayashi Y., Hotta T., Saito H., Kinoshita T. (2000) Cancer Genet. Cytogenet. 122, 37–42 [DOI] [PubMed] [Google Scholar]
- 22.Lai J. S., Herr W. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 6958–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig E., Zhang Z. K., Davies K. P., Kalpana G. V. (2002) EMBO J. 21, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stommel J. M., Marchenko N. D., Jimenez G. S., Moll U. M., Hope T. J., Wahl G. M. (1999) EMBO J. 18, 1660–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McHugh M. M., Woynarowski J. M., Sigmund R. D., Beerman T. A. (1989) Biochem. Pharmacol. 38, 2323–2328 [DOI] [PubMed] [Google Scholar]
- 26.Wei Y., Qu M. H., Wang X. S., Chen L., Wang D. L., Liu Y., Hua Q., He R. Q. (2008) PLoS ONE 3, e2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandegraaff N., Devroe E., Turlure F., Silver P. A., Engelman A. (2006) Virology 346, 415–426 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.