Abstract
Site-directed mutagenesis of MCT1 was performed on exofacial lysines Lys38, Lys45, Lys282, and Lys413. K38Q-MCT1 and K38R-MCT1 were inactive when expressed at the plasma membrane of Xenopus laevis oocytes, whereas K45R/K282R/K413R-MCT1 and K45Q/K282Q/K413Q-MCT1 were active. The former exhibited normal reversible and irreversible inhibition by DIDS, whereas the latter showed less reversible and no irreversible inhibition. K45Q/K413Q-MCT1 retained some irreversible inhibition, whereas K45Q/K282Q-MCT1 and K282Q/K413Q-MCT1 did not. These data suggest that the two DIDS SO3− groups interact with positively charged Lys282 together with Lys45 and/or Lys413. This positions one DIDS isothiocyanate group close to Lys38, leading to its covalent modification and irreversible inhibition. Additional mutagenesis revealed that DIDS cross-links MCT1 to its ancillary protein embigin using either Lys38 or Lys290 of MCT1 and Lys160 or Lys164 of embigin. We have modeled a possible structure for the outward facing (open) conformation of MCT1 by employing modest rotations of the C-terminal domain of the inner facing conformation modeled previously. The resulting model structure has a DIDS-binding site consistent with experimental data and locates Lys38 in a hydrophobic environment at the bottom of a substrate-binding channel. Our model suggests a translocation cycle in which Lys38 accepts a proton before binding lactate. Both the lactate and proton are then passed through the channel via Asp302− and Asp306+, an ion pair already identified as important for transport and located adjacent to Phe360, which controls channel selectivity. The cross-linking data have also been used to model a structure of MCT1 bound to embigin that is consistent with published data.
Monocarboxylate transporter 1 (MCT1)3 is a member of the monocarboxylate transporter family (SLC16) of which there are 14 known members encoded by both the human and mouse genomes (1). All of the members of this family are thought to have 12 transmembrane alpha helices (TMs) with a large loop between TMs 6 and 7 and the C and N termini facing the cytosol (2, 3). The only members of the MCT family that have been shown to catalyze transport of monocarboxylates such as l-lactate across the plasma membrane are isoforms 1–4 (4–8). This transport is proton-linked and leads to the net uptake or release of lactic acid from cells, which is critical for metabolic pathways such as anaerobic glycolysis, gluconeogenesis, and lactate oxidation (9). MCT8 is a high affinity thyroid hormone transporter (10), whereas MCT10 (TAT1) is an aromatic amino acid transporter (11). The other members of the MCT family remain to be characterized.
MCT1 is the most widely distributed member of the MCT family and was first identified as the lactate transporter present in red blood cells where its kinetics and substrate and inhibitor specificity were investigated in detail (9, 11, 12). These studies revealed that MCT1 can be inhibited by stilbene disulfonate derivatives such as DIDS and 4,4′-dibenzamido-stilbene-2,2′-disulfonate (DBDS). DIDS was shown to exhibit a rapid reversible inhibition of transport that was competitive with respect to l-lactate. This is followed by a slowly developing irreversible inhibition that is not exhibited by DBDS and is thought to be caused by one of the isothiocyanate groups of DIDS attacking a lysine residue on MCT1 (13–15). Prolonged incubation with DIDS also led to a fraction of the MCT1 becoming cross-linked to a 70-kDa glycoprotein that was identified as embigin, also known as gp70 (16). Embigin has a short intracellular C terminus, a single TM sequence containing a glutamic acid residue, and a large extracellular N terminus containing two immunoglobulin domains (17, 18). Subsequent studies revealed that either embigin or, more frequently, the homologous protein basigin (also known as CD147) is required as a chaperone to take MCT1 to the membrane (19) where the two proteins must remain associated for transport activity to be maintained (20, 21).
Expression of MCTs 1, 2, and 4 in Xenopus laevis oocytes has enabled their further characterization and the effects of site-directed mutagenesis to be investigated (4, 5, 7, 8, 22–24). Such studies, together with homology modeling have enabled us to propose a three-dimensional structure of MCT1 based around the published structure of the Escherichia coli glycerol-3-phosphate transporter (Protein Data Bank 1PW4) (24). This model can account for the effects of mutating a range of amino acids, including some that disrupt the interaction with basigin, and has led to the proposal that the single TM of basigin or embigin lies between TMs 3 and 6 of MCT1. The model also reveals exofacial lysines that are present in MCT1 that might be responsible for the irreversible inhibition of MCT1 by DIDS and the cross-linking of MCT1 to embigin. In rat MCT1 these residues are Lys38, Lys45, Lys282, Lys284, Lys290, and Lys413. In this paper, we use site-directed mutagenesis of these lysine residues to identify which of them are involved in DIDS binding to MCT1. In addition we use site-directed mutagenesis of embigin to demonstrate that Lys160 and Lys164 are involved in its cross-linking to MCT1. Our new data allow us to propose a modified structural model of MCT1 in its outward facing conformation that binds DIDS. This model is consistent with the site-directed mutagenesis data and also suggests a mechanism for the translocation cycle that involves Lys38 as well as Asp302 and Arg306 that have already been identified as important for transport (23, 24). We have also been able to model a structure of MCT1 bound to embigin that is consistent with published data.
EXPERIMENTAL PROCEDURES
Materials
All of the reagents were obtained from Sigma unless otherwise stated. Polyclonal antibodies against the C-terminal 16 amino acids of rat MCT1, rat basigin, and a peptide within the loop of TM7 and TM8 of rat MCT1 were raised in rabbits as described previously (2, 21). Polyclonal antibodies were also raised against the cytoplasmic C-terminal region of rat embigen (gp70, amino acids 287–328) using glutathione S-transferase conjugated proteins overexpressed in E. coli using the pGEX-4T-3 vector as described previously (10, 21). The anti-(MCT1-embigin conjugate) antibody (BP) was raised to the MCT1-embigin DIDS-crossed linked product (16). Anti-rabbit secondary antibodies for Western blotting were from Amersham Biosciences. The coding regions of rat MCT1 and embigin were subcloned into the EcoRI site of the pCI-neo mammalian expression vector (Promega) and the Xenopus oocyte vector pGHJ as described previously (8, 21).
Site-directed mutagenesis of MCT1 and embigin within the relevant vector was performed using a QuikChange kit (Stratagene), and the presence of the correct mutation was confirmed by sequencing. Primers containing the desired mutation were designed to be between 25 and 45 bases in length with a melting point greater that 78 °C. The sequences are given in supplemental Table S1. Typically, for mutant MCT1, thermocycling was performed using the following parameters: 30 s at 95 °C, 30 s at 55 °C, and a 4.5- or 6.9-min extension time at 68 °C depending on expression vector. PCR conditions for creating mutant embigin were similar but with an extension time of 4.1 or 6.5 min.
Measurement of MCT1 Transport Activity in Xenopus Oocytes
cRNA was prepared by in vitro transcription from the appropriate linearized pGHJ plasmid (mMessage mMachine; Ambion) and 20 ng injected into X. laevis oocytes prepared from mature adult female toads as described previously (24). Controls received the equivalent volume (9.2 nl) of water. The oocytes were then cultured in OR3 medium for 72 h with fresh medium each day. For the determination of MCT1 activity the uptake l-[14C]lactate into oocytes was measured as described previously (24). In brief, 20 oocytes were placed in a Petri dish containing 2 ml of uptake buffer (75 mm NaCl, 2 mm KCl, 0.82 mm MgCl2, 1 mm CaCl2, 20 mm MES, pH 6.0) and allowed to equilibrate for 5 min. Five oocytes were removed and placed in 50 μl of uptake buffer containing l-[14C]lactate (0.5 mm, 7.4 MBq/ml) with or without 1.0 mm DIDS as required. Following incubation at room temperature (22–25 °C) for 5 min, over which period uptake was linear with time, the oocytes were rapidly washed five times with ice-cold uptake buffer. After the final wash each egg was transferred into a scintillation vial and homogenized in 100 μl of 2% (w/v) SDS by vigorous vortex mixing prior to the addition of 10 ml of scintillation fluid (Emulsifier-Safe; PerkinElmer Life Sciences) and an assay of [14C] by scintillation counting. For irreversible inhibition by DIDS, the oocytes were equilibrated in incubation buffer (75 mm NaCl, 2 mm KCl, 0.82 mm MgCl2, 1 mm CaCl2, 20 mm Tris/HEPES, pH 7.4) and then incubated for 4 h at 18 °C in incubation buffer plus 1 mm DIDS. The oocytes were then washed three times in uptake buffer, and transport was measured as above. For reversible inhibition by DIDS, equilibrated oocytes were placed in incubation buffer plus 1 mm DIDS for 5 min at room temperature (22–25 °C). Five oocytes were removed and placed in 50 μl of uptake buffer containing l-[14C]lactate (0.5 mm, 7.4 MBq/ml) with 1.0 mm DIDS, and transport was measured as above. MCT1 expression at the plasma membrane was confirmed by both SDS-PAGE and Western blotting of a crude oocyte membrane preparation and by immunofluorescence microscopy as described previously (10, 24).
Transfection of COS-7 Cells
COS-7 cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 2 mm l-glutamine (Invitrogen), penicillin 100 units/ml, and streptomycin 100 μg/ml (Sigma). The cells were cultured to 60–70% confluence in 10-cm3 Petri dishes and transfection carried out essentially as described previously (19). Briefly, DNA (11.6 μg) was mixed with 35 μl of FuGENE 6 (Roche Applied Science) in 300 μl of serum-free medium and incubated for 20 min before adding to the cells and incubating for 24 h. At this point the medium was replaced and DIDS-mediated cross-linking of MCT1 to embigin carried out after a further 24 h.
Detection of DIDS-mediated Cross-linking of MCT1 to Embigin
Transfected COS-7 cells were harvested by scraping and washed in phosphate-buffered saline (pH 7.2). The cells were resuspended in citrate buffer (84 mm sodium citrate, 1 mm EGTA, pH 7.4) plus or minus 0.2 mm DIDS. The cells were incubated at 37 °C for 1 h, after which they were washed once in citrate buffer. The cells were lysed at room temperature with 50 μl of lysis buffer (0.15 m NaCl, 5 mm EDTA, 1% (w/v) Triton X-100, 10 mm Tris-HCl, pH 7.4) containing the proteinase inhibitors phenylmethylsulfonyl fluoride (0.5 mm), benzamidine (0.5 mm), leupeptin (4 μg/ml), antipain (4 μg/ml), and pepstatin (4 μg/ml) for 5 min. The insoluble fraction was removed by centrifugation for 10 min at 16,000 × g. The supernatant was assayed for protein using Bradford reagent and 5 μg of protein subjected to SDS-PAGE and Western blotting.
Western Blotting
Following SDS-PAGE, Western blotting was performed using the appropriate polyclonal antibodies at 1 in 1000, and blots were developed with enhanced chemiluminescence (ECL detection kit; Amersham Biosciences) using anti-rabbit secondary antibody conjugated to horseradish peroxidase.
Erythrocyte Preparation and DIDS Labeling
The preparation and labeling with DIDS of erythrocytes from rat and rabbit blood was carried out as described by Poole and Halestrap (15, 16). Erythrocytes were collected in citrate buffer (84 mm citrate, 1 mm EGTA, pH 7.4) and washed once in bicarbonate-buffered saline buffer (120 mm NaCl, 25 mm NaHCO3, equilibrated with 95% O2, 5% CO2) and then twice more in citrate buffer. The cells were resuspended to 10% hematocrit and incubated plus or minus 100 μm DIDS for 1 h at 37 °C. The cells were collected by centrifugation and washed once in citrate buffer containing 0.5% (w/v) bovine serum albumin and once in citrate buffer.
Erythrocyte Ghost Preparation, Trypsin Digestion, and Deglycosylation
Erythrocyte ghosts were prepared from both control and DIDS-treated cells as described previously (25) without phenylmethylsulfonyl fluoride in the lysis buffer. The ghosts were incubated at a protein concentration of 10 mg/ml for 10 min at 37 °C in phosphate-buffered saline, pH 7.3, plus or minus trypsin (50 μg/μl; Worthington Biochemical Corporation). After incubation, 1 mm phenylmethylsulfonyl fluoride was added, and the resulting digests were subjected to SDS-PAGE and Western blotting. For deglycosylation N-glycanase F (from New England Biolabs UK Ltd) was used according to the manufacturer's instructions. The ghosts were solubilized in SDS-PAGE sample buffer and mixed with an equal volume of 2.5% Nonidet P-40 (w/v) in 125 mm sodium phosphate buffer (pH 7.5) before the addition of 5000 units of N-glycanase F and incubation for 1 h at 37 °C. The resulting digests were subjected to SDS-PAGE and Western blotting as above.
Modeling the Open Structure of MCT1 and DIDS Binding
The recently reported model of the closed MCT1 transporter (24), built on the E. coli glycerol-3-phosphate transporter, GlpT (1PW4), was the starting point for the model of the open channel conformation described here. Details of how the modeling was performed are described under “Discussion,” where the model is presented. InsightII-2005 and Discover 2.98 (Accelrys) were the tools used for this modeling.
RESULTS
Lys38 is Essential for MCT1 Activity
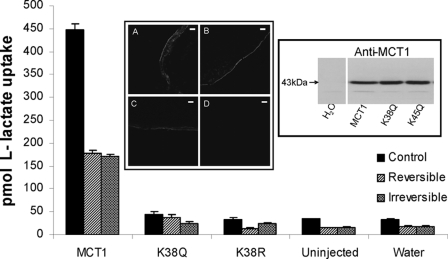
Fig. 1 shows that the mutation of Lys38 to arginine totally abolished lactate transport by MCT1 expressed in oocytes. Mutation to glutamine had a similar effect, although a very small residual transport activity was maintained in some experiments. Neither of these mutations prevented expression of MCT1 at the plasma membrane. Taken together these data imply that Lys38 plays a critical role in the transport mechanism. Fig. 1 also shows that under the conditions used 0.2 mm DIDS caused ∼60% reversible and irreversible inhibition of wild type MCT1 activity. We have previously shown that reversible inhibition is immediate and dependent only on the Ki and inhibitor concentration, whereas irreversible inhibition develops over 60 min (14). We purposefully chose conditions that gave incomplete inhibition so that either an increase or a decrease in inhibitor affinity caused by subsequent mutations could be readily detected.
FIGURE 1.
Lys38 is essential for MCT1 activity. Xenopus oocytes were injected with water or cRNA for wild type MCT1 or the mutant shown, and after expression for 3 days the uptake of l-[14C]lactate was determined as describe under “Experimental Procedures.” Where indicated the oocytes were treated with 0.2 mm DIDS under conditions designed to reveal reversible or irreversible inhibition. The conditions were chosen that gave incomplete inhibition so that either increases or decreases in inhibitor affinity caused by subsequent mutations could be readily detected. The data are shown as the means ± S.E. of 30–180 separate oocytes. The insets show a Western blot of crude membrane fractions (equal protein loading) and sections of oocytes subject to immunofluorescence microscopy to confirm that the lack of lactate transport by the K38Q (inset B) or K38R (inset C) mutants is not due to reduced MCT1 expression compared with wild type MCT1 (inset A). Inset D is water-injected. Scale bar, 20 μm.
Effects of Single Lysine Mutations of MCT1 on DIDS Inhibition of Transport Activity
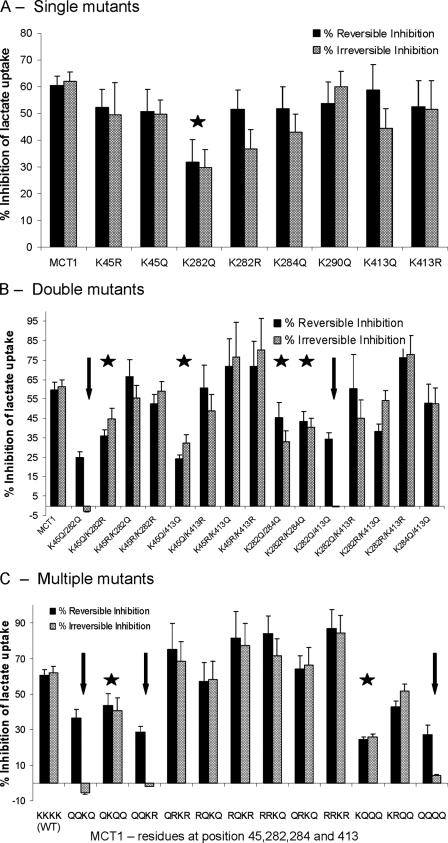
The data of Fig. 2A show the effects of mutating Lys45, Lys282, Lys284, Lys290, and Lys413 to either glutamine or arginine on reversible and irreversible inhibition of transport by DIDS. This should reveal whether any of these residues is critical for the irreversible inhibition of MCT1 by DIDS because neither arginine nor glutamine is capable of reacting covalently with the isothiocyanate group of DIDS. However, the substitution of lysine with arginine, but not glutamine, maintains the positive charge of the residue and thus might reveal whether this is important for either reversible or irreversible inhibition. It was confirmed that all of these mutants were expressed at the cell surface and gave rates of lactate transport similar to wild type MCT1 (supplemental Table S2). Thus none of these residues, unlike Lys38, is essential for MCT1 activity. Nor did any of the mutations alone prevent either reversible or irreversible inhibition of transport by DIDS, although the K282Q mutation gave a slight reduction in inhibition, suggesting that this residue might play some role in inhibitor binding.
FIGURE 2.
The effect of lysine mutations of MCT1 on the reversible and irreversible inhibition of lactate transport by DIDS. Lactate transport by oocytes expressing MCT1 with single (A) double (B) or multiple (C) mutations of lysines to glutamines or arginines was measured in the absence or presence of DIDS under conditions designed to reveal reversible (solid bars) or irreversible inhibition (shaded bars) as described under “Experimental Procedures.” The data are expressed as percentages of inhibition of transport by DIDS and as the means ± S.E. of at least 12 oocytes. All of these mutant forms of MCT1 were active and supported rates of lactate transport within 45–120% of wild type (WT) MCT1 as reported in supplemental Table S2. Mutations showing a significant reduction in reversible and irreversible inhibition (p < 0.05 by Student's t test) are indicated with a star, whereas those in which irreversible inhibition is abolished are indicated with an arrow.
Effects of Multiple Lysine Mutations of MCT1 on DIDS Inhibition of Transport Activity
The data of Fig. 2B show the effects of mutating combinations of two lysines to either glutamine or arginine. The double mutations K282Q/K284Q, K282R/K284Q, K45Q/K282Q, and K45Q/K413Q gave significant reductions in both reversible and irreversible inhibition, similar to that observed with the K282Q single mutation. The double mutations K45Q/K282Q and K282Q/K413Q also gave significant decreases in reversible inhibition, but in these cases the irreversible inhibition was totally lost. The data of Fig. 2B also show that when these lysine residues were mutated to arginine rather than glutamine, there was no reduction in either reversible or irreversible inhibition; if anything inhibition was greater. These data suggest that it is the positive charge of Lys45, Lys282, and Lys413 that plays an important role in DIDS binding. This is likely to be critical for the correct orientation of the isothiocyanate group of DIDS to react with another lysine residue such as Lys38 and so cause irreversible inhibition. This conclusion was further strengthened by making triple mutants as shown in Fig. 2C. Thus the mutation of all three residues (Lys45, Lys282, and Lys413) to arginine maintained reversible and irreversible inhibition with a trend toward enhancement of inhibition as more lysine residues were replaced by arginines. However, replacement of these three lysine residues with glutamines caused a reduction in reversible inhibition while abolishing irreversible inhibition as predicted from the double mutants of Fig. 2B.
Although rat MCT1 has a lysine at position 284, human MCT1 has a glutamine at the equivalent position, and thus our initial studies focused on Lys45, Lys282, and Lys413. However, for completion we included mutations of Lys284, and in most cases mutation of this residue had little additional effect. Thus, as predicted, when Lys45, Lys282, Lys284, and Lys413 were all mutated to glutamine, irreversible inhibition was abolished, and reversible inhibition was greatly reduced (Fig. 2C). However, unexpectedly the K282Q/K284Q/K413Q triple mutation did show some irreversible inhibition unlike the K282Q/K413Q double mutation that did not.
The Effects of MCT1 Lysine Mutations on DIDS Cross-linking to Embigin
The irreversible inhibition of lactate transport by DIDS is an indicator of the covalent binding of DIDS to MCT1. The ability to maintain irreversible binding even when Lys45, Lys282, Lys284, and Lys413 are mutated to arginine implies that these residues are not involved in the covalent modification of MCT1. Rather their importance would seem to lie in their ability to mediate binding through their positive charges interacting with the negative charges of the sulfonate groups of DIDS. This suggests that Lys38 may be the lysine that covalently binds DIDS. However, because this residue is essential for transport activity (Fig. 1), it was not possible to confirm this conclusion using irreversible inhibition of transport by DIDS as a surrogate marker for covalent binding to Lys38. We attempted to use anti-DIDS antibodies (15, 16), with or without immunoprecipitation of MCT1, to detect covalent binding of DIDS to wild type and mutant MCT1 expressed in COS-7 cells but found the antibody to lack sufficient potency and specificity to provide reliable data.
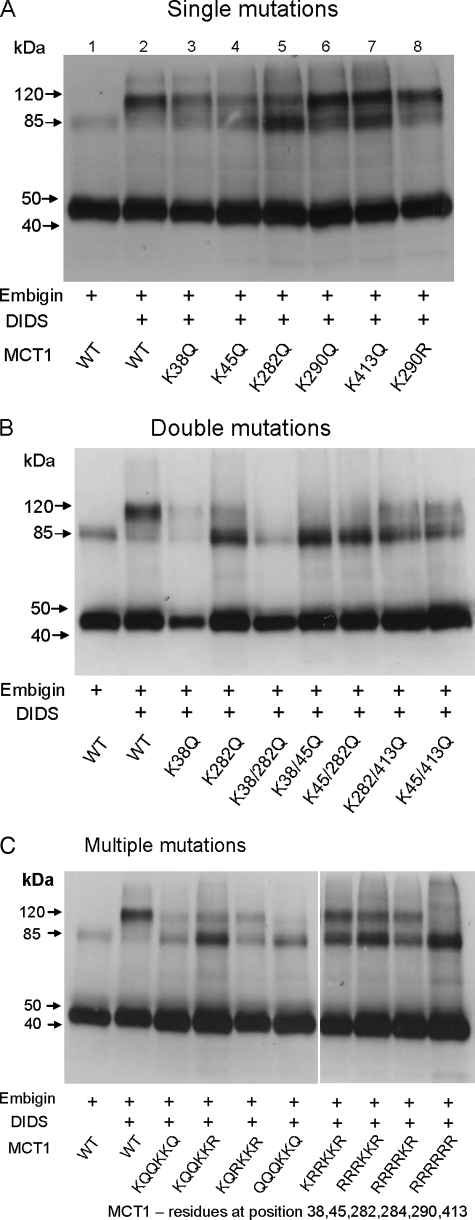
As an alternative strategy we used the ability of DIDS to cross-link MCT1 to embigin to establish the involvement of Lys38 in covalent binding of DIDS. These experiments are reported in Fig. 3. Where cross-linking to embigin occurs, a band of 120 kDa is seen with the MCT1 antibody. It should be noted that this is distinct from the MCT1 dimer that is observed at ∼85 kDa. Such dimers occur even in the absence of DIDS, and we have shown previously that this occurs as a result of aggregation of MCT1 in detergent (16). Our data suggest that the extent of this dimerization is variable and difficult to control. However, two factors appear to increase the intensity of the dimer band: the overall expression level of MCT1 and the absence of embigin. We suspect that excess MCT1 expression overloads the endogenous basigin, and thus in the absence of embigen this may lead to more incorrectly folded MCT1 that has a greater tendency to dimerize. We were unable to confirm the identity of the MCT1-embigin cross-linked product using the embigin antibody because of the presence of broad bands between 110 and 150 kDa induced by DIDS that overlapped the MCT1-embigin band. These bands probably correspond to dimers of embigin caused by DIDS cross-linking.
FIGURE 3.
The effect of lysine mutations of MCT1 on the DIDS-mediated cross-linking of MCT1 to embigin. COS cells were transfected with the indicated pCIneo MCT1 and embigin constructs and harvested after 48 h before incubating with or without 0.2 mm DIDS at 37 °C for 1 h. The cells were then washed and lysed to produce crude lysates, which were subject to SDS-PAGE and Western blotting with anti-MCT1 antibody as described under “Experimental Procedures.” The bands at ∼45 and 85 kDa represent monomeric and dimeric MCT1, whereas the band at ∼120 kDa represents the cross-linked product of MCT1 with embigin. The data shown are representative of two or three experiments that gave similar results. WT, wild type.
In Fig. 3A we show that mutation of Lys38, Lys45, or Lys282 to glutamine gave a considerable reduction in DIDS-mediated cross-linking of MCT1 to embigin, whereas mutation of Lys290 and Lys413 had little or no effect. No additional reduction in cross-linking was observed when the double mutants K45Q/K413Q and K282Q/K413Q were employed, but the K38Q/K45Q, K38Q/K282Q and K45Q/282Q double mutants almost totally abolished cross-linking (Fig. 3B) as did the triple mutants K45Q/K282Q/K413Q, K45Q/K282Q/K413R, and K45Q/K282R/K413R and the quadruple mutant K38Q/K45Q/K282Q/K413Q (Fig. 3C). Interestingly, the decrease in cross-linking was much less when lysines were mutated to arginines rather than glutamine. Thus the multiple mutants K45R/K282R/K413R, K38R/K45R/K282R/K413R, and K38R/K45R/K282R/K284R/K413R all maintained significant cross-linking, implying that rather than forming the cross-link themselves, it is the positive charge of these residues that plays an important role in DIDS binding prior to cross-linking. Only with the hextuple mutant K38R/K45R/K282R/K284R/K290R/K413R was cross-linking totally prevented. Taken together our cross-linking data suggest that DIDS may bind in two ways. Consistent with the inhibition data (Fig. 2) DIDS may bind using noncovalent interaction between its two sulfonate groups with the positive charges of Lys45, Lys282, and Lys413 leaving one isothiocyanate group available to bind covalently to Lys38. The second isothiocyanate may then bind to embigin. However, there would appear to be a second mode of cross-linking that involves Lys290 rather than Lys38. Thus the K38R/K45R/K282R/284R/K413R mutant still shows significant, although reduced, cross-linking, whereas the K38R/K45R/K282R/K284R/K290R/K413R mutant does not (Fig. 3C). Further evidence to support the likelihood of DIDS cross-linking involving two different lysine residues on MCT1 is provided by studies on cross-linking of endogenous MCT1 and embigin in rat erythrocytes treated with DIDS.
DIDS Cross-linking of MCT1 to Embigin in Rat Erythrocytes
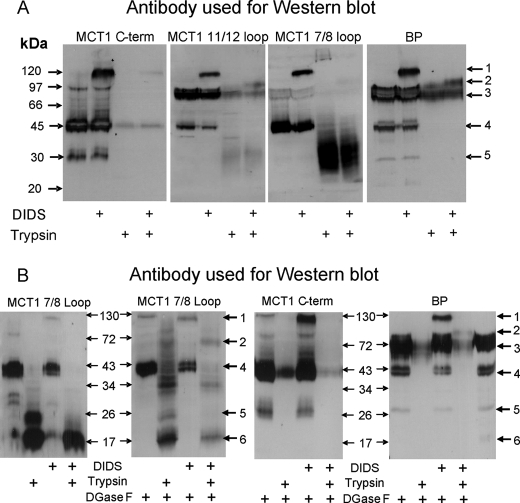
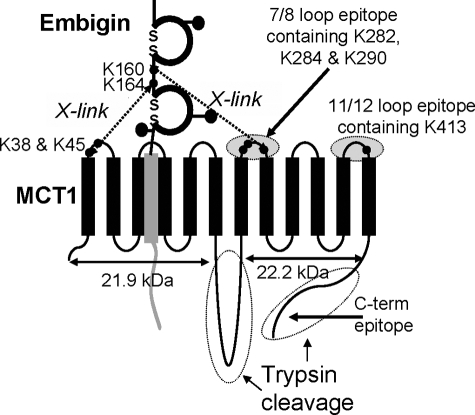
The data of Fig. 4 confirm the cross-linking of MCT1 to embigin in rat erythrocytes using four different antibodies: the normal C-terminal antibody, an antibody to residues 278–293 of the 7/8 extracellular loop of MCT1, an antibody to residues 403–418 of the 11/12 extracellular loop of MCT1, and an antibody raised against the purified MCT1-embigin cross-linked product (BP antibody). The latter detects both MCT1 at 45 kDa (Fig. 4, arrow 4) and embigin at ∼70 kDa (Fig. 4, arrow 3), with the cross-linked product running at ∼120 kDa (Fig. 4, arrow 1). The identity of the band at ∼75 kDa detected with the 11/12 loop antibody (Fig. 4B) is uncertain, although it runs at a similar mobility to both embigin and dimeric MCT1. All four antibodies detected the cross-linked product between MCT1 and embigin at 120 kDa, albeit with different sensitivities. However, after trypsin treatment the picture was more complex. As shown schematically in Fig. 5, trypsin will cleave at lysines within the 6/7 loop splitting the MCT1 into N- and C-terminal fragments of 21–25 and 22–28 kDa, the exact size depending on the extent of additional cleavage at lysines and arginines in the 6/7 loop and C terminus. This accounts for the range of products, seen with the 7/8 antibody following trypsin cleavage.
FIGURE 4.
The effects of trypsin digestion of DIDS-treated rat erythrocyte ghosts on the cross-linked product of MCT1 with embigin. In panel A, rat erythrocytes were incubated in the absence or presence of 100 μm DIDS for 1 h at 37 °C prior to preparation of ghosts and, where required, treatment with trypsin as described under “Experimental Procedures.” The proteins were subjected to SDS-PAGE and Western blotting using the antibodies against the C terminus or 7/8 loop of MCT1 or an antibody raised against the cross-linked product of MCT1 with embigin (16). In B data are shown for samples that have been treated with N-glycanase F (DGaseF) as described under “Experimental Procedures.” The data shown are representative of two or three experiments that gave similar results.
FIGURE 5.
Schematic representation of trypsin cleavage and antibody binding sites on MCT1 and the probable locus of cross-linking between MCT1 and embigin.
Additional complexity may be caused by the presence of lysines within the peptides against which the antibodies were raised. This could lead to the destruction of the antibody epitope during trypsin treatment. Nevertheless, following trypsin hydrolysis of the DIDS-treated cells, the 11/12, 7/8, and BP antibodies all detected a cross-linked product of ∼100 kDa (Fig. 4, arrow 2) consistent with the cross-linking between embigin and Lys290 in the C-terminal fragment of MCT1. In Fig. 4B we show that detection of this band was enhanced if samples were partially deglycosylated using N-glycanase F, perhaps indicating that in the cross-linked product access of the antibody to the 7/8 loop of the MCT1 was impeded by glycosylation of the embigin. Trypsin treatment prevented the C-terminal antibody from detecting any MCT1 band consistent with the predicted proteolysis of the C-terminal tail of MCT1. Trypsin treatment also abolished MCT1 detection with the BP antibody, suggesting that the epitopes for this antibody are contained within the C terminus and/or 6/7 loop degraded by trypsin. The loss of the majority of the embigin detected by the BP antibody probably reflects the almost total digestion of both the extracellular and intracellular domains by trypsin because both are predicted to be accessible to the enzyme and contain many lysines and arginine residues. Unfortunately, we have no antibody that can detect a cross-linked product between the N terminus of MCT1 and embigin, and thus we cannot use erythrocytes to confirm the cross-link between Lys38 of MCT1 and embigin deduced from the experiments of Fig. 3.
Lys160 and Lys164 in Embigin Are Involved in DIDS-mediated Cross-linking to MCT1
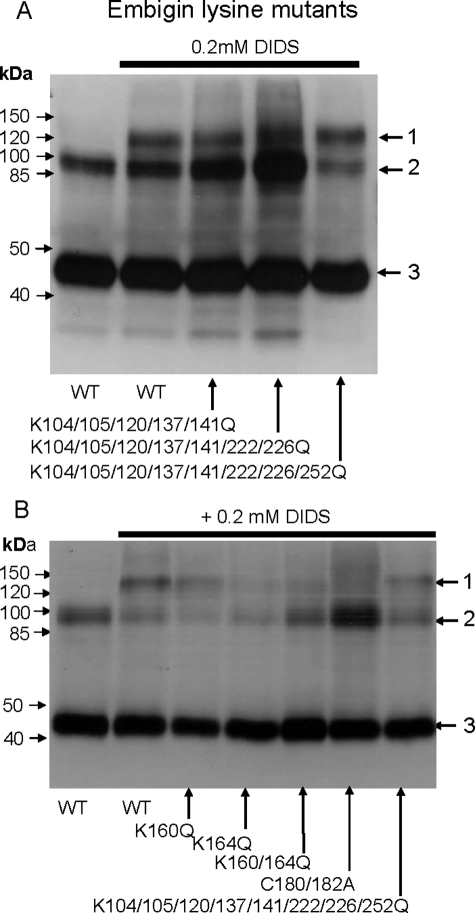
To identify the residues in embigin involved in the DIDS-mediated cross-link with MCT1, we made a range of site-directed mutants of embigin and investigated their cross-linking with wild type MCT1 in COS-7 cells. Previous studies had shown that DIDS produced no cross-linked product in rabbit erythrocytes that contain basigin rather than embigin (15, 21). The inability of DIDS to cross-link MCT1 to basigin is confirmed in the data presented in supplemental Fig. S1. Thus initially we focused our mutagenesis on lysine residues in embigin that were not present in equivalent positions in basigin. We first mutated Lys263, Lys252, Lys226, and Lys224 that would be predicted to be quite close to the TM region of embigin (residues 265–282) and thus likely to be within cross-linking range of the surface of MCT1. However, there was no diminution of cross-linking when any or all of these lysines were mutated to glutamine (data not shown). We then mutated additional lysine residues to glutamine, alone or in combination, but even when eight lysines in the immunoglobulin domains were mutated to glutamine (residues 104, 105, 120, 137, 141, 222, 226, and 252) cross-linking was still observed (Fig. 6A). However, when we mutated either Lys160 or Lys164 to glutamine DIDS-mediated cross-linking was reduced with the greatest effect observed with K164Q, whereas the K160Q/K164Q double mutant showed no detectable cross-linking (Fig. 6B). These two lysine residues are in the region of embigin that links the two immunoglobulin domains. Protein sequence alignment revealed that in rat basigin, the residues equivalent to Lys160 and Lys164 of embigin were Arg106 and Gly110, consistent with the inability of DIDS to cross-link MCT1 to basigin. The choice between Lys160 and Lys164 might depend on whether the cross-linking involves Lys38 or Lys290 on MCT1 as shown schematically in Fig. 5.
FIGURE 6.
Lysine residues 160 and 164 of embigin are involved in DIDS-mediated cross-linking to MCT1. COS cells were transfected as detailed under “Experimental Procedures” with the following pCIneo vector constructs. A, first and second lanes, MCT1 + embigin; third lane, MCT1 + K104/105/120/137/141Q embigin; fourth lane, MCT1 + K104/105/120/137/141/222/226Q embigin; fifth lane, MCT1 + K104/105/120/137/141/222/226/252Q embigin. Arrows 1–3 indicate the positions of MCT1-embigin cross-linked product, MCT1 dimer, and MCT1 monomer, respectively. B, MCT1 plus the embigin mutant indicated. COS cells were harvested 48 h after transfection and treated ± 0.2 mm DIDS, washed, and lysed. SDS-PAGE and Western blots were performed on the crude lysates using anti-MCT1 antibody. WT, wild type.
The data of Fig. 6B also demonstrate that we were able to abolish DIDS-mediated cross-linking of MCT1 to embigin by mutating cysteines at position 180 and 182 of embigin to alanine. This would be predicted to disrupt the disulfide bond stabilizing the second immunoglobulin domain, implying that correct folding of this domain is essential to bring the reactive lysine in close proximity to the DIDS. By contrast, disruption of the first immunoglobulin domain by mutation of Cys144 that forms the disulfide bond had no effect on cross-linking (supplemental Fig. S2). This domain is likely to be further from the membrane and so not involved in locating Lys160 and Lys164 close to DIDS.
DISCUSSION
The Role of Lysine Residues in the Inhibition of MCT1 by DIDS
We have used of a range of single and multiple mutations of MCT1 in which the extracellular facing lysines (Lys38, Lys45, Lys282, Lys284, and Lys413) were replaced by glutamine or arginine to determine the role of these residues in the reversible and irreversible inhibition of MCT1 by DIDS. Our data suggest that the positive charges of Lys45, Lys282, and Lys413 are important for DIDS to bind in an orientation that allows the covalent modification of Lys38 required for irreversible inhibition. Thus MCT1 in which all these three residues have been mutated to arginine is inhibited both reversibly and irreversibly even better than the wild type MCT1 (Fig. 2C). Mutation of Lys282 but not Lys45 or Lys413 to glutamine reduces both types of inhibition (Fig. 2A), suggesting that Lys282 is the most critical residue. This is confirmed by the observation that both the K45Q/K282Q and K282Q/K413Q mutants show no irreversible inhibition, whereas the K45Q/K413Q mutant retains reduced irreversible inhibition (Fig. 2B). Taken together our data imply a DIDS-binding site on MCT1 that involves the two sulfonate groups interacting with the positive charge of Lys282 together with Lys45 and/or Lys413. This brings one of the isothiocyanate groups into close proximity with Lys38, with which it can react covalently to produce irreversible inhibition. This Lys38 residue appears to be essential for transport activity. The second isothiocyanate group is predicted to face outwards toward embigin, with which it can form a cross-link. With this information, we sought to model the DIDS-binding site of MCT1.
Molecular Modeling of MCT1 with the Substrate and Inhibitor-binding Site Facing Outward
We have previously proposed a molecular model of MCT1 that was based upon the published structure of the E. coli glycerol-3-phosphate transporter (24). This model could account for the available site-directed mutagenesis data and provided the starting point for identifying lysine residues involved in DIDS binding. However, our published model structure represents the conformation in which the substrate is bound on the intracellular side, whereas DIDS would be predicted to bind when the substrate-binding site is extracellular. Indeed we have shown previously that DIDS binding and inhibition is greater when cells are subject to a pH gradient (alkaline inside) (16, 27). This is consistent with the proposed kinetic model, which predicts that such a pH gradient would favor protonation of the transporter prior to monocarboxylate binding (9). Thus to model DIDS binding, we needed to generate a molecular model of MCT1 in the conformation with an extracellular binding site.
MCT1 is composed of a six-helix N-terminal domain and a similar six-helix C-terminal domain linked by a 30-residue loop of unknown, or no, structure. Hence it is reasonable to assume that modest reorganization of the interface between these domains is possible. Indeed, just such rearrangements have been suggested for the Lac-Permease during its catalytic cycle (28), and recent data provide further confirmation of this (29–31). A similar rearrangement has also been suggested for MCT1, although no detailed modeling was performed (23). Our own modeling strategy was to juxtapose the N- and C-terminal halves to maximize the interactions between them while ensuring that the transmembrane channel can accommodate a small 2-hydroxy acid. This was achieved as follows. Initially, the six-helix C-terminal domain of the closed model was transformed relative to the six-helix N-terminal domain by two rotations around its center of mass, in the Cartesian axis system shown in Fig. 7. The transformations are a rotation of 20° in z followed by a rotation of −20° in y and then translations of −3 and −4 Å in y and z, respectively. This brings the helices in both domains approximately parallel to the z axis orientation. The second stage of the modeling is to straighten helix 11 at its kink point (characterized by a PP motif) and to remodel the loop connecting helix 11 to helix 12. Hydrogen atoms were added to this model corresponding to pH 7, a layer of water molecules was added around the structure to a depth of 10 Å, and the whole system was relaxed by energy minimization while weakly tethering the backbone atoms to their initial positions, for a total of 3000 conjugate-gradient steps. Structures of DIDS and analogues were built with each sulfonate group carrying a single negative charge and then docked into the cavity on the extracellular face of the open MCT1 model by inspection, having removed the water molecules. A new 10-Å-thick layer of water was added over the surface of the complex, and the whole system energy was minimized as described above. Coordinates of the resulting models presented below are provided in the supplemental material.
FIGURE 7.
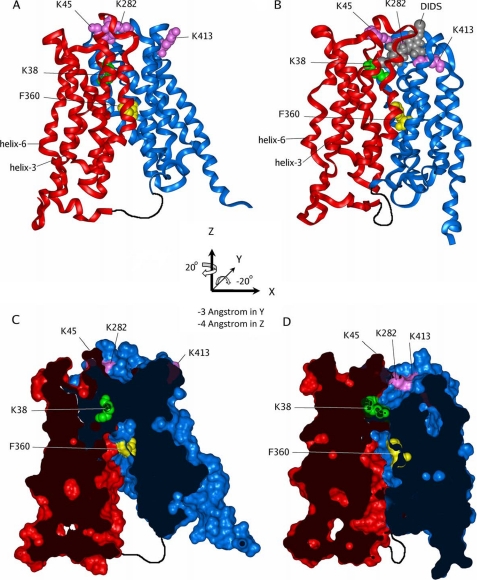
MCT1 models. A and C, closed; B and D, open. The N-terminal domain is colored red, and the C-terminal domain colored blue; Lys38 is green, and Lys45, Lys282, and Lys413 are pink, and Phe360 is yellow, shown in space-filling representation. The 30-residue loop connecting the N- and C-terminal domains is indicated by the sketched black line (not modeled). A and B are ribbon diagrams of the closed and open models, respectively. A space-filling model of DIDS in gray shows the proposed binding site in the open conformation (B). C and D show the same structures as A and B but are cross-sections of the models rendered with a solvent-accessible surface. Hence C shows how the TM channel is closed on the extracellular (upper) side, whereas D show the open TM channel, gated by Lys38 with the DIDS-binding site on the extracellular side of this gate. The position of the specificity filter (Asp302, Arg306, and Phe360) is indicated by the position of Phe360 (Asp302 and Arg306 are not shown for clarity, but line the channel next to Phe360). The axis system used for the C-terminal domain rotations and translations to generate the open model is shown in the center of the figure.
The new outward facing model structure is shown in Fig. 7 (B and D) alongside the inward facing model structure (A and C) published previously (24) and shows how the new orientation of helices 5 and 8 is such that they lie anti-parallel with an interhelical contact surface along their length. This seals the farthest seam of the TM channel. In the inward facing structure helix 11 is bent (as it is in the homology modeling template 1PW4), but after applying the above transformation, the intracellular half of helix 11 is juxtaposed with the corresponding half of helix 2, sealing the lower half of the nearest seam of the TM channel. Helix 11 bends at a PP motif, and these helix-breaking residues can form a hinge between the two halves of helix 11. Straightening helix 11 allows the extracellular half to pack against the corresponding half of helix 2, completing the nearest seam and forming the whole TM channel. Following this manipulation, the loop connecting helix 11 to helix 12 requires remodeling. This loop contains Lys413, which is involved in DIDS binding; however, it points away from the TM channel in the closed MCT1 model. Remodeling the helix 11 to helix 12 loop allows Lys413 to access the extracellular entrance of the TM channel.
The New Molecular Model Suggests a Potential Mechanism for the Translocation Cycle of MCT1
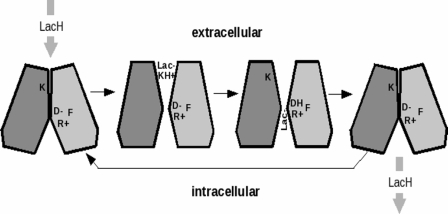
The open and closed conformations of MCT1 that we have modeled are entirely consistent with the “rocker switch” mechanism of substrate translocation suggested for other members of major facilitator superfamily (32). In this model it is proposed that the N- and C-terminal halves of the protein rock back and forth against each other along an axis that runs along the interface between the domains within the membrane. This allows the substrate-binding site to have exclusive access to either side of the membrane in an alternating fashion. For MCT1 our model allows us to suggest critical residues that may be involved in binding the lactate and proton as the protein moves between the two conformations. The cross-section in Fig. 7C shows the model of the closed channel. The upper (extracellular) part of the channel has collapsed, removing the ability of the protein to bind and transport lactate. We might hypothesize that this is the resting state, prevalent at higher extracellular pH that inhibits MCT1 transport (9), and that in this state the buried lysine (Lys38), which is essential for activity, is neutral (unprotonated). Switching into the active state (Fig. 7D) may be driven by protonation at lower pH. The active state has a complete TM channel, blocked only by Lys38 itself. We suggest that the positively charged (protonated) Lys38 binds a lactate ion from the extracellular medium and “passes” lactic acid onto the aspartate Asp302− and arginyl Arg306+ ion pair that have been implicated in proton translocation by Bröer and co-workers (23). Interestingly, the model places Phe360 adjacent to this couple, and it has been shown that mutagenesis of this Phe to Cys allows the channel to transport the larger 2-hydroxymonocarboxylate, mevalonate (33). This is consistent with the idea that the putative binding site formed by Asp302, Arg306, and Phe360 controls channel selectivity (24). Once Lys38 is deprotonated (by passing lactic acid on to D302/R306) MCT1 relaxes back to the closed state, opening the Asp302/Arg306 site to the intracellular medium and allowing the bound lactic acid to diffuse into the intracellular medium and the transport cycle to repeat. A schematic of this hypothetical mechanism is shown in Fig. 8.
FIGURE 8.
Cartoon illustrating the proposed mechanism of lactic acid transport by MCT1. Lactic acid protonates Lys38 (K) causing the channel to open. Lactate then moves into the open extracellular side of the pore and forms an ion pair with Lys38. In the next step the proton on Lys38 is transferred to aspartate 302 (D-) neutralizing the aspartate side chain (DH). This is followed by migration of lactate through the pore where it forms an ion pair with R306 (R+). The size of the adjacent side chain of residue 360 (F) governs the ability of this site to bind the α-hydroxy acid. Once Lys38 is deprotonated, and lactate is occupying the specificity filter, the transporter relaxes back toward the closed state and releases lactic acid into the intracellular space.
On the basis of their mutagenesis studies Bröer and co-workers (23) proposed that in the outward facing conformation of MCT1 there might be an ion pair between Arg143 and Glu369 that was broken in the translocation cycle. Although our new model reduces the distance between Arg143 and Glu369 from 22 to 17 Å, the residues still remain too far apart to form a salt bridge. To bring them close enough to form such a salt bridge would require reregistering the residues on each helix and another 3 Å movement between the N- and C-terminal domains. This would be hard to reconcile with all our other data.
The New Molecular Model Defines a Potential Binding Site for DIDS That Is Consistent with the Experimental Data
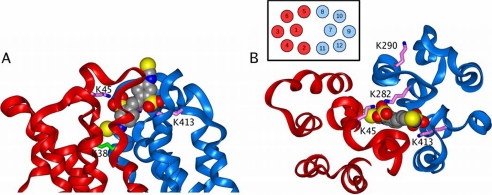
This is illustrated in Fig. 9 where views are given looking down from the extracellular surface (Fig. 9A) and in cross-section (Fig. 9B). The model allows one of the two sulfonate groups on DIDS to bind to Lys45 and Lys282 and the other to bind to Lys413. This orients the DIDS such that one of the two isothiocyanate groups is brought into close proximity with Lys38, enabling it to react covalently (irreversible inhibition). Because the major role of Lys45, Lys282, and Lys413 is to orient the DIDS through their positive charges interacting with the negative charges of the sulfonate, their mutation to arginine would not be expected to prevent binding, and this agrees with our experimental data. By contrast, the removal of the positive charge by mutation to glutamine would impair binding as we observed. An unexpected experimental finding was that both the K45Q/K282Q/K284Q and K282Q/K284Q/K413Q triple mutants showed some irreversible inhibition by DIDS (Fig. 3C), whereas neither the K45Q/K282Q nor the K282Q/K413Q double mutant did (Fig. 3B). This might be explained if the positive charge on Lys284 can act as an alternative binding site for the sulfonate in the absence of the positive charge of Lys282, causing preferential binding of DIDS in a different orientation such as that proposed below for cross-linking of MCT1 to embigin via Lys290 of MCT1.
FIGURE 9.
The binding of DIDS to the open conformation of MCT1. The figure shows a side (A) and top (B) view of the extracellular part of the open MCT1 model. The color scheme is the same as that in Fig. 7, except that DIDS is colored according to atom type with the carbon atoms colored gray. The figure shows how one isothiocyanate group of DIDS is in close contact with Lys38, whereas the two sulfonate groups are bound to Lys45, Lys282, and Lys413. The inset in B shows the numbering of the TM helices in this view.
It should be noted that the isothiocyanate group is not required for reversible inhibition by a variety of DIDS analogues in which the isothiocyanate is replaced by bulkier hydrophobic groups. Thus DBDS and TBenzDS are even more potent reversible inhibitors of MCT1 than is DIDS (14, 34). The model allows for this because Lys38 is contained within a hydrophobic cavity that could accommodate these groups. By contrast, DADS and DNDS in which the isothiocyanate group is replaced by an amino or nitro group, respectively, are much poorer reversible inhibitors. This behavior is consistent with one of the N,N′ substituents binding into the hydrophobic cavity above Lys38. This favors bulky hydrophobic substituents over small hydrophilic substituents and is consistent with the observed potency of DIDS analogues as inhibitors of MCT1 activity (i.e. TBenzDS > DBDS > DIDS > DADS, DNDS) (14, 34).
DIDS Cross-linking
Our data imply that cross-linking between MCT1 and embigin by DIDS can occur in two ways. The first is by DIDS binding in the manner shown in Fig. 8, allowing one isothiocyanate group on DIDS to bind to Lys38 of MCT1 and the other to project out of the substrate-binding site sufficiently to interact with embigin. However, cross-linking is not totally abolished in the K38Q mutants, implying that some DIDS must be binding more promiscuously to other surface lysines. Lys290 appears to be the residue that is covalently modified by the isothiocyanate group of DIDS under these conditions because mutation of lysine residues 38, 45, 282, 284, and 413 to arginines maintained some cross-linking, but this was prevented when Lys290 was also mutated to arginine. We have modeled a possible alternative binding site that can account for our data (see supplemental Fig. S3), and in this model the free isothiocyanate group of DIDS emerges from the surface of MCT1 displaced by 14.5 Å from its location in the primary binding site.
To identify the residue(s) on embigin involved in the cross-linking, we employed lysine and cysteine mutants of embigin (Fig. 6). These data implicate Lys160 and Lys164 in the cross-linking and suggest that the Ig-like V domain of embigin nearest to the membrane may bend over toward the extracellular surface of MCT1. This would bring Lys160 and Lys164 into close proximity with the mouth of the substrate-binding pocket in which DIDS binds. The equivalent residues in rat basigin are Arg106 and Gly110, consistent with the inability of DIDS to cross-link MCT1 to basigin. Furthermore, we found that C180A/C182A embigin also failed to cross-link with MCT1 consistent with disruption of the disulfide bridge in the V domain, preventing its correct folding such that Lys160 and Lys164 are no longer correctly located for cross-linking.
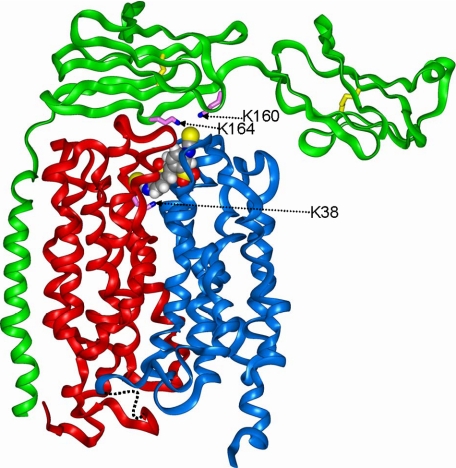
On the basis of site-directed mutagenesis studies, we have previously proposed that the single transmembrane helix of basigin or embigin interacts with transmembrane helices 3 and 6 of MCT1 (21, 24). The conformational changes we have predicted to occur in MCT1 during the translocation cycle are consistent with this model because TMs 3 and 6 remain on the outer face of the N-terminal helical bundle and would only be required to tilt slightly in the membrane. Recently, the crystal structure of the extracellular domains of basigin has been published (34), and we have used this structure to model the extracellular domains of embigin attached to the TM helix lying adjacent to TMs 3 and 6 of MCT1 (Fig. 10). The embigin model was constructed according to the published sequence alignment (34). Subunit C was chosen for the model template because the nearly linear arrangement of domains in this structure allows the N-terminal domain to remain in solution after docking. The rat embigin model was built and refined in the standard manner (24). The embigin model was docked onto the MCT1 model by inspection, and the complex was relaxed by energy minimization as above. The second domain and TM helix of embigin fit snugly against the N-terminal domain of MCT1, and the model illustrates how Lys160 and Lys164 are located over the face of MCT1 where the isothiocyanate group of DIDS emerges and thus would be available for cross-linking. Coordinates of the resulting model are provided in the supplemental material.
FIGURE 10.
Structural model of MCT1 associated with embigin illustrating the site of DIDS cross-linking. Details of the modeling strategy are given in the text. The proteins are represented as ribbons (red, N-MCT1; blue, C-MCT1; green, embigin). The lysine residues involved in cross-linking (Lys38 of MCT1, and Lys164 and Lys160 of embigin) are indicated as magenta sticks, whereas the cysteine disulfide bridges of the two immunoglobulin domains of embigin are shown as yellow sticks. DIDS is shown as a space-filling representation, with standard atom coloring.
Supplementary Material
Acknowledgment
We thank Agnieszka Bierzynska for skilled technical support
This work was supported by Project Grant 079792/z/06 from the Wellcome Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- MCT
- monocarboxylate transporter
- DIDS
- di-isothiocyanostilbene disulfonate
- DBDS
- 4,4′-dibenzamidostilbene-2,2′-disulfonate
- TM
- transmembrane domain
- MES
- 4-morpholineethanesulfonic acid
- TBenzDS
- N,N,N′,N′-tetrabenzyl-4,4′-diamino-2,2′-stilbenedisulfonate
- DADS
- 4,4′-diamino-2,2′-stilbenedisulfonate
- DNDS
- 4,4′-dinitro-2,2′-stilbenedisulfonate.
REFERENCES
- 1.Halestrap A. P., Meredith D. (2004) Pflugers Arch. 447, 619–628 [DOI] [PubMed] [Google Scholar]
- 2.Poole R. C., Sansom C. E., Halestrap A. P. (1996) Biochem. J. 320, 817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halestrap A. P., Price N. T. (1999) Biochem. J. 343, 281–299 [PMC free article] [PubMed] [Google Scholar]
- 4.Bröer S., Schneider H. P., Bröer A., Rahman B., Hamprecht B., Deitmer J. W. (1998) Biochem. J. 333, 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bröer S., Bröer A., Schneider H. P., Stegen C., Halestrap A. P., Deitmer J. W. (1999) Biochem. J. 341, 529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grollman E. F., Philp N. J., McPhie P., Ward R. D., Sauer B. (2000) Biochemistry 39, 9351–9357 [DOI] [PubMed] [Google Scholar]
- 7.Dimmer K. S., Friedrich B., Lang F., Deitmer J. W., Bröer S. (2000) Biochem. J. 350, 219–227 [PMC free article] [PubMed] [Google Scholar]
- 8.Manning Fox J. E., Meredith D., Halestrap A. P. (2000) J. Physiol. 529, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole R. C., Halestrap A. P. (1993) Am. J. Physiol. 264, C761–782 [DOI] [PubMed] [Google Scholar]
- 10.Friesema E. C., Ganguly S., Abdalla A., Manning Fox J. E., Halestrap A. P., Visser T. J. (2003) J. Biol. Chem. 278, 40128–40135 [DOI] [PubMed] [Google Scholar]
- 11.Kim D. K., Kanai Y., Chairoungdua A., Matsuo H., Cha S. H., Endou H. (2001) J. Biol. Chem. 276, 17221–17228 [DOI] [PubMed] [Google Scholar]
- 12.Deuticke B. (1982) J. Membr. Biol. 70, 89–103 [DOI] [PubMed] [Google Scholar]
- 13.Poole R. C., Halestrap A. P. (1990) Biochem. Soc. Trans. 18, 1245–1246 [DOI] [PubMed] [Google Scholar]
- 14.Poole R. C., Halestrap A. P. (1991) Biochem. J. 275, 307–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poole R. C., Halestrap A. P. (1992) Biochem. J. 283, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poole R. C., Halestrap A. P. (1997) J. Biol. Chem. 272, 14624–14628 [DOI] [PubMed] [Google Scholar]
- 17.Guenette R. S., Sridhar S., Herley M., Mooibroek M., Wong P., Tenniswood M. (1997) Dev. Genet. 21, 268–278 [DOI] [PubMed] [Google Scholar]
- 18.Muramatsu T., Miyauchi T. (2003) Histol. Histopathol. 18, 981–987 [DOI] [PubMed] [Google Scholar]
- 19.Kirk P., Wilson M. C., Heddle C., Brown M. H., Barclay A. N., Halestrap A. P. (2000) EMBO J. 19, 3896–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson M. C., Meredith D., Halestrap A. P. (2002) J. Biol. Chem. 277, 3666–3672 [DOI] [PubMed] [Google Scholar]
- 21.Wilson M. C., Meredith D., Fox J. E., Manoharan C., Davies A. J., Halestrap A. P. (2005) J. Biol. Chem. 280, 27213–27221 [DOI] [PubMed] [Google Scholar]
- 22.Rahman B., Schneider H. P., Bröer A., Deitmer J. W., Bröer S. (1999) Biochemistry 38, 11577–11584 [DOI] [PubMed] [Google Scholar]
- 23.Galić S., Schneider H. P., Bröer A., Deitmer J. W., Bröer S. (2003) Biochem. J. 376, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manoharan C., Wilson M. C., Sessions R. B., Halestrap A. P. (2006) Mol. Membr. Biol. 23, 486–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poole R. C., Halestrap A. P. (1988) Biochem. J. 254, 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deleted in proof
- 27.Carpenter L., Halestrap A. P. (1994) Biochem. J. 304, 751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. (2003) Science 301, 610–615 [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y., Guan L., Freites J. A., Kaback H. R. (2008) Proc. Nat. Acad. Sci. U.S.A. 105, 3774–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smirnova I., Kasho V., Choe J. Y., Altenbach C., Hubbell W. L., Kaback H. R. (2007) Proc. Nat. Acad. Sci. 104, 16504–16509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan L., Mirza O., Verner G., Iwata S., Kaback H. R. (2007) Proc. Nat. Acad. Sci. 104, 15294–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Law C. J., Maloney P. C., Wang D. N. (2008) Annu. Rev. Microbiol. 62, 289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia C. K., Goldstein J. L., Pathak R. K., Anderson R. G., Brown M. S. (1994) Cell 76, 865–873 [DOI] [PubMed] [Google Scholar]
- 34.Poole R. C., Cranmer S. L., Holdup D. W., Halestrap A. P. (1991) Biochim. Biophys. Acta 1070, 69–76 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.