Abstract
Methotrexate is a slow, tight-binding, competitive inhibitor of human dihydrofolate reductase (hDHFR), an enzyme that provides key metabolites for nucleotide biosynthesis. In an effort to better characterize ligand binding in drug resistance, we have previously engineered hDHFR variant F31R/Q35E. This variant displays a >650-fold decrease in methotrexate affinity, while maintaining catalytic activity comparable to the native enzyme. To elucidate the molecular basis of decreased methotrexate affinity in the doubly substituted variant, we determined kinetic and inhibitory parameters for the simple variants F31R and Q35E. This demonstrated that the important decrease of methotrexate affinity in variant F31R/Q35E is a result of synergistic effects of the combined substitutions. To better understand the structural cause of this synergy, we obtained the crystal structure of hDHFR variant F31R/Q35E complexed with methotrexate at 1.7-Å resolution. The mutated residue Arg-31 was observed in multiple conformers. In addition, seven native active-site residues were observed in more than one conformation, which is not characteristic of the wild-type enzyme. This suggests that increased residue disorder underlies the observed methotrexate resistance. We observe a considerable loss of van der Waals and polar contacts with the p-aminobenzoic acid and glutamate moieties. The multiple conformers of Arg-31 further suggest that the amino acid substitutions may decrease the isomerization step required for tight binding of methotrexate. Molecular docking with folate corroborates this hypothesis.
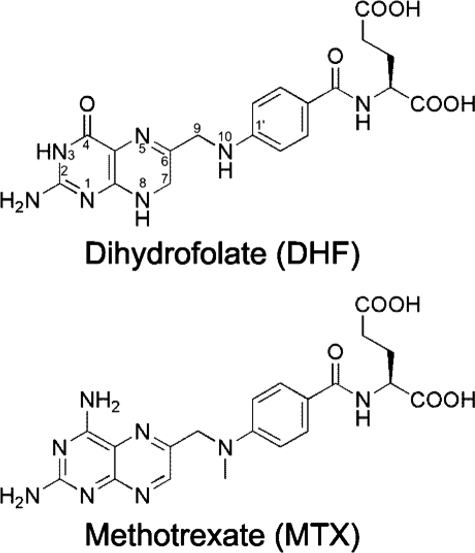
Human dihydrofolate reductase (hDHFR)6 catalyzes the reduction of 7,8-dihydrofolate (DHF) to 5,6,7,8-tetrahydrofolate in a NADPH-dependent manner. 5,6,7,8-Tetrahydrofolate is a cofactor in purine and thymidylate biosynthesis, which are essential metabolites in cell division and proliferation. As a consequence of its essential role in nucleoside biosynthesis, hDHFR has been extensively exploited as a drug target. Inhibition with folate antagonists, or antifolates, arrests cell proliferation. The most effective clinical antifolate to date is methotrexate (MTX (Fig. 1)), a slow, tight-binding competitive inhibitor that displays high affinity for hDHFR (KiMTX = 3.4 pm). MTX is currently used to treat a variety of diseases, including cancer (1–3), and autoimmune diseases such as juvenile idiopathic arthritis (4). A number of resistance mechanisms to MTX have been observed in cancer patients, including impaired transport of MTX to the cytoplasm (5) and decreased retention of MTX in the cell (6). Numerous ex vivo studies have reported mutations in the hDHFR gene resulting in an enzyme variant with decreased affinity for MTX (7, 8). These have contributed to increase our understanding of the molecular basis for active-site discrimination between the substrate, DHF, and its competitive inhibitor, MTX. Understanding the molecular interactions that affect tight binding of MTX to the active site of DHFR will contribute to our understanding of antifolate binding to DHFR, which can in turn contribute to the design of more efficient inhibitors.
FIGURE 1.
Chemical structures of hDHFR ligands. Atom numbering is shown on DHF.
A considerable number of DHFR active-site variants have been identified in MTX-resistant cancer cell lines (although never in patients) (9) or engineered in vitro to elucidate the role of active site residues in the binding of MTX. Amino acid substitutions at residues Ile-7 (10), Leu-22 (11, 12), Phe-31 (13), Phe-34 (14), Arg-70 (15), and Val-115 (16) have yielded MTX-resistant variants. These residues are all present in the folate-binding pocket (17). Because MTX and DHF bind to the active site of hDHFR in a similar manner, all known substitutions causing a decrease in MTX affinity also decrease DHF affinity and overall catalytic efficiency (7, 16, 18). However, the loss of DHF affinity and catalytic efficiency is generally smaller than the loss of MTX affinity. This is often attributed to formation of different contacts with either ligand due to the 180° inversion of the pterin ring of bound DHF relative to MTX (17, 19).
Crystal structures of MTX-resistant point mutants have offered insight into the causes of decreased binding of MTX or other antifolates (17, 20–24). To this day, crystal structures of MTX-resistant hDHFR variants L22F, L22R, and L22Y (12), as well as F31G and F31S (25), complexed to various antifolates, have been reported. Only the L22Y variant has been co-crystallized with MTX. Despite its decreased affinity for MTX (L22Y KiMTX = 11 nm versus WT KiMTX < 31 pm (18)), the inhibitor in the variant structure was bound in the same way as in the native enzyme, making interpretation of decreased affinity difficult to assess. Nonetheless, the low probability conformation of residue Tyr-22 suggested that the presence of a bulky aromatic residue in this area of the folate-binding pocket generated unfavorable hydrophobic interactions with the 2,4-diaminopterin moiety of the inhibitor (12). This is also expected to reduce DHF substrate binding. Structures of MTX-resistant variants F31G and F31S were obtained complexed to N-[4-[(2,4-diaminofuro[2,3-d]pyrimidin-5-yl)methyl]methylamino]benzoyl]-l-glutamate (MTXO) (25), a MTX analog in which the 2,4–2,4-diaminopterin moiety is replaced by a 2,4-diaminofuropyrimidine moiety. Superposition of MTXO-bound variants with MTX-bound WT hDHFR revealed that the ligands bind to the active site in an analogous manner. It was suggested that decreased MTX binding in the substituted variants resulted from the loss of van der Waals and hydrophobic contacts established between the native Phe-31 and the p-ABA and 2,4-diaminopterin moieties of MTX. F31G and F31S display a 10-fold decrease in affinity for MTX relative to WT hDHFR (KiMTX < 31 pm (18)). Further Phe-31 variants (i.e. F31R; KiMTX = 7 nm, 200-fold decrease in MTX affinity) (10) display larger decreases in affinity relative to F31G and F31S. This cannot be rationalized by reduction of side-chain contacts with the inhibitor due to the presence of a smaller side chain.
These results illustrate the difficulty of gaining insight into the molecular causes for altered MTX binding. This may be partly attributed to the very tight binding of MTX to the native enzyme, such that binding to resistant variants often remains in the sub-nanomolar or low nanomolar range, where the general mode of ligand binding has not changed appreciably relative to the native enzyme. Combining active-site mutations in hDHFR by protein engineering has been shown to generate variants with greatly decreased affinity to MTX (18, 26). Studying the molecular interactions in highly MTX-resistant hDHFR variants offers the possibility of capturing more important changes in enzyme-ligand interactions.
Here, we report detailed observations for the mode of MTX resistance in the combinatorial variant F31R/Q35E. Variant F31R/Q35E is a relevant candidate for better understanding the specific interactions that govern ligand recognition in the folate binding site, because it displays a >650-fold decrease in MTX affinity (KiMTX = 21 nm) accompanied by a modest, 9-fold decrease of affinity for the substrate DHF relative to WT hDHFR (18). In addition, we have recently shown that this variant is an efficient selectable marker for various mammalian cell types, including murine hematopoietic stem cells (18).7 Because mutations giving rise to MTX resistance are not observed in mammals, and because MTX is approved for human treatment, engineered resistant DHFRs offer great potential as human selective markers ex vivo or in vivo (10, 27, 28). To better understand the effect of either amino acid substitution on each ligand, a kinetic double mutant cycle was constructed with the simple variants F31R and Q35E. The crystal structure of the F31R/Q35E variant was obtained with bound MTX at 1.7-Å resolution, to elucidate the structural basis of MTX resistance in this variant. In addition, molecular docking was performed with the F31R/Q35E structure to evaluate the role of the two substitutions toward folate binding. Overall, the results reveal synergistic effects of the combined substitutions toward loss of MTX binding, characterized by increased disorder of specific residues throughout the active site of the highly MTX-resistant F31R/Q35E variant.
EXPERIMENTAL PROCEDURES
Construction of Vectors hDHFR F31R/Q35E-pET24, hDHFR F31R-pQE32, and hDHFR Q35E-pQE32
The hDHFR F31R/Q35E gene was amplified by PCR using the following primer set: forward (5′-CACACACCATATGGTTGGTTCGCTAAACTG-3′, NdeI restriction site in italics) and reverse (5′-GTTCTGAGGTCATTACTGG-3′, external primer) from the hDHFR F31R/Q35E-pQE32 template (18). The amplified gene was subcloned in the modified pET24 vector (29) between the NdeI and HindIII restriction sites using T4 DNA ligase, and the ligation mixture was transformed into electrocompetent BL21(DE3) cells. The expected sequence was confirmed by DNA sequencing.
The F31R and Q35E substitutions were created by megaprimer PCR using external primer set 2 described in a previous study (18) and the mutagenic primers 5′-TCTCTGGAAATATCTACGTTCGTTCCTTAAGG (F31R, reverse) and 5′-GTTGTGGTCATTTCTTTCGAAATATCTAAATTCGT (Q35E, reverse), respectively. The amplified gene was subcloned in the pQE32 vector between the BamHI and HindIII restriction sites using T4 DNA ligase, and the ligation mixture was transformed into electrocompetent SK037 cells (18). Expression, purification, and determination of kinetic and inhibitory constants was performed as previously described (18). Briefly, kinetic and inhibition assays were conducted in MATS buffer (25 mm MES, 25 mm acetate, 50 mm Tris, 100 mm sodium acetate, and 0.02% (w/v) sodium azide) (pH 7.6) at 23 °C, by monitoring the NADPH and DHF depletion (Δϵ340 nm = 12 800 m−1 cm−1). All assays were performed in at least four independent experiments, and the average values are reported. The initial rates during the first 15% of substrate conversion were recorded for all assays. Kinetic and inhibition parameters were obtained from a non-linear regression fit to the Henri Michaelis-Menten equation using Prism (GraphPad Software, San Diego, CA). The kcat values were determined in the presence of saturating substrate concentrations (100 μm each of DHF and NADPH) in 1-cm cells according to kcat = Vmax/[E]. KmDHF values were obtained using 10-cm cells containing 1 nm enzyme, 10 μm NADPH and a range of DHF concentrations (0.05 μm to 10 μm). IC50MTX was determined in the presence of saturating substrate concentrations and variable MTX concentrations (0.025 μm to 100 μm). Inhibition constants for MTX (KiMTX) were calculated from IC50MTX according to the equation for competitive inhibitor binding (30).
Expression and Purification of hDHFR F31R/Q35E
An overnight culture of BL21(DE3)/hDHFR F31R/Q35E-pET24 was used to inoculate 1 liter of LB medium. The culture was grown at 37 °C until the A600 nm reached ≈0.7. Protein expression was induced with the addition of 1 mm of isopropyl 1-thio-β-d-galactopyranoside, after which the cells were grown for 16 h at 22 °C. Induced cells were harvested by centrifugation (4000 × g for 30 min at 4 °C). The cell pellet was resuspended in 10 mm Tris-HCl, pH 8.3, at 4 °C. The cells were lysed on ice using a Branson sonicator (four pulses at 200 watts for 30 s with a tapered micro-tip). The cellular debris was pelleted by centrifugation (4000 × g for 30 min at 4 °C), and the supernatant was filtered through a 0.2-μm filter before purification.
Purification was performed following a two-step purification protocol on an AKTA fast-protein liquid chromatography (Amersham Biosciences) at 5 °C. First, the supernatant was applied to an anion-exchange DEAE-Sepharose column (1.6 × 30 cm) followed by a 3-column volume wash with 10 mm Tris-HCl, pH 8.3, at 2 ml/min. A linear gradient of 5 column volumes with NaCl (0–200 mm) in 10 mm Tris-HCl, pH 8.3, was used to elute the F31R/Q35E variant. hDHFR activity was monitored in MATS buffer, pH 7.6, in the presence of 100 μm each of NADPH and DHF. Activity was measured in flat-bottom plates (Costar #3595) by monitoring concurrent depletion of NADPH and DHF (Δϵ340 nm = 12 800 m−1 cm−1) on a FLUOstar OPTIMA UV-visible plate reader (BMG Laboratories, Offenburg, Germany). Active fractions were pooled and dialyzed overnight at 4 °C against 50 mm phosphate buffer, pH 7.5. Following dialysis, the sample (45 ml) was concentrated to 1.5 ml using an Amicon concentrator (molecular weight cut-off 10000, Millipore), for injection on a Superose 12 column (1.6 × 55 cm). The sample was eluted with 50 mm phosphate buffer, pH 7.5, at a flow rate of 1.5 ml/min. hDHFR activity was monitored as described above. Enzyme purity was evaluated using separation by SDS-PAGE (15% (w/v) polyacrylamide gel) stained by the zinc-imidazole method (31) and quantified using the public domain image analysis software Scion Image (NIH, rsb.info.nih.gov/nih-image). Protein concentration was quantified using the Bradford assay (Bio-Rad).
Crystallization and X-ray Data Collection of hDHFR F31R/Q35E
Purified hDHFR F31R/Q35E enzyme was concentrated to 10 mg/ml using an Amicon concentrator (molecular weight cut-off 10000). MTX and NADPH were prepared as described previously (18) and were added at a final concentration of 2 mm each (5-fold molar excess) to the protein sample. Crystallization experiments were set up using hanging drop, vapor-diffusion experiments, with a reservoir volume of 1 ml and a drop size of 4 μl of equal volumes of protein and reservoir solutions. A reservoir solution containing 0.2 m cadmium phosphate and 2.2 m ammonium sulfate yielded crystal-like formations that diffracted poorly. These crystals were crushed using a Hampton Seed Bead Kit and used as seeds (1/10 dilution). Rod-shaped crystals were obtained from crystallization experiments with 0.2 m sodium phosphate and 2.2 m ammonium sulfate as the reservoir solution and with drops containing 1.5 μl of protein, 2 μl of reservoir solution, and 0.5 μl of seeding solution. The crystals were soaked in the mother liquor supplemented with 15% glycerol as a cryoprotectant and frozen in a nitrogen cryostream (model X-stream 2000). Data were collected using a Rigaku RU-H3R generator, equipped with Osmic focusing mirrors, and an R-axis IV++ image plate detector, and processed using HKL-2000 (32) (Table 1).
TABLE 1.
Crystallographic data
| Data collection statistics | |
| Space group | P212121 |
| Number of molecules per asymmetric unit | 1 |
| a (Å) | 42.348 |
| b (Å) | 47.868 |
| c (Å) | 90.715 |
| α = β = γ (°) | 90 |
| Wavelength (Å) | 1.5418 |
| Resolution range (Å)a | 1.70–10.52 (1.70–1.76) |
| Completeness (%)a | 97.2 (93.9) |
| Redundancya | 11.7 (9.5) |
| Rmerge (%)a | 6.4 (47.3) |
| Refinement statistics | |
| Total number of reflections (reflections in R-free set) | 20,304 (2,066) |
| Rfactor (%) | 17.93 |
| Rfree (10% free test set) (%) | 22.30 |
| Number of atoms | 1719 |
| Protein | 1507 |
| Water | 138 |
| Ions | 41 |
| Inhibitor | 33 |
| r.m.s.d. | |
| Bond length (Å) | 0.012 |
| Bond angle (°) | 1.417 |
| Average atomic B-factor (Å2) | 16.372 |
| Protein (Å2) | 14.99 |
| Water (Å2) | 30.57 |
| Ions (Å2) | 35.20 |
| Inhibitor (Å2) | 15.78 |
| Wilson B-factor (Å2) | 21.585 |
| Luzzati sigma A coordinate error (observed) (Å) | 0.12 |
| Luzzati sigma A coordinate error (R-free set) (Å) | 0.11 |
| Ramachandran plot (non-Gly, non-Pro residues) | 159 (100%) |
| Residues in favored positions | 146 (91.8%) |
| Residues in allowed positions | 13 (8.2%) |
| Residues in disallowed positions | 0 (0%) |
a Items in parentheses refer to the highest resolution shell.
Structure Determination and Refinement
The structure was determined by molecular replacement using Phaser (33), which found a single protein molecule in the asymmetric unit (Resolution Range Used: 1.70–25.99; Log Likelihood Gain (refined): 726.123). A lower quality His-tagged F31R/Q35E-MTX-NADPH structure (see supplemental materials) was used as a molecular replacement model. Reciprocal-space refinement was performed using Refmac (34) and included individual isotropic B-factor refinement as well as TLS refinement in the final stages of refinement. Manual model building was performed periodically using Coot (35) (Table 1).
In Silico Automated Docking of Folate
The ligands were prepared as pdb files using ChemDraw 8.0 and Chem3D 8.0 (CambridgeSoft, Cambridge, MA). Energy minimization of ligands was performed with the integrated MM2 energy minimization script in Chem3D. Automated docking experiments were performed using the Autodock 4 software package (Scripps Research Institute, La Jolla, CA). Macromolecules PDB ID 1U72 and 3EIG were stripped of all ligands and heteroatoms, with the exception of the highly conserved active site water molecule (H2O #216 in 1U72, H2O #244 in 3EIG), and were prepared using default settings. A box covering the entire folate binding site and more than half the NADPH binding site was generated as a docking grid. 50 runs of a Lamarckian genetic algorithm using default settings were performed. Following docking, clusters were evaluated according to total binding energies calculated by Autodock 4, and the minimal energy conformation within the lowest energy cluster was retained for comparison with crystal structures.
RESULTS
Kinetic and Inhibition Double Mutant Cycle of hDHFR Variant F31R/Q35E
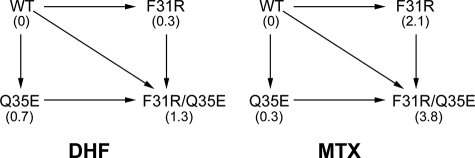
To determine the effect of each constituent amino acid substitution of the F31R/Q35E variant on DHF and MTX binding, the singly-substituted F31R and Q35E were created and their kinetic and inhibition parameters were determined (Table 2). The reactivity (kcat) of F31R and Q35E are 5-fold lower (1.9 ± 0.3 s−1) and 2-fold lower (4.6 ± 0.2 s−1) than WT hDHFR, respectively. The kcat of F31R/Q35E is similar to that of the F31R variant, indicating that loss of reactivity in F31R/Q35E is primarily due to the F31R substitution. The Michaelis constants (KmDHF) of variants F31R and Q35E are 110 ± 60 and 250 ± 90 nm, respectively, illustrating a slight decrease in DHF affinity (1.5- and 3-fold, respectively) relative to the WT. The high % error on KmDHF results from the low values of KmDHF and the correspondingly low spectrophotometric signal, which was enhanced by the use of 10-cm path length cuvettes. Kinetic data for the F31R variant had previously been reported (10), and compares well with our data, although we determined a KmDHF value that is 6-fold lower than previously reported. Because we expected a low value for KmDHF and a correspondingly low spectrophotometric signal, we used 10-cm cuvettes in KmDHF determination, rather than 1-cm cuvettes (10). This enabled a more precise measurement in the target range. Inhibition constants for MTX (KiMTX) revealed that the F31R substitution (KiMTX = 1.1 nm) confers a 35-fold loss in MTX affinity, whereas the Q35E substitution (KiMTX = 0.048 nm) displays a modest decrease of MTX affinity relative to WT (1.5-fold decrease). Like the F31R variant, the F31R/Q35E variant displayed larger decreases in MTX affinity than DHF affinity relative to the WT. However, the Q35E substitution modestly decreased DHF affinity, while having a negligible effect on MTX affinity. The F31R/Q35E variant presented both of these features, as DHF affinity decreased 9-fold relative to WT, whereas MTX affinity decreased >650-fold relative to the WT. Thus, addition of the Q35E substitution to variant F31R increased the KmDHF of F31R 6-fold while increasing the KiMTX of F31R nearly 20-fold.
TABLE 2.
Kinetic and inhibitory parameters of WT hDHFR and hDHFR variant F31R/Q35E
| Variant | kcat | KmDHF | kcat/KmDHF | ΔΔGDHFa | IC50MTX | KiMTX | ΔΔGMTXb |
|---|---|---|---|---|---|---|---|
| s−1 | nm | s−1 μm−1 | kcal/mol | nm | nm | kcal/mol | |
| WTc | 10 ± 2 | <75 | >130 | 0 | 41 ± 14 | <0.031 | 0 |
| F31R | 1.9 ± 0.3 | 110 ± 60 | 17 ± 12 | 0.3 | 1100 ± 600 | 1.1 ± 0.6 | 2.1 |
| Q35E | 4.6 ± 0.2 | 250 ± 90 | 18 ± 7 | 0.7 | 19 ± 4 | 0.048 ± 0.009 | 0.3 |
| F31R/Q35Ec | 1.3 ± 0.2 | 690 ± 13 | 1.9 ± 0.2 | 1.3 | 3100 ± 1600 | 21 ± 11 | 3.8 |
To better quantify the effect of the combined substitutions on MTX and DHF binding, we calculated the loss of binding free energy (ΔΔG) for each variant relative to WT hDHFR. For KmDHF, the sum of the ΔΔGF31R and ΔΔGQ35E values was comparable to the value of ΔΔGF31R/Q35E (Table 2 and Fig. 2), indicating additive effects of the two substitutions toward loss of DHF affinity. However, for KiMTX, ΔΔGF31R/Q35E was considerably greater than the sum of ΔΔGF31R and ΔΔGQ35E, indicating a synergistic effect of the two substitutions on loss of MTX affinity (Table 2 and Fig. 2). These data demonstrate that the F31R substitution is the most important contributor to the binding properties of the F31R/Q35E variant, while the addition of the Q35E substitution synergistically decreases MTX affinity with only a modest, additive reduction of DHF affinity.
FIGURE 2.
Double mutant cycle of F31R/Q35E for DHF and MTX affinity. Numbers in parentheses are ΔΔG values (in kilocalories/mol) for each variant relative to the WT, for KmDHF and for KiMTX, respectively.
Overall Structure and General Characteristics of hDHFR F31R/Q35E Complexed to MTX
To further understand the structural basis of the important loss in MTX affinity, we obtained two crystal structures of variant F31R/Q35E. The first structure had bound NADPH and MTX and contained six hDHFR molecules within the asymmetric unit (supplemental Fig. S1). However, considerable twinning of the crystal lattice resulted in poor data quality, leading to an inability to refine the crystal structure to acceptable Rfactor and Rfree values (supplemental Table S1). This precluded detailed analysis of molecular contacts; however, we used this structure to corroborate observations in the second structure. The full-length of the hDHFR backbone was modeled from electron density map in the second structure, with several side chains exhibiting multiple conformations. Some side chains did not display well defined electron density, and thus were excluded from the model. A large region of very well defined electron density was modeled as MTX in the DHF-binding site (supplemental Fig. S2). There was no electron density in the NADPH-binding site with which to model in NADPH, although two peaks modeled in as sulfate ions were visible where the phosphates of NADPH have been observed in other structures. Six other sulfates, as well as a cadmium ion, which originates from the crystal seed stabilization solution, were also built into the model.
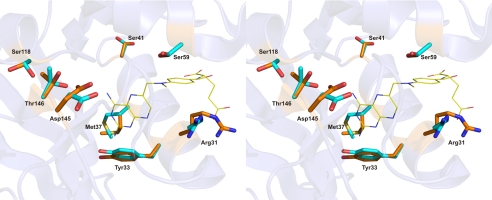
The overall fold and tertiary structure of variant F31R/Q35E are very similar to those reported for other hDHFRs (12, 17, 19–25, 36, 37). hDHFR F31R/Q35E is composed of a central β-sheet containing seven parallel and one anti-parallel strands and four α-helices interconnected by a series of loops (supplemental Fig. S3). Density surrounding the side chains of residues Arg-32, Arg-36, Glu-78, Glu-81, His-87, Arg-91, Lys-98, Glu-101, and Glu-161 was either absent or poorly defined. Those side chains, which are all at the surface of the protein, were not included in the model. Nonetheless, a number of residues were revealed to have two conformers that were clearly visible. Interestingly, these residues are clustered either in the folate (F31R, Tyr-33, Met-37, and Ser-41) or NADPH (Ser-59, Ser-118, Asp-145, and Thr-146) binding sites (Fig. 3). Other high resolution structures have reported two conformers at Ser-41, Ser-118, and Thr-146 when NADPH was not present in the active site (36), but the presence of two conformers at the other residues (F31R, Tyr-33, Met-37, Ser-59, and Asp-145) has never been previously reported and suggests that this is a consequence of the substitutions at residues 31 and/or 35.
FIGURE 3.
Active site residues observed as two or more conformers in the crystal structure of the F31R/Q35E mutant. MTX is shown in line representation and relevant residues are shown as sticks, colored by atom (C: yellow (MTX), orange (conformers ‘a’), and cyan (conformers ‘b’); O: red; N: blue). Residues Ser-118, Thr-145, and Asp-146 belong to the NADPH-binding sub-site.
Interactions between F31R/Q35E and Bound MTX at the Active Site
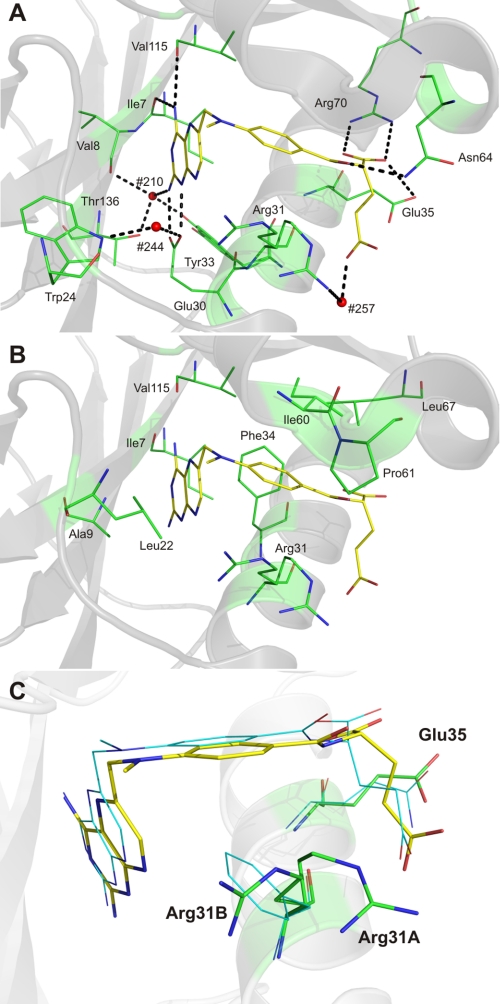
To identify structural changes in variant F31R/Q35E complexed with MTX, the 1.7-Å resolution structure was superimposed onto WT human DHFR complexed with NADPH and MTX (1U72, 1.9-Å resolution) (17) and WT human DHFR complexed with folate (1DRF, 2.0-Å resolution) (19) (r.m.s.d. 0.66 and 0.69 Å, respectively). Those structures were selected because they contain structurally similar ligands in the folate binding site, and because 1DRF lacked NADPH, which enabled identification of structural differences originating from NADPH binding. In contrast to the two WT structures, variant F31R/Q35E displays a considerable shift of loop 17–27 toward the active site (discussed below), in addition to the residues present as two conformers in the NADPH-binding cleft. The low r.m.s.d. value (0.304 Å) between 1U72 and 1DRF was also an incentive for comparison with variant F31R/Q35E. MTX binds in a similar fashion in the F31R/Q35E active site (Fig. 4) as in WT hDHFR (1U72). There was little difference in the orientation of the side chains of residues involved in binding, with the exception of two active site residues, Arg-31 and Tyr-33, which were each present in two distinct conformations. Therefore, as in the WT enzyme, residues 31 and 35 in variant F31R/Q35E do not form specific contacts with the pterin moiety of MTX.
FIGURE 4.
Bound MTX in hDHFR variant F31R/Q35E. Polar (A) and non-polar interactions (B). MTX is shown in stick representation, and relevant residues are shown as lines, colored by atom (C: yellow (MTX) and green (active-site residues); O: red; N: blue). In A, H-bonds and salt bridges are shown as dashed black lines, while active-site water molecules #210, #244, and #257 are shown as red spheres. The backbone carbonyls of Ile-7 and Val-115 are within H-bonding distance of the pterin 4-amino group, as is the hydroxyl group of Tyr-121. The carboxylate of the catalytic Glu-30 residue forms a salt bridge with the pterin N1 and 2-amino group. A conserved active-site water molecule (H2O #257), coordinated via H-bonding interactions with the indole ring of Trp-24 and the Glu-30 carboxylate group, is within H-bonding distance of the pterin N8. Another highly conserved water molecule (H2O #210) present in the active site can H-bond with the pterin 2-amino group, the backbone carbonyl of Val-8, and the hydroxyl group of Thr-136. The p-ABA moiety of MTX interacts mainly with residues Phe-34, Ile-60, Pro-61, and Leu-67 through hydrophobic contacts. A sole H-bond is formed between the carbonyl group of the p-ABA moiety and the γ-amino group of residue Asn-64. C, position of MTX, Arg-31, and Glu-35 in F31R/Q35E relative to the position observed in WT hDHFR (1U72). Residues and MTX from the F31R/Q35E structure are shown in stick representation, whereas residues and MTX from superposed WT hDHFR (1U72) are shown as lines, colored by atom (C: green (F31R/Q35E), cyan (WT hDHFR), and yellow (MTX from 3EIG); O: red; N: blue). Superposition was performed by Cα alignment of the crystal structures.
Binding of the pterin ring involves characteristic H-bonding with specific amino acids and with conserved water molecules (Fig. 4A). H2O #210 is also H-bonded to the hydroxyl group of one of the two distinct conformers of Tyr-33 observed in the F31R/Q35E structure (Fig. 4A). To our knowledge, this is the first hDHFR structure that reports two distinct Tyr-33 conformers in the same macromolecule, identified as Tyr-33A and Tyr-33B (supplemental Fig. S4). The Tyr-33A conformer is homologous to that observed in all but one structure of hDHFR. It is positioned to form the same close ring stacking interaction with Phe-179 that has been previously described for murine DHFR (38) and that exists in WT hDHFR. A ∼30° rotation around the Cα–Cβ bond toward the active site slides the hydroxyl group 2 Å away from its initial conformation, slightly increasing the ring-stacking distance with Phe-179 while bringing the hydroxyl within H-bonding distance of H2O #210 (Tyr-33B (Fig. 4A)); this may result in the Tyr-33B conformer being slightly less stabilized than the Tyr-33A. A similar H-bond is observed in WT hDHFR complexed with NADPH and PT523 (PDB ID 1OHK), a MTX-like inhibitor (24). Hydrophobic and van der Waals interactions are also formed between the pterin moiety of MTX and variant F31R/Q35E (Fig. 4B). The side chains of Ile-7, Ala-9, Leu-22, Phe-34, and Val-l15 are all within van der Waals distance of the pterin ring.
Hydrophobic contacts are predominant in binding of the p-aminobenzoic acid (p-ABA) moiety of MTX, via residues Phe-34, Ile-60, Pro-61, and Leu-67 (Fig. 3B). The two side-chain conformers of the mutated Arg-31 (Arg-31A and Arg-31B; Fig. 3C and supplemental Fig. S4B) are also within van der Waals distance of the p-ABA phenyl ring. However, Arg-31 cannot establish the hydrophobic and possibly edge-to-face aromatic contacts occurring between with WT residue Phe-31 and the p-ABA phenyl ring, the consequences of which will be discussed below. Importantly, the side-chain conformation adopted by four of the six Arg-31 residues from the lower resolution structure we obtained clustered about conformation Arg-31B (supplemental Fig. S5A). The other two Arg-31 residues from the lower resolution structure clustered together in a new conformation, whereas none adopted the Arg-31A conformation (supplemental Fig. S5B). The high conformational variation of this residue provides evidence that the conformers result from the amino acid substitutions, rather than from crystallization artifacts.
The glutamate moiety of MTX is mostly solvent-exposed and interacts mainly via polar contacts with residues of the active site and water molecules at the surface of the protein. The most characteristic contact is the salt bridge formed between the guanidinium group of Arg-70 in variant F31R/Q35E and the α-carboxylate of the glutamate moiety. Arg-70 is strictly conserved in DHFRs from all species, and this interaction is present in all crystal structures complexed with ligands containing a glutamate moiety. In the F31R/Q35E variant, the α-carboxylate of MTX is also within H-bonding distance of a network involving three water molecules (H2O #232, 256, and 257) and the carbonyl group of the p-ABA moiety. This H-bond network has not been observed in other hDHFR structures. Molecule H2O #257 may also H-bond with the γ-carboxylate of MTX and the backbone carbonyl of Arg-28. The backbone nitrogen of Arg-32 is within H-bonding distance of the γ-carboxylate group of MTX as is the ϵNH group of mutated Arg-31A. The Arg-31A conformer places the side chain in proximity to the glutamate portion of MTX and enables H-bonding with H2O #257 and the MTX γ-carboxylate.
Differences in MTX Binding Resulting from the F31R and Q35E Substitutions
Despite similar interactions formed with MTX in variant F31R/Q35E and WT hDHFR (1U72), differences were observed that are consistent with weakened MTX binding in the variant (Fig. 4C). Table 3 lists the distances for apparent polar contacts between the enzyme and the inhibitor, in F31R/Q35E and 1U72. With the exception of unique contacts related to each structure, which mainly involve H-bonds mediated with H2O #210 and 257, the most important differences were observed in the Trp-24-H2O #244 H-bond (0.8 Å shorter) and the Glu-30 ϵO1-N8 of MTX H-bond (0.6 Å shorter). The change in distance of the Trp-24-H2O #244 H-bond can be attributed to the shift of residues 17–27 (Fig. 5). This shift enables closer contacts between the Trp-24 indole ring and the conserved water molecule in variant F31R/Q35E. Another difference results from a specific rotation around the C6–C9 of the pterin ring of MTX in F31R/Q35E relative to 1U72. The slight rotation (∼7°) around the C6–C9 bond brings the pterin 2-amino group slightly closer (0.6 Å) to the catalytic Glu-30 residue. These structural changes in residues that interact with the pterin-moiety appear to be induced by the substitutions at positions 31 and 35, which do not form direct contacts with the pterin moiety.
TABLE 3.
Polar interactions in F31R/Q35E and WT hDHFR (1U72) complexed with MTX
| Polar contact | Distance F31R/Q35E | Distance 1U72 (17) |
|---|---|---|
| Å | ||
| Ile-7 O–4-NH2 MTX | 2.9 | 2.7 |
| Val-8 N–HOH#210 | 4.1 | |
| HOH1#210–2-NH2 MTX | 3.1 | |
| Trp-24 N–HOH#244 | 3.1 | 3.9 |
| HOH#244–N8 MTX | 3.2 | 3.2 |
| Arg-28 O–HOH#257 | 2.8 | |
| HOH#257–Oϵ1 Glu MTX | 3.2 | |
| Glu-30 Oϵ1–2-NH2 MTX | 2.7 | 3.3 |
| Glu-30 Oϵ2–N1 MTX | 2.8 | 2.9 |
| Glu-30 Oϵ2–HOH#244 | 2.8 | 2.6 |
| Arg-31A NH1–HOH#257 | 3.3 | |
| Arg-32 N–Oϵ2 Glu MTX | 3.2 | |
| Tyr-33B OH–HOH#210 | 2.7 | |
| Gln-35 Oϵ–O1 Glu MTX | 3.3 | |
| Asn-64 Nδ–O p-ABA MTX | 2.8 | 2.8 |
| Arg-70 NH1–O2 Glu MTX | 3.0 | 2.4 |
| Arg-70 NH2–O1 Glu MTX | 2.8 | 2.7 |
| Val-115 O–4-NH2 MTX | 2.9 | 3.2 |
| Tyr-121 O–4-NH2 MTX | 3.3 | 3.3 |
| Thr-136 OH–HOH#210 | 2.7 | |
FIGURE 5.
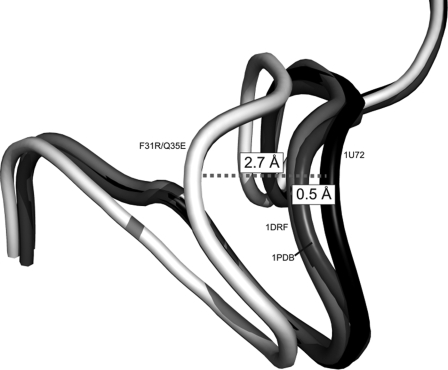
Shift of loop 17–27 in hDHFR variant F31R/Q35E. Loops are shown in schematic representation. Residues 17–27 are shown for variant F31R/Q35E (white), and WT hDHFR from three structures: 1U72 (complexed with NADPH and MTX; black), 1DRF (complexed with folate; light gray), and 1PDB (apoenzyme; dark gray). Distances were calculated between the 1U72 Cα of Gly-20 of 1U72 and the respective Cα of Gly-20 for the three structures. Superposition was performed by Cα alignment of the crystal structures.
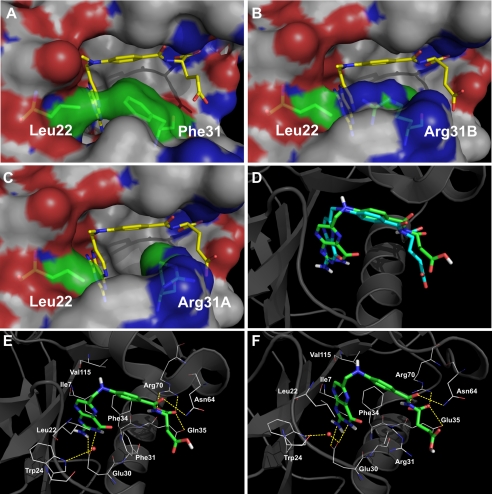
The p-ABA portion of MTX is shifted by ∼0.6 Å in F31R/Q35E relative to WT hDHFR, bringing this moiety closer to the mutated Arg-31, for which two conformers were resolved. Arg-31B (supplemental Fig. S4B) points in roughly the same direction as the WT Phe-31 residue. Its χ1 angle (−81°) is similar to the WT residue (−84°), such that the guanidinium side chain occupies the area of the active site normally occupied by Phe-31. However, a rotation of ∼90° about the Cβ–Cγ bond of Arg-31B takes the δCH2 and ϵNH groups out of the plane of the Phe-31 phenyl group (supplemental Fig. S4B). This results in a loss of hydrophobic and van der Waals contacts, consistent with decreased MTX affinity in variant F31R/Q35E. Nonetheless, the bulky Arg side chain conserved some van der Waals contacts with the p-ABA moiety of the inhibitor, and so this may not be sufficient to rationalize the large decrease in MTX affinity. The shift of the p-ABA moiety also brings the MTX N10-methyl group closer to residue 22 in F31R/Q35E as a result of a 7° rotation about the N10–C1′ bond. Leu-22 belongs to the 17–27 loop, which displays an important shift toward the active site in the variant structure. This loop shift may be partly attributable to the change of position of residue Leu-22 (0.8 Å Cα–Cα distance, relative to 1U72) to maximize van der Waals interactions with MTX, and also with the side chain or Arg-31B. In variant F31R/Q35E, one of the ηNH groups of Arg-31B is in close proximity to the side chain of Leu-22 (Fig. 6B), mimicking an interaction observed with Phe-31 in WT hDHFR (Fig. 6A). Considering that the van der Waals radius for nitrogen is 0.2 Å smaller than for carbon, Leu-22 must therefore be closer to the Arg-31 side chain to optimize van der Waals interactions.
FIGURE 6.
Comparison of WT hDHFR and variant F31R/Q35E by modeling. A–C, surface representation of the contacts established between residues 22 and 31 in WT hDHFR (A, 1U72), variant F31R/Q35E with Arg-31B (B) or with Arg-31A (C) bound to MTX. MTX and residues 22 and 31 are in stick representation, colored by atom (C: yellow (MTX) and green (residues 22 and 31), O: red, N: blue). Surface is colored by atoms (C: white, O: red, N: blue). D–F, docking of folate onto WT hDHFR and variant F31R/Q35E. D, superposition of the original crystal structure 1DRF (WT DHFR bound to folate in blue) and the docking model of 1U72 (WT DHFR) docked with folate (in green). Results for the minimum energy binding conformers are shown for folate (E) docked onto WT hDHFR (1U72), as well as for folate (F) docked onto F31R/Q35E with Arg-31A conformer. The ligands are shown in stick representation while the residues are shown as lines, colored by atom (C: green (ligands) and white (residues), O: red, N: blue). Superposition was performed by Cα alignment of the crystal structures.
The second Arg-31 conformer, Arg-31A, is rotated about Cα–Cβ by >70° relative to Arg-31B and the native Phe-31. This rotation brings the guanidinium side chain closer to the γ-carboxylate group of MTX, where it H-bonds with the γ-carboxylate via a water molecule. As shown in Fig. 6C, considerable hydrophobic and van der Waals contacts with MTX are lost when Arg-31 adopts this second conformation, again consistent with the important decrease of MTX affinity in the variant.
The Gln-35 → Glu substitution also appears to contribute, however slightly, to the decreased MTX affinity. Residue 35 is located at the surface of the protein. In the WT, the side chain of Gln35 H-bonds with the MTX α-glutamate, but in F31R/Q35E, the Glu-35 side chain is slightly shifted away from the glutamate moiety of MTX relative to the WT 1U72 structure. As a result, the Glu-35 side chain points away from the MTX α-glutamate group such that H-bonding would be weakened or absent. An important rotation around the N–Cα bond of the MTX glutamate moiety (34° relative to 1U72), which tilts the α-carboxylate group closer to helix α1, could be caused by unfavorable electrostatic interactions due to two close negative charges (the Glu-35 side chain and the glutamate moiety of MTX). This tilt of the MTX glutamate portion results in the lengthening of the salt bridge between Arg-70 ϵNH1 and αO2 of MTX (0.6 Å difference) in variant F31R/Q35E.
A further potential role of the Q35E substitution in reducing MTX affinity may be a reorientation of Glu-35 in the absence of MTX or other ligands, to form a salt bridge with the guanidinium group of Arg-70. This speculation is based on the presence of a H-bond between Gln-35 and Arg-70 in the WT hDHFR apoenzyme structure following repositioning of the Gln-35 side chain relative to WT hDHFR bound to MTX (21). An intramolecular salt bridge between Glu-35 and Arg-70 would hinder the binding of the α-carboxylate group of MTX or DHF, thereby decreasing affinity for either compound. However, the kinetic data obtained for the Q35E variant suggests that, if these effects are present, they are not the predominant cause for important loss of binding of either ligand.
Docking of Folate upon the Crystallized F31R/Q35E Structure
One of the key features of variant F31R/Q35E is the more important decrease in MTX affinity relative to DHF affinity. This may be attributed to the flip of the pterin ring of folate relative to MTX, resulting in establishment of different, specific contacts of either ligand with the active-site residues. As mentioned above, the variant residues 31 and 35 do not interact directly with the pterin moiety of MTX. It is thus unlikely that the selective decrease in MTX affinity relative to DHF could result from different contacts with the variant residues. The crystal structure presented herein shows that the substitutions mainly perturb interactions with the p-ABA and glutamate portions of the bound inhibitor. WT hDHFR structures complexed with folate, the more oxidized form of the substrate, have shown that the p-ABA and glutamate portions bind similarly to those of MTX. Therefore, loss of contacts with these portions due to substitutions would be expected to reduce the affinity by a similar factor for either ligand. As our attempts to obtain high quality crystals of F31R/Q35E with folate have been unsuccessful thus far, molecular docking studies were performed with variant F31R/Q35E to provide insight into the potential binding mode of folate in the variant enzyme, using the WT hDHFR (PDB ID 1U72) as a reference. Because NADPH was not present in the structure of variant F31R/Q35E, it was removed from the 1U72 structure. For F31R/Q35E, two independent structures, containing either of the observed conformers at Arg-31 (supplemental Fig. S4B), were created. Because the minimal energy conformers for all tested ligands were indistinguishable for the two conformers of Arg-31, we report only the results obtained for F31R/Q35E Arg-31A.
As a control for the docking protocol, MTX was docked onto 1U72 and F31R/Q35E structures. The docking results confirmed that the protocol enabled good prediction of interactions between MTX and the macromolecules. The minimal energy conformers of docked MTX for WT hDHFR or F31R/Q35E closely resembled the binding observed in the respective crystal structures (r.m.s.d. MTX to MTX = 0.9 Å, results not shown). Furthermore, the minimal energy conformer of folate docked onto the 1U72 structure was superimposable with the crystallized folate molecule contained in the 1DRF crystal structure (r.m.s.d. folate to folate = 1.2 Å, results not shown). The orientation of the pterin ring of folate and MTX was correctly predicted in all the minimal energy conformers obtained. Additionally, all known contacts (except when involving non-active-site water molecules) were present, including those of the p-ABA and glutamate moieties (Fig. 6, D–F). The docking studies with folate suggest that the p-ABA and glutamate moieties of folate and MTX bind similarly to the active site of variant F31R/Q35E. If the loss of MTX affinity in this variant were uniquely attributable to loss of interactions with residues 31 and 35, we would expect the MTX and DHF affinities to decrease by a similar factor. It thus appears that the main effect of substitutions F31R and Q35E is to decrease local order within the active-site area, as evidenced by the number of residues observed as more than one conformer. The resulting active-site composition is more detrimental to MTX affinity than to DHF affinity, resulting in reduced discrimination between these two ligands.
DISCUSSION
We report the structure of the highly MTX-resistant, doubly substituted hDHFR variant F31R/Q35E bound to MTX. A major obstacle to effective gene therapy for treatment of hematologic disorders is the low transduction efficiency that reduces effectiveness of bone marrow repopulation with cells carrying the desired gene. We have shown that this variant of hDHFR allows rapid and efficient selection of hematopoietic cells, offering the potential to address this issue.7 The distinguishing feature of this variant, relative to others that have previously been tested toward this goal (11, 26, 28), is the much larger decrease in MTX affinity (>650-fold decrease of MTX affinity relative to WT hDHFR) than DHF affinity (9-fold relative to WT hDHFR). When performing directed evolution of this enzyme, we had selected for a decrease in MTX affinity and concomitant retention of catalytic activity (hence, substrate binding) (18). This is a particularly difficult task, considering the structural similarity between the substrate and the inhibitor, and the observation (from a number of DHFR mutants) that a decrease in affinity for the one has generally been accompanied by a similar decrease in affinity for the other (7). Nonetheless, it has been recently suggested that the native function of enzymes tend to be more resistant to mutations than drug binding, which is a promiscuous (secondary) property of enzymes (39).
In this variant, the mutations resulting in the desired phenotype provided weaker discrimination between the ligands. The MTX resistance appears to be attributed to loss of van der Waals contacts between Arg-31 and the MTX p-ABA moiety resulting in part from multiple conformations adopted by this side chain, as well as unfavorable electrostatic contacts between Glu-35 and the glutamate portion of MTX. These differences could also account for the decrease of DHF affinity, which also contains p-ABA and glutamate moieties. Kinetic characterization of the simple F31R and Q35E variants showed that the decrease of MTX affinity in variant F31R/Q35E was attributable to a synergistic effect of the combined substitutions, while the decrease of DHF affinity was attributable to an additive effect. Docking studies predicted that folate would bind to variant F31R/Q35E in a similar manner to what is observed in WT hDHFR crystal structures bound to folate or MTX (19, 37), and that loss of contacts with the p-ABA and glutamate moieties of MTX should also prevail with DHF or folate. This suggests that the larger decrease in MTX affinity is not solely caused by loss of contacts between the enzyme and inhibitor.
In addition to the broad distribution of conformers observed for our two structures for the mutated F31R residue, seven non-mutated residues in proximity to the folate-binding site were observed in two distinct conformers (Fig. 3). Such a far-reaching disruption of active-site order has never been observed in any other vertebrate DHFR structure reported to date, where few or no secondary conformers are observed even at high resolution. This observation is striking because two neighboring mutations on a single α-helix (F31R/Q35E) have led to apparent disorder throughout the active-site region. This suggests that variant F31R/Q35E has a more dynamic character than the WT hDHFR and suggests the mode by which the increased ligand discrimination operates. There has been a great amount of work reported over the past 10 years relating protein dynamics and function (reviewed by Doucet (40)), including human immunodeficiency virus protease and antibodies and, more specifically, with drug binding (reviewed by Teague (41)). Further work has shown that variations of ligand binding and catalytic activity can be related to amino acid substitutions that change the dynamics of an enzyme (29, 42, 43). Here, putative dynamic effects throughout the active-site area as a result of two substitutions appear to provide a selective decrease of inhibitor binding. The structural resolution of hDHFR variant F31R/Q35E provides insight into an unsuspected path to drug resistance, representing a significant advancement into our understanding of active-site mutations on drug binding. Although our structural data provide information on the MTX-bound enzyme, we may assume that yet more active-site disorder would be observed in the free enzyme. Ligand binding would occur at the expense of greater entropy, which would be reflected in the binding constants; nonetheless, the data currently available does not allow us to provide an entropic basis for ligand discrimination.
As indicated by the inhibition constants, the F31R substitution is the main contributor to decreased MTX affinity. Structural data has suggested that the decrease in MTX affinity for variants F31A, F31S, and F31G could be due to the loss of van der Waals interactions (13). Although some loss of contacts between MTX and residue 31 is apparent in the F31R/Q35E structure, it is hard to reconcile with the fact that variants F31R or F31R/Q35E display larger decreases in MTX affinity than the F31G variant (13), where no contacts can be formed between the ligand and residue 31. This suggests that the basis for the greater decrease of MTX affinity relative to DHF affinity is attributable to a feature that specifically promotes MTX affinity.
It has been shown experimentally that an isomerization step following initial cofactor and MTX binding increases MTX affinity >60-fold in the native enzyme, leading to a non-dissociating hDHFR·NADPH·MTX complex (44). It has been suggested that Phe-31 is a key residue in this isomerization (13). This assumption was based on two observations: the presence of a second conformer at the homologous Tyr-31 residue in chicken DHFR bound to NAPD+ and biopterin (45) and a non-native-like Phe-31 conformer present in one of the two macromolecules observed in WT hDHFR complexed with folate (37). This suggested a possible readjustment of this residue following ligand binding, which would be required for isomerization.
The multiple conformers at residue 31 when MTX is bound to variant F31R/Q35E suggest two possibilities. The substitution of Phe-31 may decrease the isomerization constant (Kiso) (44), altering the dynamic process required for tight MTX binding. This isomerization effect has never been reported in the binding of DHF or folate; amino acid substitutions causing perturbation of Kiso would not affect the binding of these ligands as much as it would MTX, consistent with the greater decreases in MTX affinity observed for variant F31R/Q35E. The F31R substitution, which results in multiple distinct conformers, may perturb Kiso, thus reducing MTX binding. Biophysical data will be required to confirm the disruption of a putative isomerization step in MTX binding in the F31R and F31R/Q35E variants.
Supplementary Material
This work was supported by grants from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada (NSERC).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
J. P. Volpato, N. Mayotte, G. Sauvageau, and J. N. Pelletier, submitted for publication.
- hDHFR
- human dihydrofolate reductase
- MES
- 4-morpholineethanesulfonic acid
- DHF
- dihydrofolate
- MTX
- methotrexate
- MTXO
- N-[4-[(2,4-diaminofuro[2,3-d]pyrimidin-5-yl)methyl]methylamino]benzoyl]-l-glutamate
- p-ABA
- p-aminobenzoic acid.
REFERENCES
- 1.Slamon D. J., Romond E. H., Perez E. A. (2006) Clin. Adv. Hematol. Oncol. 4, Suppl. 1, 4–9 [PubMed] [Google Scholar]
- 2.Daw N. C., Billups C. A., Rodriguez-Galindo C., McCarville M. B., Rao B. N., Cain A. M., Jenkins J. J., Neel M. D., Meyer W. H. (2006) Cancer 106, 403–412 [DOI] [PubMed] [Google Scholar]
- 3.Strojan P., Soba E., Budihna M., Auersperg M. (2005) J. Surg. Oncol. 92, 278–283 [DOI] [PubMed] [Google Scholar]
- 4.Ramanan A. V., Whitworth P., Baildam E. M. (2003) Arch. Dis. Child 88, 197–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flintoff W. F., Sadlish H., Gorlick R., Yang R., Williams F. M. (2004) Biochim. Biophys. Acta 1690, 110–117 [DOI] [PubMed] [Google Scholar]
- 6.Takemura Y., Kobayashi H., Miyachi H. (1999) Anticancer Drugs 10, 677–683 [DOI] [PubMed] [Google Scholar]
- 7.Blakley R. L., Sorrentino B. P. (1998) Hum. Mutat. 11, 259–263 [DOI] [PubMed] [Google Scholar]
- 8.Volpato J. P., Pelletier J. N. (2009) Drug Resistance Updates 12, 28–41 [DOI] [PubMed] [Google Scholar]
- 9.Spencer H. T., Sorrentino B. P., Pui C. H., Chunduru S. K., Sleep S. E., Blakley R. L. (1996) Leukemia 10, 439–446 [PubMed] [Google Scholar]
- 10.Patel M., Sleep S. E., Lewis W. S., Spencer H. T., Mareya S. M., Sorrentino B. P., Blakley R. L. (1997) Hum. Gene Ther. 8, 2069–2077 [DOI] [PubMed] [Google Scholar]
- 11.Ercikan-Abali E. A., Waltham M. C., Dicker A. P., Schweitzer B. I., Gritsman H., Banerjee D., Bertino J. R. (1996) Mol. Pharmacol. 49, 430–437 [PubMed] [Google Scholar]
- 12.Lewis W. S., Cody V., Galitsky N., Luft J. R., Pangborn W., Chunduru S. K., Spencer H. T., Appleman J. R., Blakley R. L. (1995) J. Biol. Chem. 270, 5057–5064 [DOI] [PubMed] [Google Scholar]
- 13.Chunduru S. K., Cody V., Luft J. R., Pangborn W., Appleman J. R., Blakley R. L. (1994) J. Biol. Chem. 269, 9547–9555 [PubMed] [Google Scholar]
- 14.Nakano T., Spencer H. T., Appleman J. R., Blakley R. L. (1994) Biochemistry 33, 9945–9952 [DOI] [PubMed] [Google Scholar]
- 15.Thompson P. D., Freisheim J. H. (1991) Biochemistry 30, 8124–8130 [DOI] [PubMed] [Google Scholar]
- 16.Fossati E., Volpato J. P., Poulin L., Guerrero V., Dugas D. A., Pelletier J. N. (2008) J. Biomol. Screen 13, 504–514 [DOI] [PubMed] [Google Scholar]
- 17.Cody V., Luft J. R., Pangborn W. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 147–155 [DOI] [PubMed] [Google Scholar]
- 18.Volpato J. P., Fossati E., Pelletier J. N. (2007) J. Mol. Biol. 373, 599–611 [DOI] [PubMed] [Google Scholar]
- 19.Oefner C., D'Arcy A., Winkler F. K. (1988) Eur. J. Biochem. 174, 377–385 [DOI] [PubMed] [Google Scholar]
- 20.Cody V., Luft J. R., Pangborn W., Gangjee A., Queener S. F. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 646–655 [DOI] [PubMed] [Google Scholar]
- 21.Cody V., Luft J. R., Pangborn W., Gangjee A. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1603–1609 [DOI] [PubMed] [Google Scholar]
- 22.Cody V., Galitsky N., Luft J. R., Pangborn W., Gangjee A. (2003) Acta Crystallogr. D. Biol. Crystallogr. 59, 654–661 [DOI] [PubMed] [Google Scholar]
- 23.Gangjee A., Vidwans A. P., Vasudevan A., Queener S. F., Kisliuk R. L., Cody V., Li R., Galitsky N., Luft J. R., Pangborn W. (1998) J. Med. Chem. 41, 3426–3434 [DOI] [PubMed] [Google Scholar]
- 24.Cody V., Galitsky N., Luft J. R., Pangborn W., Rosowsky A., Blakley R. L. (1997) Biochemistry 36, 13897–13903 [DOI] [PubMed] [Google Scholar]
- 25.Cody V., Galitsky N., Luft J. R., Pangborn W., Blakley R. L., Gangjee A. (1998) Anticancer Drug Des. 13, 307–315 [PubMed] [Google Scholar]
- 26.Ercikan-Abali E. A., Mineishi S., Tong Y., Nakahara S., Waltham M. C., Banerjee D., Chen W., Sadelain M., Bertino J. R. (1996) Cancer Res. 56, 4142–4145 [PubMed] [Google Scholar]
- 27.Allay J. A., Persons D. A., Galipeau J., Riberdy J. M., Ashmun R. A., Blakley R. L., Sorrentino B. P. (1998) Nat. Med. 4, 1136–1143 [DOI] [PubMed] [Google Scholar]
- 28.Flasshove M., Banerjee D., Mineishi S., Li M. X., Bertino J. R., Moore M. A. (1995) Blood 85, 566–574 [PubMed] [Google Scholar]
- 29.Doucet N., Savard P. Y., Pelletier J. N., Gagné S. M. (2007) J. Biol. Chem. 282, 21448–21459 [DOI] [PubMed] [Google Scholar]
- 30.Segel I. H. (1993) in Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems, pp. 100–120, Wiley Classics Library Ed John Wiley and Sons, New York [Google Scholar]
- 31.Fernandez-Patron C., Castellanos-Serra L., Rodriguez P. (1992) BioTechniques 12, 564–573 [PubMed] [Google Scholar]
- 32.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 286–306 [DOI] [PubMed] [Google Scholar]
- 33.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 34.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36.Klon A. E., Héroux A., Ross L. J., Pathak V., Johnson C. A., Piper J. R., Borhani D. W. (2002) J. Mol. Biol. 320, 677–693 [DOI] [PubMed] [Google Scholar]
- 37.Davies J. F., 2nd, Delcamp T. J., Prendergast N. J., Ashford V. A., Freisheim J. H., Kraut J. (1990) Biochemistry 29, 9467–9479 [DOI] [PubMed] [Google Scholar]
- 38.Cody V., Pace J., Chisum K., Rosowsky A. (2006) Proteins 65, 959–969 [DOI] [PubMed] [Google Scholar]
- 39.Nobeli I., Favia A. D., Thornton J. M. (2009) Nat. Biotechnol. 27, 157–167 [DOI] [PubMed] [Google Scholar]
- 40.Doucet N., Pelletier J. N. (2009) in Protein Engineering Handbook (Lutz S., Bornscheuer U. eds) pp. 187–211, Wiley-VCH, Weinheim, Germany [Google Scholar]
- 41.Teague S. J. (2003) Nat. Rev. Drug Discov. 2, 527–541 [DOI] [PubMed] [Google Scholar]
- 42.Watt E. D., Shimada H., Kovrigin E. L., Loria J. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11981–11986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisenmesser E. Z., Millet O., Labeikovsky W., Korzhnev D. M., Wolf-Watz M., Bosco D. A., Skalicky J. J., Kay L. E., Kern D. (2005) Nature 438, 117–121 [DOI] [PubMed] [Google Scholar]
- 44.Appleman J. R., Prendergast N., Delcamp T. J., Freisheim J. H., Blakley R. L. (1988) J. Biol. Chem. 263, 10304–10313 [PubMed] [Google Scholar]
- 45.McTigue M. A., Davies J. F., 2nd, Kaufman B. T., Kraut J. (1992) Biochemistry 31, 7264–7273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.