Abstract
Cadherin 23 (CDH23), a transmembrane protein localized near the tips of hair cell stereocilia in the mammalian inner ear, is important for delivering mechanical signals to the mechano-electric transducer channels. To identify CDH23-interacting proteins, a membrane-based yeast two-hybrid screen of an outer hair cell (OHC) cDNA library was performed. EHD4, a member of the C-terminal EH domain containing a protein family involved in endocytic recycling, was identified as a potential interactor. To confirm the interaction, we first demonstrated the EHD4 mRNA expression in hair cells using in situ hybridization. Next, we showed that EHD4 co-localizes and co-immunoprecipitates with CDH23 in mammalian cells. Interestingly, the co-immunoprecipitation was found to be calcium-sensitive. To investigate the role of EHD4 in hearing, compound action potentials were measured in EHD4 knock-out (KO) mice. Although EHD4 KO mice have normal hearing sensitivity, analysis of mouse cochlear lysates revealed a 2-fold increase in EHD1, but no increase in EHD2 or EHD3, in EHD4 KO cochleae compared with wild type, suggesting that a compensatory increase in EHD1 levels may account for the absence of a hearing defect in EHD4 KO mice. Taken together, these data indicate that EHD4 is a novel CDH23-interacting protein that could regulate CDH23 trafficking/localization in a calcium-sensitive manner.
Hair cells located in the mammalian inner ear transform mechanical stimuli into electrical signals that in turn facilitate neurotransmitter release onto auditory neurons. The key element in the transduction process is the mechano-electric transducer (MET)2 apparatus located near the top of the stereocilium. CDH23 is a single pass transmembrane protein with 27 extracellular cadherin repeats. It is one of the components of the tip-link (1, 2), which connects the top of a shorter stereocilium to the side of its taller neighbor (3). Vibrations of the basilar membrane of the inner ear ultimately result in deflection of the hair bundles, which modulates tension on the tip-link, thereby controlling the opening probability of cation-selective MET channels (3, 4). Cations, principally K+ and Ca2+, flow through the MET channels and ultimately change the membrane potential. A mutation in the gene encoding CDH23, the Usher syndrome type 1D factor (USH1D), causes deaf-blindness in humans (5). Several interacting partners of CDH23 have been reported and include another tip-link protein protocadherin 15 (6), a multi-PDZ domain-containing scaffold protein harmonin (7) and a stereociliary scaffolding protein MAGI-1 (8). Protocadherin 15 binds to CDH23 through its extracellular domains (6), whereas the cytoplasmic region of CDH23 interacts with MAGI-1 and harmonin through its PDZ domain-binding interfaces (PBI). Harmonin also associates with other USH1 factors like myosin VIIa, protocadherin 15, and sans (9). These findings indicate that harmonin bridges CDH23 to the cytoskeletal actin core of the stereocilium and is probably essential for the developmental differentiation of stereocilia (10–12). However, it is currently unknown how CDH23 is transported to the tip of stereocilia. To search for additional interacting partners of CDH23, we performed a membrane-based yeast two-hybrid assay, which identified EHD4 as a potential binding partner (13).
EHD4 belongs to an evolutionarily conserved EH (Eps 15 homology) domain-containing protein family involved in endocytic trafficking and recycling. Four highly homologous members of this family, EHD1–4, are expressed in mammalian cells. They contain a single C-terminal EH domain, an N-terminal nucleotide-binding loop and a coiled-coil region responsible for oligomerization (14–16). Of the four EHD proteins EHD1 is the best characterized and is involved in regulating the recycling of membrane receptors including the transferrin receptor and the major histocompatibility complex class I (17, 18). EHD1 is also involved in controlling cholesterol recycling and homeostasis (19) and in facilitating endosome to Golgi retrieval (20). EHD3 appears to regulate receptor movements from the early endosome (EE) to the endocytic recycling compartment (ERC) and Golgi (21, 22). EHD2 was isolated from GLUT4-enriched fractions of adipocytes and was shown to regulate insulin-mediated translocation of GLUT4 to the plasma membrane (23, 24). Additionally, EHD2 is involved in the regulation of transferrin receptor internalization (23), recycling (25), and actin cytoskeleton rearrangement (23). EHD4, also called Pincher, was first reported as an extracellular matrix protein (26). Subsequent studies have shown this intracellular protein to be involved in the regulation of neurotrophin receptor TrkA endocytosis in pheochromocytoma (PC12) cells (27). It is also involved in interactions with the cell fate determinant, NUMB, and co-localizes with the small GTP-binding protein, Arf6 (28). Recently, Sharma et al. (29) showed that EHD4 regulates the exit of endocytic cargo from the early endosome toward both the recycling compartment and the late endocytic pathway. They also indicated that EHD4 and EHD1 interact transiently as most of the EHD4 resides on peripheral early endosomes, while EHD1 resides primarily on tubular recycling compartments. This partial overlap/association might be necessary for the transport of proteins through the early endosome to the ERC. Previously, George et al. (25) had also demonstrated that EHD4 interacts with EHD1 and its paralogs, which suggests cooperation and partial overlap of function between EHD4 and EHD1.
Unlike other CDH23-binding proteins, EHD4 does not contain a PDZ domain that could bind to the PBI located in the cytoplasmic tail of CDH23. In addition, the cytoplasmic tail of CDH23 lacks an Asn-Pro-Phe (NPF) motif that could mediate an interaction with the EH domain of EHD4. Therefore, we proceeded to characterize the authenticity of interaction between EHD4 and CDH23 identified in yeast and mammalian cells, using both in vitro and in vivo methods. We verified the expression of EHD4 mRNA in mouse cochlea and investigated the physiological role of EHD4 protein in the cochlea using EHD4-KO mice.
EXPERIMENTAL PROCEDURES
DNA Constructs
Constructs encoding Myc-tagged and GFP-tagged full-length EHD4 and EHD4ΔEH are described previously (25). Otocdh23 DF-pFLAG-CMV-1 (kindly provided by Dr. James R. Bartles) contains a FLAG tag, several extracellular cadherin repeats (domains 14–27), the transmembrane domain, and the cytoplasmic tail. Constructs encoding GFP-tagged prestin and V5-tagged prestin were described previously (30).
Antibodies
Primary antibodies were monoclonal anti-V5 antibody and monoclonal anti-Myc-HRP-conjugated antibody from Invitrogen (Carlsbad, CA); monoclonal anti-FLAG antibody from Sigma, rabbit polyclonal anti-FLAG and anti-Hsc70 antibodies from Santa Cruz Biotechnology, rabbit polyclonal EHD1, EHD2, EHD3, and EHD4 antibodies were custom-made as described before (25), and goat polyclonal EHD4 was purchased from Abcam (Cambridge, MA). Anti-digoxigenin-AP Fab fragments antibody was purchased from Roche Applied Science (Indianapolis, IN). Secondary antibodies include goat anti-mouse IgG-AlexaFluor 546 and goat anti-rabbit IgG-AlexaFluor 488 (Molecular Probes, Eugene, OR); protein A-HRP (Zymed Laboratories Inc., San Francisco, CA), and donkey anti-goat IgG-HRP and goat anti-mouse IgG-HRP (Jackson ImmunoResearch, West Grove, PA).
Yeast Two-hybrid Analyses
Otocdh23 DF (CDH23) was inserted into the bait expression vector pTMBV4 (DUALsystems Biotech, Switzerland) with CUB-LexA-VP16 downstream of and in-frame with CDH23. The bait vector carries the LEU2 gene for auxotrophic selection. The sequence of the CDH23-bait vector was confirmed through DNA sequencing. Expression of the CDH23-CUB-LexA-VP16 fusion protein was further verified by Western blot analysis with anti-FLAG antibody. Part of EHD4 (1408–1727 bp) was inserted into the prey-expressing vector pDL2-Nx (DUALsystems Biotech) with NubG upstream of and in-frame with EHD4. The prey vector carries the TRP1 gene for auxotrophic selection. pMBV-Alg5 is a negative control bait construct, which expresses the Cub-LexA-VP16 fusion protein in the correct orientation in the yeast membrane. The CDH23-bait construct and the negative control, pMBV-Alg5, were transformed into yeast strain NMY51 (MATa his3Δ200 trp1−901 leu2−3, 112 ade2 LYS2::(lexAop)4- HIS3 ura3::(lexAop)8-lacZ ade2::(lexAop)8-ADE2 GAL4) (DUALsystems Biotech, Switzerland) and grown on leucine selective plates (SD-L), respectively. EHD4 prey constructs were transformed into CDH23-bait-expressing yeast or pMBV-Alg5 expressing yeast and grown on leucine-tryptophan double selective plates (SD-LT). Positive interactions were identified by the ability of yeast to grow on leucine-tryptophan-histidine-alanine selective plates (SD-LTHA) in the presence of 2 mm 3-aminotriazole, and by β-galactosidase expression, indicated by the blue color observed in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal).
In Situ Hybridization
All surgical and experimental procedures were conducted in accordance with the policies of Northwestern University Animal Care and Use Committee and the National Institutes of Health. The detailed procedures for producing whole mount cochlear samples were described in Judice et al. (31). Briefly, adult wild type and EHD4-KO mice were cardiac-perfused, first with heparinized saline and then with 4% formaldehyde, followed by at least 48-h postfixation. Cochleae were dissected and decalcified in 10% EDTA for at least 24 h. A 319-bp fragment corresponding to the EHD4 cDNA (nucleotides 1408–1727 bp) was cloned into pGEM-7Z. Antisense and sense mRNAs transcribed from the T7 and SP6 promoters, respectively, were labeled with Dig-UTP using the Dig RNA labeling kit (Roche Applied Science). After purification of RNA probes with ChromaSpin-30 columns (Clontech), the Dig-labeled probe was used to hybridize cochlear tissue. Samples were mounted on glass slides and viewed with a standard microscope. Images were captured with a CCD camera.
Cell Culture and Immunofluorescence
Plasmids encoding GFP-tagged EHD4 or GFP-tagged EHD4ΔEH were transiently co-transfected with that of CDH23 or prestin in opossum kidney (OK) cells according to the protocols described in Zheng et al. (30). Approximately 46-h post-transfection, the transiently transfected cells were fixed with 1% formaldehyde in PBS for 10 min at room temperature. Cells were blocked in PBS with 5% BSA and 0.1% saponin for 1 h at room temperature, and then incubated with monoclonal anti-FLAG (1:1000), anti-V5 (1:1600), or rabbit polyclonal anti-FLAG (1:200) antibodies in PBS with 5% BSA and 0.1% saponin for 2 h at room temperature. Following brief washing, secondary antibodies, goat anti-mouse IgG-Alexa Fluor 546 (1:400) and/or goat anti-rabbit IgG-Alexa Fluor 488 (1:400) were incubated in PBS with 5% BSA, 0.1% saponin, and 10% normal goat serum for 1 h at room temperature. The samples were mounted on glass slides with Fluoromount-G (Southern Biotechnology Associates, Inc., Birmingham, AL) and observed using a Leica confocal system with a standard configuration DMRXE7 microscope.
Generating EHD4 Knock-out Mice
Details on the generation and characterization of Ehd4-deficient mice are described elsewhere.3 In brief, a conditional gene knock-out targeting construct was generated using the “recombineering” method described by Liu et al. (32). C57BL/6-derived ES cells were used for gene targeting, and the loxP-flanked first exon of Ehd4 was deleted by crossing the mice with B6.FVB-Tg (EIIa-Cre) C5379Lmgd/J mice, which express Cre recombinase in a wide range of tissues including the germ cells. All mice used in this study have been maintained on the C57BL/6J background.
Protein Expression from Native Tissue and Western Blot
Cochlea and lung from either wild type or EHD4-KO mice were collected in CelLytic mammalian tissue lysis/extraction reagent (Sigma) supplemented with a combination of inhibitors (1:100 mixture protease inhibitor (Sigma), 100 μg/ml phenylmethylsulfonyl fluoride, and 10 units/ml DNase). After homogenization, samples were centrifuged (800 × g for 10 min) to separate the nuclei, unlysed cells, and bone structures. The protein concentration was measured using the Bio-Rad Protein Assay and bovine serum albumin as a standard. Human lung epithelial cell line HBE135 lysate was used as an additional control.
For Western blots, lysates were mixed with 2× LDS Laemmli sample buffer combined with 100 mm dithiothreitol, 50 mm β-mercaptoethanol, and boiled for 5 min. Aliquots of 90 μg of protein lysate were separated by 7.5% SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with 1:2000 dilutions of primary antibodies (anti EHD1, EHD2, EHD3, EHD4, or Hsc70), followed by 1:20,000 dilution of protein A-HRP. Signals were detected using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences, Boston, MA) and Kodak X-Omat Blue XB-1 film (PerkinElmer Life Sciences).
Hearing Sensitivity Studies
Gross cochlear potentials were acquired at the round window membrane using a silver wire ball electrode in young mice (P33-P60) anesthetized with sodium pentobarbital (80 mg/kg, IP). A traditional ventro-lateral approach was used, and the headholder was heated to prevent cooling of the cochlea. Compound action potential (CAP) thresholds were obtained for a 10 μV N1/P1 criterion voltage (33). All procedures were approved by the National Institutes of Health and by Northwestern's Institutional Review Board.
Preparation of Cell Extracts and Co-immunoprecipitation
For co-immunoprecipitation of epitope-tagged proteins, HEK293T cells were transiently co-transfected with plasmids encoding FLAG-CDH23 and either with Myc-EHD4 or Myc-EHD4ΔEH using ProFection Mammalian Transfection System-Calcium Phosphate (Promega Corp., Madison, WI). Approximately 42-h post-transfection, cells were harvested and lysed in cold lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 0.1% Nonidet P-40, 1 mm EDTA, pH 7.6) supplemented with a mixture of protease inhibitor (1:100) and 100 μg/ml phenylmethylsulfonyl fluoride. Cells were lysed either in cold lysis buffer plus 1 mm EDTA or in cold lysis buffer plus 1 mm CaCl2 to assess calcium sensitivity of the co-immunoprecipitation. Insoluble material was removed by centrifugation at 10,000 × g for 15 min.
Co-IPs were carried out by rocking 1.5 mg of aliquots of protein lysate, 2 μg of anti-FLAG antibody, and protein A-Sepharose beads at 4 °C overnight. Cell lysate transfected with either Myc-EHD4 or Myc-EHD4ΔEH only was used as a negative control. Beads were washed three times with cold lysis buffer, and bound proteins were eluted in Laemmli buffer with 100 mm dithiothreitol. The proteins were resolved on a 10% NEXT gel (AMRESCO, Solon, OH) followed by immunoblotting. Myc-EHD4 was detected with polyclonal goat anti-EHD4 antibody at 1:2000 and Donkey anti-goat IgG-HRP at 1:5000, whereas Myc-EHD4ΔEH was detected by using a monoclonal anti-Myc-HRP-conjugated antibody at 1:2000. CDH23 protein was detected with monoclonal anti-FLAG antibody at 1:1000 followed by a Goat anti-mouse IgG-HRP at 1:5000. Signals were detected using Enhanced Chemiluminescent (ECL) substrate (Pierce).
RESULTS
Yeast Two-hybrid Analyses Isolate EHD4 as a Novel Interacting Partner for CDH23
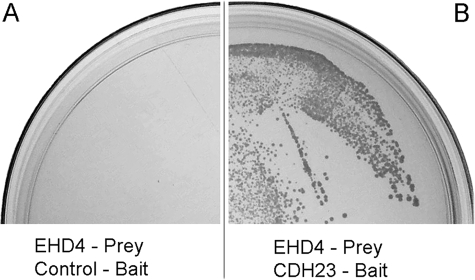
A membrane-based yeast two-hybrid screen was performed to identify potential CDH23 partners from an OHC cDNA library (13). This screening method is a highly sensitive genetic approach, suitable for identifying low abundance protein partners whether they are present in the cytoplasm and/or on the cell membrane (34). By using partial CDH23 as the bait, two independent clones encoding EHD4 were isolated (13). The sequences of these clones mapped to the EH domain of EHD4. The interaction between CDH23-bait and EHD4-prey was further tested using SD-LTHA selective plates to select yeast expression of CDH23 and EHD4, as well as interaction between them. As shown in Fig. 1, A and B, the yeast co-expressing EHD4-prey and CDH23-bait grew on SD-LTHA-selective plates (Fig. 1B) and turned blue when tested for the activation of the lacZ gene (data not shown). However, yeast expressing control-bait and EHD4-prey showed no growth (Fig. 1A) indicating that EHD4 prey could interact with CDH23 bait but not with control bait. These data suggest that EHD4 is a potential interacting partner of CDH23 in yeast.
FIGURE 1.
EHD4 interacts with CDH23 in yeast. A, when a prey plasmid encoding EHD4 cDNA was transformed into the control-bait yeast, there was no growth on SD-LTHA plates because EHD4 and the control-bait proteins failed to interact. B, when the EHD4-prey plasmid was transfected into the CDH23-bait yeast, there was a bait/prey interaction, and histidine was produced, as evidenced by the growth on SD-LTHA-selective plates.
EHD4 mRNA Is Expressed in Hair Cells
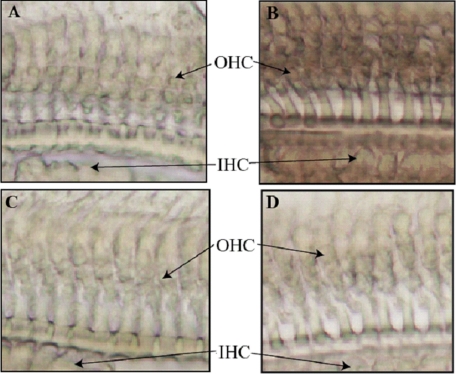
CDH23 is expressed in hair cells and Reissner's membrane of the cochlea (35, 36). To examine cochlear EHD4 mRNA distribution in hair cells, in situ hybridization was performed to detect EHD4 mRNA at the cellular level. When WT mouse cochlear tissue was stained with either antisense or sense EHD4 RNA probes (see “Experimental Procedures”), only the antisense EHD4 RNA probe showed hybridization signals in OHCs and IHCs (brown color in Fig. 2B). The sense probe, which was used as a negative control, produced no signal (Fig. 2A). As expected, no hybridization signal was observed when EHD4-KO mouse cochlear tissue was stained with either the antisense EHD4 RNA probe (Fig. 2D) or the sense EHD4 RNA probe (Fig. 2C). These data demonstrate that EHD4 mRNA is present in both populations of sensory receptor cells in WT mouse cochleae, where CDH23 is known to be expressed. The co-existence of EHD4/CDH23 mRNA in hair cells is consistent with the possibility that CDH23 and EHD4 proteins could interact in vivo.
FIGURE 2.
Expression of EHD4 mRNA in the cochlea. In situ hybridization was used to detect the expression of EHD4 mRNA in the cochlea. WT and EHD4-KO mouse cochlear tissues were stained with the sense (A for WT and C for EHD4-KO) or antisense EHD4 RNA probes (B for WT and D for EHD4-KO). Only the antisense EHD4 RNA probe stained OHCs, as well as IHCs in WT, but not in EHD4-KO mouse cochlear tissue, thereby demonstrating that EHD4 is present in both populations of sensory receptor cells.
EHD4 Co-localizes with CDH23
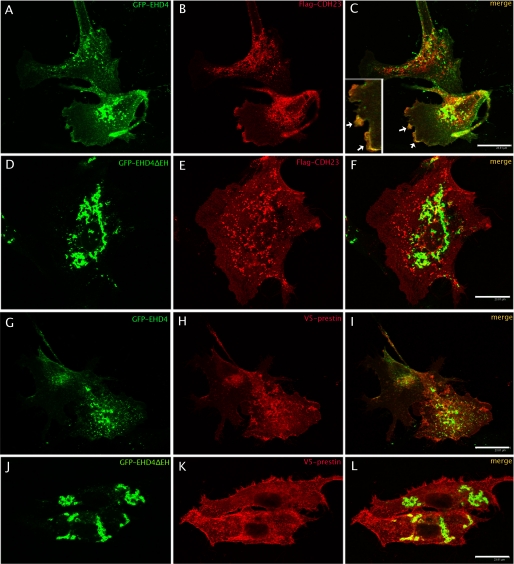
We further investigated the localization of EHD4 protein in hair cells. However, anti-EHD4 antibodies, both custom-made (25) and purchased (from Abcam), cross-react with other EHD (1–3) proteins apparently due to high homology among these proteins. Therefore, it was not feasible to selectively detect EHD4 protein expression in the cochlea. To investigate the co-localization between CDH23 and EHD4 proteins, plasmids containing GFP-EHD4 and GFP-EHD4ΔEH (EHD4 protein without EH domain) were co-transfected with a FLAG-CDH23 plasmid into opossum kidney (OK) cells. Distinct co-localization was observed between GFP-EHD4 and CDH23 (Fig. 3C) near the plasma membrane. CDH23 is a single domain transmembrane protein whereas EHD4 does not contain any transmembrane domain. However, EHD4 has been shown to associate with membranes either through a direct interaction with lipids or with lipid-embedded proteins (37, 38).
FIGURE 3.
A–F, co-localization of CDH23 and EHD4 in OK cells. OK cells were transiently co-transfected with GFP-EHD4+FLAG-CDH23 (A and B), or GFP-EHD4ΔEH+FLAG-CDH23 (D and E). After 48 h, cells were fixed and incubated with mouse anti-FLAG followed by the corresponding secondary antibody. Yellow images (right column) are superimposed from green and red images, indicating the co-localization of CDH23 and EHD4 (C) at the plasma membrane as indicated by the arrow. No co-localization was found for GFP-EHD4ΔEH and FLAG-CDH23 (F). For better visualization of the co-localization, a portion of panel C is shown in the left corner of panel C at higher magnification. G–L, no co-localization between prestin and EHD4 in OK cells. EHD4 (G–I) did not co-localize with prestin, which was used as a negative control. Bar, 23.8 μm.
The co-localization of EHD4 with CDH23 in the plasma membrane could reflect a direct or indirect interaction between these proteins. It has been shown previously that EHD proteins localize to membranes of tubules and loss of the EH domain (EHDΔEH) results in their re-localization exclusively in punctuate structures (17, 25, 29, 37). Like Sharma et al. (29), we also observed that EHD4ΔEH no longer associated with tubules but localized to large punctate structures (Fig. 3D). We could not find any co-localization between EHD4ΔEH and CDH23 (Fig. 3F), which supports the conclusion that interaction with CDH23 requires the EHD4 EH domain or the EH domain-dependent membrane localization. When prestin, a transmembrane protein (30) (used as a negative control) was co-expressed with EHD4, no co-localization was found with either EHD4 or EHDΔEH (Fig. 3, G–L). Thus, the co-localization between EHD4 and CDH23 is specific.
EHD4 Co-immunoprecipitates with CDH23, and the EH Domain of EHD4 Is Required for This Interaction
To directly assess if EHD4 and CDH23 interact, co-immunoprecipitation experiments were performed. HEK293T cells were co-transfected with plasmids encoding FLAG-CDH23 and Myc-EHD4/Myc-EHD4ΔEH. HEK293T cells transfected only with the plasmid encoding Myc-EHD4/Myc-EHD4ΔEH served as a negative control. Forty-eight hours after transfection, cells were lysed in a buffer containing 1 mm EDTA, and cell lysates were subjected to anti-FLAG antibody IP followed by anti-EHD4 Western blotting. Myc-EHD4ΔEH was detected by using a monoclonal anti-Myc-HRP-conjugated antibody, while Myc-EHD4 was detected with polyclonal goat anti-EHD4 antibody plus secondary donkey anti-goat IgG-HRP. As shown in Fig. 4A, EHD4 co-immunoprecipitated with CDH23 and in contrast, no EHD4 signal was observed in IPs of EHD4-only expressing lysate. These data confirm the interaction between EHD4 and CDH23. Interestingly, no co-IP was observed when 1 mm EDTA was omitted from the lysis buffer (data not shown) or when buffer with 1 mm CaCl2 was used (Fig. 4A). As shown in Fig. 4A, eluted proteins contain CDH23 as expected under both EDTA and CaCl2 conditions, and EHD4 is present in the flow-through, indicating that the lack of co-IP under these conditions is not due to loss of proteins. In contrast, Fig. 4B shows that EHD4ΔEH was not co-immunoprecipitated with CDH23 either in the presence of 1 mm EDTA or 1 mm CaCl2. It appears that the EH domain of EHD4 is involved in the CDH23-EHD4 interaction. Taken together, these data demonstrate that EHD4 binds to CDH23 in a calcium-sensitive manner through the EH domain.
FIGURE 4.
EHD4 directly binds to CDH23 in a Ca2+-sensitive fashion, and this binding is dependent on the EH domain of EHD4. HEK293T cell lysates from Myc-EHD4-expressing (EHD4) and Myc-EHD4+FLAG-CDH23-expressing (EHD4+CDH23)/Myc-EHD4ΔEH-expressing (EHD4ΔEH) and Myc-EHD4ΔEH+FLAG-CDH23-expressing cells were subjected to co-immunoprecipitation using 2 μg of anti-FLAG antibody and protein A-Sepharose (beads) by rotating at 4 °C for overnight. A, co-IP of Myc-EHD4 and CDH23 where EHD4 was visualized using a goat anti-EHD4 antibody. Lanes 1 and 2 show proteins from the flow-through fraction. Lanes 3 and 4 are eluates from CDH23-anti-FLAG-beads. EHD4 protein was detected by anti-EHD4 antibody and co-immunoprecipitated with CDH23 only in the no Ca2+ condition (1 mm EDTA), i.e. not in 1 mm Ca2+. The lysate containing only overexpressed Myc-EHD4 was used as a negative control. B, co-IP of Myc-EHD4ΔEH and CDH23 shows no EHD4ΔEH protein in the eluates (lane 3) from CDH23-anti-FLAG-beads when blotted with anti-Myc-HRP-conjugated antibody. The lysate containing only overexpressed Myc-EHD4ΔEH was used as a negative control (lane 4). Lanes 1 and 2 show proteins from the flow-through fraction. EHD4ΔEH protein was not co-immunoprecipitated either in the presence of 1 mm EDTA or 1 mm CaCl2 demonstrating that the EH domain of EHD4 is required for the Ca2+-sensitive CDH23-EHD4 binding.
EHD4 Knock-out Mice Have Normal Hearing Ability
To assess the potential role of EHD4 in CDH23-dependent hair cell function, CAP thresholds were measured on EHD4-KO versus wild type mice. As shown in Fig. 5, the sound pressure level necessary to obtain the 10 μV criterion voltage was similar for wild type (n = 7) and KO (n = 4) mice. The increase in thresholds below ∼5 kHz reflects the fact that mice have very poor low frequency hearing. The shift at high frequencies above ∼32 kHz is due to age-related hearing loss, which is observed in mice of the C57BL/6 strain used here. Thus, mice lacking EHD4 protein expression have normal cochlear sensitivity.
FIGURE 5.
CAP thresholds indicate that EHD4-KO mice have normal sensitivity. Recordings from the round window membrane were made in 4 KO mice (-- -- --) and compared with an average threshold curve obtained in wild type controls (———).
EHD1 May Functionally Compensate for Loss of EHD4 in the Cochlea
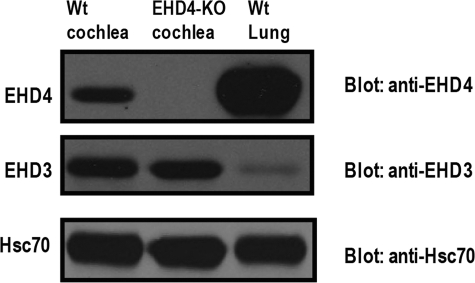
Because mammalian EHD4 has three closely related homologs (EHD1–3), it is possible that EHD4 function in EHD4-KO mice is compensated by other EHD proteins present in the cochlea. Previously, using a complementation assay in a Caenorhabditis elegans mutant lacking a functional worm EHD ortholog, Rme-1, it had been shown that all four human EHD proteins could rescue the endocytic recycling defect in the intestine of the mutant worm (25), suggesting that EHD proteins can function in a redundant manner. Also, a co-operation between EHD4 and EHD1 in terms of regulation of endocytic trafficking has been reported (29, 39). Therefore, we examined the expression of EHD1, EHD2, EHD3, and EHD4 proteins in WT and EHD4-KO cochleae using Western blot analysis to verify if other EHD protein levels are elevated in the absence of EHD4. Interestingly, the level of expression of EHD1 was elevated in EHD4-KO cochleae compared with that of WT (Fig. 6A). The graphical presentation (Fig. 6B) of the quantified EHD bands shows that EHD1 protein level in EHD4-KO cochleae is increased by nearly 2-fold compared with that in WT cochlea. These data indicate that EHD1 is up-regulated in the EHD4-KO cochlea. On the other hand, there was no considerable change in EHD2 protein levels in WT and EHD4-KO cochleae (Fig. 6A). Similar to EHD2, EHD3 expression was unchanged between WT and EHD4-KO cochleae (Fig. 7). These results are consistent with the likelihood that an increase in EHD1 levels may compensate for loss of EHD4 function, thereby leading to a normal hearing phenotype in EHD4-null mice. This possibility is in agreement with results in cellular models where functional overlap between EHD4 and EHD1 has been demonstrated (29, 39). Our result is the first report suggesting that EHD1 could functionally compensate for the lack of EHD4 with respect to a physiological function in mouse models.
FIGURE 6.
A, EHD1 possibly compensates for the loss of EHD4 in EHD4-KO cochleae. Cochlear lysates from EHD4-KO and WT mice were mixed with SDS-Laemmli sample buffer and run on 7.5% SDS-PAGE, then probed with anti-EHD4, anti-EHD1, and anti-EHD2 antibodies, respectively. EHD1 expression in EHD4-KO cochlea is higher than EHD1 expression in WT cochlea but the expression of EHD2 is similar in WT and EHD4-KO cochlea. Hsc70 was used as a loading control. The lane between EHD4-KO cochleae and WT lung is empty. B, semi-quantitative analysis of EHD proteins. Densitometric analysis of (upper image) EHD4 and EHD1 proteins and (lower image) EHD4 and EHD2 proteins in WT (n = 4) and EHD4-KO cochleae (n = 4) blotted with (upper image) anti-EHD1 antibody and (lower image) anti-EHD2 antibody, respectively. EHD1 expression in EHD4-KO cochleae is more than double the EHD1 expression in a WT cochlea. In contrast, EHD2 expression is similar in both WT and EHD4-KO cochleae.
FIGURE 7.
EHD3 protein levels do not change in EHD4-KO cochleae. Cochlear lysates from EHD4-KO and WT mice were mixed with SDS-Laemmli sample buffer and run on 7.5% SDS-PAGE, then probed with anti-EHD4, anti-EHD3, and anti-Hsc70 antibodies sequentially. EHD3 protein level is similar in EHD4-KO and WT cochleae. Hsc70 was used as a loading control. Lysates from EHD4 WT lung were used as an additional control.
DISCUSSION
CDH23, which is essential for auditory signal transduction, is expressed in cochlear hair cells and in Reissner's membrane. It has been previously shown that CDH23 interacts with protocadherin 15, harmonin, and MAGI-1 in cochlear hair cells. In this report, we identify EHD4 as an additional, novel interacting partner for CDH23. Although this interaction was found in yeast, it was verified in mammalian cells using co-localization and co-immunoprecipitation experiments.
The yeast two-hybrid screen identified two independent fragments corresponding to the EH domain of EHD4 as interaction partners for CDH23, suggesting a possible interaction between the two proteins via the EH domain. This finding was further supported by co-localization experiments where EHD4 and CDH23 were co-localized in the plasma membrane, while an EHD4 mutant lacking the EH domain (EHD4ΔEH) was not. In addition, co-immunoprecipitation experiments demonstrate that EHD4 binds to CDH23, and this interaction is abolished in the absence of the EH domain of EHD4. Therefore, we conclude that the EH domain of EHD4 is responsible for the interaction between CDH23 and EHD4.
Previous studies have identified at least 20 direct or indirect binding partners for EHD proteins. EH domains of EHD proteins bind to sequences with a core NPF motif in target proteins. This interaction is facilitated by oligomerization through the coiled-coil region of EHD proteins (21, 40). Interestingly, our results show strong binding (as shown by both yeast two-hybrid and mammalian cell co-IP analyses) between EHD4 and CDH23, which disappears when the EH domain of EHD4 is deleted, suggesting that the binding is unlikely to be mediated by other domains in EHD4. Yet, no apparent NPF motif is present in CDH23. These results suggest the likelihood that the EHD4 EH domain binds to CDH23 through a non-NPF motif. The alternative possibility is that an unknown NPF-containing adaptor protein links the EH domain of EHD4 with CDH23; this is unlikely given their interaction in the yeast two-hybrid system. Harmonin and MAGI-1, two PDZ domain-containing proteins, are reported to bind to two PBIs in the cytoplasmic region of CDH23 (41). Besides these PBIs, no other binding motifs in the cytoplasmic region of CDH23 are known. Here we show for the first time that EHD4, which is not a PDZ domain-containing protein, is able to bind to CDH23. It is possible that inside the cell, interacting partners for CDH23 are not simply restricted to PDZ domain-containing proteins. Hence, our data suggest the presence of novel binding motifs in the cytoplasmic domain of CDH23 as well as the potential recognition of a non-NPF motif by the EH domain of EHD4.
The calcium-sensitive nature of the EHD4-CDH23 interaction was unexpected. At present, the mechanisms of this interaction and its functional significance are speculative. Calcium is known to play several important roles in cochlear hair cell functions, ranging from triggering neurotransmitter release to mechanical signal amplification (for review, see Refs. 42–44). To perform these functions, calcium concentration inside hair cells is tightly regulated using a number of Ca2+-binding/regulator proteins (45). As calcium has also been known to regulate protein trafficking in hair cells (46–48), it is conceivable that the transport of CDH23 in hair cells is regulated by calcium through its modulation of binding between CDH23 and the endocytic trafficking regulatory protein, EHD4.
As mutations in the human CDH23 gene cause Usher syndrome, which is associated with hearing loss, the interaction between EHD4 and CDH23 suggested that hearing may be impaired in EHD4-KO mice if EHD4 plays an important role in the trafficking/localization or function of CDH23. However, electrophysiological measurements in EHD4-KO mice did not reveal any loss of hearing when compared with WT mice. Notably, the level of the closely related family member EHD1 in EHD4-KO mice cochlea was nearly twice as much as in WT controls. This result suggests the likelihood of a functional compensation by up-regulation of EHD1 with respect to CDH23 trafficking/localization as well as cochlear function. This possibility is supported by previous results where EHD4 has been shown to be capable of interacting with EHD1 (25), and co-operating with EHD1 in endocytic trafficking (29). EHD1 KO mice with a truncation of the EHD1 protein have been reported and we have now established a complete EHD1 knock-out mouse.4 Analyses of these mice as well as EHD1/4 double-KO mice should help assess if indeed EHD1 and 4 function in a redundant manner downstream of CDH23 in the cochlea.
In conclusion, our data demonstrate that EHD4 is a novel interacting partner for CDH23 and show that the EHD4-CDH23 interaction represents a novel mode of EH domain binding to a non-NPF motif on an EHD partner. Further studies of the EHD-CDH23 interaction and analyses of EHD KO mice should help determine the biochemical basis and the physiological significance of this novel interaction.
Acknowledgments
We thank Dr. Jaime Garcia-Anoveros, Dr. Lili Zheng, as well as Dr. James R. Bartles of Northwestern University for providing the CDH23 cDNA plasmid. We also thank Dr. W. Russin at the Biological Imaging Facility of Northwestern University for his help in image processing.
This work was supported, in whole or in part, by National Institutes of Health Grants DC00089 (to P. D.), DC006412 (to J. Z.), CA87986, CA105489, CA99900, CA116552, and CA99163 (to H. B.) and The Hugh Knowles Center Leadership Fund (to J. Z.).
M. George, M. Naramura, M. A. Rainey, G. Ying, L. Doglio, M. H. Vitaterna, S. E. Crawford, V. Band, and H. Band, submitted for publication.
M. A. Rainey, M. George, M. Naramura, G. Ying, R. Akakura, L. Doglio, S. E. Crawford, R. A. Hess, V. Band, and H. Band, submitted for publication.
- MET
- mechano-electric transducer
- OHC
- outer hair cell
- IHC
- inner hair cell
- CDH23
- cadherin 23
- PBI
- PDZ-binding interface
- EH
- Eps 15 homology
- KO
- knock-out
- WT
- wild type
- EHD
- EH domain-containing protein
- USH1D
- Usher syndrome type 1D factor
- EE
- early endosome
- ERC
- endocytic recycling compartment
- GFP
- green fluorescence protein
- HRP
- horseradish peroxidase
- EHD4ΔEH
- EHD4 protein without the EH domain
- OK
- opossum kidney
- HEK
- human embryonic kidney
- HSC70
- heat shock cognate protein 70
- Co-IP
- co-immunoprecipitation
- CAP
- compound action potential
- Nub
- the N-terminal domain of ubiquitin
- NubG
- the mutant form of Nub
- Cub
- the C-terminal domain of ubiquitin
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Siemens J., Lillo C., Dumont R. A., Reynolds A., Williams D. S., Gillespie P. G., Müller U. (2004) Nature. 428, 950–955 [DOI] [PubMed] [Google Scholar]
- 2.Söllner C., Rauch G. J., Siemens J., Geisler R., Schuster S. C., Müller U., Nicolson T. (2004) Nature. 428, 955–959 [DOI] [PubMed] [Google Scholar]
- 3.Pickles J. O., Comis S. D., Osborne M. P. (1984) Hear Res. 15, 103–112 [DOI] [PubMed] [Google Scholar]
- 4.Howard J., Hudspeth A. J. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 3064–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Amraoui A., Petit C. (2005) J. Cell Sci. 118, 4593–4603 [DOI] [PubMed] [Google Scholar]
- 6.Kazmierczak P., Sakaguchi H., Tokita J., Wilson-Kubalek E. M., Milligan R. A., Müller U., Kachar B. (2007) Nature. 449, 87–91 [DOI] [PubMed] [Google Scholar]
- 7.Boëda B., El-Amraoui A., Bahloul A., Goodyear R., Daviet L., Blanchard S., Perfettini I., Fath K. R., Shorte S., Reiners J., Houdusse A., Legrain P., Wolfrum U., Richardson G., Petit C., Weil D. (2002) EMBO J. 21, 6689–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z., Peng A. W., Oshima K., Heller S. (2008) J. Neurosci. 28, 11269–11276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adato A., Michel V., Kikkawa Y., Reiners J., Alagramam K. N., Weil D., Yonekawa H., Wolfrum U., El-Amraoui A., Petit C. (2005) Hum. Mol. Genet. 14, 347–356 [DOI] [PubMed] [Google Scholar]
- 10.Frolenkov G. I., Belyantseva I. A., Friedman T. B., Griffith A. J. (2004) Nat. Rev. Genet. 5, 489–498 [DOI] [PubMed] [Google Scholar]
- 11.Michel V., Goodyear R. J., Weil D., Marcotti W., Perfettini I., Wolfrum U., Kros C. J., Richardson G. P., Petit C. (2005) Dev. Biol. 280, 281–294 [DOI] [PubMed] [Google Scholar]
- 12.Petit C. (2006) Trends Mol. Med. 12, 57–64 [DOI] [PubMed] [Google Scholar]
- 13.Zheng J., Anderson C. T., Miller K. K., Cheatham M., Dallos P. (2009) BMC Genomics. 10, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D. W., Zhao X., Scarselletta S., Schweinsberg P. J., Eisenberg E., Grant B. D., Greene L. E. (2005) J. Biol. Chem. 280, 17213–17220 [DOI] [PubMed] [Google Scholar]
- 15.Mintz L., Galperin E., Pasmanik-Chor M., Tulzinsky S., Bromberg Y., Kozak C. A., Joyner A., Fein A., Horowitz M. (1999) Genomics. 59, 66–76 [DOI] [PubMed] [Google Scholar]
- 16.Pohl U., Smith J. S., Tachibana I., Ueki K., Lee H. K., Ramaswamy S., Wu Q., Mohrenweiser H. W., Jenkins R. B., Louis D. N. (2000) Genomics. 63, 255–262 [DOI] [PubMed] [Google Scholar]
- 17.Caplan S., Naslavsky N., Hartnell L. M., Lodge R., Polishchuk R. S., Donaldson J. G., Bonifacino J. S. (2002) EMBO J. 21, 2557–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin S. X., Grant B., Hirsh D., Maxfield F. R. (2001) Nat. Cell Biol. 3, 567–572 [DOI] [PubMed] [Google Scholar]
- 19.Naslavsky N., Rahajeng J., Rapaport D., Horowitz M., Caplan S. (2007) Biochem. Biophys. Res. Commun. 357, 792–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gokool S., Tattersall D., Seaman M. N. (2007) Traffic. 8, 1873–1886 [DOI] [PubMed] [Google Scholar]
- 21.Naslavsky N., Rahajeng J., Sharma M., Jovic M., Caplan S. (2006) Mol. Biol. Cell. 17, 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naslavsky N., McKenzie J., Altan-Bonnet N., Sheff D., Caplan S. (2009) J. Cell Sci. 122, 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guilherme A., Soriano N. A., Bose S., Holik J., Bose A., Pomerleau D. P., Furcinitti P., Leszyk J., Corvera S., Czech M. P. (2004) J. Biol. Chem. 279, 10593–10605 [DOI] [PubMed] [Google Scholar]
- 24.Park S. Y., Ha B. G., Choi G. H., Ryu J., Kim B., Jung C. Y., Lee W. (2004) Biochemistry. 43, 7552–7562 [DOI] [PubMed] [Google Scholar]
- 25.George M., Ying G., Rainey M. A., Solomon A., Parikh P. T., Gao Q., Band V., Band H. (2007) BMC Cell Biol. 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo H. J., Tran N. T., Clary S. A., Morris N. P., Glanville R. W. (2001) J. Biol. Chem. 276, 43103–43110 [DOI] [PubMed] [Google Scholar]
- 27.Shao Y., Akmentin W., Toledo-Aral J. J., Rosenbaum J., Valdez G., Cabot J. B., Hilbush B. S., Halegoua S. (2002) J. Cell Biol. 157, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith C. A., Dho S. E., Donaldson J., Tepass U., McGlade C. J. (2004) Mol. Biol. Cell. 15, 3698–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma M., Naslavsky N., Caplan S. (2008) Traffic. 9, 995–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng J., Long K. B., Shen W., Madison L. D., Dallos P. (2001) Neuroreport. 12, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 31.Judice T. N., Nelson N. C., Beisel C. L., Delimont D. C., Fritzsch B., Beisel K. W. (2002) Brain Res. Brain Res. Protoc. 9, 65–76 [DOI] [PubMed] [Google Scholar]
- 32.Liu P., Jenkins N. A., Copeland N. G. (2003) Genome. Res. 13, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheatham M. A., Huynh K. H., Gao J., Zuo J., Dallos P. (2004) J. Physiol. 560, 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stagljar I., Fields S. (2002) Trends Biochem. Sci. 27, 559–563 [DOI] [PubMed] [Google Scholar]
- 35.Wilson S. M., Householder D. B., Coppola V., Tessarollo L., Fritzsch B., Lee E. C., Goss D., Carlson G. A., Copeland N. G., Jenkins N. A. (2001) Genomics. 74, 228–233 [DOI] [PubMed] [Google Scholar]
- 36.Lagziel A., Ahmed Z. M., Schultz J. M., Morell R. J., Belyantseva I. A., Friedman T. B. (2005) Dev. Biol. 280, 295–306 [DOI] [PubMed] [Google Scholar]
- 37.Blume J. J., Halbach A., Behrendt D., Paulsson M., Plomann M. (2007) Exp. Cell Res. 313, 219–231 [DOI] [PubMed] [Google Scholar]
- 38.Naslavsky N., Rahajeng J., Chenavas S., Sorgen P. L., Caplan S. (2007) J. Biol. Chem. 282, 16612–16622 [DOI] [PubMed] [Google Scholar]
- 39.Grant B. D., Caplan S. (2008) Traffic 9, 2043–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rotem-Yehudar R., Galperin E., Horowitz M. (2001) J. Biol. Chem. 276, 33054–33060 [DOI] [PubMed] [Google Scholar]
- 41.Siemens J., Kazmierczak P., Reynolds A., Sticker M., Littlewood-Evans A., Müller U. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14946–14951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fettiplace R., Ricci A. J. (2003) Curr. Opin. Neurobiol. 13, 446–451 [DOI] [PubMed] [Google Scholar]
- 43.LeMasurier M., Gillespie P. G. (2005) Neuron. 48, 403–415 [DOI] [PubMed] [Google Scholar]
- 44.Vollrath M. A., Kwan K. Y., Corey D. P. (2007) Annu. Rev. Neurosci. 30, 339–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hackney C. M., Mahendrasingam S., Penn A., Fettiplace R. (2005) J. Neurosci. 25, 7867–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beurg M., Safieddine S., Roux I., Bouleau Y., Petit C., Dulon D. (2008) J. Neurosci. 28, 1798–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beutner D., Voets T., Neher E., Moser T. (2001) Neuron. 29, 681–690 [DOI] [PubMed] [Google Scholar]
- 48.Seiler C., Nicolson T. (1999) J. Neurobiol. 41, 424–434 [PubMed] [Google Scholar]