Abstract
Forkhead transcription factors (FoxOs) play a pivotal role in controlling cellular proliferation and survival. The cellular level of these factors is tightly regulated through the phosphoinositide 3-kinase/Akt and ubiquitin-mediated degradation. However, the ubiquitin ligases responsible for the degradation of FoxO1 and the relevance of this regulation to smooth muscle cell (SMC) proliferation and survival have not been fully identified. Here we showed that overexpression of C terminus of Hsc70-interacting protein (CHIP) promoted ubiquitination and degradation of FoxO1 in SMCs in response to tumor necrosis factor-α. Both the U-box (containing ubiquitin ligase activity) and the charged (essential for FoxO1 binding) domains within CHIP were required for CHIP-mediated FoxO1 down-regulation. Moreover, interaction and ubiquitination of FoxO1 by CHIP depended on phos pho ryl a tion of FoxO1 at Ser-256. Furthermore, overexpression of CHIP repressed FoxO1-mediated transactivation and its proapo pto tic function following tumor necrosis factor-α treatment. In contrast, knockdown of CHIP by small interfering RNA enhanced FoxO1-mediated transactivation and its effect on SMC proliferation and survival. Taken together, our data indicate that CHIP is a negative regulator of FoxO1 activity through ubiquitin-mediated degradation, and inhibition of CHIP may serve as a potential therapeutic target for reducing proliferative arterial diseases.
The atherosclerotic lesion is characterized by the endothelial dysfunction, inflammation, and accumulation of vascular smooth muscle cells (SMCs),3 foam cells, and matrix protein and lipids in the intima (1, 2). Although the pathogenic mechanisms of atherosclerosis and restenosis are complex, the balance between proliferation and apoptosis of vascular SMCs seems to be a major factor in the progression of these diseases (1, 2). Various growth factors and cytokines are known to be involved in these processes (3–6). One important pleiotropic cytokine is tumor necrosis factor-α (TNF-α), which is believed to play a key role in modulating SMC proliferation, migration, survival, or apoptosis (3–6). TNF-α has been shown to promote cellular proliferation and survival via phosphoinositide 3-kinase (PI3K)/Akt and nuclear factor-κB signaling pathways in several cell types (7–10). Activated Akt phosphorylates and regulates a number of downstream proapoptotic proteins, among which are the forkhead factors FoxO1, FoxO3a, and FoxO4 (formally known as FKHR, FKHRL1, and AFX, respectively) (11).
The FoxO proteins are multifunctional transcription factors, which have important roles in regulating cellular differentiation, proliferation, survival in various cell lines, including cancer cells, fibroblasts, myoblasts, endothelial cells, and SMCs (3–6). Activated FoxO proteins modulate apoptosis through regulation of a number of proapoptotic proteins, inducing Bim, the TNF-related death inducing ligand, Fas ligand, and TNF-R1-associated death domain, which all are involved in apoptotic signaling (11–14). Recent studies demonstrate that FoxOs are expressed in the vasculature and exert a growth-suppressive effect in endothelial cells and SMCs via p27Kip1-dependent cycle arrest and apoptosis, thereby inhibiting neointimal formation (4, 6, 15).
FoxO1 proteins lie downstream of TNF-α, platelet-derived growth factor-BB, and insulin-like growth factor 1 signaling pathways in SMCs in vitro, and treatment with these growth factors stimulates Akt-dependent phosphorylation and nuclear exclusion of FoxOs to the cytoplasm where they are inactive through the ubiquitin-proteasome system (16–20). Importantly, FoxO1 has been shown to be targeted by the F-box protein Skp2 for proteasomal degradation, and this effect of Skp2 requires Akt-specific phosphorylation of FoxO1 (18), indicating that protein phosphorylation and subsequent proteasomal degradation are key steps for FoxO1 inactivation in SMCs. However, little information is known about the ubiquitin ligase necessary for FoxO1 degradation in SMCs in response to TNF-α.
C terminus of Hsc70-interacting protein (CHIP) was recently identified as a dual-function cochaperone/ubiquitin ligase that is highly expressed in the heart, vascular cells, and other cells (21–23). CHIP is demonstrated to target chaperone-bound client proteins such as p53 (24), ErbB2 (25), cystic fibrosis transmembrane conductance regulator (26, 27), Tau (28, 29), ASK1 (30), and ataxin-1(31, 32) for the ubiquitin-mediated degradation. Recently, CHIP has been reported to play an important role in preventing cell apoptosis. CHIP-deficient mice undergo temperature-sensitive apoptosis after environmental stress (33). CHIP also displays a critical cardioprotective effect in response to ischemia/reperfusion injury (34). Furthermore, CHIP inhibits ASK1-mediated apoptosis via its degradation (30). More recently, we found that CHIP promotes myocardin ubiquitin-mediated degradation and attenuates myocardin-dependent SMC differentiation (35). However, the role of CHIP in TNF-α-induced SMC proliferation and survival has not been examined. In the present study, we showed that CHIP interacted with and promoted ubiquitin-dependent degradation of FoxO1, thereby repressing FoxO1-induced anti-proliferation and apoptosis in SMCs following TNF-α treatment. In contrast, depletion of CHIP by small interfering RNA attenuated the inhibitory effect of FoxO1 on SMC proliferation and survival. Thus, CHIP appears to have an important role in regulating SMC phenotype modulation and apoptosis.
EXPERIMENTAL PROCEDURES
Plasmids
Mammalian expression plasmids Myc-CHIP wild type and mutants, His- or HA-ubiquitin, and Bim luciferase reporter, FLAG-FoxO1 wild-type and mutants, HA-FoxO3a and FLAG-FoxO4 have been described previously (18, 21, 33, 36, 37). The pEGFP vector was purchased from BD Biosciences (Clontech). siRNA sequences for CHIP was described as previously (30). All mutated clones were verified by fully sequencing.
Antibodies and Chemicals
The following antibodies were used: anti-CHIP (H-231) and anti-Myc (clone 9E10, Santa Cruz Biotechnology), anti-FLAG (M2, Sigma-Aldrich), anti-HA (clone 12CA5, Roche Diagnostics), anti-GST (Amersham Pharmacia Biosciences), anti-ubiquitin and anti-His (Chemicon International Inc.), anti-AKT, anti-phospho-AKT (Ser-473), anti-FoxO1, anti-phospho-FoxO1 (Ser-256), anti-FoxO3a, anti-FoxO4, anti-GAPDH, and horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology). TNF-α was purchased from Sigma-Aldrich. Cycloheximide, wortmannin, and MG-132 were purchased from Calbiochem.
Cell Culture, Transfection, and Luciferase Assays
293T, rat aortic smooth muscle cells A10 and human umbilical endothelial cells (human umbilical endothelial cells) were obtained from ATCC (Manassas, Virginia) and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C as described (38). Rat neonatal cardiomyocytes were isolated by enzymatic disassociation of 1- to 2-day-old neonatal rat hearts as described previously (38). Recombinant adenoviruses vector control and Myc-tagged CHIP were generated as described previously (35).
Cycloheximide, MG132, or TNF-α was added at a final concentration of 10 μg/ml, 10 μm, or 10 ng/ml, respectively. The luciferase reporter constructs were cotransfected using Lipofectamine 2000 (Invitrogen) with expression vectors carrying CHIP wild type or mutants, FoxO1 expression vectors into 293T cells or with siRNA CHIP into A10 cells, and luciferase activity was measured as described previously (38). The data represent means ± S.E. of three independent experiments in duplicate and normalization for β-galactosidase activity.
Cell Viability, Apoptosis, and Proliferation Assays
Cell viability was determined by Trypan blue exclusion assay. Cell apoptosis was detected by TUNEL assay using an in situ apoptosis detection kit (Roche Applied Science) following the manufacturer's instructions. Percentages of positive-stained cells were determined by counting the numbers of labeled and total cells. Cell proliferation was determined by WST-1 assay (Roche Applied Science) as suggested by the manufacturer. SMCs were seeded in 24-well plates and transfected with the indicated plasmids. At 24 h after transfection, cells were starved for overnight and treated with TNF-α for 24 h, and then incubated with WST-1 for 2 h at 37 °C. Formation of formazan was directly quantitated with an enzyme-linked immunosorbent assay reader at 460 and 690 nm. The data represent the means ± S.E. of three independent experiments in duplicate.
Immunocytochemistry
SMCs were processed for immunofluorescence as described previously (36). The visualization of CHIP and FoxO1 location was performed by immunostaining with CHIP or FoxO1 primary antibodies and appropriate secondary antibodies. Fluorescent images were collected on Nikon Eclipse E800 microscope.
Immunoprecipitations, Western Blotting, and GST Pulldown Assays
Immunoprecipitation and Immunoblotting were performed as described previously (38). Briefly, 293 cells were cotransfected with expression vectors for Myc-CHIP, FLAG-FoxO1. Tagged proteins were immunoprecipitated for 2 h at 4 °C with the appropriate antibody (anti-Myc). The beads were washed and analyzed by immunoblotting. GST pulldown assays were performed as described (38). Briefly, 293 cells were transfected with FLAG-FoxO1 expression plasmid for 36 h, and cells were lysed for 30 min in lysis buffer. Lysates were precleared with GST beads for 1 h and incubated with GST or GST-CHIP fusion proteins for 1 h at 4 °C. The bound beads were washed four times with lysis buffer and analyzed by immunoblotting.
Pulse-Chase Assays
To analyze the degradation kinetics of FoxO1, 293T cells were seeded in 6-well plates and were transfected with FLAG-FoxO1 and Myc-CHIP or vector as indicated. Protein lysates were prepared at indicated time points after addition of cycloheximide (10 μg/ml). Cells were washed twice in phosphate-buffered saline and lysed in immunoprecipitation lysis buffer. Equal amounts of protein were separated by SDS-PAGE. Levels of FoxO1 proteins were determined by immunoblotting and quantified at indicated time points.
In Vivo Ubiquitination Assays
To assess ubiquitination in vivo, 293 cells were transfected with His-Ub or HA-Ub, FLAG-FoxO1, and Myc-CHIP and treated with TNF-α for 24 h. Lysate proteins were precipitated and analyzed by immunoblotting using appropriate antibodies as previously described method (38).
RNA Analysis
Total RNA was purified from cultured cells with TRIzol (Invitrogen). Reverse transcription-PCR analysis of transcripts of FoxO1, FoxO3a, FoxO4, Bim, p27kip1, GADD45, and GAPDH (as a control) was performed with primers designed to detect rat gene products as described previously (37, 38).
Carotid Artery Ligation and Immunohistochemistry
The carotid artery was isolated from anesthetized 2-month-old male rat by using an approved institutional protocol according to Peking Union Medical College guidelines. Carotid artery ligation was performed as described previously (39). 24 days after ligation, the left and right carotid arteries were harvested, fixed, and stained using anti-CHIP or anti-FoxO1 antibody (1:50) following standard protocol.
Statistical Analysis
Data are presented as means ± S.E. Differences between groups were evaluated for statistical significance using Student's t test. p values <0.05 were regarded as significant.
RESULTS
CHIP Down-regulates FoxO1 Expression
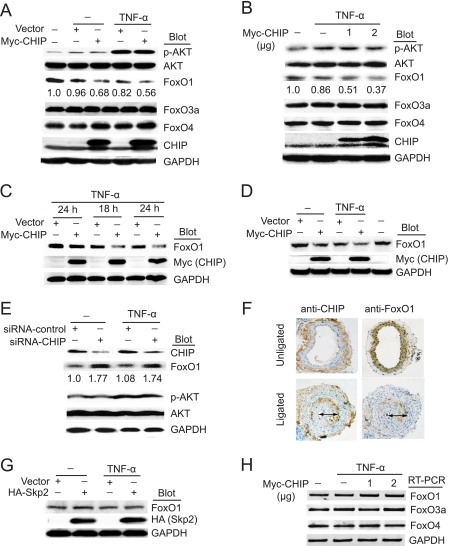
It has been demonstrated previously that FoxO1, FoxO3a, and FoxO4 can be degraded by the ubiquitin-proteasome pathway in several cell types (16–20). We hypothesized that CHIP as an E3 ligase might be involved in this proteolysis, thereby modulating SMC proliferation and apoptosis. To test this hypothesis, we first investigated whether CHIP affected the expression of Akt and FoxO proteins in SMCs without or with TNF-α stimulation. As shown in Fig. 1A, overexpression of CHIP decreased the level of endogenous FoxO1 protein, and this effect was further enhanced by TNF-α. Moreover, CHIP had a dose-dependent inhibitory effect on the level of endogenous FoxO1 protein (Fig. 1B). In contrast, CHIP was unable to down-regulate the expression of FoxO3a and FoxO4 (Fig. 1, A and B). Furthermore, overexpression of CHIP was found to decrease expression of endogenous FoxO1 protein in human umbilical endothelial cells and rat neonatal cardiomyocytes, although to varying degrees (Fig. 1, C and D). Interestingly, Akt was activated by TNF-α, but no significant change in Akt phosphorylation was observed in CHIP-transfected cells compared with vector transfection (Fig. 1, A and B). To further test the involvement of endogenous CHIP in regulating FoxO proteins, we performed gene knockdown experiments with CHIP siRNA. As shown in Fig. 1E, depletion of endogenous CHIP resulted in an increase of endogenous FoxO1 protein, whereas transfection with nonspecific siRNA had no effect. Collectively, these results indicate that the inhibitory effect of CHIP on the expression of FoxO1 was specific.
FIGURE 1.
CHIP decreases FoxO1 protein level. A, SMCs were transfected with Myc-CHIP or vector and treated with TNF-α (10 ng/ml) for 24 h. Whole cell extracts were subjected to immunoblot analysis using antibodies against Akt, FoxO1, FoxO3a, FoxO4, CHIP, and GAPDH (as a loading control). B, SMCs were transfected with increasing amounts of Myc-CHIP and treated with TNF-α (10 ng/ml) for 24 h. Whole cell extracts were prepared for Western blot analysis as in A. C and D, human umbilical endothelial cells or cardiomyocytes were infected with adenovirus Myc-CHIP or vector control and treated with TNF-α (10 ng/ml) for the indicated time points. Whole cell extracts were subjected to immunoblot analysis using antibodies against FoxO1, Myc, and GAPDH. A representative blot is shown (n = 3). E, SMCs were transfected with siRNA-control or siRNA-CHIP and treated with TNF-α (10 ng/ml) for 24 h. Whole cell extracts were prepared for Western blot analysis as in A. F, the left common carotid artery was ligated for 24 days; both right and left common carotid arteries were harvested, fixed, and stained with anti-CHIP or anti-Foxo1 antibody (arrow indicates neointimal formation in vessels, original magnification, ×100). G, SMCs were transfected with Skp2 and treated as in A. Whole cell extracts were subjected to immunoblot analysis using antibodies against FoxO1, HA, and GAPDH. A representative blot is shown (n = 3). H, SMCs were transfected and treated as in B. Total RNA was isolated, and the expression levels of transcripts for FoxO1, FoxO3a, and FoxO4 were determined by reverse transcription-PCR analysis, normalized to GAPDH. A representative blot is shown (n = 3).
Importantly, immunohistochemical staining for CHIP and FoxO1 was performed in the neointimal layer of the rat carotid artery following flow cessation injury. As shown in Fig. 1F, CHIP expression was up-regulated, whereas FoxO1 was decreased compared with the control uninjured artery.
Because Skp2 also targets FoxO1 for proteasomal degradation in LNCaP, NIH 3T3, and COS7 cell (18), we further investigated whether Skp2 down-regulated FoxO1 level in SMCs. Overexpression of Skp2 did not significantly decrease FoxO1 protein in SMCs without or with TNF-α stimulation (Fig. 1G). Thus, it appears that Skp2 has a very minimal effect on FoxO1 protein level in SMCs.
To elucidate the underlying mechanism, we examined whether CHIP regulated the expression of FoxO1 at the messenger level by using reverse transcription-PCR. CHIP did not affect the mRNA level of FoxO1, FoxO3a, and FoxO4 (Fig. 1H). Taken together, these results suggest that CHIP specifically down-regulates FoxO1 expression in SMCs at protein level.
CHIP Targets FoxO1 for Ubiquitination-mediated Degradation
To determine whether CHIP as an E3 ligase promotes ubiquitination and degradation of FoxO1, we performed a pulse-chase assay and found that overexpression of CHIP resulted in a rapid decrease in the FoxO1 protein compared with vector control (Fig. 2A). In contrast, treatment with proteasome inhibitor MG132 significantly blocked CHIP-induced down-regulation of FoxO1 protein despite of TNF-α treatment (Fig. 2B). Furthermore, CHIP markedly promoted ubiquitination and degradation of FoxO1, and this effect was enhanced markedly by TNF-α (Fig. 2, C and D). These results demonstrate that CHIP targets FoxO1 for proteasomal degradation, which is triggered by TNF-α.
FIGURE 2.
CHIP promotes ubiquitination and degradation of FoxO1. A, 293 cells were cotransfected with FLAG-FoxO1, vector control, or Myc-CHIP and treated with cycloheximide (Chx, 10 μg/ml) for the indicated time points. Equal amounts of cell lysates were subjected to SDS-PAGE. The level of protein was detected by Western blotting with anti-FLAG, anti-Myc, or anti-GAPDH antibodies (top). The amount of FoxO1 in presence of CHIP (squares) and absence of CHIP (triangle) was quantified. Values shown are the means ± S.E. of three independent experiments (bottom). B, SMCs cotransfected with Myc-CHIP or vector were treated with TNF-α (10 ng/ml) in the absence or presence of MG132 for 6 h. Whole cell extracts were prepared for Western blot analysis using FoxO1, Myc, and GAPDH antibodies (top). Values shown are the means ± S.E. of three independent experiments (bottom). *, p < 0.01 versus vector. C, 293 cells were cotransfected with His-ubiquitin (His-Ub), FLAG-FoxO1, and Myc-CHIP and treated with TNF-α (10 ng/ml) for 24 h and MG132 for 6 h. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-His (top) or anti-FLAG (bottom) antibodies. D, 293 cells were cotransfected and treated as in C. Whole cell extracts were subjected to direct immunoblot analysis using anti-FLAG or anti-Myc antibodies, and GAPDH as loading controls.
CHIP Interacts with FoxO1
Because one of the characteristics for an E3 ligase is the recognition of specific substrates, it will be interesting to test whether CHIP is physically associated with FoxO1. GST pulldown assays showed that FoxO1 (but not FoxO3a or FoxO4) more strongly bound to GST-CHIP (Fig. 3A). Coimmunoprecipitation assays confirmed that anti-CHIP precipitated FoxO1, and this interaction was enhanced by MG132 (Fig. 3B). Moreover, FLAG-FoxO1 was detected in the Myc-CHIP immune complex, whereas no FLAG-FoxO1 was found in the immune complex precipitated with a nonspecific IgG (Fig. 3C). Finally, immunostaining demonstrated that CHIP was colocalized with FoxO1 in cytoplasm of SMCs (Fig. 3D). These data indicate that CHIP specifically interacts with FoxO1 in vivo and in vitro.
FIGURE 3.
CHIP interacts with FoxO1. A, lysates of 293T cells transfected with FLAG-FoxO1, HA-FoxO3a, and FLAG-Foxo4 were subjected to GST pulldown assay by GST-CHIP or GST alone fusion protein, and bound proteins were analyzed by immunoblotting using anti-FLAG or anti-HA antibody after binding reactions. B, SMCs treated without or with MG132 for 12 h were immunoprecipitated with anti-CHIP antibody or control IgG, and the immunoprecipitates were subjected to SDS-PAGE and immunoblotted with anti-FoxO1 antibody. C, SMCs were cotransfected with the indicated plasmids, and whole cell lysates were immunoprecipitated (IP) with anti-Myc antibody or control IgG. Immunoprecipitates were analyzed by immunoblotted with anti-FLAG and anti-Myc antibodies. D, immunostaining analysis of endogenous CHIP and FoxO1 in SMCs showed that CHIP colocalized with FoxO1.
Both the U-box and Charged Domains Are Essential for CHIP-induced Ubiquitination and Degradation of FoxO1
To map the FoxO1 binding region of CHIP, we performed GST pulldown assays using a series of GST-fusion CHIP deletion mutants and FoxO1 expressed in 293T cells. Full-length CHIP efficiently interacted with FoxO1, and deletion of the U-Box domain (Δ198–303aa) had less effect on CHIP binding to FoxO1. However, deletion of the charged domain (Δ143–197aa) completely abolished the binding of CHIP to FoxO1 in this assay (Fig. 4, A and B), indicating that residues between 143 and 197 of CHIP are sufficient to bind FoxO1.
FIGURE 4.
Both the U-Box and charged domains are required for CHIP-induced FoxO1 ubiquitination and down-regulation. A, the residues of CHIP required for binding to FoxO1 were determined with GST pulldown assays. GST-CHIP fusion proteins were purified from bacteria and analyzed by immunoblotting with GST antibody (top). The ability of the truncated CHIP fusion proteins to bind to FLAG-FoxO1 expressed in 293T cells was analyzed by blotting with anti-FLAG antibody (middle). 10% of input was shown in the bottom panel. B, schematic representation of CHIP truncations that interact with FoxO1. TPR, tetratricopeptide repeat domain; +/−, charged region; U-Box, U-box domain. C, 293T cells were transfected with HA-ubiquitin, FLAG-FoxO1, CHIP Wt, or mutants plasmids in the presence of TNF-α (10 ng/ml) for 24 h and MG132 for 6 h. Cell extracts were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-HA (top) or FLAG (bottom) antibodies. D, 293T cells were transfected with the indicated plasmids and treated with TNF-α. Whole cell extracts were subjected to direct immunoblot analysis using anti-FLAG or anti-Myc antibodies, and GAPDH as loading controls.
To further examine whether the ubiquitin ligase activity and FoxO1 interaction domain are required for CHIP-mediated FoxO1 degradation, three CHIP mutants were used. As shown in Fig. 4 (C and D), both CHIP Wt and ΔTPR (Δ1–142aa) significantly increased ubiquitination of FoxO1 and down-regulated its protein levels. In contrast, neither CHIP ΔU-box (Δ198–303aa) nor Δ±(Δ143–197aa) mutant had any effect, indicating that both U-box and charged domains in CHIP are involved in ubiquitination and degradation of FoxO1 proteins.
CHIP-induced Ubiquitination of FoxO1 Requires Phosphorylation of FoxO1 at S256
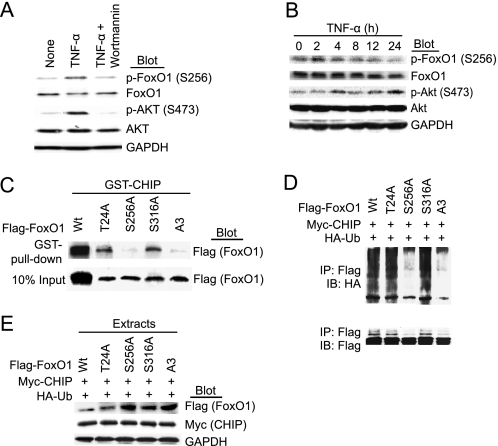
Because PI3K/Akt-dependent phosphorylation plays a key role in the proteasomal degradation of FoxO1 (17–19, 40), we determined whether TNF-α modulates FoxO1 phosphorylation through PI3K/Akt signaling pathway in SMCs. As shown in 5A, incubation of SMCs with TNF-α for 30 min resulted in markedly phosphorylation of Akt and FoxO1, and this effect was completely blocked by pretreatment with the PI3K inhibitor wortmannin. This is consistent with the notion that FoxO1 acts downstream of Akt phosphorylation in the PI3K/Akt pathway.
CHIP has been known to preferentially recognize phosphorylated substrates such as Tau (28, 29), we therefore examine whether ubiquitination of FoxO1 by CHIP requires its phosphorylation in vivo. As shown in Fig. 5B, treatment with TNF-α increased the level of Akt phosphorylation and decreased total and phosphorylated FoxO1 protein in a time-dependent manner, indicating that TNF-α induced degradation of FoxO1 via Akt activation. We next examined whether mutation of the Akt-phosphorylation sites at Thr-24, Ser-256, and/or Ser-319 to alanine affects the interaction of FoxO1 with CHIP. All of the mutant forms of FoxO1 were expressed at comparable levels in 293T cells (Fig. 5C, bottom). Both T24A and S319A mutants were able to form a complex with CHIP. However, the interaction between FoxO1 and CHIP was almost abolished by a point mutation at S256A or triple mutations A3 at T24A, S256A, and S319A (Fig. 5C, top). Moreover, ubiquitination was observed on the FoxO1 or mutants at position either T24A or S319A, whereas ubiquitination of FoxO1 was diminished by the single mutation at S256A or triple mutations A3 (Fig. 5D).
FIGURE 5.
CHIP-induced ubiquitination of FoxO1 requires phosphorylation of FoxO1 at Ser-256. A, SMCs were pretreated with wortmannin (200 nm) for 30 min and then incubated with TNF-α (10 ng/ml) for further 30 min. Whole cell extracts were subjected to direct immunoblot analysis using anti-FoxO1, anti-Akt, and anti-GAPDH antibodies. B, SMCs were treated with TNF-α (10 ng/ml) for 0–24 h. Whole cell extracts were detected as in A. C, lysates of 293T cells transfected with FLAG-FoxO1 Wt or mutants (T24A, S256A, S316A, or A3) were subjected to GST pulldown by GST-CHIP purified from bacteria (top). 10% of input is shown in the bottom panel. D, 293T cells were cotransfected with HA-ubiquitin, CHIP, and FoxO1 mutants plasmids and treated with TNF-α for 24 h and MG132 for 6 h. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-HA (top) or anti-FLAG (bottom) antibodies. E, 293T cells were transfected with the indicated plasmids and treated with TNF-α for 24 h. Whole cell extracts were subjected to direct immunoblot analysis using anti-FLAG or anti-Myc antibodies, and GAPDH as loading controls.
We next investigated the effect of CHIP on the protein stability of FoxO1 Wt and its mutants. Cells were transfected with CHIP and with FoxO1 Wt or mutants with TNF-α stimulation for 24 h. Overexpression of CHIP markedly decreased FoxO1 Wt, T24A, and S319A protein levels, whereas the CHIP had no effect on FoxO1 S256A and A3 following TNF-α treatment (Fig. 5E). Together, these findings demonstrate that interaction, ubiquitination, and degradation of FoxO1 by CHIP depend on its phosphorylation at Ser-256.
CHIP Inhibits FoxO1-mediated Gene Transactivation
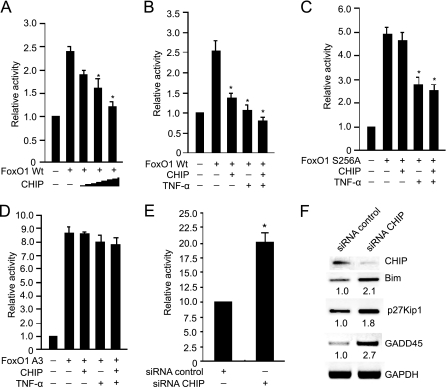
After having established a role for CHIP in FoxO1 protein turnover, we next examined the effect of CHIP on FoxO1-mediated gene transactivation. A luciferase assay was performed using Bim reporter, FoxO1, and CHIP. As shown in Fig. 6(A and B), CHIP inhibited the transcriptional activity of FoxO1 in a dose-dependent manner. TNF-α treatment alone also attenuated the transcriptional activity of FoxO1. Moreover, the inhibition of FoxO1 transactivation was enhanced by the combination of CHIP and TNF-α. Interestingly, the single mutation of FoxO1 at Ser-256 resulted in a higher transactivation activity of FoxO1. In contrast to the wild-type FoxO1, there was no effect of CHIP expression on transcriptional activity of FoxO1 S256A. However, treatment with TNF-α decreased the activity of FoxO1 S256A (Fig. 6C). This is presumably due to activation of Akt by TNF-α, resulting in FoxO1 phosphorylation at Thr-24 and Ser-319 and inactivation. Interestingly, expression of FoxO1 A3 caused an over 3-fold increase in its transactivation when compared with the wild-type FoxO1, and FoxO1 A3 mutant was resistant to CHIP- or TNF-α-induced down-regulation of transactivation (Fig. 6D).
FIGURE 6.
CHIP inhibits FoxO1-mediated transcriptional activity. A, 293T cells were transfected with Bim luciferase reporter, FLAG-Foxo1, and increasing amounts of Myc-CHIP (0.25, 0.5, and 1.0 μg). 24 h after infection, Cells were harvested and luciferase activity was measured (n = 3, means ± S.E.). *, p < 0.01 versus FoxO1 alone. B, 293T cells were transfected with Bim luciferase reporter, FLAG-FoxO1, and Myc-CHIP, and cells were treated without or with TNF-α (10 ng/ml) for 24 h. The luciferase analysis was performed as in A. *, p < 0.01 versus FoxO1 alone. C, 293T cells were transfected with Bim luciferase reporter, FoxO1 S256A, and Myc-CHIP, and cells were treated without or with TNF-α for 24 h. The luciferase analysis was performed as in A. *, p < 0.01 versus FoxO1 S256A alone. D, 293T cells were transfected with Bim luciferase reporter, FoxO1 A3, and Myc-CHIP, and cells were treated without or with TNF-α for 24 h. The luciferase analysis was performed as in A. E, SMCs were transfected with Bim luciferase reporter, siRNA-CHIP, or siRNA-control. The luciferase activity was determined (n = 3, means ± S.E.). *, p < 0.01 versus siRNA-control. F, SMCs cells were transfected with siRNA-CHIP or siRNA-control vector and treated with TNF-α (10 ng/ml) for 24 h. Total RNA was isolated and the expression levels of transcripts for CHIP, Bim, p27Kip1, and GADD45 were determined by reverse transcription-PCR analysis, normalized to GAPDH (n = 3).
To further investigate the functional role of endogenous CHIP in FoxO1-mediated transactivation in SMCs, we depleted endogenous CHIP protein using siRNA and performed luciferase assay with Bim reporter. As shown in Fig. 6E, transcriptional activity of the Bim reporter by FoxO1 was increased ∼2-fold by siRNA-CHIP than scrambled siRNA-control. Furthermore, the mRNA expression of known FoxO target genes, including Bim, p27kip1, and GADD45 was concomitantly increased in siRNA-CHIP transfected cells. Quantitative analysis indicated that the expression of each of these target genes was increased by ∼1.8-fold or more in siRNA-CHIP-transfected cells compared with siRNA-control cells (Fig. 6F). Taken together, these data suggest that CHIP negatively regulates the transcriptional activity of FoxO1 through proteasomal degradation.
CHIP Blocks FoxO1-mediated Apoptosis and Growth Arrest of SMCs
To investigate a functional role of CHIP in FoxO1-mediated cell growth arrest and apoptosis, we performed cell viability and proliferation assays. As shown in Fig. 7 (A and B), similar to previous data (4), overexpression of FoxO1 Wt and S256A mutant resulted in a significant decrease of viability and proliferation in SMCs, and the effect of FoxO1 Wt was markedly abolished by cotransfection of CHIP. However, no action was observed when cells were cotransfected with CHIP and FoxO1 S256A mutant. Moreover, TNF-α treatment increased CHIP-mediated SMC viability and proliferation in the presence of FoxO1 Wt and S256A mutant. Furthermore, depletion of endogenous CHIP by siRNAs markedly increased protein level of FoxO1, cleaved caspase-3 and poly(ADP-ribose) polymerase compared with siRNA-control (Fig. 7C), and this effect was further enhanced with TNF-α stimulation. More importantly, CHIP knockdown promoted FoxO1-induced cell apoptosis (Fig. 7D) and decreased cell proliferation in SMCs (Fig. 7E). These results indicate that CHIP is a negative regulator for FoxO1-induced growth arrest and apoptosis in SMCs.
FIGURE 7.
CHIP antagonizes the proapoptotic and anti-proliferative function of FoxO1. A, SMCs cells were cotransfected with vector pcDNA-GFP, GFP-FoxO1 wt, or GFP-FoxO1 S256A mutant in the presence or absence of CHIP and treated without or with TNF-α (10 ng/ml) for 24 h. Transfected viable cells were photographed under both UV and transmitted light. Quantitative data of viable cells were determined using trypan blue assay from three independent experiments in A. *, p < 0.01 versus vector. B, SMCs were transfected and treated as in A, cell proliferation was performed using WST-1 assay. *, p < 0.01 versus vector control. C, cells were transfected with siRNA-CHIP or siRNA-control and treated without or with TNF-α for 24 h. The protein levels of CHIP, FoxO1, cleaved caspase 3, and poly(ADP-ribose) polymerase (PARP) were examined by immunoblotting with antibodies as indicated. D, SMCs were cotransfected with control vector or wild-type FoxO1 in the presence of siRNA-control or siRNA-CHIP and treated with TNF-α for 24 h. To quantify apoptotic cells, cells were fixed and stained with TUNEL. At least 120 cells per dish were counted. Results were expressed as means ± S.E. for three independent experiments. #, p < 0.05 or *, p < 0.01 versus siRNA-control. E, SMCs were transfected and treated as in D, and cell proliferation analysis was performed using WST-1 assay. Results were expressed as means ± S.E. of three separate experiments. #, p < 0.05 or *, p < 0.01 versus siRNA-control. F, proposed mechanisms for CHIP to regulate SMC proliferation and survival through ubiquitination and degradation of FoxO1 by the proteasome.
DISCUSSION
It is now recognized that the balance between proliferation and apoptosis is linked to various vascular diseases, such as atherosclerosis and neointimal hyperplasia after vascular intervention. FoxO1, a main member of the forkhead transcription factor family, has been shown to play an important role in cell cycle arrest and apoptosis of various cell lines (41). In the present study, we demonstrated that the protein level of FoxO1 is tightly controlled by CHIP through ubiquitin-mediated degradation following TNF-α treatment, and this effect of CHIP requires phosphorylation of FoxO1 at Ser-256. Importantly, overexpression of CHIP represses FoxO1-mediated transactivation and its inhibitory function on SMC survival and proliferation. In contrast, deletion of CHIP by siRNA had opposite effects. Thus, we have identified a previously uncharacterized mechanism of inactivation of FoxO1 by CHIP in SMCs.
The significance of the FoxO family members to diverse physiological events in the transcription-based function is extensively investigated (41). Activation of Akt regulates activity of FoxO1 factor by phosphorylating it at three conserved serine/threonine residues (Thr-24, Ser-256, and Ser-319). This leads to the release of FoxO1 from DNA and translocation to the cytoplasm. In the absence of growth or survival signal stimulation, inactivation of Akt in the quiescent cells results in nuclear retention of FoxO factors, thereby enhancing expression of its downstream target genes by binding to other transcription factors or acting as an activator that modulate the metabolic state, DNA repair (e.g. GADD45), cell cycle transitions (e.g. p27kip1), or cellular apoptosis (e.g. Fas ligand, TNF-R1-associated death domain, and Bim) (41). Recent studies have demonstrated that expression of FoxOs inhibits SMC growth and survival via up-regulation of p27Kip1, leading to inhibition of neointimal hyperplasia (4, 15, 19). These data indicate that FoxO transcription factors play a pivotal role in regulating SMC proliferation and survival.
FoxO transcription factors are controlled by post-translational modifications. In addition to phosphorylation, FoxO factors are also regulated by ubiquitination (41). It has been shown that steady-state levels of FoxO1 are reduced in several cell lines in response to growth and survival factors such as insulin, insulin-like growth factor 1, and platelet-derived growth factor, and this effect is inhibited by proteasome inhibitors, suggesting that proteasome-mediated degradation of FoxO1 depends on Akt activation (4, 15–19). Recently, Abid et al. have shown that TNF-α induces phosphorylation of FoxO1 proteins on conserved Akt phosphorylation site Ser-256 in human coronary artery SMCs and induces nuclear exclusion of FoxO1 (4). However, it is not studied whether TNF-α-induced phosphorylation is required for the ubiquitination of FoxO1, and the E3 ligase responsible for this event is also unknown.
CHIP as ubiquitin ligase comprises three functional domains: a tetratricopeptide repeat (TPR) at the N terminus, a U-box domain at the C terminus, and a highly charged region (21). CHIP has been shown to play a critical regulatory function in the protein quality control machinery of the cell lines (21–24, 42) and implicated in the pathology of several neurological disorders characterized by protein misfolding and aggregation (29, 32). It is believed that CHIP exerts biological effects via inducing chaperone client proteins for proteasomal degradation. Recently, we reported that CHIP inhibited myocardin-induced SMC differentiation via ubiquitin-proteasome system (35). In the present study, we found that CHIP interacted and down-regulated FoxO1 protein level through ubiquitin-mediated degradation, and this effect is enhanced by TNF-α treatment (Figs. 1–3). Moreover, CHIP-induced ubiquitination and degradation of Foxo1 requires its E3 ligase activity (U-box) and FoxO1 binding domain (charged region) (Fig. 4, C and D). In accordance with the ubiquitination data in vivo, we further demonstrated that CHIP overexpression dramatically decreases FoxO1-mediated transactivation that is further enhanced by TNF-α (Fig. 6, A and B). In contrast, knockdown of CHIP increased FoxO1-mediated transactivation and the expression of FoxOs target genes (Fig. 6, E and F). These findings demonstrate that the cellular protein level and activity of FoxO1 are tightly controlled by CHIP.
Phosphorylation is required for the ubiquitination of many transcriptional factor proteins, and in several cases the target proteins are first marked by phosphorylation for ubiquitination. For example, Tau is first phosphorylated by glycogen synthase kinase 3β and then targeted to proteasomal degradation by CHIP (28, 29, 43). Indeed, it is evident that FoxO proteins are regulated by the PI3K/Akt pathway, and the phosphorylation of FoxO proteins by Akt results in their cytoplasmic retention and proteasomal degradation (4, 16–19). More recently, Skp2, an oncogenic subunit of the Skp1/Cul1/F-box protein ubiquitin complex, was shown to interact with and promote the degradation of Foxo1 that requires Akt-induced phosphorylation of FoxO1 at Ser-256 (18). Consistent with these findings, our study demonstrated that TNF-α can induce PI3K-dependent activation of Akt, leading to FoxO1 phosphorylation and degradation (Fig. 5, A and B). Importantly, phosphorylation at position Ser-256 of FoxO1 was required for its interaction with CHIP and ubiquitin-mediated degradation (Fig. 5, C–E). Furthermore, TNF-α treatment alone markedly decreased FoxO1 S256A-mediated transactivation. However, CHIP had no this effect (Fig. 6C). These data indicate that CHIP selectively promotes ubiquitination and degradation of phosphorylated FoxO1 in vivo.
Recent studies demonstrate that CHIP-deficient mice develop apoptosis in cardiomyocytes and especially endothelial cells of intramural vessels in response to ischemia-reperfusion injury (33, 34). CHIP degrades ASK1 and inhibits JNK activation in response to oxidative stress (30). Furthermore, CHIP cooperates with E2 enzymes of the Ubc4/5 family to mediate ubiquitin-mediated proteasomal degradation of chaperone-bound p53 (24). These observations led us to search for a mechanism whereby CHIP might be directly involved in apoptosis in the face of cellular stress. Indeed, the present studies extend these observations by demonstrating a functional role of endogenous CHIP in FoxO1-mediated apoptotic signaling and cell proliferation. Overexpression of CHIP significantly attenuated the effect of FoxO1 Wt on viability and proliferation of SMCs without or with TNF-α treatment (Fig. 7, A and B). However, no effect was observed when cells were cotransfected with CHIP and FoxO1 S256A mutant (Fig. 7, A and B). In contrast, depletion of CHIP increased the level of cleaved caspase-3 and poly(ADP-ribose) polymerase (Fig. 7C), resulting in a significant increase of apoptosis and decrease of proliferation in SMCs via activation of FoxO1 (Fig. 7, D and E), indicating that CHIP participates in inhibiting FoxO1-mediated apoptosis and growth arrest in SMCs.
In conclusion, our data strongly indicate that CHIP regulates phenotypic modulation of SMC through ubiquitination and degradation of FoxO1. Based on these findings, we propose a novel model to explain the involvement of CHIP in enhancing proliferation and survival of SMCs in response to TNF-α (Fig. 7F). TNF-α binds its specific receptor and activates PI3K/Akt signaling (16, 18), resulting in FoxO1 phosphorylation. CHIP promotes the ubiquitin-mediated degradation of phosphorylated FoxO1 proteins, leading to consistently decreased protein levels and inactivation of FoxO1. Consequently, TNF-α stimulates SMC proliferation and survival, resulting in neointimal formation. This study reveals the importance of CHIP in proliferation and survival of SMCs and suggests that it may serve as a target for interventions in pathological states. Future studies are required to confirm the molecular mechanisms by which CHIP promotes degradation of FoxO1 and regulates phenotypic modulation of vascular SMCs in animal model.
This work was supported by the China Natural Science Foundation (Grants 2006CB910306, 2006CB503805, and 30670860 to H. L.) and by The Ph.D. Programs Foundation of Ministry of Education of China (Grant 20060023051 to H. L.).
- SMCs
- smooth muscle cells
- FoxOs
- forkhead transcription factors
- CHIP
- C terminus of Hsc70-interacting protein
- TPR
- a tetratricopeptide repeat
- TNF-α
- tumor necrosis factor-α
- PI3K
- phosphoinositide 3-kinase
- GST
- glutathione S-transferase
- TUNEL
- terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- siRNA
- small interference RNA
- HA
- hemagglutinin
- aa
- amino acid(s)
- Ub
- ubiquitin
- E2
- ubiquitin carrier protein
- E3
- ubiquitin-protein isopeptide ligase
- Wt
- wild type
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 2.Owens G., Emons M. F., Christian-Herman J., Lawless G. (2007) Dis. Manag. 10, 74–82 [DOI] [PubMed] [Google Scholar]
- 3.Potente M., Urbich C., Sasaki K., Hofmann W. K., Heeschen C., Aicher A., Kollipara R., DePinho R. A., Zeiher A. M., Dimmeler S. (2005) J. Clin. Invest. 115, 2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abid M. R., Yano K., Guo S., Patel V. I., Shrikhande G., Spokes K. C., Ferran C., Aird W. C. (2005) J. Biol. Chem. 280, 29864–29873 [DOI] [PubMed] [Google Scholar]
- 5.Abid M. R., Shih S. C., Otu H. H., Spokes K. C., Okada Y., Curiel D. T., Minami T., Aird W. C. (2006) J. Biol. Chem. 281, 35544–35553 [DOI] [PubMed] [Google Scholar]
- 6.Park K. W., Kim D. H., You H. J., Sir J. J., Jeon S. I., Youn S. W., Yang H. M., Skurk C., Park Y. B., Walsh K., Kim H. S. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 742–747 [DOI] [PubMed] [Google Scholar]
- 7.Ozes O. N., Akca H., Mayo L. D., Gustin J. A., Maehama T., Dixon J. E., Donner D. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4640–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozes O. N., Mayo L. D., Gustin J. A., Pfeffer S. R., Pfeffer L. M., Donner D. B. (1999) Nature 401, 82–85 [DOI] [PubMed] [Google Scholar]
- 9.Osawa Y., Banno Y., Nagaki M., Brenner D. A., Naiki T., Nozawa Y., Nakashima S., Moriwaki H. (2001) J. Immunol. 167, 173–180 [DOI] [PubMed] [Google Scholar]
- 10.Osawa Y., Nagaki M., Banno Y., Brenner D. A., Asano T., Nozawa Y., Moriwaki H., Nakashima S. (2002) Infect. Immun. 70, 6294–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Der Heide L. P., Hoekman M. F., Smidt M. P. (2004) Biochem. J. 380, 297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modur V., Nagarajan R., Evers B. M., Milbrandt J. (2002) J. Biol. Chem. 277, 47928–47937 [DOI] [PubMed] [Google Scholar]
- 13.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 14.Gilley J., Coffer P. J., Ham J. (2003) J. Cell Biol. 162, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H. Y., Chung J. W., Youn S. W., Kim J. Y., Park K. W., Koo B. K., Oh B. H., Park Y. B., Chaqour B., Walsh K., Kim H. S. (2007) Circ. Res. 100, 372–380 [DOI] [PubMed] [Google Scholar]
- 16.Hu M. C., Lee D. F., Xia W., Golfman L. S., Ou-Yang F., Yang J. Y., Zou Y., Bao S., Hanada N., Saso H., Kobayashi R., Hung M. C. (2004) Cell 117, 225–237 [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaki H., Daitoku H., Hatta M., Tanaka K., Fukamizu A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11285–11290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H., Regan K. M., Wang F., Wang D., Smith D. I., van Deursen J. M., Tindall D. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plas D. R., Thompson C. B. (2003) J. Biol. Chem. 278, 12361–12366 [DOI] [PubMed] [Google Scholar]
- 20.Brenkman A. B., de Keizer P. L., van den Broek N. J., Jochemsen A. G., Burgering B. M. (2008) PLoS ONE 3, e2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999) Mol. Cell. Biol. 19, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) Nat. Cell Biol. 3, 93–96 [DOI] [PubMed] [Google Scholar]
- 23.Esser C., Alberti S., Höhfeld J. (2004) Biochim. Biophys. Acta 1695, 171–188 [DOI] [PubMed] [Google Scholar]
- 24.Esser C., Scheffner M., Höhfeld J. (2005) J. Biol. Chem. 280, 27443–27448 [DOI] [PubMed] [Google Scholar]
- 25.Galigniana M. D., Harrell J. M., Housley P. R., Patterson C., Fisher S. K., Pratt W. B. (2004) Brain Res. Mol. Brain Res. 123, 27–36 [DOI] [PubMed] [Google Scholar]
- 26.Alberti S., Böhse K., Arndt V., Schmitz A., Höhfeld J. (2004) Mol. Biol. Cell 15, 4003–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. (2001) Nat Cell Biol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- 28.Shimura H., Schwartz D., Gygi S. P., Kosik K. S. (2004) J. Biol. Chem. 279, 4869–4876 [DOI] [PubMed] [Google Scholar]
- 29.Dickey C. A., Kamal A., Lundgren K., Klosak N., Bailey R. M., Dunmore J., Ash P., Shoraka S., Zlatkovic J., Eckman C. B., Patterson C., Dickson D. W., Nahman N. S., Jr., Hutton M., Burrows F., Petrucelli L. (2007) J. Clin. Invest. 117, 648–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang J. R., Zhang C., Patterson C. (2005) Cell Stress Chaperones 10, 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Ramahi I., Lam Y. C., Chen H. K., de Gouyon B., Zhang M., Pérez A. M., Branco J., de Haro M., Patterson C., Zoghbi H. Y., Botas J. (2006) J. Biol. Chem. 281, 26714–26724 [DOI] [PubMed] [Google Scholar]
- 32.Choi J. Y., Ryu J. H., Kim H. S., Park S. G., Bae K. H., Kang S., Myung P. K., Cho S., Park B. C., Lee do H. (2007) Mol. Cell Neurosci. 34, 69–79 [DOI] [PubMed] [Google Scholar]
- 33.Dai Q., Zhang C., Wu Y., McDonough H., Whaley R. A., Godfrey V., Li H. H., Madamanchi N., Xu W., Neckers L., Cyr D., Patterson C. (2003) EMBO J. 22, 5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C., Xu Z., He X. R., Michael L. H., Patterson C. (2005) Am. J. Physiol. Heart Circ Physiol. 288, H2836–2842 [DOI] [PubMed] [Google Scholar]
- 35.Xie P., Fan Y., Zhang H., Zhang Y., She M., Gu D., Patterson C., Li H. (2009) Mol. Cell. Biol. 29, 2398–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H. H., Willis M. S., Lockyer P., Miller N., McDonough H., Glass D. J., Patterson C. (2007) J. Clin. Invest. 117, 3211–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z. P., Wang Z., Yanagisawa H., Olson E. N. (2005) Dev. Cell 9, 261–270 [DOI] [PubMed] [Google Scholar]
- 38.Li H. H., Kedar V., Zhang C., McDonough H., Arya R., Wang D. Z., Patterson C. (2004) J. Clin. Invest. 114, 1058–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar A., Hoover J. L., Simmons C. A., Lindner V., Shebuski R. J. (1997) Circulation 96, 4333–4342 [DOI] [PubMed] [Google Scholar]
- 40.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 41.Huang H., Tindall D. J. (2007) J. Cell Sci. 120, 2479–2487 [DOI] [PubMed] [Google Scholar]
- 42.Jiang J., Ballinger C. A., Wu Y., Dai Q., Cyr D. M., Höhfeld J., Patterson C. (2001) J. Biol. Chem. 276, 42938–42944 [DOI] [PubMed] [Google Scholar]
- 43.Kosik K. S., Shimura H. (2005) Biochim. Biophys. Acta 1739, 298–310 [DOI] [PubMed] [Google Scholar]