Abstract
Berberine bridge enzyme (BBE) is a member of the recently discovered family of bicovalently flavinylated proteins. In this group of enzymes, the FAD cofactor is linked via its 8α-methyl group and the C-6 atom to conserved histidine and cysteine residues, His-104 and Cys-166 for BBE, respectively. 6-S-Cysteinylation has recently been shown to have a significant influence on the redox potential of the flavin cofactor; however, 8α-histidylation evaded a closer characterization due to extremely low expression levels upon substitution. Co-overexpression of protein disulfide isomerase improved expression levels and allowed isolation and purification of the H104A protein variant. To gain more insight into the functional role of the unusual dual mode of cofactor attachment, we solved the x-ray crystal structures of two mutant proteins, H104A and C166A BBE, each lacking one of the covalent linkages. Information from a structure of wild type enzyme in complex with the product of the catalyzed reaction is combined with the kinetic and structural characterization of the protein variants to demonstrate the importance of the bicovalent linkage for substrate binding and efficient oxidation. In addition, the redox potential of the flavin cofactor is enhanced additively by the dual mode of cofactor attachment. The reduced level of expression for the H104A mutant protein and the difficulty of isolating even small amounts of the protein variant with both linkages removed (H104A-C166A) also points toward a possible role of covalent flavinylation during protein folding.
Since the discovery of the first known example of a covalent bond between a flavin cofactor and an amino acid side chain occurring in enzymes in the 1950s (1), a number of different types of linkages have been identified: 8α-histidylation (either to N1 or to N3), 8α-O-tyrosylation, 8α-S-cysteinylation, and 6-S-cysteinylation. For current reviews relating to these modes of flavin attachment, see Refs. 2 and 3. Recently, another way of covalent tethering of FAD to proteins was discovered in x-ray crystallographic studies on glucooligosaccharide oxidase (GOOX)4 from Acremonium strictum (4). The mode of flavin linkage observed in this case employs both 8α-histidylation and 6-S-cysteinylation to form a bicovalently attached cofactor. Representative members of all these groups have been studied in detail, and several explanations for the role of the covalent flavinylation have been put forward. Some of the suggestions tend to be rather specific for the system being studied, e.g. prevention of cofactor inactivation at the C-6 position for trimethylamine dehydrogenase (5) or facilitation of electron transfer from the flavin to the cytochrome subunit for p-cresol methylhydroxylase (6). Other explanations including the increase of the flavin redox potential due to the covalent linkage (7–9) and the prevention of cofactor dissociation (10, 11) were found for several enzymes also harboring different types of cofactor attachments. Taking into account that protein stability (12) and optimal binding of substrate molecules (11, 13) are also positively influenced by covalent tethering of the flavin, one might speculate that no generally applicable explanation for the covalent attachment of flavins to proteins exists. Therefore, it seems likely that the large variety of systems operating with one of the above mentioned modes of cofactor tethering might have evolved to also adapt to a diversity of enzymatic challenges.
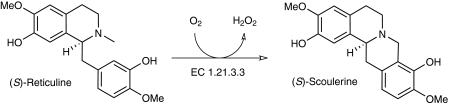
Berberine bridge enzyme (BBE) from Eschscholzia californica is a plant enzyme involved in alkaloid biosynthesis, catalyzing the challenging oxidative cyclization of (S)-reticuline to (S)-scoulerine (Scheme 1). This enzyme was recently shown to belong to the group of flavoenzymes with a bicovalently attached FAD (14). After the discovery of this unusual mode of linkage in the crystal structure of GOOX (4), several members of this group, all belonging to the vanillyl-alcohol oxidase family (15), were identified by biochemical methods (16–18) and also structural studies (19). Because some of the suggested benefits of a covalent cofactor attachment can easily be brought about by a single linkage, e.g. prevention of cofactor dissociation or stabilization of the tertiary structure, the two amino acids attached to FAD might have different and individual functions as well as an additive effect on physicochemical properties such as redox potentials or substrate binding and oxidation. To elucidate the relative importance for the overall enzymatic functioning of members of this group, more detailed studies have been performed on GOOX (11), chito-oligosaccharide oxidase (ChitO) from Fusarium graminearum (17), and BBE (20). Common results of these analyses show that the bicovalent FAD has a redox potential of about +130 mV, which is among the highest potentials reported for flavoenzymes. Replacement of one of the amino acids involved in anchoring of the cofactor generally reduces the rate of cofactor reduction and the steady-state turnover rate, but whether this can be directly linked to reduced redox potentials of these mutant proteins has been under debate (11).
SCHEME 1.
Overall reaction catalyzed by BBE.
To address these issues further, we report the expression of the H104A mutant protein of BBE. A biochemical characterization of this protein variant with respect to the redox potential, transient kinetics, and steady-state analysis is combined with the structural analysis of both the H104A and the C166A mutant proteins. In addition, a structure of wild type (WT) BBE in complex with the product of the enzyme-catalyzed reaction is presented, which provides further insights toward the involvement of active site amino acids during the course of the reaction. Together with the recently reported x-ray crystal structure of WT BBE with and without substrate bound (21) and the biochemical characterization of the C166A mutant protein (20), these results provide interesting insights into the role of bicovalent FAD attachment in enzymes.
EXPERIMENTAL PROCEDURES
Optimization of Heterologous Expression
To overcome the low level of expression for BBE H104A and H104A-C166A reported previously (20), we attempted to improve the level of secretory expression in Pichia pastoris by co-overexpression of protein disulfide isomerase (PDI), a protein reported to have a general positive effect on the secretory machinery in yeasts (22, 23). The plasmid containing the coding sequence of PDI from P. pastoris in the vector pPICK was a generous gift of Prof. A. Glieder and Dr. S. Abad. Transformation of this plasmid into P. pastoris KM71H was performed as described in Ref. 24 using the condensed protocol. 20 differently sized transformants grown on YPD plates containing Geneticin were pooled and used for preparation of competent cells for another round of transformation using the plasmid pPICZα containing various BBE coding sequences (WT, H104A, and H104A-C166A (20)). To screen a larger amount of transformants for expression analysis, we employed deep well plates for the cultivation of about 30–50 clones of each protein variant as described in Ref. 25. These plates were harvested by centrifugation after 72 h of induction with methanol, and the supernatants were analyzed for BBE expression by dot-blot assays. Therefore, 100 μl of the supernatant were applied onto a nitrocellulose membrane according to the instructions of the Bio-Dot® microfiltration apparatus (Bio-Rad) manual. A qualitative analysis of BBE expression on the membrane was performed according to standard Western blot procedures. Briefly, the membrane was blocked with 15% milk powder in TTBS (0.1 m Tris, 150 mm NaCl, 0.1% Tween 20, pH 7.5) for 1 h at room temperature. After a brief washing step with TTBS, the nitrocellulose membrane was incubated with the primary antibody targeted against BBE (anti-BBE 1:500; donated by Prof. T. M. Kutchan) at 4 °C overnight. Excess first antibody was removed by washing the membrane with TTBS three times, 10 min each, followed by incubation with the secondary antibody (anti-rabbit IgG (peroxidase-coupled) 1:15000; Sigma-Aldrich) for 1 h at 4 °C. After this, the membrane was again washed three times with TTBS, and detection of the protein dots was carried out by incubating the membrane with 4 ml of SuperSignal® West Pico luminol/enhancer solution and 4 ml of SuperSignal® West Pico stable peroxidase solution (both from Thermo Scientific) and exposing the membrane to an Eastman Kodak film for various time periods depending on the level of BBE expression.
Protein Expression and Isolation
The best producing colonies identified by the deep well plate assay were used to start large scale fermentations as described in Ref. 14. Purification of the secreted enzymes was performed as described for the WT enzyme (14), but due to the low level of protein expression in the case of BBE H104A, the purification procedure included an additional anion exchange step. Fractions from the gel filtration step showing activity were pooled and loaded onto a MonoQTM 5/50 GL column (GE Healthcare) equilibrated with 50 mm Tris/HCl, pH 9.0 (buffer A). Although most of the impurities bound strongly to the column, BBE eluted with the breakthrough due to limited capacity of the column. This flow-through was then diluted 4-fold with distilled water and rechromatographed on the same column. Bound BBE was eluted during a 1-h gradient from 100% buffer A to 70% buffer A and 30% 50 mm Tris/HCl, 1 m NaCl, pH 9.0 (buffer B).
Kinetic Analysis
Steady-state and transient kinetic parameters were determined using the substrate (S)-reticuline as described previously (14). Substrate concentrations ranging from 2 to 350 μm reticuline were covered to study effects on Km and Ki because for C166A BBE, a considerable amount of substrate inhibition was observed. For the comparison of turnover rates in Table 1, a substrate concentration of 100 μm reticuline was used, which is ∼30 times the reported Km of WT BBE (26).
TABLE 1.
Summary of kinetic properties and redox potentials
Turnover rates are stated as mean ± maximal error of two independent measurements. Transient kinetic terms were measured four or five times and are shown as mean ± standard deviation. The redox potential given is the mean with standard deviation of three experimental setups.
Transient kinetics experiments for determination of the reductive and oxidative rate constants were performed as described recently (20). The term reductive rate refers to the apparent rate of cofactor reduction under saturating concentrations of (S)-reticuline determined in single turnover experiments. Observed rate constants at substrate concentrations ranging from 10 to 300 μm were fit with a hyperbolic function to obtain the parameters Kd and kred. For studying reoxidation of the cofactor, the protein sample was reduced with 0.9 equivalents of substrate and mixed with an air-saturated buffer solution (243 μm O2).
Redox Potential Determination
The redox potential of the H104A mutant protein was determined as published previously for the WT enzyme and the C166A variant (20) based on the dye-equilibration method originally described by Massey (27). The experimental setup was slightly modified and included an automated shutting unit to reduce the possibility of photochemical reactions in the flow cell during the long incubation times. Reductions of the enzyme and reference dye were carried out in 50 mm potassium phosphate buffer, pH 7.0, at 25 °C and contained benzyl viologen (5 μm) as mediator, 500 μm xanthine, and catalytic traces of xanthine oxidase (∼20 nm). Used reference dyes included thionine acetate (EM = +64 mV) and toluidine blue (EM = +34 mV), and the reactions were carried out over a time period of about 2 h. The redox potentials were estimated from plots of log([ox]/[red]) of H104A mutant protein versus log([ox]/[red]) of the dye toluidine blue according to Minnaert (28).
Crystallization, Data Collection, and Structure Elucidation
BBE WT crystals grown with the sitting drop vapor diffusion method as described in Ref. 21 were used for seeding of crystallization setups with protein variants H104A and C166A. For this, drops of 1 μl of enzyme solution (∼25 mg ml−1 in 50 mm Tris/HCl, 150 mm NaCl, pH 9.0) were mixed with 1 μl of reservoir solution (0.2 m MgCl2 and 30% (w/v) polyethylene glycol 4000 in 0.1 m Tris/HCl, pH 8.5) and equilibrated for 2 h prior to streak seeding. Tetragonal crystals appeared overnight and reached final dimension in 3–4 days for both mutant proteins. Crystals for native structures were directly transferred to liquid nitrogen for flash-freezing, whereas substrate soaks were performed by incubating the crystals for 60 min in drops containing 20 mm (S)-reticuline in the reservoir solution prior to flash-cooling.
Data sets for H104A, C166A, C166A_reticuline, and WT_ scoulerine were collected at beamline X06DA at the Swiss Light Source in Villigen, Switzerland. A summary of data collection, processing, and structure refinement is presented in supplemental Table S1. Processing of all data sets was carried out using the XDS software package (29). Due to the isomorphicity of all crystals (supplemental Table S1), the structures could be solved by rigid body fitting using the program PHENIX (30) based on the available model of WT BBE. The structures were further refined with the same software. Model building and fitting steps involved the graphics program Coot (31) using σA-weighted 2Fo − Fc and Fo − Fc electron density maps. Rfree values (32) were computed from 5% randomly chosen reflections not used throughout the refinement. Ligands were excluded from the initial refinement process and inserted only in the final round when clear electron density was visible for (S)-scoulerine (WT_scoulerine). In the case of C166A_ reticuline, two conformations of reticuline were included to explain the electron density. Water molecules were placed automatically into difference electron density maps and were retained or rejected based on geometric criteria as well as on their refined B-factors. In all structures, residues Gly-178, Phe-392, and Arg-409 were found in the disallowed region of the Ramachandran plot. Their electron density, however, was well defined in each case. In the structure of the H104A variant, two residues (Lys-350 and Phe-351) of the poorly defined surface loop were found in the less favored region.
RESULTS
Optimization of Expression and Protein Purification
Expression levels of the H104A single and H104A-C166A double mutant proteins of BBE with the standard expression system (20) were too low to allow a biochemical characterization of these variants. As shown in supplemental Fig. S1, co-overexpression of PDI reduced this problem. Analysis of the protein secretion level in dot-blot assays demonstrated that WT BBE was expressed much better than the H104A mutant protein, and this in turn was significantly stronger than the H104A-C166A protein variant. Analysis of several transformants for each gene construct resulted in relatively similar expression levels for all protein variants, whereas a comparison with the strains previously used for protein expression (20) without cotransformation of PDI showed a significant improvement. Transformants with the H104A-C166A bbe gene, however, showed only a very weak positive signal when compared with the negative controls. The transformant producing the most intense signal for each protein variant was then used for scale up to fermentation and protein expression in 5-liter fermenters. Purification of WT BBE allowed the recovery of about 500 mg of protein from a single fermenter, which is about twice the amount we obtained without coexpression of PDI (14). For the H104A protein variant, we were able to isolate 6 mg of pure enzyme from two fermenters. The H104A-C166A protein variant, however, could not be detected in the fermentation supernatant in quantities suitable for an initial characterization. Due to the low level of expression observed in the initial screening procedure (supplemental Fig. S1), this was not unexpected taking into account the still relatively low expression level of the H104A mutant protein.
Spectral Analysis of Cofactor Attachment
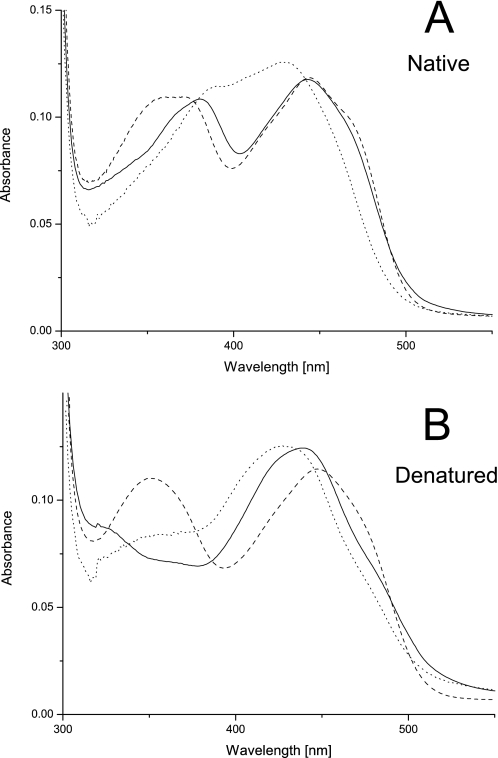
A first indication toward the mode of cofactor attachment for H104A BBE comes from the comparison of absorption spectra under native and denatured conditions (Fig. 1). As reported recently (14), the near UV absorbance band of the flavin cofactor in WT BBE at ∼380 nm disappears upon denaturation. By comparison with the spectral properties of the C166A variant, this feature was attributed to the covalent linkage between C-6 of FAD and the cysteine residue (20). In the case of the H104A mutant protein, this absorption around 380 nm is slightly shifted to higher wavelength in the native state. After denaturation, this spectral feature completely vanishes (Fig. 1B), and the overall spectrum approaches that of free 6-S-cysteinyl FMN (33). Therefore, it can be concluded that 6-S-cysteinylation of FAD also occurs without 8α-histidylation being present.
FIGURE 1.
Spectral properties of WT, C166A, and H104A BBE. All spectra are normalized to a protein concentration of 10 μm. BBE WT is shown as a solid line, C166A is shown as a dashed line, and H104A is shown as a dotted line. A represents the spectra under native conditions in 50 mm Tris/HCl, 150 mm NaCl, pH 9.0, and B represents the spectra after denaturation at a final SDS concentration of 0.5%.
Structure Elucidation
We determined the x-ray crystal structures of both the H104A and the C166A protein variants, as well as of complexes of the C166A variant with (S)-reticuline and of WT BBE with (S)-scoulerine. While crystals of the C166A variant grew to comparable dimensions as for WT enzyme, crystals obtained with the H104A mutant protein were generally significantly smaller and diffracted to a comparatively lower resolution.
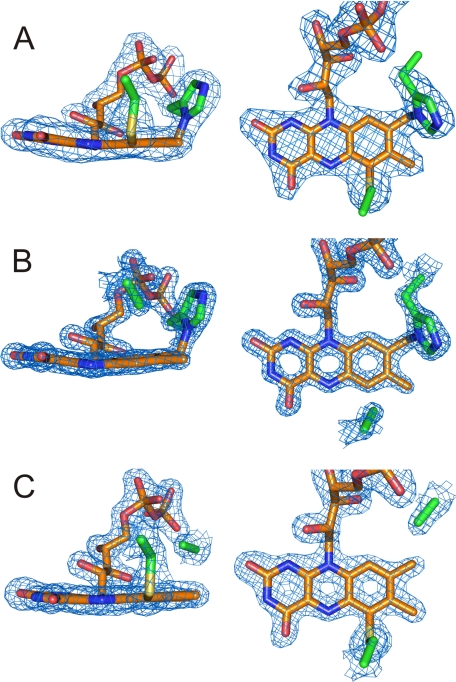
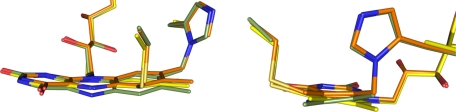
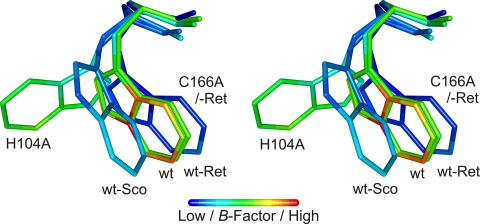
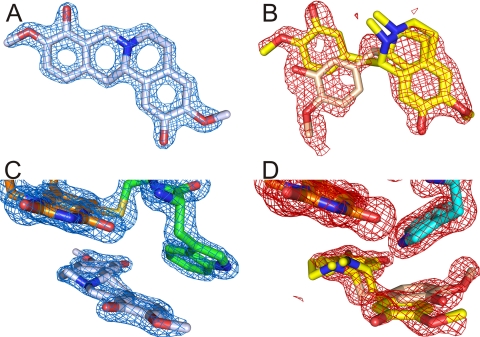
Initial electron density maps showed the missing covalent linkages according to the amino acid substitutions and confirmed the retention of the respective other FAD attachment for both H104A and C166A (Fig. 2), as suspected from the spectral characteristics (Fig. 1) (20). Apart from these differences, the overall structural changes are relatively small with root mean square deviations for Cα atoms on the order of 0.15 Å. The removal of the covalent linkages has only subtle impacts on the geometry of the isoalloxazine ring system (Fig. 3). Apart from the relaxing movement of FAD in the regions around C-6 and the 8α-methyl group away from the substituted amino acid due to removal of the covalent bond by 0.5 and 0.8 Å, respectively, only a slightly more pronounced bending around the N-5–N-10 axis in the case of C166A is observed. More pronounced are the structural influences on some of the neighboring amino acids. Removal of the sulfur atom of Cys-166 causes the disordered Trp-165 to adopt a well defined orientation. This conformation resembles that observed in the structure with bound substrate (21). Although His-104 is relatively distant to Trp-165, the latter residue is also affected by the deletion of 8α-histidylation. Similar to observations in the WT structure, this side chain is not very well defined in H104A BBE. The conformation best representing the electron density, however, corresponds to an ∼100° rotation around the Cβ–Cγ bond when compared with the conformation found in the WT structure (Fig. 4). The removal of 8α-histidylation additionally causes Gln-59 to attain a new conformation, occupying the space created by the His-104 to Ala substitution. This in turn breaks the interaction of the glutamine with Lys-350 (supplemental Fig. S2), which is part of a poorly defined surface loop becoming better ordered upon substrate binding (21). Although in the structures of WT and C166A BBE, residues 349–353 already have a significantly higher B-factor than main chain atoms of well defined regions, this loop is completely disordered in the case of the H104A mutant protein (supplemental Fig. S2).
FIGURE 2.
Densities of the FAD moieties in WT, C166A, and H104A BBE. Each panel shows two perpendicular views of the electron density of the cofactor including the amino acids 104 and 166 observed in the three BBE variants. A, Fo − Fc density map of WT BBE contoured at 5.0 σ. B, 2Fo − Fc density map of C166A BBE contoured at 3.0 σ. C, 2Fo − Fc density map of H104A BBE contoured at 1.5 σ.
FIGURE 3.
Comparison of the isoalloxazine ring systems of WT (orange), H104A (yellow), and C166A BBE (olive). The flavin cofactors were superimposed by aligning the two protein variants H104A and C166A to the WT BBE structure.
FIGURE 4.
Stereo representation of the different Trp-165 conformations observed for various BBE structures. The alignment was performed using the full-length structures reported in this study combined with native and substrate-bound WT BBE structures (Protein Data Bank (PDB): 3D2J and 3D2D, respectively). Ret, reticuline; Sco, scoulerine;
Previous short term soaks with (S)-reticuline resulted in electron density corresponding to the substrate in the active site (21). In the present study, we also tested better diffracting crystals with longer incubation times. We observed a time-dependent change in the electron density corresponding to C–C bond formation during the conversion of (S)-reticuline to (S)-scoulerine (Scheme 1). As a representative data set, we show a 1-h soak of WT BBE crystals where the substrate is completely converted to product (Fig. 5A). Comparing the substrate- and product-bound structures of WT BBE shows that the same polar interactions are employed for binding. In addition to the active site base Glu-417 (21), also Asp-352 from the surface loop and Trp-165 are involved in binding of (S)-reticuline as well as (S)-scoulerine (supplemental Fig. S3). Analogous soaks with C166A BBE crystals yielded no clear electron density for neither substrate nor product, and we modeled the electron density as a superposition of two different substrate binding modes (Fig. 5B). Because both data sets could be refined to 1.5 Å, however, the significantly increased B-factor of the substrate molecules also indicates a lower total occupancy when compared with the WT BBE product complex (occupancy 84%). When analyzing the active sites of the two complex structures, significant differences are evident for Trp-165 and the loop at the entrance to the active site (residues 349–353). Most importantly, Trp-165 of the C166A BBE soak remains unchanged when compared with the unbound mutant protein structure. In contrast, this residue adopts a completely different position in the case of the WT soak when compared with the native or substrate-bound structures (21). A representation of the various Trp-165 conformations is presented in Fig. 4.
FIGURE 5.
Comparisons of 60-min substrate soak densities for WT and C166A BBE. A, 2Fo − Fc density map at 1.75 σ of the compound bound to the active site of WT BBE modeled as (S)-scoulerine, the product of the enzyme-catalyzed reaction. B, 2Fo − Fc density map contoured at 1.0 σ of the same area as in panel A as observed in the C166A protein variant. A mixture of two differently oriented substrate molecules is modeled into the obtained electron density. The light pink orientation of the benzyl moiety corresponds to an unproductive binding of (S)-reticuline. C and D, the same density maps as in panels A and B for WT and C166A BBE, respectively, including the isoalloxazine ring system and the different orientations of Trp-165.
Steady-state Kinetics
Due to significant changes in the affinity of substrates to protein variants with a modified FAD attachment observed in both GOOX and ChitO (11, 17) we analyzed the steady-state kinetics of WT BBE in comparison with both mutant proteins over a substrate concentration range of 2–350 μm (Table 1). The highest value represents about 100 times the reported Km for WT BBE (26). Although the kcat values for WT and H104A BBE differ substantially, we observed comparable affinities for (S)-reticuline lying in the same range as the Kd values obtained from pre-steady-state experiments (see below). In contrast to the C166A mutant protein, where we observed significant substrate inhibition, only a minor decrease in the rate of substrate conversion at higher reticuline concentrations was observed for both WT and H104A BBE. However, a detailed evaluation of substrate inhibition in the case of the C166A mutant protein was not possible due to limitations of the high pressure liquid chromatography-based activity assay at substrate concentrations below 2 μm. At the latter concentration, we obtained the highest turnover rates (about 1.5 times the value obtained at 100 μm, Table 1) in the concentration range covered, and the activity decreased continuously down to about 30% of the initial rate at 350 μm (S)-reticuline. As far as the turnover rates at 100 μm substrate are concerned, a significant reduction can be observed for both mutant proteins when compared with WT enzyme. Because the altered reactivity of the C166A variant could previously be correlated with the change in the rate of cofactor reduction under anaerobic conditions (20), the cause for the similar reduction of kcat for the H104A mutant protein was further examined by also studying the transient kinetics of both the reductive and the oxidative half-reaction.
Pre-steady-state Kinetics of the Reductive and Oxidative Half-reaction
Reduction of the FAD cofactor of this protein variant was studied under anaerobic conditions over a similar substrate concentration range as for the WT and C166A proteins (20). The obtained transients showed a biphasic characteristic with the substrate-dependent fast phase contributing to more than 90% of the change in absorbance at 430 nm. The Kd value shown in Table 1 is not very well represented by data points, but it still illustrates that there are no major alterations of this value in comparison with WT BBE and the C166A mutant protein. Under pseudo first order conditions (more than 20-fold excess of substrate), a limiting reductive rate of 3.4 ± 0.3 s−1 is observed for the H104A variant, with the spectral course of flavin reduction resembling that of WT BBE (supplemental Fig. S4) (20). This is substantially higher than the steady-state turnover rate, indicating that another step during catalysis is the limiting factor for catalysis. To see whether regeneration of the cofactor might be limiting, we also determined the observed rate of flavin reoxidation at a single oxygen concentration. Comparison of the obtained 0.8 ± 0.1 m−1 s−1 with the recently determined kox of WT and C166A BBE showed that it is in the same range and, hence, cannot explain the reduced rate observed during steady-state turnover.
Redox Potential Determination
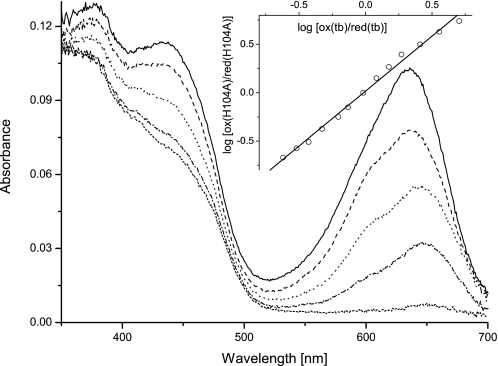
We determined the redox potential of H104A BBE employing the xanthine/xanthine oxidase system (27) in the presence of the redox indicators toluidine blue (tb, EM = +34 mV) and thionine acetate (EM = +64 mV). As depicted in Fig. 6, a direct 2-electron reduction without any indication of radical intermediates could be observed during reductions typically lasting for 2 h. The slope of the linear fit in the Nernst plot of the inset is 1.1, which is close to the theoretical value of unity expected for a simultaneous 2-electron transfer for both systems occurring in equilibrium. The estimated potential obtained from this plot is 28 ± 4 mV. Because this value is significantly lower than reported redox potentials of similar protein variants of GOOX and ChitO (11, 17), we also tested the higher potential redox indicator thionine acetate. This dye, however, was reduced to more than 50% before significant changes in the flavin absorption could be monitored even under very slow reductions (>2 h), indicating that the redox potential of H104A BBE is significantly lower than that of the reference dye.
FIGURE 6.
Redox potential determination of H104A BBE. Selected spectra of the reductive titration of H104A BBE in the presence of toluidine blue (tb) are shown (ranging from solid lines (both oxidized) to short dashed lines (fully reduced)). The inset shows the Nernst plot used for estimation of the redox potential, showing only every fifth measurement for clarity. Data points for evaluation were extracted at 436 and 636 nm for protein and dye, respectively, where no significant contributions to the absorbance are caused by the other chromophore.
DISCUSSION
The major objective of this study was to elucidate the structural and functional importance of bicovalent flavinylation for BBE. Due to the significant influence of both 8α-histidylation and 6-S-cysteinylation on the kinetic properties (Table 1), we addressed the relative contributions of physicochemical properties and structural differences to the overall catalytic efficiency of the enzyme.
In a previous study, we have correlated the change in redox potential for the C166A mutant protein directly to the change in the limiting rate of cofactor reduction (20). Taking into account the results obtained in this study, however, we think that this assumption cannot be generalized. Because for the H104A variant of BBE an even larger decrease in the redox potential was observed, although the reductive rate remained more than 10-fold higher, there clearly are additional factors contributing to the differences in reactivity. The fact that reoxidation of the cofactor is not significantly altered and that the Kd values are only slightly affected by the removal of the covalent linkages suggests a rather subtle change in the interaction of the substrate with FAD to cause the altered kinetic parameters. Similar conclusions were also put forward by Lim et al. (34) stating that a change in the redox potential need not quantitatively correlate with the differences in rates. Other factors such as the geometry of the transition state or the mode of orbital reorganization might also strongly affect the kinetics of the reaction.
Comparing x-ray crystal structures of both mutant proteins with that of the WT enzyme, however, shows that the influence of the removed covalent bonds on the geometry of the isoalloxazine ring system is also rather small (Fig. 3). The overall positioning of FAD is nearly unchanged with the ribityl side chains perfectly overlapping in all three structures. In contrast, neighboring amino acids seem to be more affected by the removal of either 8α-histidylation or 6-S-cysteinylation, which rather supports the idea of substrate-interacting residues playing an important role for the catalytic efficiency. Most notably, the conformation of Trp-165 in the case of the C166A mutant protein (but also to some extent in the H104A variant) as well as a stretch of amino acids (residues 349–353), which seems to play a role in substrate binding/release (supplemental Figs. S2 and S3) for the H104 mutant protein, is substantially changed. Similar observations, where the effect on substrate-interacting amino acids was more pronounced than on the flavin cofactor, were recently described for the analogous protein variant of GOOX where 6-S-cysteinylation was removed (11). The issue of the interaction of the enzyme with substrate or product of the catalyzed reaction was further addressed by soaking crystals of all BBE variants and by comparing the resulting structures with the recently reported structure of the WT BBE substrate complex (21). While crystals of the H104A mutant protein did no longer diffract well enough after soaking, we were able to obtain a structure of WT BBE in complex with the product (S)-scoulerine (Fig. 5, A and C). For the C166A variant, we obtained a difficult to interpret electron density in the active site (Fig. 5, B and D). When compared with the observations of short term substrate soaks (21), the major alterations in the WT_scoulerine complex again involve the amino acid Trp-165 and the surface loop including residues 349–353 (supplemental Fig. S3). In this case, Trp-165 adopts an additional conformation not yet observed in any of the other structures (Figs. 4 and 5C), most importantly also different from the one observed in the short term soaks. In the WT complex, the B-factors of all product atoms are not significantly higher than those of surrounding amino acid atoms, indicating a high occupancy (refined to 84%). This is also one of the most striking differences in comparison with the soak of C166A BBE crystals. As can be seen in Fig. 5B, the electron density clearly differs from that observed in WT crystals. Combined with steady-state kinetics data indicating significant substrate inhibition for this protein variant, we think that these observations are best represented by a superposition of productive and unproductive substrate binding modes. The additional electron density extending from the methoxy group of the benzyl moiety in the unproductive orientation, however, could also indicate complete rotation of the substrate in the active site (changing the position of the isoquinoline and benzyl ring systems). This would be in keeping with the rather poor representation of the N-methyl groups in both alternate conformations of the bound reticuline. On the other hand, this could also be caused by constant inversion of the nitrogen if the attaining of a conformation capable of C–C bond formation is prevented by the active site environment. The unfavorable active site architecture could be linked to the high degree of order of Trp-165 for C166A BBE, which remains as in the native crystals (compare Figs. 4 and 5D) and might prevent binding of the substrate in a way allowing efficient catalysis. Interestingly, the position equivalent to Trp-165 is always occupied by a bulky aromatic amino acid (Trp or Tyr) in all BBE homologs reported to catalyze the reaction shown in Scheme 1.
Substrate binding could additionally be affected by the changed electrostatic potential of the isoalloxazine ring system as shown in computational studies of 6-S-cysteinylation in trimethylamine dehydrogenase (13). Because the nitrogen atom of reticuline is positioned close to the C-6 atom of the cofactor (4.0 Å), changes in the surrounding electrostatic environment could negatively affect positioning of the substrate. As shown recently, (S)-reticuline in WT BBE is perfectly oriented for the simultaneous formation of the C–C bond and substrate oxidation (21). Therefore, subtle changes in the positioning of the substrate would negatively affect the rate of FAD reduction, which is supported by our experimental results.
In contrast, for H104A BBE, the retained electrostatic potential due to 6-S-cysteinylation combined with a flexible Trp-165 could account for the more than 10-fold higher rate of cofactor reduction when compared with C166A BBE, although the redox potential is 20 mV lower than that of the C166A mutant protein. These significant differences caused by subtle changes in the active site architecture might also be related to the rate-limiting step of catalysis for the H104A mutant protein. Neither substrate oxidation nor cofactor regeneration is limiting (Table 1), leaving product dissociation as a plausible limiting step. Although polar interactions with the active site include the same amino acids for both substrate and product, only the conformation of Trp-165 is significantly altered in the WT enzyme (supplemental Fig. S3). This structural change is unlikely to occur synchronously with the enzymatic reaction and therefore suggests that product release is affected by the positioning of Trp-165. In the case of H104A BBE, the conformation of this residue significantly differs from the WT structure. Therefore, changes in the relative energies of substrate- and product-bound Trp-conformations could affect the release of product prior to binding of the next (S)-reticuline molecule during turnover. With the high degree of disorder of the loop involved in substrate binding and product release also pointing in this direction, product dissociation is apparently the rate-limiting step during catalysis in the case of H104A BBE. Understanding that bicovalent flavinylation affects several properties of BBE with quite complex implications for the catalytic functioning of the enzyme, it would be interesting to see whether these observations can be extended to other members in the group of flavoprotein oxidases sharing the same type of cofactor attachment.
Interestingly, most properties of the cofactor for the WT enzymes and the single mutant proteins addressing the bicovalent attachment of FAD are in relatively good agreement between the systems studied in more detail (GOOX, ChitO, and BBE (11, 14, 17, 20)). The absorbance spectra of WT and the single mutant proteins with one covalent linkage removed show characteristic differences (Fig. 1), but these changes are retained among the various systems. This shows that although the electronic properties of the FAD cofactor are strongly affected by the bicovalent attachment, substantial differences in the surrounding active site amino acid composition in these different enzymes seem to have only a minor influence. Interestingly, the characteristic spectral properties of 6-S-cysteinylation as observed in trimethylamine dehydrogenase (33) are only observed upon denaturation of the WT enzyme and the H104A variant. Similar observations have also been made for ChitO (17), and they demonstrate that the sole absorbance maximum around 435 nm is caused by the 6-S-cysteinyl FAD linkage. With the spectral properties of the C166A mutant protein being characteristic for 8α-histidylation (20), these results demonstrate that both types of covalent linkage can be formed independently.
Similar to the spectral properties, the redox potentials of the WT and mutant enzymes, which have been studied for the systems mentioned above, also correlate relatively well. For all three bicovalently modified FAD cofactors, an unusually high redox potential of about +130 mV was found, and removal of 6-S-cysteinylation is accompanied by a decrease of around 70 mV (11, 17, 20). For the protein variants with a substitution corresponding to H104A in BBE, however, controversial results have been obtained (11, 17). Although for ChitO an increase in the redox potential of about 30 mV was measured (17), the analogous GOOX mutant protein instead showed a decrease of around 60 mV (11). As pointed out by the authors of these studies, however, the measurements were erratic for the ChitO system, and in the case of GOOX, the evaluations of data were not straightforward (11, 17). We did not experience any difficulties, and data interpretation yielded a slope close to unity in the Nernst plots as expected for a simultaneous 2-electron reduction of cofactor and reference dye (Fig. 6). In combination with the results of our measurement with a higher potential reference dye, this indicates that no limiting kinetic effects account for the low midpoint potential. The observed reduction of more than 100 mV for the removal of 8α-histidylation in BBE is also in good agreement with similar systems where the redox potential in the absence of the covalent bond has been analyzed. For both cholesterol oxidase and vanillyl-alcohol oxidase, substitution of the corresponding histidine residues with alanine has led to a decrease in the midpoint potential of 100–110 mV (7, 8). Therefore, we believe that the high redox potential observed in the group of bicovalently flavinylated oxidases is caused by the cumulative increase of the potential due to both linkages observed in the WT enzymes.
Because the changes in redox potentials, however, cannot easily be correlated with the kinetic rates, the most important conclusions from our steady-state turnover measurements are not the significant deteriorations of catalytic efficiency but rather the occurrence of substrate inhibition or the changes in the rate-limiting step. These results are generally related to structural changes of active site amino acids, and hence, a comparison of effects on Km and kcat between the studied systems of bicovalent flavoenzymes is quite complex as expected from the differences in active site composition and substrate spectra (11, 17, 20) and shows almost no general trends (11). Other interesting differences are related to the expression level of different mutant proteins addressing bicovalent flavinylation. Optimization of the secretion level of BBE in P. pastoris by co-overexpression of PDI was necessary to obtain amounts sufficient for biochemical and structural characterization of the H104A mutant protein. When compared with the WT enzyme and the C166A protein variant (20) the optimized expression level of H104A BBE is, however, still relatively low (about 1% when compared with WT BBE), and for the double mutant protein H104A-C166A, the level of secretion is even further decreased. The inability to isolate the double mutant variant was also reported for ChitO (17); however, there are also other examples where the corresponding double mutant protein could be isolated (11, 18). Assuming that this problem is specific for a given system, one could argue that it might be related to the amino acid content of the relatively large solvent-accessible active site. Systems with mainly apolar residues such as BBE (21) could have more difficulties adopting a stable fold without the covalently attached FAD cofactor than, for example, GOOX, which contains mostly polar amino acids for the interaction with its sugar substrate (4).
In conclusion, our results show that the bicovalent attachment of FAD in BBE strongly affects the physicochemical properties of the enzyme. In addition to a cumulative increase of the redox potential, both types of linkage, 8α-histidylation and 6-S-cysteinylation, are also responsible for fine tuning of the active site geometry. Appropriate positioning of catalytically important residues accompanied by providing the electrostatic potential for binding of the substrate in the correct conformation for efficient catalysis are facilitated by this recently discovered mode of cofactor attachment.
Supplementary Material
Acknowledgments
We are grateful to Prof. Toni M. Kutchan (The Donald Danforth Plant Science Center, St. Louis, MO) for donating the substrate of the BBE catalyzed reaction (S)-reticuline. We also thank Prof. Anton Glieder and Dr. Sandra Abad (Institute of Molecular Biotechnology, Graz University of Technology, Graz, Austria) for providing the plasmid containing the PDI coding sequence and the input for optimization of enzyme expression in P. pastoris. The support of Dr. M. Oberer (Institute of Molecular Biosciences, University of Graz, Graz, Austria) for the refinement of product density in the long time substrate soaks for WT BBE is highly appreciated. In addition, we are grateful for the help of staff scientists at the Swiss Light Source in Villigen, Switzerland.
This work was supported by the Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung (FWF)) through the Doktoratskolleg “Molecular Enzymology” Grant W901-B05 (to K. G. and P. M.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and supplemental Figs. S1–S4.
The atomic coordinates and structure factors (codes 3FW7, 3FW8, 3FW9, and 3FWA) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GOOX
- glucooligosaccharide oxidase
- BBE
- berberine bridge enzyme
- ChitO
- chito-oligosaccharide oxidase
- PDI
- protein disulfide isomerase
- WT
- wild type.
REFERENCES
- 1.Singer T. P., Kearney E. B., Massey V. (1956) Arch. Biochem. Biophys. 60, 255–257 [DOI] [PubMed] [Google Scholar]
- 2.Mewies M., McIntire W. S., Scrutton N. S. (1998) Protein Sci. 7, 7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edmondson D. E., Newton-Vinson P. (2001) Antioxid. Redox. Signal. 3, 789–806 [DOI] [PubMed] [Google Scholar]
- 4.Huang C. H., Lai W. L., Lee M. H., Chen C. J., Vasella A., Tsai Y. C., Liaw S. H. (2005) J. Biol. Chem. 280, 38831–38838 [DOI] [PubMed] [Google Scholar]
- 5.Huang L., Scrutton N. S., Hille R. (1996) J. Biol. Chem. 271, 13401–13406 [DOI] [PubMed] [Google Scholar]
- 6.Kim J., Fuller J. H., Kuusk V., Cunane L., Chen Z. W., Mathews F. S., McIntire W. S. (1995) J. Biol. Chem. 270, 31202–31209 [DOI] [PubMed] [Google Scholar]
- 7.Fraaije M. W., van den Heuvel R. H. H., van Berkel W. J. H., Mattevi A. (1999) J. Biol. Chem. 274, 35514–35520 [DOI] [PubMed] [Google Scholar]
- 8.Motteran L., Pilone M. S., Molla G., Ghisla S., Pollegioni L. (2001) J. Biol. Chem. 276, 18024–18030 [DOI] [PubMed] [Google Scholar]
- 9.Efimov I., Cronin C. N., McIntire W. S. (2001) Biochemistry 40, 2155–2166 [DOI] [PubMed] [Google Scholar]
- 10.Hassan-Abdallah A., Zhao G., Jorns M. S. (2006) Biochemistry 45, 9454–9462 [DOI] [PubMed] [Google Scholar]
- 11.Huang C. H., Winkler A., Chen C. L., Lai W. L., Tsai Y. C., Macheroux P., Liaw S. H. (2008) J. Biol. Chem. 283, 30990–30996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caldinelli L., Iametti S., Barbiroli A., Bonomi F., Fessas D., Molla G., Pilone M. S., Pollegioni L. (2005) J. Biol. Chem. 280, 22572–22581 [DOI] [PubMed] [Google Scholar]
- 13.Trickey P., Basran J., Lian L. Y., Chen Z., Barton J. D., Sutcliffe M. J., Scrutton N. S., Mathews F. S. (2000) Biochemistry 39, 7678–7688 [DOI] [PubMed] [Google Scholar]
- 14.Winkler A., Hartner F., Kutchan T. M., Glieder A., Macheroux P. (2006) J. Biol. Chem. 281, 21276–21285 [DOI] [PubMed] [Google Scholar]
- 15.Leferink N. G., Heuts D. P., Fraaije M. W., van Berkel W. J. (2008) Arch. Biochem. Biophys. 474, 292–301 [DOI] [PubMed] [Google Scholar]
- 16.Rand T., Qvist K. B., Walter C. P., Poulsen C. H. (2006) FEBS J. 273, 2693–2703 [DOI] [PubMed] [Google Scholar]
- 17.Heuts D. P., Winter R. T., Damsma G. E., Janssen D. B., Fraaije M. W. (2008) Biochem. J. 413, 175–183 [DOI] [PubMed] [Google Scholar]
- 18.Li Y. S., Ho J. Y., Huang C. C., Lyu S. Y., Lee C. Y., Huang Y. T., Wu C. J., Chan H. C., Huang C. J., Hsu N. S., Tsai M. D., Li T. L. (2007) J. Am. Chem. Soc. 129, 13384–13385 [DOI] [PubMed] [Google Scholar]
- 19.Alexeev I., Sultana A., Mäntsälä P., Niemi J., Schneider G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6170–6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler A., Kutchan T. M., Macheroux P. (2007) J. Biol. Chem. 282, 24437–24443 [DOI] [PubMed] [Google Scholar]
- 21.Winkler A., Lyskowski A., Riedl S., Puhl M., Kutchan T. M., Macheroux P., Gruber K. (2008) Nat. Chem. Biol. 4, 739–741 [DOI] [PubMed] [Google Scholar]
- 22.Robinson A. S., Hines V., Wittrup K. D. (1994) Biotechnology 12, 381–384 [DOI] [PubMed] [Google Scholar]
- 23.Inan M., Aryasomayajula D., Sinha J., Meagher M. M. (2006) Biotechnol. Bioeng. 93, 771–778 [DOI] [PubMed] [Google Scholar]
- 24.Lin-Cereghino J., Wong W. W., Xiong S., Giang W., Luong L. T., Vu J., Johnson S. D., Lin-Cereghino G. P. (2005) BioTechniques 38, 44–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weis R., Luiten R., Skranc W., Schwab H., Wubbolts M., Glieder A. (2004) FEMS Yeast Res. 5, 179–189 [DOI] [PubMed] [Google Scholar]
- 26.Dittrich H., Kutchan T. M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9969–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massey V. (1990) in Flavins and Flavoproteins (Curti B., Zanetti G., Ronchi S. eds) pp. 59–66, Walter de Gruyter, Como, Italy [Google Scholar]
- 28.Minnaert K. (1965) Biochim. Biophys. Acta 110, 42–56 [DOI] [PubMed] [Google Scholar]
- 29.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 30.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 31.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32.Kleywegt G. J., Brünger A. T. (1996) Structure. 4, 897–904 [DOI] [PubMed] [Google Scholar]
- 33.Steenkamp D. J., McIntire W., Kenney W. C. (1978) J. Biol. Chem. 253, 2818–2824 [PubMed] [Google Scholar]
- 34.Lim L., Molla G., Guinn N., Ghisla S., Pollegioni L., Vrielink A. (2006) Biochem. J. 400, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.