Abstract
Many agents that activate hematopoietic cells use phos pha tidyl ino si tol 3,4,5-trisphosphate (PtdIns 3,4,5-P3) to initiate signaling cascades. The SH2 domain-containing inositol 5′ phosphatase, SHIP1, regulates hematopoietic cell function by opposing the action of phos pha tidyl ino si tol 3-kinase and reducing the levels of PtdIns 3,4,5-P3. Activation of the cyclic AMP-de pend ent protein kinase (PKA) also opposes many of the pro-inflammatory responses of hematopoietic cells. We tested to see whether the activity of SHIP1 was regulated via phos pho ryl a tion with PKA. We prepared pure recombinant SHIP1 from HEK-293 cells and found it can be rapidly phos pho ryl a ted by PKA to a stoichiometry of 0.6 mol of PO4/mol of SHIP1. In 32P-labeled HEK-293 cells transfected with SHIP1, stimulation with Sp-adenosine 3′,5′-cyclic monophosphorothioate triethylammonium salt hydrate (Sp-cAMPS) or activation of the β-adrenergic receptor increased the phos pho ryl a tion state of SHIP1. Inhibition of protein phosphatase activity with okadaic acid also increased the phos pho ryl a tion of SHIP1. Phosphorylation of SHIP1 in vitro or in cells by PKA increased the 5′ phosphatase activity of SHIP1 by 2–3-fold. Elevation of Ca2+ in DT40 cells in response to B cell receptor cross-linking, an indicator of PtdIns 3,4,5-P3 levels, was markedly blunted by pretreatment with Sp-cAMPS. This effect was absent in SHIP−/− DT40 cells showing that the effect of Sp-cAMPS in DT40 cells is SHIP1-de pend ent. Sp-cAMPS also blunted the ability of the B cell receptor to increase the phos pho ryl a tion of Akt in DT40 and A20 cells. Overall, activation of G protein-coupled receptors that raise cyclic AMP cause SHIP1 to be phos pho ryl a ted and stimulate its inositol phosphatase activity. These results outline a novel mechanism of SHIP1 regulation.
Activation of phosphatidylinositol 3-kinase (PtdIns 3-kinase)2 is central to regulation of multiple cell functions including cell shape changes, cell migration, cell activation, and proliferation (1). PtdIns 3-kinase phosphorylates phosphatidylinositol 4,5-bisphosphate in the inner leaflet of the plasma membrane to generate phosphatidylinositol 3,4,5-trisphosphate (PtdIns 3,4,5-P3) (2). PtdIns 3,4,5-P3 then activates downstream signaling pathways by interacting with pleckstrin homology domain-containing proteins, such as phosphoinositide-dependent kinase 1 and the serine-threonine kinase Akt (3). The finding of abnormal activation of the PtdIns 3-kinase pathway in cancer cells has led to interest in the development of inhibitors for PtdIns 3-kinase (4).
The level of PtdIns 3,4,5-P3 is stimulated by multiple members of the PtdIns 3-kinase family (2) and is opposed by two phosphatidylinositol phosphatases: the Src homology 2 (SH2) domain-containing inositol 5′ phosphatase (SHIP) and the 3′ inositol phosphatase, phosphatase and tensin homolog (PTEN) (5). PTEN removes phosphate from the 3′ position in the inositol ring of PtdIns 3,4,5-P3 and converts it to phosphatidylinositol 4,5-bisphosphate (6). PTEN has a C2 domain, a PDZ-binding motif, and a N-terminal phosphatidylinositol 4,5-bisphosphate binding motif essential for translocation to the membrane and interaction with other regulatory proteins (7). There are serine and threonine residues in PTEN that have been found to be phosphorylated, but their role in regulating the activity of the enzyme is not clear (8). Mutations in the PTEN protein have been observed in many tumors, suggesting a role for this enzyme in cancer (9).
In contrast, SHIP dephosphorylates the 5′ position on the inositol ring and produces phosphatidylinositol 3,4-bisphosphate (10). There are three isoforms of SHIP: the 145-kDa hematopoietic cell restricted SHIP (also known as SHIP1); the 104-kDa stem cell-restricted SHIP, sSHIP; and the more widely expressed 150-kDa SHIP2 (11). SHIP1 is the major inositol phosphatase regulating PtdIns 3,4,5-P3 in monocytes, macrophages, B cells, and T cells (11). SHIP1 has three known structural features: the N-terminal SH2 domain, the central inositol 5′ phosphatase domain, and two NPXY sequences in the C-terminal region. The currently accepted model for regulation of PtdIns 3,4,5-P3 levels by SHIP1 envisions translocation of SHIP1 from the cytosol to the membrane. Upon stimulation by growth factors, cytokine receptors, or immunoreceptors, SHIP1 is recruited via its N-terminal SH2 domain to phosphorylated tyrosine residues in receptor kinases and degrades the elevated levels of PtdIns 3,4,5-P3 near the activated receptor (12). During this translocation process, SHIP1 is not thought to change its 5′ phosphatase activity (13). Although it is known that SHIP1 can be phosphorylated on tyrosine residues by the lyn cytoplasmic kinase (12) or following the activation of the T cell receptor (14), neither event appears to influence the 5′ phosphatase activity. To date, direct regulation of SHIP1 activity by serine/threonine kinases has not been studied.
Activation of G protein-coupled receptors that raise cAMP (i.e. β-adrenergic receptors or adenosine A2a receptors) is known to blunt the pro-inflammatory responses generated by receptors that raise the level of PtdIns 3,4,5-P3 (15). Therefore, we investigated the possibility that phosphorylation of SHIP1 by cyclic AMP-dependent protein kinase (PKA) might regulate the activity of SHIP1. We found that SHIP1 can be phosphorylated by PKA both in vitro and in cells leading to a stimulation of SHIP1 activity. Activation of PKA in DT40 and A20 cells blunted indicators of the PtdIns 3,4,5-P3 response to B cell receptor stimulation. These results indicate that SHIP1 activity can be regulated both in vitro and in cells by activation of the cyclic AMP-dependent protein kinase and highlight a new mode of SHIP regulation by G protein-coupled receptors.
EXPERIMENTAL PROCEDURES
Materials
Sp-cAMPS, isoproterenol, myristoylated PKA inhibitor fragment 14–22 (mPKI), Anti-FLAG M2 monoclonal antibody, anti-FLAG M2 affinity gel, and anti-hemagglutinin antibody were purchased from Sigma. 32P and [γ-32P]ATP were obtained from PerkinElmer Life Sciences. The PKA inhibitor peptide (fragment 6-22), wortmannin, and okadaic acid were purchased from Calbiochem. Anti-SHIP antibody (P1C1) and anti-PKA α catalytic subunit (C-20) were from Santa Cruz Biotechnology (Santa Cruz, CA). Phospho-Akt (Thr308) and Akt antibody were from Cell Signaling Technology (Danvers, MA). FLAG peptide was synthesized by the Biomolecular Research Facility at the University of Virginia. DiC8-PtdIns 3,4,5-P3, inositol 1,3,4,5-tetrakisphosphate (Ins-1,3,4,5-P4), and Malachite Green solution were purchased from Echelon Biosciences (Salt Lake City, UT). The λ-phosphatase was prepared as described (16). The purified catalytic subunit α of PKA and the plasmid encoding the catalytic subunit α (17) were kind gifts of Dr. Susan Taylor (UCSD). The expression vector encoding a full-length mouse SHIP1 has been described (18). Rabbit anti-mouse IgG (F(ab′)2 fragment) was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Rabbit anti-mouse IgM (F(ab′)2 fragment and intact antibody) were from Invitrogen. Murine monoclonal anti-chicken IgM (M4) antibody was obtained from SouthernBiotech (Birmingham, AL). All other materials were reagent grade.
Cell Culture
The murine B cell lymphoma line, A20, was cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, 2 mm l-glutamine, and 50 μm 2-mercaptoethanol. The SHIP-deficient and parental DT40 cell lines were obtained from Dr. Tomo Kurosaki (Japan) (19). The DT40 cell suspension was grown in RPMI 1640 medium supplemented with 1% chicken serum (Sigma). HEK-293 cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum.
Purification of SHIP1
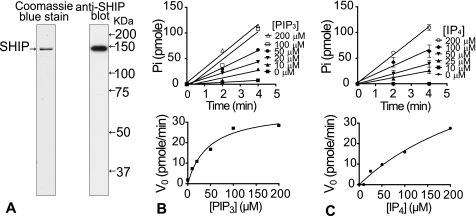
A FLAG epitope-tagged recombinant SHIP1 protein was overexpressed in HEK-293 cells and purified using immunoaffinity chromatography. The cDNA encoding mouse SHIP1 was subcloned into a pCMVTag2C vector (Stratagene), which encodes a FLAG antibody epitope at the N terminus of the expressed protein. To purify recombinant SHIP1, four 15-cm dishes of 70% confluent HEK-293 cells were transfected with the SHIP1 plasmid, and after 48 h the cells were lysed in 4 ml of a buffer containing 25 mm Hepes, pH 7.4, 150 mm NaCl, 3 mm MgCl2, 1% (v/v) Triton X-100, and a mix of protease and phosphatase inhibitors. This mix contained: 200 nm microcystin, 10 mm NaF, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml pepstatin, 100 μg/ml benzimadine, and 100 μg/ml Pefabloc SC. The cell lysate was centrifuged at 150,000 × g for 30 min at 4 °C, and the supernatant was incubated with 160 μl of anti-FLAG beads at 4 °C for 1 h in a 6-ml column. After incubation, the beads were allowed to settle in the column until the cell extract drained and then washed with 5 ml of buffer containing 25 mm Hepes, pH 7.4, 500 mm NaCl, 3 mm MgCl2, 0.5% (v/v) Triton X-100 containing the above protease inhibitor mix (wash buffer A). The column was then immediately washed with 5 ml of 25 mm Hepes, pH 7.4, 500 mm NaCl, 3 mm MgCl2, 0.1% (v/v) Tween 20, and the above protease inhibitor mix (wash buffer B). Following the wash, 500 μl of elution buffer containing 20 mm Hepes, pH 7.4, 150 mm NaCl, 3 mm MgCl2, 100 μg/ml FLAG peptide, and the above protease inhibitor mix was added to the column. The column was incubated with the elution buffer for 10 min at 4 °C to elute SHIP1. The entire 500 μl was collected, and the elution procedure was repeated six times to collect all of the bound SHIP1. Approximately 3 ml of 70 ng/ml SHIP1 was obtained. To purify SHIP1 from agonist-treated HEK-293 cells, protein from one 10-cm dish was harvested in 0.5 ml of lysis buffer and quickly purified as above using 20 μl of anti-FLAG beads in a 0.8-ml centrifuge column (Thermo Scientific, Rockford, IL) using a one-step elution of 80 μl. To determine the yield and purity of each preparation, fractions collected from the FLAG column were loaded on an 8% SDS gel and stained with Coomassie Blue (Invitrogen, “Simply Blue”). The concentration of SHIP1 was determined from the optical density of the bands by comparison to a standard curve of β-galactosidase ranging from 0–500 ng/lane. The pure protein was stored in small aliquots in elution buffer (with protease inhibitors) at −80 °C and used only once.
Assay of SHIP1 Activity
The published Malachite Green assay (20) was modified to use a short chain diC8-PtdIns 3,4,5-P3 as substrate, and the reaction was stopped by heating each reaction tube to 80 °C for 5 min. This procedure reduced the background caused by using EDTA or N-ethylmaleimide to stop the reaction and made the assay more reproducible. The assay was carried out in 20 μl of a mixture consisting of 20 mm Hepes, pH 7.4, 10 mm MgCl2, and 100 μm diC8-PtdIns 3,4,5-P3. The reaction was started by the addition of SHIP1, incubated for 2–5 min at 30 °C, stopped by heating to 80 °C for 5 min, cooled to 4 °C, and centrifuged at 18,000 × g for 15 min. The amount of inorganic phosphate released in the supernatant was determined by adding 100 μl of Malachite Green reagent, incubating for 15 min at room temperature, and measuring the absorbance at 660 nm. For the kinetic experiments, the inorganic phosphate released in the reaction was measured every minute for 4 min using concentrations of PtdIns 3,4,5-P3 ranging from 10 to 200 μm.
In Vitro Phosphorylation of SHIP1
Recombinant SHIP1 (70 ng) was incubated with 0–1200 units of PKA catalytic subunit (see figure legends) in 10 μl of buffer containing 20 mm Hepes, pH 7.4, 200 nm microcystin, 0.1 mg/ml bovine serum albumin, 12.5 mm magnesium acetate, and 1.25 mm EGTA. The reaction was started by the addition of 1 μl of 1.25 mm ATP containing 2.2 × 106 dpm [γ-32P]ATP/tube and incubated for 0–30 min at 30 °C with frequent gentle mixing. The reactions were terminated by adding SDS sample buffer and heating to 100 °C. Aliquots containing ∼70 ng of SHIP1 were run on an 8% SDS gel, stained with Coomassie Blue, and dried, and an autoradiograph was prepared with Kodak X-Omat LS film. The bands identified were cut from the gel and counted. The stoichiometry was calculated from the known specific activity of [γ-32P]ATP and the radioactivity in each gel slice.
Assay of Phosphorylated SHIP1
SHIP1 was treated with 300 units of catalytic subunit as described above for 10 min without radiolabeling. Control reactions had an identical amount of PKA storage buffer (20 mm Hepes, pH 7.4, 100 mm KCl, 1 mm dithiothreitol, 10% glycerol) added. To compare the activity of “native” SHIP1 with PKA-treated SHIP1, the PKA-treated SHIP1 was diluted 10-fold into the SHIP1 activity assay to achieve a final SHIP1 concentration of 10 ng/reaction in a volume of 20 μl.
Dephosphorylation of SHIP1
To prepare 32P-labeled SHIP1 for dephosphorylation with the λ-phosphatase, 70 ng of SHIP1 was first phosphorylated by 300 units of PKA in the presence of [32P]ATP for 30 min, and the reaction was stopped by adding the PKA inhibitor to a final concentration of 170 nm. 32P-Labeled SHIP1 was incubated with λ-phosphatase at a 1:1 or 1:10 mol ratio of SHIP1/λ-phosphatase for 0–30 min at 30 °C in a buffer containing 40 mm Tris, 0.1 mm EDTA, 2 mm MnCl2, 0.01% Brij 35, 5 mm dithiothreitol (dephosphorylation buffer). The reactions were terminated by adding SDS sample buffer and heating to 100 °C. Protein samples were resolved on an 8% SDS gel, and the relative amounts of 32P remaining in SHIP1 were determined by autoradiography.
Purification of Dephosphorylated SHIP1
As described under “Results,” the SHIP1 expressed in HEK-293 cells appeared to be partially phosphorylated. Purification of dephosphorylated SHIP1 was achieved by treating SHIP1 bound to the anti-FLAG affinity column with the highly active, nonspecific λ-phosphatase (21). SHIP1 was extracted from HEK-293 cells, bound to the anti-FLAG resin, and washed as described above. After the second wash, ∼160 μl of SHIP containing resin was separated into two equal parts, one fraction as a control treated with dephosphorylation buffer only and another fraction for treatment with λ-phosphatase at a concentration of 50 ng/μl. Both columns were warmed to 30 °C for 30 min, then cooled to 4 °C, and washed with 5 ml of wash buffer B at of 4 °C. The dephosphorylated SHIP1 was eluted with six 500-μl aliquots of elution buffer as described above. The protein was stored in small aliquots of elution buffer at −80 °C and used only once. The activity of control and dephosphorylated SHIP1 was compared using the Malachite Green assay as described above.
Cell Transfection and Metabolic Labeling
HEK-293 cells were seeded in 6-well plates, 10-cm dishes, or 15-cm dishes. Twenty-four hours later, the cells were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen) at a 1:1 ratio (w/v DNA/Lipofectamine). The cells were serum-deprived overnight prior to treatment with agonists or harvested and SHIP1 purified as described above. To label the ATP pools of the cell, HEK cells were transfected with FLAG-tagged SHIP1 for 24 h in 6-well plates, serum-starved overnight, and incubated with 0.2 mCi/well of 32PO4 at 37 °C for 1 h. The cells were then treated as described under “Results.” SHIP1 was extracted as described above and immunoprecipitated using 10 μl of anti-FLAG antibody beads, and the beads were washed with 500 μl of wash buffer A in an Eppendorf tube. SHIP1 was eluted from the beads by boiling with 10 μl of 2× Laemmli sample buffer for 5 min and resolved on an 8% gel, and the amount of 32PO4 incorporated into SHIP1 identified by autoradiography. The autoradiographs were scanned with a Bio-Rad GS-800 Densitometer, and the optical density of the bands were quantified with Quantity One Image Analysis Software.
Intracellular Ca2+ Measurement
Changes in intracellular Ca2+ levels have been demonstrated to reflect PtdIns 3,4,5-P3 levels in DT40 cells and are blocked by pretreatment with wortmannin (22). To initiate measurement of intracellular Ca2+, 3 × 106 DT40 cells were incubated in 1 μg/ml Indo-1 (Molecular Probes, Eugene, OR) in the culture medium for 20 min in a humidified incubator at 37 °C. The cells were washed twice in a medium consisting of 150 mm NaCl, 5 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 10 mm Hepes, pH 7.4, 0.1% glucose, and 1% fetal calf serum and resuspended in 1 ml of the buffer, and 300 μl was transferred to a cuvette. Secondary antibodies, 2 μg/ml F(ab′)2 fragment of rabbit anti-mouse IgM (for B cell receptor (BCR) cross-linking alone) or 4 μg/ml of intact rabbit anti-mouse IgM (for B cell receptor plus FcγRIIB (FcR) cross-linking) were added prior to measurement of Ca2+. The ratio of Indo-1 fluorescence at 398 and 480 nm was recorded using a Hitachi Model 2500 fluorescence spectrometer. The background was recorded for 20 s, followed by adding a mouse anti-chicken IgM (M4) antibody at 1 μg/ml to stimulate the B cell receptor, and calcium flux was recorded for the next 180 s. For Sp-cAMPS treatment, the cells were treated with 100 μm Sp-cAMPS for 15 min prior to BCR or BCR+FcR stimulation.
Measurement of Akt Phosphorylation in DT40 and A20 Cells
Suspension cultures of DT40 cells were washed and resuspended at 2 × 107 cells/ml in serum-free RPMI 1640. Then cells were incubated at 37 °C for 15 min during which time the cells were treated with vehicle or Sp-cAMPS prior to stimulation of the BCR. After the 15-min incubation, the secondary antibody (2 μg/ml F(ab′)2 fragment of rabbit anti-mouse IgM) was added for 5 min before adding a mouse anti-chicken IgM (M4) antibody at 1 μg/ml to stimulate the BCR. After 0, 1, 3, and 5 min, aliquots containing 1 × 107 cells were withdrawn, pelleted in a microcentrifuge for 5 s and solubilized in 200 μl of lysis buffer containing 25 mm Hepes, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, and the above protease and phosphatase inhibitors. The mixture was centrifuged at 20,000 × g for 15 min at 4 °C. Ten μl of the supernatant was resolved on a 12% SDS-polyacrylamide gel, transferred to nitrocellulose, and probed for p-Akt (T308) with Cell Signaling antibodies.
A20 cells were cultured as described above, washed, and resuspended at 1 × 107 cells/ml in serum-free RPMI 1640. The cells were preincubated at 37 °C with 100 μm Sp-cAMPS for 15 min or pretreated with 10 μm mPKI for 10 min before adding Sp-cAMPS. Following the preincubations, the cells were stimulated with 1.3 μg/ml F(ab′)2 fragment of rabbit anti-mouse IgG to cross-link the BCR. At the indicated times, aliquots containing 5 × 106 cells were removed and processed for electrophoresis and immunoblotting as described above.
Data Presentation and Statistical Analysis
Representative experiments were repeated three to five times. The averaged data are presented as the means ± S.E. Control SHIP1 activity presented in Figs. 3–5 is normalized to 100%. Actual rates of PtdIns 3,4,5-P3 hydrolysis for control samples shown in Figs. 2–5 ranged between 0.7–3 pmol of Pi/min/ ng SHIP1. Statistical differences between multiple means were analyzed via the analysis of variance method using Statview software (SAS Institute Inc.). The difference between two individual treatments was examined using unpaired t tests.
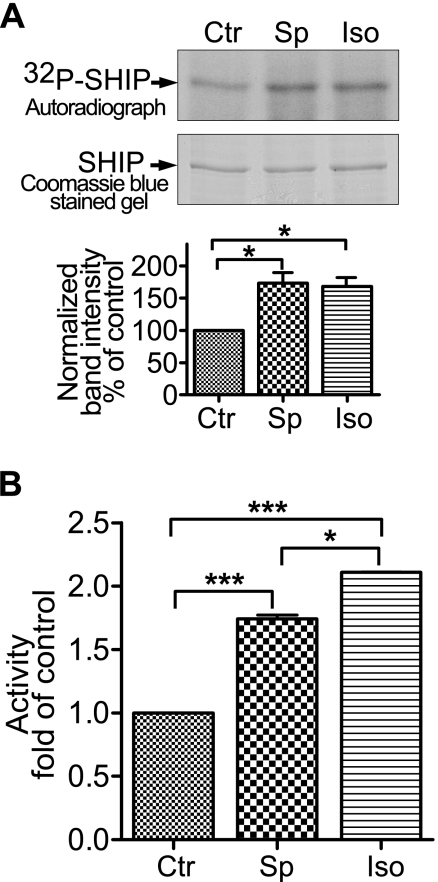
FIGURE 3.
SHIP1 is phosphorylated in HEK-293 cells. HEK-293 cells were transfected with SHIP1, labeled with 32P, and stimulated with 100 μm Sp-cAMPS (Sp) for 15 min or 10 μm isoproterenol (Iso) for 5 min. The FLAG-tagged SHIP1 was immunoprecipitated from cell extracts, resolved on a SDS gel, and subjected to autoradiography. A, upper panel presents the autoradiograph showing the effect of Sp-cAMPS or isoproterenol on phosphorylation of SHIP1. The lower panel presents the amount of 32P incorporated into SHIP1 based on the integrated band intensity normalized for protein expression. The data are expressed as percentages of control (Ctr). *, p < 0.05 (n = 3). B, treating cells with Sp-cAMPS or isoproterenol stimulates the activity of immunopurified SHIP1. HEK-293 cells were transfected with SHIP1, and the cells were incubated with 100 μm Sp-cAMPS for 15 min or 10 μm isoproterenol for 5 min before lysis. Both Sp-cAMPS and isoproterenol significantly increased activity. ***, p < 0.001 (n = 6).
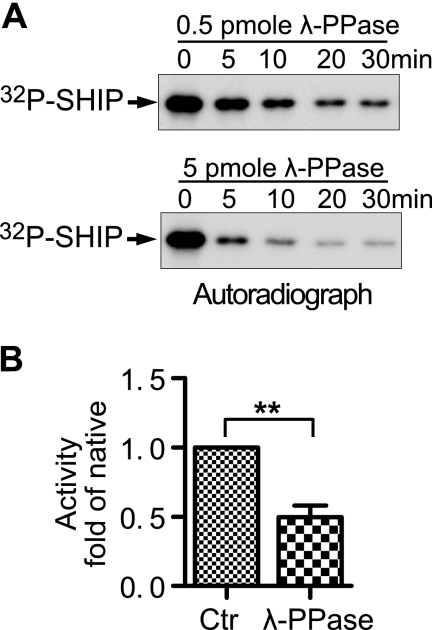
FIGURE 4.
Dephosphorylation of SHIP1 with λ-phosphatase decreases its activity. A, 70 ng of native SHIP1 was phosphorylated by 300 units of PKA in the presence of [32P]ATP before incubation with 0.5 pmol of λ-phosphatase (upper panel) or 5 pmol of λ-phosphatase (lower panel) to dephosphorylate 32P-labeled SHIP1 over a 30-min incubation. B, activity of native SHIP1 and SHIP1 treated with λ-phosphatase for 30 min were measured with 100 μm PtdIns 3,4,5-P3 as substrate. Treatment with λ-phosphatase significantly lowered activity. **, p < 0.01 (n = 6). Ctr, control.
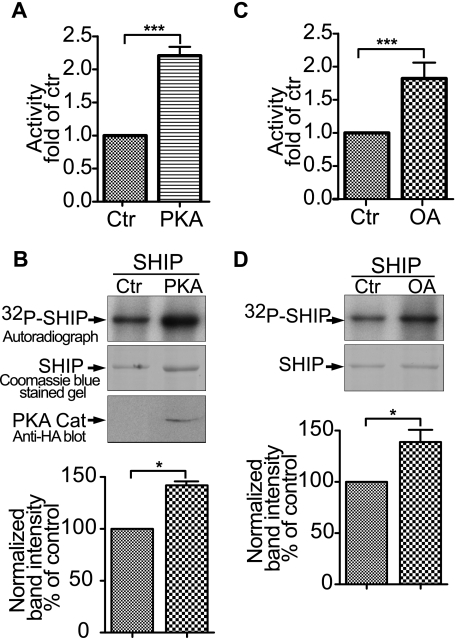
FIGURE 5.
Phosphorylation regulates SHIP1 activity in HEK-293 cells. A, cells were transfected with SHIP1 with or without PKA catalytic subunit for 40 h. Recombinant SHIP1 was immunopurified, the protein concentration was determined, and the activity of 10 ng of SHIP1 was measured. The effect of PKA on activity was significant. ***, p < 0.001 (n = 6). B, upper panel presents an autoradiograph showing the effect of co-expression of the PKA catalytic subunit on the phosphorylation of SHIP1. An immunoblot with the anti-hemagglutinin antibody shows the expression of hemagglutinin-tagged PKA catalytic subunit. The lower panel presents the amount of 32P incorporated into SHIP1 based on band intensity normalized for protein expression. The data appear below the respective band and are expressed as the percentages of control (Ctr). *, p < 0.05 (n = 3). C, HEK-293 cells were transfected with SHIP1 for 40 h and cells were treated with or without 100 nm okadaic acid (OA) for 2 h. SHIP1 was immunopurified and assayed for activity. ***, p < 0.001 (n = 6). D, upper panel presents an autoradiograph showing the effect of okadaic acid on phosphorylation of SHIP1. The lower panel presents the amount of 32P incorporated into SHIP1 based on band intensity normalized for protein expression. *, p < 0.05 (n = 3).
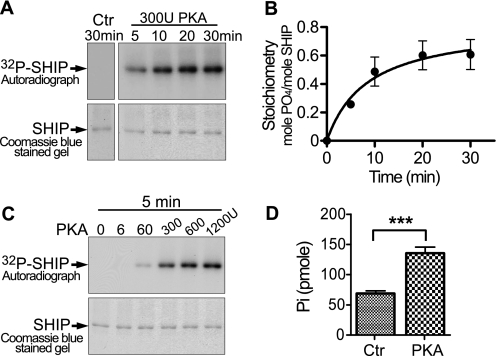
FIGURE 2.
PKA can phosphorylate SHIP1 in vitro and increase its activity. A, upper panel, autoradiograph showing time-dependent phosphorylation of SHIP1 by PKA. Each reaction contained 70 ng of SHIP1 and was incubated with 300 units of PKA for the indicated time. Lower panel, Coomassie Blue-stained 8% SDS gel used to make autoradiograph. B, the bands cut from the gel in A were counted in a scintillation counter. The stoichiometry was calculated from the known specific activity of [32P]ATP used in the reaction mixture and the amount of 32P in each gel slice. C, PKA-dependent phosphorylation of SHIP1. Pure SHIP1 was phosphorylated with the indicated concentration of PKA for 5 min. D, activity of native SHIP1 and PKA-treated SHIP1 was compared following treatment with 300 units of PKA for 10 min. PKA significantly increased activity. ***, p < 0.001 (n = 6). Ctr, control.
RESULTS
Purification of Recombinant SHIP1
Recombinant SHIP1 was purified as a 145-kDa protein that corresponds to the full-length SHIP1 molecule. An example of the purity of the protein is shown in Fig. 1A (left panel) along with immunoblotting for SHIP1 (right panel). Previous investigators have experienced difficulty purifying full-length SHIP1 because the protein has a C-terminal protease site near Ser942 (23); cleavage at this site leads to truncated SHIP1 molecules of 100–120 kDa (23). Our optimized protocol provides highly pure SHIP1 that is free of proteolytic breakdown products. This result is likely due to rapid extraction of the protein from the cell lysate, use of a high affinity FLAG epitope tag that allows extensive washing of the column, and use of multiple protease inhibitors during the entire protocol.
FIGURE 1.
Purification and kinetics of recombinant SHIP. A, left panel, 100 ng of SHIP1 purified from HEK-293 cells, resolved on an 8% SDS gel and stained with Coomassie Blue. The right panel presents an immunoblot with a SHIP1 antibody. B, upper panel, time-dependent phosphatase activity of SHIP1 with the indicated concentrations of PtdIns 3,4,5-P3 (PIP3); lower panel, plot of initial rate against PtdIns 3,4,5-P3 concentration. Vmax and Km were estimated as 1.8 pmol Pi/min/ng and 46.0 μm, respectively. C, upper panel, time-dependent phosphatase activity of SHIP1 with different concentrations of Ins-1,3,4,5-P4 (IP4) as the substrate. Vmax and Km were estimated as 3.8 pmol Pi/min/ng and 351.4 μm, respectively.
Kinetics of SHIP1 Activity
The catalytic activity of SHIP1 displayed conventional Michaelis-Menten kinetics with two substrates as shown in Fig. 1 (B and C). The Km using diC8-PtdIns 3,4,5-P3 as substrate was 46.0 μm, and the Vmax was 1.8 pmol Pi/min/ng. No published data exist on the Km of SHIP1 toward PtdIns 3,4,5-P3, but our result is similar to the value of 50 μm reported for PTEN using PtdIns 3,4,5-P3 as substrate (24). SHIP1 is also capable of dephosphorylating Ins-1,3,4,5-P4, but with a much higher Km value (351.4 μm) and Vmax of 3.8 pmol Pi/min/ng. The 7-fold selectivity of SHIP1 for PtdIns 3,4,5-P3 over Ins-1,3,4,5-P4 is much greater than the 2-fold difference observed with PTEN (24), suggesting that PtdIns 3,4,5-P3 is the preferred substrate of SHIP1. Based on the data shown in Fig. 1 (B and C), we measured SHIP1 activity in subsequent experiments using 100 μm of diC8-PtdIns 3,4,5-P3 as the substrate and at 2–4 min of reaction time to keep the assay within the linear range.
PKA Can Phosphorylate SHIP1 in Vitro and Increase Its Catalytic Activity
To assess whether SHIP1 can serve as a target for PKA-mediated phosphorylation, we incubated pure SHIP1 with the catalytic subunit of PKA and [γ-32P]ATP. Recombinant SHIP1 is rapidly phosphorylated when incubated with 300 units of PKA at 30 °C (Fig. 2A). The stoichiometry of phosphorylation was estimated to be 0.53 mol PO4/mol SHIP1 at 10 min and 0.6 mol PO4/mol SHIP1 at 30 min (Fig. 2B). Control reactions revealed that this preparation of SHIP1 does not undergo phosphorylation in the absence of added kinase in vitro (Fig. 2A, far left panel). When SHIP1 was treated with increasing concentrations of PKA (0–1200 units) for 5 min, we found that SHIP1 can be efficiently phosphorylated with 300 units of PKA (Fig. 2C). Higher concentrations of PKA (600–1200 units) were only slightly more effective. Thus, in the subsequent experiments we used 300 units PKA and a 10-min incubation time to phosphorylate SHIP1.
We next investigated the effect of SHIP1 phosphorylation on its activity toward PtdIns 3,4,5-P3. Phosphorylation of SHIP1 with PKA increased SHIP1 activity ∼2-fold (p < 0.001) (Fig. 2D). Overall, these findings indicate that SHIP1 is a substrate for PKA-mediated phosphorylation in vitro and that the activity of SHIP1 is increased after phosphorylation by PKA.
Phosphorylation of SHIP1 in HEK-293 Cells
To determine whether SHIP1 could be phosphorylated by elevating cAMP levels in intact cells, HEK-293 cells were transfected with SHIP1, serum-starved, labeled with [32P]orthophosphate, and treated with agents expected to activate PKA, and the SHIP1 was immunopurified. The phosphorylation state of SHIP1 is increased following activation of the endogenous PKA with 100 μm Sp-cAMPS for 15 min or by activation of the β-adrenergic receptor of the cells with 10 μm isoproterenol for 5 min (Fig. 3A). Treatment of the cells with either Sp-cAMPS or isoproterenol increases the incorporation of 32P into SHIP1 ∼2-fold. Interestingly, a high basal level of 32P incorporation into SHIP1 is observed in untreated cells (see below).
To address how phosphorylation of SHIP1 in intact cells affects its activity, experiments analogous to those shown in Fig. 3A were performed with unlabeled HEK-293 cells. As expected, SHIP1 immunopurified from cells treated with Sp-cAMPS or isoproterenol is activated ∼2-fold (Fig. 3B). These results clearly indicate that SHIP1 is a substrate for PKA in HEK cells and that phosphorylation of SHIP1 stimulates its activity.
In Vitro Dephosphorylation of SHIP1 with λ-Phosphatase Decreases Its Activity
Because the activity of SHIP1 is increased by phosphorylation, it is likely that SHIP1 is a substrate for a protein phosphatase. To test this possibility, we phosphorylated 70 ng of SHIP1 with [32P]ATP and PKA for 30 min, stopped the kinase reaction, added 0.5 pmol or 5 pmol of the highly active λ-phosphatase (21) to the reaction, and removed aliquots for gel electrophoresis at various times over a 30-min incubation. Fig. 4A shows the autoradiographs made from the time course of dephosphorylation. Note that labeled SHIP1 was rapidly dephosphorylated by both concentrations of λ-phosphatase (Fig. 4A). In pilot experiments, we found that SHIP1 dephosphorylated by the λ-phosphatase can be rephosphorylated to a stoichiometry of 0.74 mol PO4/mol SHIP1, ∼0.15 mol PO4/mol SHIP1 higher than that of the SHIP purified directly from cells. From this observation, we believe that recombinant SHIP1 from HEK-293 cells may be partially phosphorylated in the “basal” state (see also Fig. 3A) and that dephosphorylation of SHIP1 should lower basal SHIP1 activity. To explore this possibility, we treated HEK cell expressed SHIP1 with the λ-phosphatase, while it was bound to the anti-FLAG column during purification to obtain dephosphorylated SHIP1 for biochemical studies. A control preparation of SHIP1 was prepared for this experiment by incubating SHIP bound to the anti-FLAG column with the λ-phosphatase buffer. Interestingly, the dephosphorylated form of SHIP1 has approximately one-half the activity of native SHIP1 (p < 0.01) (Fig. 4B). Taken together, the data in Figs. 2–4 indicate that the activity of SHIP1 is directly regulated by its phosphorylation state.
Phosphorylation Regulates SHIP1 Activity in Cells
We next explored how different manipulations of the kinase and phosphatase activity in HEK-293 cells affected the activity of SHIP1. First, we tested the direct involvement of PKA by co-transfecting cells with SHIP1 and a plasmid expressing the PKA catalytic subunit, immunopurified the expressed SHIP1, and measured its activity. Fig. 5A shows that expression of an exogenous PKA catalytic stimulated SHIP1 activity over 2-fold. HEK cells transfected with the plasmid expressing the catalytic subunit were labeled with 32PO4 and assessed for SHIP1 phosphorylation. SHIP1 phosphorylation was increased following co-transfection with the catalytic subunit (Fig. 5B).
To inhibit the native protein phosphatases in the HEK-293 cells, we treated them with okadiac acid, an inhibitor of both protein phosphatase 1 and protein phosphatase 2A (25). Fig. 5C shows that SHIP1 immunopurified from cells treated with 100 nm okadiac acid is activated ∼2-fold, and Fig. 5D shows that okadiac acid also increases the phosphorylation state of immunoprecipitated SHIP1.
Regulation of SHIP1 Activity in B Cells
Previous studies in B lymphocytes have shown a rapid calcium flux induced by cross-linking of the BCR that is dependent on PtdIns 3,4,5-P3 generation (26). This response can be blocked by wortmannin, making the Ca2+ flux a convenient reflection of PtdIns 3,4,5-P3 levels (22). Attenuation of BCR-mediated downstream responses by the inhibitory FcγRIIB receptor depends on the recruitment of SHIP1 to the membrane by the FcγRIIB receptor and the subsequent degradation of PtdIns 3,4,5-P3 by SHIP1 (19). Some level of SHIP1 is also recruited to the membrane during BCR stimulation alone, and this provides a dampening of the BCR-induced calcium flux in B cells (27, 28). SHIP1 also provides a basal attenuation of the BCR-mediated calcium flux as indicated by a greatly enhanced calcium flux following BCR cross-linking in SHIP1 deficient DT40 cells (22). Accordingly, DT40 cells provide an attractive system to test whether the PKA-mediated activation of the SHIP1 inositol phosphatase could inhibit the BCR induced calcium flux. We used parental DT40 B cells and DT40 cells that have been made SHIP-deficient by targeted gene deletion to examine the effect of Sp-cAMPS on the intracellular Ca2+ response to BCR stimulation. Importantly, DT40 cells do not express SHIP2, and the responses studied are entirely dependent on SHIP1 expression and function (29).
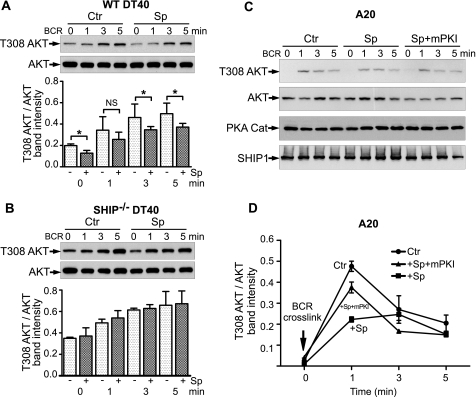
As shown in Fig. 6A, cross-linking of the BCR on wild type DT40 cells induces a large rise in intracellular calcium; this event is inhibited by pretreatment of the DT40 cells with Sp-cAMPS. The inhibition is equal to that seen after co-cross-linking of the BCR with the inhibitory FcγRIIB receptor. To confirm that the calcium response was induced by a rise in PtdIns 3,4,5-P3, we pretreated the cells with wortmannin. As expected (22), the rise in intracellular Ca2+ was blocked (not shown). In contrast, DT40 cells lacking SHIP1 expression are not inhibited by Sp-cAMPS or by BCR+FcR co-cross-linking (Fig. 6B). This result suggests that the inhibition of BCR induced calcium flux by Sp-cAMPS is due to an effect on SHIP1 activity.
FIGURE 6.
The B cell receptor induced calcium flux in DT40 cells is blocked by Sp-cAMPS in a SHIP-dependent manner. The upper panel shows experiments performed using wild type DT40 cells (A), and the lower panel shows experiments with DT40 cells with harboring a targeted disruption of SHIP1 expression (SHIP-deficient) (B). The level of intracellular Ca2+ was measured in DT40 cells under four conditions: DT40 cells stimulated by cross-linking the B cell receptor alone (BCR, closed black circles); DT40 cells pretreated with Sp-cAMPS for 15 min and then stimulated by cross-linking the B cell receptor alone (Sp-cAMPS+BCR, closed red triangles); DT40 cells in which both the BCR and FcR were cross-linked (BCR+FcR, blue closed squares); and DT40 cells pretreated with Sp-cAMPS for 15 min in which both the BCR and FcR were cross-linked (Sp-cAMPS+BCR+FcR, open green circles). WT, wild type.
Another convenient indicator of PtdIns 3,4,5-P3 levels in B cells following BCR engagement is the phosphorylation of Akt (30). Akt activation is dependent on generation of PtdIns 3,4,5-P3 in the membrane, leading to phosphorylation on Thr308 and Ser473 by phosphoinositide-dependent kinases 1 and 2 (31). Accordingly, we measured the p-Akt in DT40 and murine A20 cells following BCR stimulation to determine whether BCR-induced Akt activation could be blocked by the application of Sp-cAMPS.
In wild type DT40 cells, BCR ligation caused a substantial increase in phosphorylation of Akt on Thr308 at 1, 3, and 5 min (Fig. 7A). BCR cross-linking also caused phosphorylation of Akt at Ser473, although phosphorylation level on Ser473 is lower than on Thr308 (data not shown). Pretreatment of cells with 100 μm Sp-cAMPS reduced the BCR-induced phosphorylation of Akt over the 5-min time course (dark bars versus light bars). The experiments with SHIP-deficient DT40 cells (Fig. 7B) provide two important observations. First, the p-Akt level was approximately twice that in the WT cells at time 0 and higher at all subsequent time points. This finding is consistent with the conclusion that SHIP1 is essential in regulating PtdIns 3,4,5-P3 levels in B cells (11). Second, Sp-cAMPS did not blunt the BCR cross-linking response in SHIP-deficient DT40 cells (dark bars versus light bars). These results suggest that activation of SHIP1 via phosphorylation is an important regulatory event in hematopoietic cells.
FIGURE 7.
Sp-cAMPS inhibits BCR-induced phosphorylation of Akt. Wild type DT40 (A) and SHIP-deficient DT40 cells (B) were treated with 100 μm Sp-cAMPS for 15 min, and the rabbit anti-mouse IgM F(ab′)2 fragment was added for 5 min and then stimulated with a murine anti-chicken IgM (M4) antibody to activate the BCR for the indicated times. Aliquots of 1 × 107 cells were removed, pelleted, and resuspended in 200 μl of lysis buffer. A 10-μl aliquot of cell lysate was analyzed for p-Akt (T308) as described under “Experimental Procedures” (upper panels). To control for total Akt content, the blots were reprobed with an Akt antibody (lower panels). The effect of Sp-cAMPS was significant. *, p < 0.05 (n = 3). In SHIP-deficient DT40 cells, Sp-cAMPS had no effect. C, A20 cells were treated with 100 μm of Sp-cAMPS for 15 min with or without 10 μm of mPKI before the BCR was stimulated with a rabbit anti-mouse IgG F(ab′)2 for the indicated times. Aliquots containing 5 × 106 cells were processed for p-Akt blotting as described. The blots were then reprobed with Akt, SHIP1, and PKA catalytic subunit (PKA Cat) antibodies. D, results were quantified and normalized for Akt expression. Ctr, control.
We also tested the effect of Sp-cAMPS on BCR-induced phosphorylation of Akt in the murine A20 B cell lymphoma line. As shown in Fig. 7 (C and D), there is a low basal phosphorylation of Akt in A20 cells, compared with DT40 cells. At 1 min of BCR cross-linking, there was a marked, 40-fold increase in phosphorylation of Akt. This increase is inhibited ∼50% by pretreatment of the cells with Sp-cAMPS (squares). When cells were treated with myristoylated PKA inhibitor peptide (mPKI), the effect of Sp-cAMPS was nearly abolished (triangles). Taken together, the data from the DT40 cells and A20 cells provide strong evidence for the cAMP-mediated phosphorylation and activation of SHIP1.
DISCUSSION
Elevation of cAMP in hematopoietic cells inhibits the inflammatory activity induced by agents that raise PtdIns 3,4,5-P3 (32–34), suggesting that PKA may regulate the synthesis or degradation of PtdIns 3,4,5-P3. Because the major inositol phosphatase thought to regulate PtdIns 3,4,5-P3 in hematopoietic cells is SHIP1 (11), we examined the ability of PKA to regulate the activity of SHIP1. Our results show SHIP1 can be phosphorylated by PKA, resulting in an increase in SHIP1 activity both in vitro and in HEK-293 cells (Figs. 2 and 3). In addition, experiments with DT40 and A20 cells demonstrate that PKA can regulate SHIP1 in hematopoietic cells (Figs. 6 and 7).
There is extensive research about the phosphorylation of SHIP by tyrosine kinases, such as Src family kinases or Syk (12), or by platelet-derived growth factor (35), but phosphorylation of SHIP1 on serine or threonine residues has not been reported. Here, we provide six lines of evidence that SHIP1 is phosphorylated by PKA and that this event stimulates the catalytic activity of SHIP1. First, the PKA catalytic subunit can rapidly phosphorylate recombinant SHIP1 in vitro (Fig. 2). Second, recombinant SHIP1 expressed in HEK-293 cells is phosphorylated following stimulation of their endogenous PKA by Sp-cAMPS or activation of the β-adrenergic receptor by isoproterenol (Fig. 3A). This suggests that the phosphorylation of SHIP1 is a physiologically relevant process. Third, the phosphorylation of SHIP1 by PKA in vitro increases its activity 2-fold (Fig. 2D); moreover, SHIP1 immunopurified from cells treated with Sp-cAMPS or isoproterenol is twice as active as SHIP1 from control cells. Fourth, overexpression of the PKA catalytic subunit in HEK-293 cells stimulated SHIP1 activity to the same extent as activating the endogenous PKA. Fifth, we show that SHIP1 is a substrate for protein phosphatases in vitro and inhibition of protein phosphatase activity in HEK293 cells with okadaic acid increased the phosphorylation state of SHIP1 and activated the enzyme to the same extent as treatment with PKA (Fig. 5, C and D). Finally, experiments with the DT40 and A20 cell lines suggest that activation of SHIP1 by Sp-cAMPS can inhibit PtdIns 3,4,5-P3-linked responses induced by B cell receptor cross-linking (Figs. 6 and 7). Taken together, these data strongly suggest that phosphorylation of SHIP1 by PKA is an important regulatory event.
The current model for SHIP1 function in various physiological situations is primarily based on the translocation of SHIP1 from the cytosol to the membrane following receptor activation. After stimulation of cells via receptor tyrosine kinases (growth factors, cytokine receptors, or immunoreceptors) SHIP1 is thought to bind via its N-terminal SH2 domain to phosphorylated tyrosines on receptors or receptor associated proteins and then degrade PtdIns 3,4,5-P3 near the activated receptor (12). In this translocation model, SHIP1 is not thought to change its 5′ phosphatase activity (12, 13, 36). In the context of B cells, where SHIP1 function has been most studied, SHIP1 can be recruited to the FcγRIIB receptor during inhibitory signaling (when BCR+FcR are co-engaged) or recruited to the plasma membrane during BCR stimulation alone via interactions yet to be described (28, 37, 38). In this context, our observation that phosphorylation of SHIP1 with PKA markedly activates its function is novel and provides insight into an important new mode of SHIP1 regulation.
Overall, the data suggest a new mode of SHIP1 regulation: phosphorylation and activation via PKA. This model of regulation of SHIP1 catalytic activity is in keeping with recent studies showing that the enzymatic activity of SHIP can be regulated by means other than recruitment to the membrane. First, it was shown that the more widely expressed SHIP2 isoform can be activated by phosphorylation on tyrosine in response to EGF and tyrosine phosphatase inhibitors (39). Although the SHIP1 isoform is not thought to be regulated by tyrosine phosphorylation (12, 13, 36), this possibility may need to be more rigorously tested. The second study found the product of SHIP1, phosphatidylinositol 3,4-bisphosphate, could bind to the C2 domain of SHIP1, leading to a 5-fold allosteric increase in the activity of SHIP1. Interestingly, Ong et al. (40) found a compound, AQX-MN100, that also bound to the C2 domain and activated the enzyme. Based on these findings, these investigators suggest that molecules that allosterically activate SHIP1 might be useful drugs to lower PtdIns 3,4,5-P3 levels, inhibit the PtdIns 3-kinase pathway, and treat hematopoietic disorders (40). Although complete details about the activation of SHIP remain to be established, the above findings suggest that regulation of SHIP1 may be more complex than originally anticipated. Moreover, the regulation of the level of the SHIP1 protein and its activity are emerging as important events in the inflammatory process. For example, Sly et al. (41) demonstrated that LPS induces an increase in the amount of SHIP1 protein in macrophages.
Although the paradigms of SHIP1 regulation have been shaped by known features of the receptor tyrosine kinases, receptors coupled to G proteins play a large role in regulating PtdIns 3,4,5-P3 levels in cells of hematopoietic origin (42). In neutrophils, mast cells, and macrophages where SHIP1 is highly expressed (11), the Gi-coupled f-Met-Leu-Phe and C5a receptors raise PtdIns 3,4,5-P3 levels via activation of the p110γ form of PtdIns 3-kinase (2); this effect is markedly blunted by stimulation of Gs-coupled receptors that raise cAMP (32, 43, 44). In particular, activation of the A2a adenosine receptor has been found to inhibit the actions of inflammatory mediators in a variety of cell types (32–34). As one example, the A2a adenosine receptor agonist, ATL146e, stimulates cAMP accumulation and inhibits TNF-α release from LPS-stimulated mouse intraperitoneal macrophages (45). In addition, Sun et al. (46) found that treatment of monocytes with either the dibutyryl cAMP or forskolin markedly inhibited LPS-induced production of TNF-α, suggesting that the anti-inflammatory response of A2a adenosine receptor agonist is mediated by cAMP and PKA. Thus, the phosphorylation of important regulatory molecules by PKA in hematopoietic cells appears to be central to the anti-inflammatory effects of these receptors (32). However, the targets for PKA are only beginning to emerge. Our findings indicate that activation of the β-adrenergic receptor (which raises cAMP) results in the phosphorylation and activation of SHIP1, and this event attenuates the PtdIns 3,4,5-P3 signal. Because activation of Gs coupled receptors inhibits many inflammatory events (32), this mode of SHIP1 regulation may be widespread and important in the overall regulation of hematopoietic cells.
In conclusion, our results provide novel insights on the regulation of SHIP1 activation. The finding that SHIP1 activity can be regulated via phosphorylation by PKA provides a logical extension of its biological role in hematopoietic cells. These additional modes of SHIP1 regulation fit with the different mechanisms used to stimulate PtdIns 3,4,5-P3 levels, allowing a single enzyme to reduce elevated PtdIns 3,4,5-P3 levels. When PtdIns 3,4,5-P3 levels are raised by receptor kinases, the SH2 binding mechanism allows SHIP1 to translocate from the cytosol to the membrane and reduce PtdIns 3,4,5-P3 levels. This mechanism is used by receptors such as cytokine receptors or immunoreceptors. In contrast, when PtdIns 3,4,5-P3 levels are raised in hematopoietic cells by G protein-coupled receptors, such as the C5a receptor in neutrophils, there is no phosphorylated tyrosine kinase receptor to cause translocation of SHIP1. In this setting, other receptors such as β-adrenergic or the adenosine A2a receptor can raise cAMP, activate SHIP1, and inhibit inflammatory signals.
Acknowledgments
We thank Dr. Susan Taylor for providing the expression vector containing the catalytic subunit of PKA and the purified protein. We also thank all the members of the Garrison laboratory for advice and support on this project.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM076236 and DK199520.
- PtdIns 3-kinase
- phosphatidylinositol 3-kinase
- PtdIns 3,4,5-P3
- phosphatidylinositol 3,4,5-trisphosphate
- Ins-1,3,4,5-P4
- inositol 1,3,4,5-tetrakisphosphate
- PKA
- cyclic AMP dependent protein kinase
- Sp-cAMPS
- Sp-adenosine 3′,5′-cyclic monophosphorothioate triethylammonium salt hydrate
- SHIP
- Src homology 2 domain-containing inositol 5′ phosphatase
- mPKI
- myristoylated protein kinase A inhibitor
- BCR
- B cell antigen receptor
- FcR
- the inhibitory receptor FcγRIIB
- SH
- Src homology
- PTEN
- phosphatase and tensin homolog.
REFERENCES
- 1.Deane J. A., Fruman D. A. (2004) Annu. Rev. Immunol. 22, 563–598 [DOI] [PubMed] [Google Scholar]
- 2.Koyasu S. (2003) Nat. Immunol. 4, 313–319 [DOI] [PubMed] [Google Scholar]
- 3.Cantley L. C. (2002) Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Echeverria C., Sellers W. R. (2008) Oncogene 27, 5511–5526 [DOI] [PubMed] [Google Scholar]
- 5.Sly L. M., Rauh M. J., Kalesnikoff J., Büchse T., Krystal G. (2003) Exp. Hematol. 31, 1170–1181 [DOI] [PubMed] [Google Scholar]
- 6.Sulis M. L., Parsons R. (2003) Trends Cell Biol. 13, 478–483 [DOI] [PubMed] [Google Scholar]
- 7.Leslie N. R., Batty I. H., Maccario H., Davidson L., Downes C. P. (2008) Oncogene 27, 5464–5476 [DOI] [PubMed] [Google Scholar]
- 8.Wang X., Jiang X. (2008) Oncogene 27, 5454–5463 [DOI] [PubMed] [Google Scholar]
- 9.Keniry M., Parsons R. (2008) Oncogene 27, 5477–5485 [DOI] [PubMed] [Google Scholar]
- 10.Krystal G. (2000) Semin. Immunol. 12, 397–403 [DOI] [PubMed] [Google Scholar]
- 11.Sly L. M., Ho V., Antignano F., Ruschmann J., Hamilton M., Lam V., Rauh M. J., Krystal G. (2007) Front. Biosci. 12, 2836–2848 [DOI] [PubMed] [Google Scholar]
- 12.Phee H., Jacob A., Coggeshall K. M. (2000) J. Biol. Chem. 275, 19090–19097 [DOI] [PubMed] [Google Scholar]
- 13.Giuriato S., Payrastre B., Drayer A. L., Plantavid M., Woscholski R., Parker P., Erneux C., Chap H. (1997) J. Biol. Chem. 272, 26857–26863 [DOI] [PubMed] [Google Scholar]
- 14.Lamkin T. D., Walk S. F., Liu L., Damen J. E., Krystal G., Ravichandran K. S. (1997) J. Biol. Chem. 272, 10396–10401 [DOI] [PubMed] [Google Scholar]
- 15.Hasko G., Linden J., Cronstein B., Pacher P. (2008) Nat. Rev. Drug Discov. 7, 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayeenuddin L. H., Garrison J. C. (2006) J. Biol. Chem. 281, 1921–1928 [DOI] [PubMed] [Google Scholar]
- 17.Sastri M., Barraclough D. M., Carmichael P. T., Taylor S. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aman M. J., Walk S. F., March M. E., Su H. P., Carver D. J., Ravichandran K. S. (2000) Mol. Cell Biol. 20, 3576–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono M., Okada H., Bolland S., Yanagi S., Kurosaki T., Ravetch J. V. (1997) Cell 90, 293–301 [DOI] [PubMed] [Google Scholar]
- 20.Maehama T., Taylor G. S., Slama J. T., Dixon J. E. (2000) Anal. Biochem. 279, 248–250 [DOI] [PubMed] [Google Scholar]
- 21.Zhuo S., Clemens J. C., Hakes D. J., Barford D., Dixon J. E. (1993) J. Biol. Chem. 268, 17754–17761 [PubMed] [Google Scholar]
- 22.Bolland S., Pearse R. N., Kurosaki T., Ravetch J. V. (1998) Immunity 8, 509–516 [DOI] [PubMed] [Google Scholar]
- 23.Horn S., Meyer J., Heukeshoven J., Fehse B., Schulze C., Li S., Frey J., Poll S., Stocking C., Jücker M. (2001) Leukemia 15, 112–120 [DOI] [PubMed] [Google Scholar]
- 24.Maehama T., Taylor G. S., Dixon J. E. (2001) Annu. Rev. Biochem. 70, 247–279 [DOI] [PubMed] [Google Scholar]
- 25.Dounay A. B., Forsyth C. J. (2002) Curr. Med. Chem. 9, 1939–1980 [DOI] [PubMed] [Google Scholar]
- 26.Hikida M., Kurosaki T. (2005) Adv. Immunol. 88, 73–96 [DOI] [PubMed] [Google Scholar]
- 27.Aman M. J., Tosello-Trampont A. C., Ravichandran K. (2001) J. Biol. Chem. 276, 46371–46378 [DOI] [PubMed] [Google Scholar]
- 28.Brauweiler A., Tamir I., Dal Porto J., Benschop R. J., Helgason C. D., Humphries R. K., Freed J. H., Cambier J. C. (2000) J. Exp. Med. 191, 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.March M. E., Lucas D. M., Aman M. J., Ravichandran K. S. (2000) J. Biol. Chem. 275, 29960–29967 [DOI] [PubMed] [Google Scholar]
- 30.Okkenhaug K., Vanhaesebroeck B. (2003) Nat. Rev. Immunol. 3, 317–330 [DOI] [PubMed] [Google Scholar]
- 31.Gold M. R., Scheid M. P., Santos L., Dang-Lawson M., Roth R. A., Matsuuchi L., Duronio V., Krebs D. L. (1999) J. Immunol. 163, 1894–1905 [PubMed] [Google Scholar]
- 32.Linden J. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 775–787 [DOI] [PubMed] [Google Scholar]
- 33.McColl S. R., St-Onge M., Dussault A. A., Laflamme C., Bouchard L., Boulanger J., Pouliot M. (2006) FASEB J. 20, 187–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan G. W., Rieger J. M., Scheld W. M., Macdonald T. L., Linden J. (2001) Br. J. Pharmacol. 132, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Artemenko Y., Gagnon A., Ibrahim S., Sorisky A. (2007) J. Cell Physiol. 211, 598–607 [DOI] [PubMed] [Google Scholar]
- 36.Blero D., De Smedt F., Pesesse X., Paternotte N., Moreau C., Payrastre B., Erneux C. (2001) Biochem. Biophys. Res. Commun. 282, 839–843 [DOI] [PubMed] [Google Scholar]
- 37.Barbour S. E., Wong C., Rabah D., Kapur A., Carter A. D. (1998) Mol. Immunol. 35, 977–987 [DOI] [PubMed] [Google Scholar]
- 38.Okada H., Bolland S., Hashimoto A., Kurosaki M., Kabuyama Y., Iino M., Ravetch J. V., Kurosaki T. (1998) J. Immunol. 161, 5129–5132 [PubMed] [Google Scholar]
- 39.Batty I. H., van der Kaay J., Gray A., Telfer J. F., Dixon M. J., Downes C. P. (2007) Biochem. J. 407, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ong C. J., Ming-Lum A., Nodwell M., Ghanipour A., Yang L., Williams D. E., Kim J., Demirjian L., Qasimi P., Ruschmann J., Cao L. P., Ma K., Chung S. W., Duronio V., Andersen R. J., Krystal G., Mui A. L. F. (2007) Blood 110, 1942–1949 [DOI] [PubMed] [Google Scholar]
- 41.Sly L. M., Rauh M. J., Kalesnikoff J., Song C. H., Krystal G. (2004) Immunity 21, 227–239 [DOI] [PubMed] [Google Scholar]
- 42.Hirsch E., Katanaev V. L., Garlanda C., Azzolino O., Pirola L., Silengo L., Sozzani S., Mantovani A., Altruda F., Wymann M. P. (2000) Science 287, 1049–1053 [DOI] [PubMed] [Google Scholar]
- 43.Giembycz M. A., Lynch O. T., De Souza P. M., Lindsay M. A. (2000) Pulm. Pharmacol. Ther. 13, 195–223 [DOI] [PubMed] [Google Scholar]
- 44.Braun M. C., Kelsall B. L. (2001) Microbes. Infect. 3, 99–107 [DOI] [PubMed] [Google Scholar]
- 45.Murphree L. J., Sullivan G. W., Marshall M. A., Linden J. (2005) Biochem. J. 391, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun W. C., Moore J. N., Hurley D. J., Vandenplas M. L., Linden J., Cao Z., Murray T. F. (2008) Vet. Immunol. Immunopathol. 121, 91–100 [DOI] [PubMed] [Google Scholar]