Abstract
Formins assemble non-branched actin filaments and modulate microtubule dynamics during cell migration and cell division. At the end of mitosis formins contribute to the generation of actin filaments that form the contractile ring. Rho small GTP-binding proteins activate mammalian diaphanous-related (mDia) formins by directly binding and disrupting an intramolecular autoinhibitory mechanism. Although the Rho-regulated activation mechanism is well characterized, little is known about how formins are switched off. Here we reveal a novel mechanism of formin regulation during cytokinesis based on the following observations; 1) mDia2 is degraded at the end of mitosis, 2) mDia2 is targeted for disposal by post-translational ubiquitin modification, 3) forced expression of activated mDia2 yields binucleate cells due to failed cytokinesis, and 4) the cytokinesis block is dependent upon mDia2-mediated actin assembly as versions of mDia2 incapable of nucleating actin but that still stabilize microtubules have no effect on cytokinesis. We propose that the tight control of mDia2 expression and ubiquitin-mediated degradation is essential for the completion of cell division. Because of the many roles for formins in cell morphology, we discuss the relevance of mDia protein turnover in other processes where ubiquitin-mediated proteolysis is an essential component.
Formin proteins play a role in diverse processes such as cell migration (1, 2), vesicle trafficking (3, 4), tumor suppression (5, 6), and microtubule stabilization (7, 8). Formins also play an essential and conserved role in cytokinesis (9–11). Proper cell division is essential in all animals to maintain the integrity of their genome. Failure to complete cytokinesis can result in genomic instability and ultimately lead to disease such as cancer (12).
The members of the mDia2 family of formins are autoregulated Rho effectors that remodel the cytoskeleton by nucleating and elongating non-branched actin filaments (13). The amino terminus of mDia contains a GTPase binding domain (GBD) that directs interaction with specific Rho small GTP-binding proteins. The adjacent Dia inhibitory domain (DID) mediates mDia autoregulation through its interaction with the carboxyl-terminal diaphanous autoregulatory domain (DAD) (14, 15). Between the DID and DAD domains lie the conserved formin homology 1 (FH1) and FH2 domains. The FH1 domain is a proline-rich region that mediates binding to other proteins such as profilin, Src, and Dia-interacting protein (16–19). In contrast, the FH2 domain binds monomeric actin to generate filamentous actin (F-actin) and can also bind microtubules directly to induce their stabilization (8, 20).
Although the mechanism of mDia activation is well characterized, little is known about its inactivation. Previous reports have suggested that formins can cycle between active, partially active, and inactive states (21, 22) due to GTP hydrolysis upon Rho binding to GTPase-activating proteins. Another formin inactivation mechanism is through mDia interactions with Dia-interacting protein (23). In the context of cortical actin assembly, Dia-interacting protein negatively regulates mDia2 actin polymerization but has no effect on mDia1 actin polymerization despite its ability to interact with both proteins directly (17). Because of the fundamental role for formins in cell division, we sought to identify how mDia2 is inactivated in mitosis.
During cell division, the expression level and activity of many proteins (e.g. cyclins and Aurora and Polo kinases) are tightly regulated (24). A unifying regulatory mechanism among these proteins is ubiquitin-mediated proteolysis. In this study we find that mDia2 protein levels are constant from S phase into mitosis and dramatically decrease at the end of mitosis due to ubiquitin-mediated degradation. Failure to inhibit mDia2 actin assembly results in multinucleation, which supports an essential role for the tight regulation of mDia2 during cell division.
EXPERIMENTAL PROCEDURES
Cells and Plasmids
HEK293T and HeLa cells were grown in Dulbecco's modified Eagle's medium containing 10% (v/v) fetal bovine serum. Cells were transfected with Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Myc-mDia2, Myc-EGFP-mDia2-GBD, Myc-mDia2-ΔDAD, Myc-EGFP-mDia2-ΔFH1, Myc-EGFP-mDia2-DAD, and Myc-EGFP-mDia2-DAD-M1041A were previously described (15, 16). EGFP-mDia2-ΔGBD/ΔDAD (AA 521–1040) and EGFP-mDia2-ΔGBD/ΔDAD-I704A were gifts from Gregg Gundersen. HA-ubiquitin was a gift from Richard Cerione. Myc-mDia2 K118R, K118R/K119R, and K493R/K494R mutants were constructed using site-directed mutagenesis (Stratagene) following the manufacturer's protocol.
Antibodies and Reagents
The following antibodies were used in this study; anti-HA (clone 12CA5) and anti-Myc (clone 9E10) were generated at the Van Andel Research Institute Monoclonal Antibody core facility, anti-cyclin E, anti-Myc, and anti-GFP were from Santa Cruz Biotechnology, and anti-GFP as well as Hoechst 33342 and Texas Red phalloidin were from Molecular Probes. Anti-β-catenin was from BD Transduction Laboratories, anti-ubiquitin (clone P4G7) was from Covance, anti-ubiquitin (clone FK1) was from BioMol, anti-β-tubulin (clone E7) was from the Developmental Studies Hybridoma Bank, anti-EF1α was from Upstate Biotechnologies, rabbit anti-mDia2 (1358) was raised against the mDia2 FH2 domain (generated as recombinant protein in Escherichia coli) as described previously (3, 17), and anti-rabbit conjugated to fluorescein isothiocyanate, anti-mouse conjugated to TRITC, and anti-rabbit and anti-mouse conjugated to horseradish peroxidase were from Jackson ImmunoResearch. MG132 proteasome inhibitor was from Calbiochem. Cell lysis and immunoprecipitation were performed as previously described (17). Immunoblots were performed using 4–20% Tris-glycine gels (Invitrogen) transferred to a 0.45-μm polyvinylidene difluoride membrane (Invitrogen).
Cell Cycle Arrest and Release
G1/S phase cell cycle arrest was performed by incubating HeLa and HEK293T cells in growth medium containing 2 mm thymidine (Sigma) for 16 h. Cells were briefly washed in 1× phosphate-buffered saline and incubated in growth medium for 10 h followed by another thymidine incubation for 16 h. Cells were released into growth medium after washing with phosphate-buffered saline. Cells were collected for flow cytometry analysis or lysed at specific time points for immunoblotting. Mitotic cell cycle arrest was performed by incubating HEK293T cells in growth medium containing 100 ng/ml nocodazole (Sigma) for 18 h. Cells were briefly washed and allowed to progress through the cell cycle by incubating in growth medium.
Flow Cytometry
HeLa and HEK293T cells were briefly washed in 1× phosphate-buffered saline. Cells were stained with a propidium iodide (Sigma) solution containing Nonidet P-40 and RNase. DNA content was acquired using a FACSCalibur flow cytometer (BD Biosciences). The percentages of cells in G0/G1, S, or G2/M phase were determined using ModFit LT software.
Microscopy and Microinjection
HeLa cells were plated onto glass coverslips as previously described (25). Cells were microinjected with plasmid DNA at a concentration of 50 μg/ml as previously described (15). Cells were fixed with 3.7% formaldehyde and permeabilized with 0.3%Triton X-100.
RESULTS
mDia2 Localization and Expression Are Cell Cycle- dependent
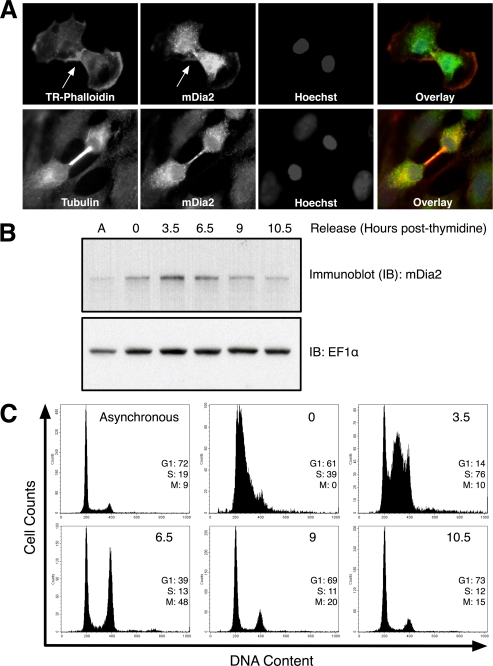
Previously, mDia2 was shown to localize to the cleavage furrow and midbody in dividing NIH 3T3 cells (26). We show a similar cell cycle-dependent localization of mDia2 in human cells. HeLa cells were grown on coverslips and stained for endogenous mDia2 as well as for F-actin or β-tubulin (Fig. 1A). We observed that mDia2 co-localized with the actin-rich cleavage furrow and with the microtubule-rich central spindle during cytokinesis. Because mDia2 localization is tightly linked to the cell cycle, we then asked whether mDia2 protein expression is also cell cycle-dependent. HeLa cells were arrested at the G1/S phase transition using a double thymidine block and allowed to progress through the remainder of the cell cycle. Immunoblots revealed that mDia2 protein expression was increased in S phase and mitotic cells compared with cells predominantly in G0/G1 phase (Fig. 1B). As cells completed mitosis and re-entered the G0/G1 phase, mDia2 protein expression decreased to levels similar to those found in asynchronously growing cells. Consistent with these results, we observed increased endogenous mDia2 fluorescence in cells undergoing mitosis (Fig. 1A, lower panel). Flow cytometry was performed in parallel to measure DNA content and quantify the percentage of cells in each cell cycle phase (Fig. 1C).
FIGURE 1.
mDia2 localization and expression is cell cycle-dependent. A, HeLa cells were stained with rabbit anti-mDia2 (fluorescein isothiocyanate (FITC)) and phalloidin (Texas Red) or mDia2 (fluorescein isothiocyanate) and tubulin (TRITC) along with DNA (Hoechst, blue). Overlay shows the merged image of all three channels. Arrows in the top panels denote the actin-rich cleavage furrow containing mDia2. B, HeLa cell lysates were collected at time points indicated after double thymidine G1/S arrest and release. A represents lysate from asynchronous population of cells. Immunoblots (IB) were probed with anti-mDia2 (1358) and anti-EF1α as a loading control. C, cells from B were labeled as described under “Experimental Procedures” to determine DNA content and analyzed on a flow cytometer. Plots show cell numbers relative to DNA content. The percentages of cells in G1, S, or M phase are shown for each time point.
Proteasome Inhibition Blocks mDia2 Degradation at the End of Mitosis
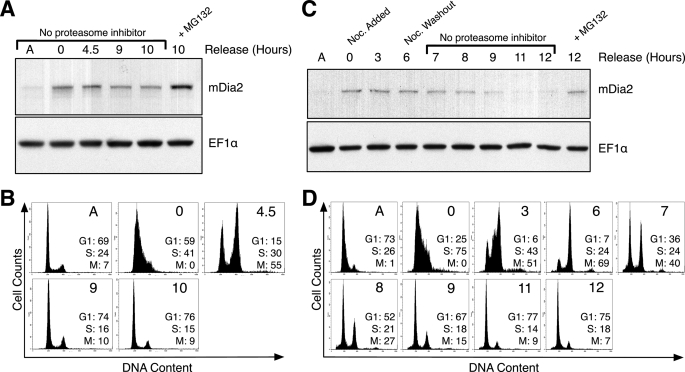
The ordered proteolysis of numerous cell cycle proteins is required for cell division to occur properly (24). We asked if the decrease in mDia2 levels was the result of proteasomal degradation during the cell cycle. HeLa cells were arrested using a double thymidine block and released into growth medium with and without the proteasome inhibitor MG132. Cells treated with MG132 maintained elevated mDia2 levels relative to untreated cells that completed mitosis (Fig. 2A). The corresponding flow cytometry analysis confirmed the G1/S phase arrest, with subsequent completion of cell division upon thymidine washout (Fig. 2B).
FIGURE 2.
Proteasome inhibition prevents mDia2 degradation. A, HeLa cells were arrested with a double thymidine block and released into growth media. MG132 proteasome inhibitor (20 μm) was added to a population of cells upon thymidine release and incubated for 10 h (lane 6). Lysates were collected at the time points indicated and immunoblotted for mDia2. EF1α was probed as a loading control. A represents cell lysate from an asynchronous population. B, cells from A were labeled as described under “Experimental Procedures” to determine DNA content and analyzed on a flow cytometer. Plots show cell numbers relative to DNA content. The percentages of cells in G1, S, or M phase are shown for each time point. C, HeLa cells were thymidine arrested as in A (t = 0 h) but released into growth media containing 100 ng/ml nocodazole for 6 h (t = 0–6 h). Cells were then rinsed and released into normal growth media. MG132 (20 μm) was added to a population of cells 1 h after the nocodazole release and incubated for 5 h (lane 10). Immunoblotting of lysates were performed as in A. D, flow cytometry profiles showing DNA content of cells examined in C.
We then wanted to address whether proteolytic degradation of mDia2 primarily occurs in late mitosis, when formins contribute to contractile ring formation or if mDia2 is degraded earlier in the cell cycle. HeLa cells were arrested in G1/S phase and released into growth medium containing nocodazole. This procedure allows cells to progress through S phase but causes arrest in early mitosis as a result of blocked microtubule assembly. Cells were then washed in normal growth medium to remove the nocodazole for the completion of cell division. Immunoblotting revealed that mDia2 protein levels remain constant from G1/S phase into mitosis (t = 0–6 h) (Fig. 2C). After nocodazole washout, mDia2 expression diminished to levels observed in asynchronously growing cells. However, cells treated with MG132 after nocodazole washout did not show diminished mDia2 expression, suggesting that mDia2 was proteolytically degraded in late mitosis. Flow cytometry confirmed the mitotic arrest in nocodazole-treated cells, with subsequent completion of mitosis upon nocodazole washout (Fig. 2D).
mDia2 Is Post-translationally Modified by Ubiquitin
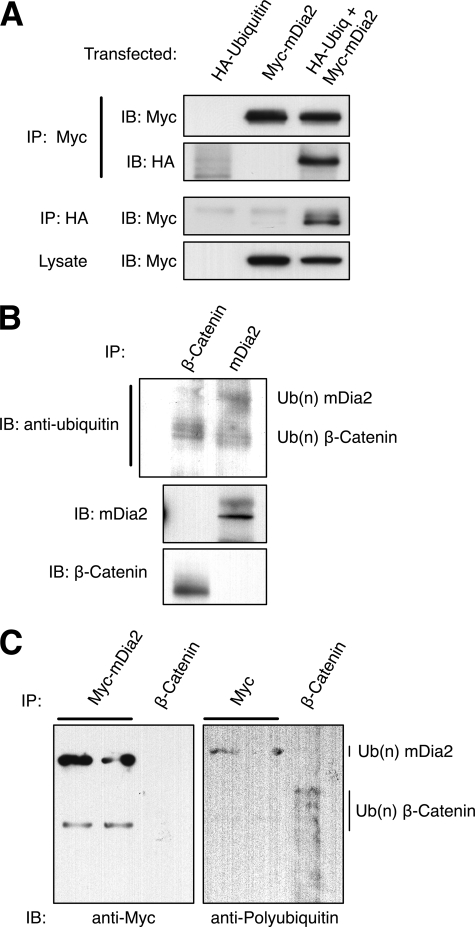
Cell cycle-dependent protein degradation is often a result of polyubiquitination, which targets proteins for degradation in the proteasome (27). We hypothesized that polyubiquitination of mDia2 may signal for its degradation during mitosis. HEK293T cells were transiently transfected with epitope-tagged ubiquitin and mDia2. After mDia2 immunoprecipitation and immunoblotting against ubiquitin, we found that ubiquitin covalently modified mDia2 (Fig. 3A). This was also confirmed by performing the reverse immunoprecipitation to pull down ubiquitin and probing for mDia2 (Fig. 3A). We often observed a “laddering” of mDia2 at increased molecular sizes on immunoblots, characteristic of polyubiquitination. Although mDia2 ubiquitination was shown to occur in cells, we wanted to verify that it was not simply an artifact of overexpressed proteins. To test this, endogenous mDia2 was immunoprecipitated in HeLa cells and blotted for endogenous ubiquitin. β-Catenin, well known for its modification by ubiquitin, was immunoblotted as a positive control. Our results confirmed that endogenous mDia2 was ubiquitinated, which supports our data using transiently expressed mDia2 (Fig. 3B).
FIGURE 3.
mDia2 is polyubiquitinated. A, HEK293T cells were transfected with Myc-mDia2, HA-ubiquitin, or co-transfected with both plasmids. Lysates were immunoprecipitated (IP) for either Myc or HA. Immunoblots (IB) of lysates and immunoprecipitations were probed with anti-Myc or anti-HA to examine the extent of ubiquitination. B, HeLa cells were incubated with 10 μm MG132 for 18 h. Lysates were immunoprecipitated using anti-β-catenin or anti-mDia2. Immunoblots were probed with anti-ubiquitin, anti-β-catenin, or anti-mDia2 (1358) to examine endogenous ubiquitination. C, HEK293T cells were co-transfected with Myc-mDia2 and HA-ubiquitin. Lysates were immunoprecipitated with anti-Myc. HeLa cells were incubated with 20 μm MG132 for 4 h. Lysates were immunoprecipitated with anti-β-catenin. Immunoprecipitations were immunoblotted with anti-Myc and an antibody specific to polyubiquitin chains.
Different types of ubiquitin modification often dictate different cellular outcomes (28). Mono-ubiquitination (that is, the attachment of a single ubiquitin to a lysine of a substrate protein) is usually associated with endocytosis and intracellular trafficking. On the other hand, polyubiquitination (that is, attaching several linked ubiquitin proteins to a lysine of the substrate) usually results in proteasomal targeting and degradation. We tested the hypothesis that mDia2 is polyubiquitinated, consistent with our finding that mDia2 is proteolytically degraded at the end of mitosis. We took advantage of an antibody that will recognize only polyubiquitin chains and not mono- or free ubiquitin. Myc-mDia2 was transfected in HEK293T cells and immunoprecipitated. Subsequent immunoblotting using the anti-polyubiquitin antibody revealed that mDia2 is modified by a polyubiquitin chain (Fig. 3C). Endogenous β-catenin was immunoprecipitated from HeLa cells and blotted with mDia2 as a positive control.
mDia2 Ubiquitination Is Cell Cycle-dependent
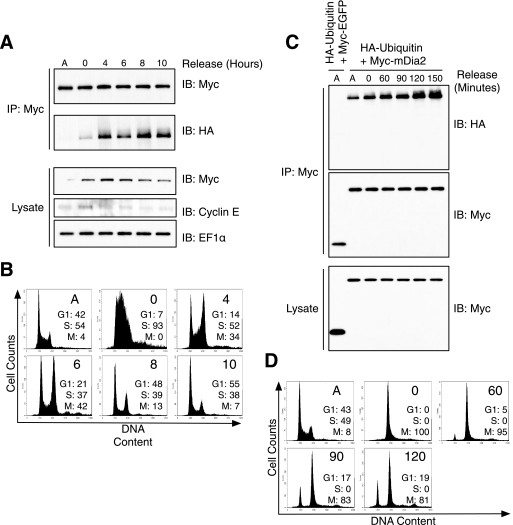
It is possible that mDia2 ubiquitination may be required for multiple cellular functions. Therefore, we asked whether mDia2 ubiquitination was regulated differently during cell division, as opposed to a constitutive process not dependent on the cell cycle. HEK293T cells expressing Myc-mDia2 and HA-ubiquitin were arrested in G1/S phase using a double thymidine block and allowed to progress through the cell cycle. Cells were collected at various time points and subjected to immunoprecipitation and immunoblotting to examine the extent of mDia2 ubiquitination. Flow cytometry was also performed to confirm progression through the cell cycle. We found that ubiquitination of mDia2 increased as cells progressed from S phase (t = 0 h, 93% of cells in S) into mitosis (t = 4–10 h) (Fig. 4A and B). To further examine the timing of ubiquitination, HEK293T cells expressing epitope-tagged mDia2 and ubiquitin were arrested in mitosis after nocodazole treatment. Cells were collected at various times after nocodazole washout and subjected to immunoprecipitation and immunoblotting. In support of our hypothesis, the levels of ubiquitinated mDia2 increased at the end of mitosis (Fig. 4, C and D), which is consistent with the timing of mDia2 degradation observed earlier (see Fig. 2). These data do not rule out the possibility that mDia2 ubiquitination occurs at other times, but it demonstrates that mDia2 modification by ubiquitin is regulated at the end of mitosis.
FIGURE 4.
mDia2 ubiquitination increases at the end of mitosis. A, HEK293T cells were co-transfected with Myc-mDia2 and HA-ubiquitin. Cells were arrested at G1/S phase with a double thymidine block. Cells were released into growth media, and lysates were collected at time points indicated and subjected to Myc immunoprecipitation (IP). Immunoblots (IB) were probed with Myc and HA to examine the extent of mDia2 ubiquitination. EF1α was blotted as a loading control, and cyclin E was blotted to verify progression through the cell cycle. B, cells from A were labeled as described under “Experimental Procedures” to determine DNA content and analyzed on a flow cytometer. Plots show cell numbers relative to DNA content. The percentages of cells in G1, S, or M phase are shown for each time point. C, HEK293T cells were co-transfected with Myc-mDia2 and HA-ubiquitin (second through seventh lanes) in addition to Myc-GFP and HA-ubiquitin as a negative control (first lane). Cells were incubated with 100 ng/ml nocodazole for 24 h to arrest cells in mitosis. Cells were rinsed, and lysates were collected at the indicated times after nocodazole washout. Immunoblots were probed with anti-Myc and anti-HA. D, flow cytometry profiles showing DNA content of cells from C.
mDia2 Is Ubiquitinated on Multiple Lysine Residues
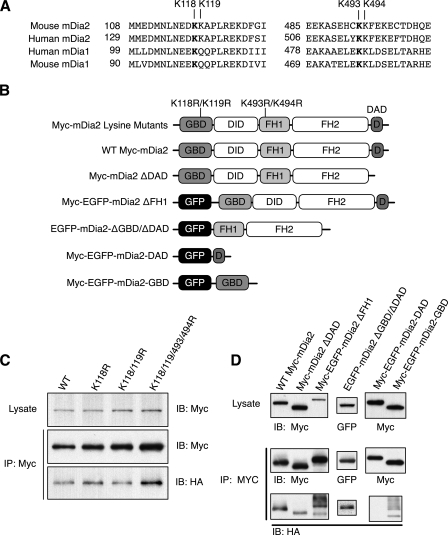
Next, we wanted to identify the region(s) of mDia2 attached to ubiquitin, which may reveal additional functions of ubiquitination beyond proteasomal degradation and help determine the functional consequence of stabilized mDia2. Mass spectrometry has been used to examine ubiquitinated substrates in MCF-7 human breast cancer cells, and that study revealed numerous proteins that were covalently modified by ubiquitin, one of which was mDia2 (29). These data not only substantiate our finding that mDia2 is ubiquitinated in cells at endogenous levels, but it also identified a specific lysine residue attached to ubiquitin. That lysine (Lys-118) is located in the mDia2 GBD. Interestingly, mDia1 was also found to contain a ubiquitinated lysine adjacent to the FH1 domain (corresponding to Lys-493 of mDia2). ClustalW protein sequence alignment showed conservation of the corresponding lysines across the mouse and human sequences of mDia1 and mDia2 (Fig. 5A). We then asked if replacing the lysines with arginines would be sufficient to prevent ubiquitination. mDia2 Lys-118, Lys-118/119, and Lys-118/119/493/494 were replaced with arginines to generate a single, double, and quadruple mutant, respectively, because E3 ligases are known to be promiscuous in their attachment of ubiquitin to substrate proteins. The altered versions of mDia2 were expressed in HEK293T cells with HA-ubiquitin to examine the levels of ubiquitination. Interestingly, mDia2 ubiquitination was not abolished in any of the Arg-substituted variants, suggesting that there are additional lysine residues that can act as acceptors for ubiquitin attachment (Fig. 5, B and C).
FIGURE 5.
Ubiquitin is attached to multiple residues on mDia2. A, ClustalW protein sequence alignment of mouse mDia1, human mDia1, mouse mDia2, and human mDia2 (accession numbers O08808, NP_005210, NP_062644, NP_001035982, respectively). Bold lysines represent ubiquitinated residues identified in Denis et al. (29). B, schematic profile of mDia2 mutants examined for ubiquitination. WT, wild type. C, lysine-to-arginine mutant versions of Myc-mDia2 were co-transfected with HA-ubiquitin in HEK293T cells. Lysates were Myc-immunoprecipitated (IP), and immunoblots (IB) were probed with anti-Myc and anti-HA. D, mDia2 mutants were co-transfected with HA-ubiquitin in HEK293T cells. Lysates were immunoprecipitated, and immunoblots were probed with anti-GFP, anti-Myc, or anti-HA to examine the extent of ubiquitination.
To better define where ubiquitin is attached, we expressed versions of mDia2 lacking certain domains or expressed specific mDia2 domains individually in HEK293T cells (Fig. 5B). Immunoblots showed that the mDia2 GBD was sufficient but not required for ubiquitination, as the ΔGBD/ΔDAD version of mDia2 could still be ubiquitinated. In addition, we found that the FH1 domain was not required for ubiquitination (Fig. 5D). These data confirm that ubiquitin can be attached to many different residues on mDia2 and is not specific to one domain or region of the protein.
Deregulated mDia2 Expression Induces Cytokinesis Failure Due to Excessive Actin Accumulation
No definitive mechanism of inactivating the mDia formins has been revealed. It is interesting to speculate that ubiquitination and proteolytic degradation of the mDias may be a general regulatory mechanism to ensure their inactivation, particularly during cell division. In this case the failure to degrade mDia efficiently would result in excessive actin accumulation.
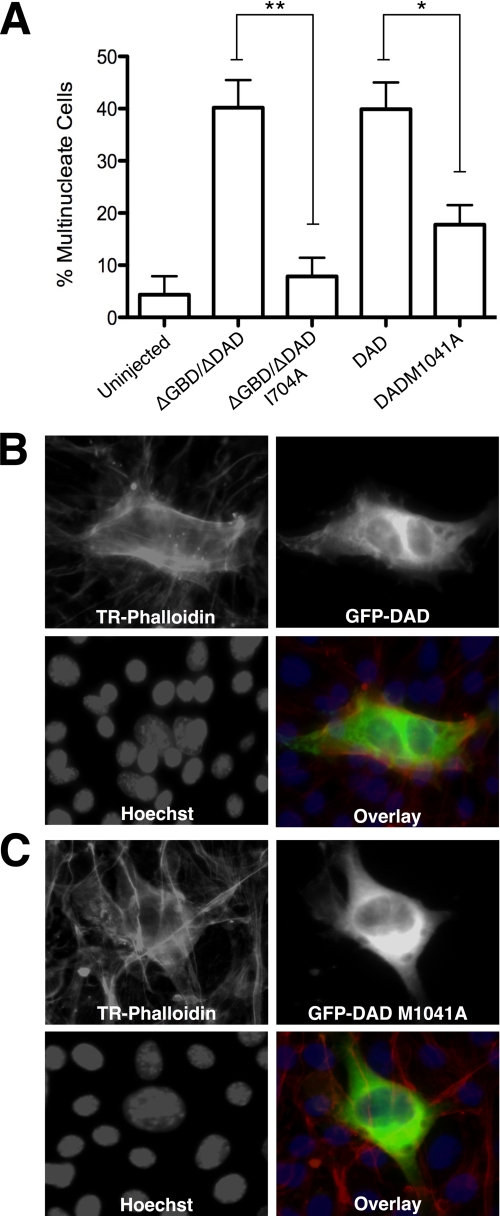
To test the biological significance of such a scenario, we microinjected HeLa cells with a ΔGBD/ΔDAD version of mDia2 (amino acids 521–1040), which lacks key autoregulatory domains. Plasmid DNA was microinjected into synchronized cells during late S phase. Cells were fixed 16 h later and stained with phalloidin and Hoechst. We found that 40% of the cells expressing deregulated mDia2 were predominantly binucleate or to a lesser extent, multinucleate, which are hallmarks of failed cytokinesis (Fig. 6A). We also showed that a version of mDia2 that can stabilize microtubules but is deficient in actin nucleation (EGFP-mDia2-ΔGBD/ΔDAD-I704A (8)) does not result in multinucleation (Fig. 6A). This important distinction shows that failed cytokinesis is due to deregulated actin polymerization and not decreased microtubule dynamics, as mDia2 was recently shown to control both properties independently (8).
FIGURE 6.
Deregulated mDia2 results in cytokinesis failure. A, bar graph showing the percent of multinucleate cells after injection with plasmids that encode EGFP-mDia2-ΔGBD/ΔDAD (amino acids 521–1040), EGFP-mDia2-ΔGBD/ΔDAD-I704A, EGFP-mDia2-DAD, or EGFP-mDia2-DAD-M1041A. Error bars represent the S.D. from three independent experiments (p < 0.01 (**) and p < 0.05 (*) using Student's t test for significance, n = 30). B, HeLa cells were microinjected with plasmid DNA encoding EGFP-mDia2-DAD, which interferes with mDia autoregulation. Cells were stained for F-actin (Texas Red) and nuclei (Hoechst, blue). C, HeLa cells were microinjected with a mutant version of the plasmid described in B that is unable to bind to DID and interfere with autoregulation (DAD-M1041A). Cells were stained as in B.
To further address whether mDia2 hyperactivation would impede the completion of cytokinesis, plasmid DNA encoding the DAD domain was microinjected into synchronized HeLa cells. Expression of this domain disrupts the formin autoregulatory mechanism in cells and results in increased actin polymerization (15, 30). DAD expression promoted multinucleation in ∼40% of cells (Fig. 6, A and B). An altered version of DAD (M1041A) that prevents binding to DID (31, 32) and does not affect formin autoregulation (15) resulted in significantly fewer multinucleate cells (Fig. 6, A and C).
DISCUSSION
The later stages of cell division are part of a highly regulated process requiring the contribution of many proteins at specific times (33). Inappropriate regulation of cytokinesis results in genetic defects (aneuploidy) that can support the progression to malignancy. Here, we report the cell cycle-dependent localization and expression of mDia2; we found that mDia2 was ubiquitinated and degraded at the end of mitosis. We propose that mDia2 degradation is a potential mechanism to inactivate the formin, thus ensuring its appropriate activity within the cell cycle. A consequence of deregulated actin polymerization during mitosis is cytokinesis failure, as activated mDia2 results in the multinucleation of cells.
Contractile ring formation during the latter stages of mitosis requires highly regulated and localized actin assembly, whereas its constriction depends upon the action of myosin (34). In both budding and fission yeast formin FH2 domains nucleate and processively elongate new actin filaments that form the contractile ring (35, 36). FH2-mediated actin polymerization is increased in the presence of the FH1 domain because of its ability to interact with profilin-bound actin monomers. Once the dense, bundled actin network of the contractile ring is finally formed (37), myosin-II pulls on the actin fibers to invaginate the plasma membrane. Soon after, components of the contractile ring were removed or degraded to allow disassembly of the ring (35).
It was recently shown that mDia2 is essential for cytokinesis of NIH 3T3 fibroblasts, presumably by acting as a scaffold for the contractile ring and maintaining furrow position (26). mDia2 is normally autoregulated in the cytosol and activated to promote actin assembly at desired locations, such as the contractile ring. But it was interesting to find coordinated mDia2 protein expression during cell division. Why are the protein levels not sustained once cell division is complete if the formin could simply become autoregulated again? Interestingly, the yeast formin cdc12p contains conserved DID and DAD domains, but its activity does not appear to be mediated by autoregulation during cytokinesis (10). This suggests that additional regulatory mechanisms mediate the specific timing of actin assembly for this formin.
Traditionally, it has been assumed that formins are activated by certain Rho GTPases and eventually become inactivated upon release of Rho or other proteins that might keep formins in an activated conformation. One way to guarantee that mDia2 (or similar formins, such as cdc12p) would no longer be activated would be to dispose of the protein entirely. Ubiquitin-mediated proteolysis is a well characterized mechanism for degrading cellular proteins via the proteasome. Protein ubiquitination has also been shown to play a crucial role in the completion of cytokinesis (38). Our discovery that mDia2 is ubiquitinated and degraded at the end of mitosis suggests a new layer of formin regulation.
In addition, we found that mDia2-mediated actin assembly will result in failed cytokinesis if not properly controlled. Deregulated versions of mDia2 also stabilize microtubules, thus making it difficult to determine whether failed cytokinesis is due to deregulated actin assembly or due to effects on microtubule stabilization. Stable microtubules play an important role in central spindle assembly (39), and the localization of mDia2 to the midbody during cytokinesis suggests that it could contribute to this process. Because mDia2-mediated actin assembly is independent of its microtubule stabilization properties, we took advantage of this distinction and used a mutant version of mDia2 (I704A) that is unable to nucleate actin but can still stabilize microtubules. We determined that failed cytokinesis is specifically the result of deregulated actin assembly.
Our findings are consistent with a similar model showing that deregulated actin polymerization by WASp (Wiskott-Aldrich syndrome protein) can lead to defective cytokinesis (40). This study showed that activating mutations in WASp, which are known to cause X-linked neutropenia, can lead to the hyperactivation and mislocalization of actin polymerization. The authors of this study concluded that failed cytokinesis was likely due to disruption of contractile ring formation and potential physical inhibition of mitosis by interfering with required signaling proteins.
It is well known that intracellular trafficking of membrane to the central spindle is crucial for cleavage furrow invagination and subsequent separation (41, 42). Our laboratory has demonstrated that constitutively active RhoB and deregulated mDia2, an effector for RhoB, slows vesicle trafficking (3). mDia2 localizes to endosomes and contributes to their motility by mediating actin dynamics. Therefore, deregulated mDia2 may also hinder the normal trafficking of membrane and other components required for cell abscission. Defective abscission results in multinucleate cells, which is what we observed in the context of deregulated mDia2. Thus, mDia2 degradation may be required for both normal actin dynamics related to contractile ring assembly and for intracellular trafficking dynamics. This is an interesting possibility considering that protein ubiquitination contributes to the normal turnover of many proteins involved in endosome trafficking (43).
The anaphase-promoting complex (APC) is an E3 ligase important for the inactivation of cytokinesis machinery by triggering its degradation (44). In budding yeast, genetic deletion of the APC activator Cdh1 results in cells that do not disassemble the contractile actin ring properly because specific proteins remain stabilized after cell contraction (45). Furthermore, a stable version of the protein IQGAP, which cross-links F-actin, can partially induce the phenotype observed in Cdh1-deficient cells. It was concluded that protein degradation of multiple contractile ring components must be responsible for efficient ring disassembly. Our findings suggest that mDia2 ubiquitination and degradation may contribute to the disassembly of the contractile ring, especially considering that mDia2 is a required component for proper actin ring assembly (26), and mDia2 was previously shown to bundle actin filaments directly (46).
A logical question is whether mDia2 can act as a substrate for APC-mediated ubiquitination. mDia2 sequence analysis shows that it contains over 10 putative destruction box motifs (RXXL or KEN) (47). We show that mDia2 deletion mutants and lysine point mutants did not prevent ubiquitination, suggesting that there are multiple residues that can attach to ubiquitin. The large number of putative APC binding sites that span mDia2 is consistent with these results. We are currently testing the hypothesis that APC is responsible for promoting mDia2 ubiquitination and degradation during cell division. A stabilized mutant version of mDia2 will be important to fully characterize the functional consequence of mDia2 ubiquitination.
Taken together, our data provide new insights for mDia2 function during cell division. Given the importance of proper cell division for genomic stability, it will be important to study the role of mDia2 ubiquitination and degradation further as well as examine other formins that might share similar properties.
Acknowledgments
We thank Rich West and Kellie Leali for assistance with flow cytometry. We also thank David Nadziejka for critical reading of the manuscript and Leanne Lash-VanWyhe for technical assistance.
This work was supported by the Van Andel Foundation.
- mDia
- mammalian diaphanous-related
- GBD
- GTPase binding domain
- DID
- Dia inhibitory domain
- DAD
- Dia autoregulatory domain
- FH
- formin homology
- APC
- anaphase-promoting complex
- TRITC
- tetramethylrhodamine isothiocyanate
- E3
- ubiquitin-protein isopeptide ligase
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP
- HA
- hemagglutinin.
REFERENCES
- 1.Eisenmann K. M., West R. A., Hildebrand D., Kitchen S. M., Peng J., Sigler R., Zhang J., Siminovitch K. A., Alberts A. S. (2007) J. Biol. Chem. 282, 25152–25158 [DOI] [PubMed] [Google Scholar]
- 2.Gupton S. L., Eisenmann K., Alberts A. S., Waterman-Storer C. M. (2007) J. Cell Sci. 120, 3475–3487 [DOI] [PubMed] [Google Scholar]
- 3.Wallar B. J., Deward A. D., Resau J. H., Alberts A. S. (2007) Exp. Cell Res. 313, 560–571 [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Borja M., Janssen L., Verwoerd D., Hordijk P., Neefjes J. (2005) J. Cell Sci. 118, 2661–2670 [DOI] [PubMed] [Google Scholar]
- 5.Peng J., Kitchen S. M., West R. A., Sigler R., Eisenmann K. M., Alberts A. S. (2007) Cancer Res. 67, 7565–7571 [DOI] [PubMed] [Google Scholar]
- 6.Kamasani U., Duhadaway J. B., Alberts A. S., Prendergast G. C. (2007) Cancer Biol. Ther. 6, 1422–1427 [DOI] [PubMed] [Google Scholar]
- 7.Palazzo A. F., Cook T. A., Alberts A. S., Gundersen G. G. (2001) Nat. Cell Biol. 3, 723–729 [DOI] [PubMed] [Google Scholar]
- 8.Bartolini F., Moseley J. B., Schmoranzer J., Cassimeris L., Goode B. L., Gundersen G. G. (2008) J. Cell Biol. 181, 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castrillon D. H., Wasserman S. A. (1994) Development 120, 3367–3377 [DOI] [PubMed] [Google Scholar]
- 10.Yonetani A., Lustig R. J., Moseley J. B., Takeda T., Goode B. L., Chang F. (2008) Mol. Biol. Cell 19, 2208–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Severson A. F., Baillie D. L., Bowerman B. (2002) Curr. Biol. 12, 2066–2075 [DOI] [PubMed] [Google Scholar]
- 12.Ganem N. J., Storchova Z., Pellman D. (2007) Curr. Opin. Genet. Dev. 17, 157–162 [DOI] [PubMed] [Google Scholar]
- 13.Goode B. L., Eck M. J. (2007) Annu. Rev. Biochem. 76, 593–627 [DOI] [PubMed] [Google Scholar]
- 14.Li F., Higgs H. N. (2005) J. Biol. Chem. 280, 6986–6992 [DOI] [PubMed] [Google Scholar]
- 15.Alberts A. S. (2001) J. Biol. Chem. 276, 2824–2830 [DOI] [PubMed] [Google Scholar]
- 16.Tominaga T., Sahai E., Chardin P., McCormick F., Courtneidge S. A., Alberts A. S. (2000) Mol. Cell 5, 13–25 [DOI] [PubMed] [Google Scholar]
- 17.Eisenmann K. M., Harris E. S., Kitchen S. M., Holman H. A., Higgs H. N., Alberts A. S. (2007) Curr. Biol. 17, 579–591 [DOI] [PubMed] [Google Scholar]
- 18.Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier M. F. (2004) Cell 119, 419–429 [DOI] [PubMed] [Google Scholar]
- 19.Higgs H. N. (2005) Trends Biochem. Sci. 30, 342–353 [DOI] [PubMed] [Google Scholar]
- 20.DeWard A. D., Alberts A. S. (2008) Curr. Biol. 18, R605–608 [DOI] [PubMed] [Google Scholar]
- 21.Martin S. G., Chang F. (2006) Curr. Biol. 16, 1161–1170 [DOI] [PubMed] [Google Scholar]
- 22.Kovar D. R. (2006) Curr. Biol. 16, R535–538 [DOI] [PubMed] [Google Scholar]
- 23.Satoh S., Tominaga T. (2001) J. Biol. Chem. 276, 39290–39294 [DOI] [PubMed] [Google Scholar]
- 24.Lindon C., Pines J. (2004) J. Cell Biol. 164, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng J., Wallar B. J., Flanders A., Swiatek P. J., Alberts A. S. (2003) Curr. Biol. 13, 534–545 [DOI] [PubMed] [Google Scholar]
- 26.Watanabe S., Ando Y., Yasuda S., Hosoya H., Watanabe N., Ishizaki T., Narumiya S. (2008) Mol. Biol. Cell 19, 2328–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed S. I. (2003) Nat. Rev. Mol. Cell Biol. 4, 855–864 [DOI] [PubMed] [Google Scholar]
- 28.Welchman R. L., Gordon C., Mayer R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 599–609 [DOI] [PubMed] [Google Scholar]
- 29.Denis N. J., Vasilescu J., Lambert J. P., Smith J. C., Figeys D. (2007) Proteomics 7, 868–874 [DOI] [PubMed] [Google Scholar]
- 30.Dong Y., Pruyne D., Bretscher A. (2003) J. Cell Biol. 161, 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallar B. J., Stropich B. N., Schoenherr J. A., Holman H. A., Kitchen S. M., Alberts A. S. (2006) J. Biol. Chem. 281, 4300–4307 [DOI] [PubMed] [Google Scholar]
- 32.Nezami A. G., Poy F., Eck M. J. (2006) Structure 14, 257–263 [DOI] [PubMed] [Google Scholar]
- 33.Glotzer M. (2005) Science 307, 1735–1739 [DOI] [PubMed] [Google Scholar]
- 34.Werner M., Glotzer M. (2008) Biochem. Soc. Trans. 36, 371–377 [DOI] [PubMed] [Google Scholar]
- 35.Pollard T. D. (2008) Biochem. Soc. Trans. 36, 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul A. S., Pollard T. D. (2008) Curr. Biol. 18, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vavylonis D., Wu J. Q., Hao S., O'Shaughnessy B., Pollard T. D. (2008) Science 319, 97–100 [DOI] [PubMed] [Google Scholar]
- 38.Mukai A., Mizuno E., Kobayashi K., Matsumoto M., Nakayama K. I., Kitamura N., Komada M. (2008) J. Cell Sci. 121, 1325–1333 [DOI] [PubMed] [Google Scholar]
- 39.Canman J. C., Cameron L. A., Maddox P. S., Straight A., Tirnauer J. S., Mitchison T. J., Fang G., Kapoor T. M., Salmon E. D. (2003) Nature 424, 1074–1078 [DOI] [PubMed] [Google Scholar]
- 40.Moulding D. A., Blundell M. P., Spiller D. G., White M. R., Cory G. O., Calle Y., Kempski H., Sinclair J., Ancliff P. J., Kinnon C., Jones G. E., Thrasher A. J. (2007) J. Exp. Med. 204, 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montagnac G., Echard A., Chavrier P. (2008) Curr. Opin. Cell Biol. 20, 454–461 [DOI] [PubMed] [Google Scholar]
- 42.Albertson R., Cao J., Hsieh T. S., Sullivan W. (2008) J. Cell Biol. 181, 777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hicke L., Dunn R. (2003) Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 44.Peters J. M. (2006) Nat. Rev. Mol. Cell Biol. 7, 644–656 [DOI] [PubMed] [Google Scholar]
- 45.Tully G. H., Nishihama R., Pringle J. R., Morgan D. O. (2009) Mol. Biol. Cell 20, 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris E. S., Rouiller I., Hanein D., Higgs H. N. (2006) J. Biol. Chem. 281, 14383–14392 [DOI] [PubMed] [Google Scholar]
- 47.King R. W., Glotzer M., Kirschner M. W. (1996) Mol. Biol. Cell 7, 1343–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]