Abstract
Differentiation of erythroid cells is regulated by cell signaling pathways including those that change the intracellular concentration of calcium. Calcium-dependent proteases have been shown previously to process and regulate the activity of specific transcription factors. We show here that the protein levels of upstream stimulatory factor (USF) increase during differentiation of murine erythroleukemia (MEL) cells. USF was subject to degradation by the Ca2+-dependent protease m-calpain in undifferentiated but not in differentiated MEL cells. Treatment of MEL cells with the specific calpain inhibitor calpeptin increased the levels of USF and strongly induced expression of the adult α- and β-globin genes. The induction of globin gene expression was associated with an increase in the association of USF and RNA po ly mer ase II with regulatory elements of the β-globin gene locus. Calpeptin also induced high level α- and β-globin gene expression in primary CD71-positive erythroid progenitor cells. The combined data suggest that inhibition of calpain activity is required for erythroid differentiation-associated increase in globin gene expression.
Calcium plays a vital role in the physiology and biochemistry of organisms and cells, and Ca2+ ions are one of the most widespread second messengers used in signal transduction pathways (1). The most prominent signal transduction pathway regulating differentiation of erythroid cells is represented by erythropoietin-induced activation of Janus kinase 2 (2). Janus kinase 2 initiates many different pathways within the cell including activation of processes mediated by phosphatidylinositol 3-kinase and phospholipase C. Phospholipase C catalyzes the generation of inositol 1,4,5-trisphosphate, which triggers intracellular calcium release (3). Furthermore phospholipase C-γ promotes calcium entry into the cells through stimulating the cell surface expression of transient receptor potential channels (TRPCs)3 such as TRPC3 (4).
Treatment of murine erythroleukemia (MEL) cells with dimethyl sulfoxide (DMSO), hexamethylene, bisacetamide, x-irradiation, or hypoxanthine results in expression of erythroid cell-specific genes and loss of cellular immortality (5). However, the mechanism(s) by which these reagents induce erythroid differentiation is (are) not known. Following exposure to inducer, a latent period of 8–12 h occurs before the cells begin to differentiate. Changes in cytosolic calcium concentration have been suggested to play a role in inducing these early changes (6). In MEL cells induced with DMSO, EGTA (a calcium chelator) blocks the commitment to differentiate, and addition of excess calcium results in the reverse of this block (6). Additionally the calcium ionophore A23187, which increases the permeability of membranes with high selectivity for calcium ions, abolishes the latent period during DMSO induction and promotes differentiation (7). However, despite these colonies being hemoglobinized, the cells do not express elevated levels of β-globin or Band3, markers for differentiated erythroid cells. Somewhat contradictory to these studies, Faletto and Macara (8) have demonstrated that DMSO decreases cellular calcium levels in MEL cells. In addition, intracellular calcium concentrations were measured in precursor erythroid cells at various stages (proerythroblast, basophil erythroblast, and normoblast erythroblast) as well as in red blood cells (9). Calcium concentration was shown to increase in cells from 0 to 24 h and then begin to decrease at 48 h until it reaches the lowest concentration in red blood cells (9).
Members of the calpain family, a heterogeneous group of cysteine proteases, are involved in a variety of calcium-regulated processes, such as signal transduction, cell proliferation and differentiation, apoptosis, membrane fusion, and platelet activation (10, 11). The proteolytic domain of calpains is configured to form an active catalytic pocket only in the presence of calcium, which is bound by the EF-hand domain. Previous studies have shown that the transcription factor USF, which has an important function during cellular differentiation, is proteolytically processed by calpain in vitro (12). USF has been shown previously to regulate gene expression in erythroid cells. For example, USF is required for the efficient recruitment of transcription complexes to the β-globin gene locus (13) where it interacts with E-box motifs (CANNTG) present in locus control region (LCR) element HS2 and in the adult β-globin gene promoter (14–16).
We demonstrate here that USF is subject to calpain-mediated proteolytic processing in undifferentiated but not differentiated erythroid cells. Treatment of DMSO-induced MEL cells with calcium ionophore led to proteolytic processing of USF and a decrease in β-globin gene expression. We further show that treatment of MEL cells with the specific calpain inhibitor calpeptin increased the level of full-length USF and induced high level globin gene expression. Calpeptin also increased globin gene expression in K562 cells as well as in primary c-Kit- and CD71-positive erythroid progenitor cells.
EXPERIMENTAL PROCEDURES
Cell Culture
K562 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. MEL cells were grown in Dulbecco's modified Eagle's medium containing 10% FBS and 1% penicillin-streptomycin. Cells were grown in 5% CO2 at 37 °C and maintained at a density between 1 × 105 and 2 × 106 cells/ml. In DMSO induction studies, MEL cells were incubated with 2% DMSO for 48 h. Calcium ionophore treatment of DMSO-induced or uninduced MEL cells was performed by incubating MEL cells with 0.3 or 0.9 μg/ml A23187 (Sigma) as described previously (7). To inhibit m-calpain, K562 and MEL cells were treated with 30 or 40 μm calpeptin (Calbiochem) for up to 7 days. In control experiments MEL cells were incubated with 0.03 or 0.04% DMSO. The starting cell density was 5 × 105 cells/ml before adding the drug. Early erythroblast c-Kit+/CD71+ cells were obtained from the bone marrow of 6-week-old C57BL/6J female mice (The Jackson Laboratory, Bar Harbor, ME). The bone marrow was obtained by flushing femurs with phosphate-buffered saline (PBS) supplemented with neonatal calf serum. Cells were centrifuged at 1100 rpm for 5 min at 4 °C. Pellets were resuspended by tapping, and red blood cells were lysed with 0.5 ml/mouse ACK buffer (0.15 m NH4Cl, 10 mm KHCO3, 0.1 mm Na2EDTA, pH 7.35) at room temperature for 5 min. 13.5 ml of PBS was added, and cells were centrifuged at 1100 rpm for 5 min at 4 °C and resuspended in 200 μl of PBS containing 10% FBS. Phycoerythrin-conjugated rat anti-mouse c-Kit (catalog number 553355) and fluorescein isothiocyanate-conjugated rat anti-mouse CD71 (catalog number 553266) antibodies (both from Pharmingen) were added at a concentration of 0.8 μg/million cells. Cells were incubated on ice for 20 min, washed, and resuspended in PBS containing 10% FBS. Fluorescence-activated cell sorting was performed, and c-Kit+/CD71+ early erythroblasts were harvested and cultured in complete Iscove's modified Dulbecco's medium containing 10% FBS, 100 units penicillin/streptomycin, 2 mm glutamine, 20 ng/ml recombinant murine interleukin-3 and interleukin-6, and 56 ng/ml recombinant murine stem cell factors for 2 days before being treated with drug. Interleukin-3, interleukin-6 (catalog number 216-16), and stem cell factors (catalog number 250-03) were purchased from PeproTech, Inc. (Rocky Hill, NJ). Cells were then incubated with 30 μm calpeptin in 0.03% DMSO or with 0.03% DMSO for 4 days with a starting density of 2.5 × 105 cells/ml.
RNA Isolation, Reverse Transcription, and Real Time PCR
RNA was isolated using the guanidine thiocyanate method as described previously (13, 17). Reverse transcription was carried out using the iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR was performed using MyiQ (Bio-Rad), and reactions were carried out using iQ SYBR Green SuperMix (Bio-Rad). Real time conditions were as described previously (13, 18). Primers for amplifying the murine βmajor (βmaj)- and human ϵ- and β-globin genes as well as that of human β-actin have been described previously (13, 18, 19). In addition, the following primer sequences were used: murine USF1: forward, 5′-GATGAGAAACGCAGGGCTCAGCATA-3′; reverse, 5′-TTAGTTGCTGTCATTCTTGATGACG-3′; α-globin: forward, 5′- CCTGGGGGAAGATTGGTG-3′; reverse, 5′GCCGTGGCTTACATCAAAGT-3′; and murine β-actin: forward, 5′-GTGGGCCGCTCTAGGCACCA-3′; reverse, 5′-TGGCCTTAGGGTGCAGGGGG-3′.
Chromatin Immunoprecipitation (ChIP)
For K562 cells ChIP assays were performed as described previously (16, 19). The following antibodies were used in this study: USF1 (H-86, sc-8983) and USF2 (N-18, sc-861) (both purchased from Santa Cruz Biotechnology) and RNA polymerase II (Pol II) (CTD45H8) (purchased from Upstate Biotechnology, Inc.).
For MEL cell lines, ChIP assays were performed as described by Dahl and Collas (20) with some minor modifications. Antibody-bead complexes were prepared as described except that the beads were diluted only 5-fold from the original stock in antibody-bead solution in 0.5-ml tubes. Antibodies used were RNA Pol II (N-20, sc-899), USF2 (C-20, sc-862), and normal rabbit IgG (sc-2027) (all purchased from Santa Cruz Biotechnology). 1 × 107 cells were collected and resuspended at 1 × 106 cells/ml in PBS. Cells were cross-linked with 1% (v/v) formaldehyde in PBS for 10 min at room temperature. After quenching the cross-linking reaction with 0.125 m glycine, cells were washed twice with ice-cold PBS, resuspended in 165 μl of lysis buffer (50 mm Tris-HCl, pH 8, 10 mm EDTA, 1% SDS, protease inhibitor mixture, 1 mm phenylmethylsulfonyl fluoride), and incubated on ice for 8 min. Cells were then sonicated using power 2 setting for 4 × 10 s with 1-min pause on ice water (Fisher Scientific Model 100 sonic dismembrator) to produce 200–500-bp chromatin fragments. 20 μl of the sonicated material was taken for fragment size examination, and the rest was centrifuged at 13,000 rpm for 10 min at 4 °C to remove cell debris. 150 μl of supernatant was transferred to a new tube and then diluted 10-fold in RIPA buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.5 mm EGTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 140 mm NaCl) with protease inhibitor mixture and 1 mm phenylmethylsulfonyl fluoride. Aliquots of 100 μl of diluted chromatin were transferred to the tube containing antibody-bead complexes after removing the original solution in the tube, and one aliquot was saved as input. Antibody-bead complexes in chromatin suspension were rotated at 4 °C overnight and then washed as described above. At the last wash in TE buffer (10 mm Tris-HCl, pH 8.0, 10 mm EDTA), the beads were transferred to a 1.5-ml tube, and the TE buffer was removed. Immunoprecipitated chromatin was eluted, reverse cross-linked, proteinase K-treated as described previously (16), and incubated with 100 μl of elution buffer (20 mm Tris-HCl, pH 7.5, 5 mm EDTA, 50 mm NaCl) containing 1% SDS, 50 μg/ml proteinase K, and 50 μg/ml RNase. The corresponding eluates were combined, and the DNA was purified using a Qiagen Miniprep kit and eluted in 100 μl of TE buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA). Real time PCR was performed using MyiQ (Bio-Rad) and iQ SYBR Green SuperMix (Bio-Rad) with 3 μl of DNA template and 6 μm forward and reverse primer mixture in a total volume of 20 μl. Human HS2 primers and human and mouse β-globin promoter primers used in this experiment have been described previously (13, 19).
Protein Isolation and Western Blotting
Proteins were isolated and analyzed by Western blotting as described by Leach et al. (16). A total of 20 μg of proteins from 2 × 106 cells were loaded onto 12% Ready Gels (Bio-Rad). After transfer proteins were detected using the ECL Plus system (Amersham Biosciences). The following antibodies were used: USF2 (c-20, sc-862), NF-E2/p45 (c-19, sc-291), USF1 (sc-229), or glyceraldehyde-3-phosphate dehydrogenase (FL-335, sc-25778) (all purchased from Santa Cruz Biotechnology).
Benzidine Staining
Cells (1 × 106) were centrifuged and resuspended in 500 μl of PBS. Benzidine working solution was prepared as described by Song et al. (21), and 100 μl was added to the cell suspension. Cells were then incubated at room temperature for 3 min, pelleted by centrifugation, resuspended in 500 μl of PBS, and counted under the microscope using a hemacytometer. The cell numbers were counted on four squares. Blue cells were calculated as percentage of total cells in each square. For taking photographs, cells were resuspended in ∼30 μl of PBS and loaded onto a glass slide.
RESULTS
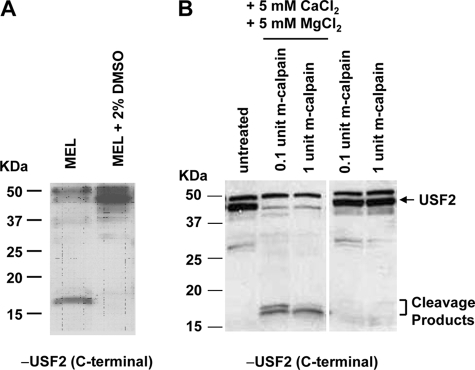
The transcription factor USF regulates many genes during the process of cellular differentiation (22). We previously demonstrated that USF is required for increased transcription of the adult β-globin gene during differentiation of MEL cells (13). During the course of these experiments we noticed that the levels of full-length USF increase during differentiation of MEL cells. In Western blotting experiments, antibodies specific for USF2 detected short protein fragments (between 15 and 20 kDa) in extracts from undifferentiated MEL cells but not in those obtained from differentiated cells (Fig. 1A). Previous studies have shown that USF is a substrate for the calcium-dependent protease m-calpain (12). Treatment of protein extracts from differentiated MEL cells with 0.1 or 1 unit of recombinant m-calpain in the presence but not in the absence of 5 mm CaCl2 and 5 mm MgCl2 led to the generation of USF2-specific cleavage products that migrate in the range between 15 and 20 kDa (Fig. 1B). We obtained similar results for USF1 (data not shown). The results suggest that USF is subject to proteolytic processing in undifferentiated erythroid cells and protected from proteolytic cleavage in differentiated cells.
FIGURE 1.
USF is subject to proteolytic cleavage by the calcium-sensitive protease m-calpain during erythroid differentiation. A, Western blot analysis of USF2 in uninduced MEL and DMSO (2%)-induced MEL cells. Whole cell protein extracts were isolated from the cells and subjected to Western blot analysis using antibodies specific for USF2. B, Western blot analysis of protein extracts from DMSO (2%)-induced MEL cells treated with recombinant m-calpain in the presence or absence of CaCl2 and MgCl2. MEL cells were incubated for 2 days in 2% DMSO to induce differentiation. Whole cell protein extracts from differentiated MEL cells were incubated in the absence or presence of different amounts of m-calpain as indicated as well as in the absence and presence of 5 mm CaCl2 and MgCl2. The protein extracts were subjected to Western blot analysis using a USF2-specific antibody.
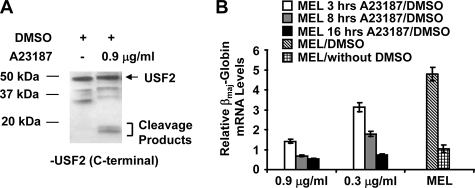
Previous studies have shown that treatment of MEL cells with calcium ionophores induced differentiation but inhibited β-globin gene expression (7). Treatment of DMSO-induced MEL cells with the calcium-specific ionophore A23187 led to the appearance of USF2-specific proteolytic fragments in the molecular mass range of 15–20 kDa (Fig. 2A), which is associated with a decline in β-globin mRNA levels in a dose- and time-dependent manner (Fig. 2B). Interestingly the treatment of differentiated MEL cells with the calcium ionophore decreased βmaj-globin gene expression to levels comparable to those detected in undifferentiated MEL cells.
FIGURE 2.
Decrease in β-globin gene expression in differentiated MEL cells exposed to calcium ionophore. A, treatment of DMSO-induced MEL cells with the calcium ionophore A23187 leads to proteolytic cleavage of USF. DMSO-treated MEL cells were incubated in the presence or absence of 0.9 μg/ml A23187 and subjected to Western blot analysis using a USF2-specific antibody. B, treatment of DMSO-induced MEL cells with the calcium ionophore A23187 reduces βmaj-globin gene expression. MEL cells were grown in the absence (MEL/without DMSO) or in the presence (MEL/DMSO) of 2% DMSO for 48 h. MEL cells grown with DMSO were incubated with 0.3 or 0.9 μg/ml A23187 for 3, 8, or 16 h as indicated. RNA was isolated from these cells, reverse transcribed, and subjected to quantitative PCR using primers specific for the βmaj-globin gene. Error bars reflect S.D. from two independent experiments.
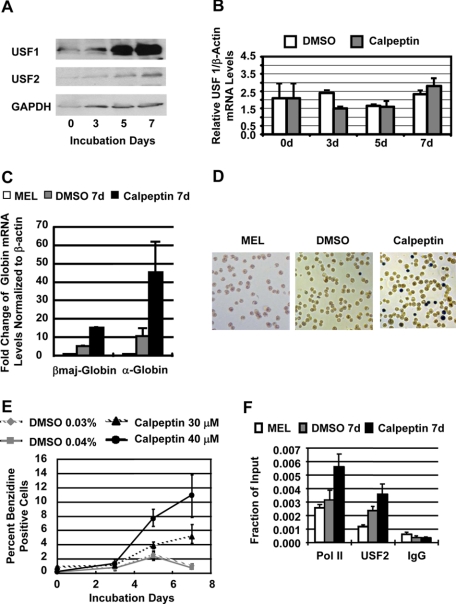
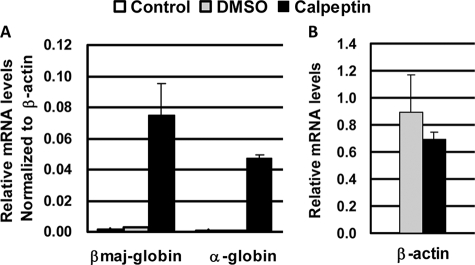
We next examined whether the specific inhibition of calpain is sufficient to induce globin gene expression in MEL cells. Treatment of uninduced MEL cells with the calpain-specific inhibitor calpeptin increased the protein levels of USF1 and USF2 (Fig. 3A). The calpeptin-induced increase in USF levels was not due to up-regulation of transcription of USF1 (Fig. 3B) or USF2 (data not shown) because the mRNA levels remained the same between untreated and treated MEL cells. The increase in the protein levels of USF was accompanied by a strong and significant increase in α- and β-globin gene expression (Fig. 3C) consistent with previous observations demonstrating that USF is required for high level β-globin gene expression in MEL cells (13). There was a more than 10-fold increase in βmaj-globin gene expression after treatment of MEL cells with 40 μm calpeptin for 7 days. We also observed an increase in expression of the α-globin gene (Fig. 3C), which was up-regulated by more than 40-fold. To confirm these results we incubated untreated and calpeptin-treated MEL cells with benzidine, which stains hemoglobin and is commonly used as an indicator of erythroid differentiation. Calpeptin treatment led to a significant increase in the number of benzidine-positive cells (Fig. 3, D and E). More than 10% of the cells stained positive for benzidine after 7-day treatment with 40 μm calpeptin. The increase in the number of hemoglobinized cells occurred in a dose- and time-dependent manner.
FIGURE 3.
Calpeptin increases the levels of USF and stimulates high level globin gene expression in MEL cells. A, Western blot analysis of USF1, USF2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in MEL cells incubated with 30 μm calpeptin for 0, 3, 5, or 7 days. B, reverse transcription-PCR analysis of USF1 mRNA levels in MEL cells treated with 0.03% DMSO only or with 30 μm calpeptin, 0.03% DMSO for 0, 3, 5, and 7 days. C, reverse transcription-PCR analysis of adult βmaj-globin and α-globin mRNA levels in MEL cells incubated for 7 days with 0.04% DMSO (DMSO 7d) or with 40 μm calpeptin, 0.04% DMSO (calpeptin 7d). RNA was isolated from the cells, reverse transcribed, and analyzed by real time PCR. Globin mRNA levels in untreated control cells (MEL) were set at 1, and -fold changes were calculated with mRNA levels of the β-actin gene serving as an internal control. D, benzidine staining of hemoglobin in MEL cells grown for 7 days in the absence (MEL), in the presence of 0.04% DMSO (DMSO), or in the presence of 40 μm calpeptin, 0.04% DMSO (Calpeptin). E, relative number of benzidine-positive MEL cells in cultures incubated with DMSO only (0.03 or 0.04%) or incubated with 30 μm calpeptin, 0.03%DMSO or 40 μm calpeptin, 0.04% DMSO for 0, 3, 5, and 7 days. F, ChIP analysis of the association of USF2 and Pol II with the βmaj-globin gene promoter in MEL cells incubated for 7 days in the absence (MEL), in the presence of 0.04% DMSO (DMSO 7d), or in the presence of 40 μm calpeptin, 0.04% DMSO (calpeptin 7d). Cells were cross-linked with formaldehyde. Chromatin was isolated, fragmented, and precipitated with antibodies against USF2 and Pol II or the unspecific IgG antibody. DNA was purified from the precipitate and analyzed by quantitative real time PCR using primers specific for the adult βmaj-globin gene promoter. Bars represent the relative enrichment over the input DNA. Error bars represent S.D. from two independent experiments. d, days.
The increase in globin gene expression was associated with an increase in the binding of USF2 and Pol II to the adult βmaj-globin gene (Fig. 3F). Treatment of MEL cells with 40 μm calpeptin significantly increased the association of both proteins with the promoter after 7 days.
The solvent for calpeptin is DMSO, and the final concentration in experiments with 30 and 40 μm calpeptin was 0.03 and 0.04%, respectively. Because 2% DMSO is an inducer of MEL cell differentiation and globin gene expression, we included control experiments in which MEL cells were incubated with DMSO only. The low concentration of DMSO alone (0.03 or 0.04%) did not significantly increase the number of benzidine-positive cells (Fig. 3, D and E) nor did it lead to an increase in globin gene expression comparable to that seen in cells treated with calpeptin (Fig. 3C). The low DMSO concentration also failed to increase the level of USF in MEL cells (data not shown).
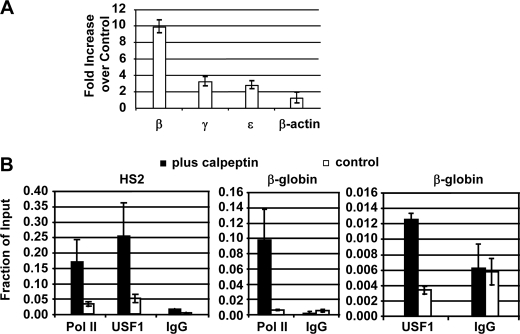
To exclude the possibility that calpeptin-induced changes in globin gene expression are an artifact associated with the MEL cell system, we repeated the experiments using human K562 cells, an erythroid cell line expressing the embryonic ϵ-globin and the fetal γ-globin genes but not the adult β-globin gene (19, 21). We treated K562 cells with the m-calpain inhibitor calpeptin and observed an 8–10-fold increase in adult β-globin gene expression in these cells (Fig. 4A). Expression of the ϵ- and γ-globin genes was increased about 2-fold, whereas expression of the control β-actin gene was not affected by the treatment. The increase in expression of the embryonic and fetal globin genes could be attributed to increased binding of USF to LCR element HS2. Indeed cells treated with the calpain inhibitor revealed an increase in RNA Pol II and USF1 loading not only to the β-globin gene promoter but also to LCR element HS2 (Fig. 4B). A similar increase in RNA Pol II and USF binding was not observed in the control β-actin gene (data not shown). These results further support the notion that USF activity is limited for the activation of the adult β-globin gene in embryonic and undifferentiated erythroid cells and that this protein plays an important role in the activation of β-globin gene expression during development and differentiation.
FIGURE 4.
Inhibition of calpain by calpeptin in K562 cells increases β-globin gene expression and Pol II interactions with the β-globin promoter and LCR core element HS2. A, relative increase in β-globin mRNA levels following treatment of K562 cells with 40 μm calpain inhibitor calpeptin for 7 days. RNA was isolated, reverse transcribed, and subjected to quantitative PCR analysis using primers specific for the human β-, γ-, and ϵ-globin genes as well as the β-actin gene. The mRNA levels are shown as -fold increase in cells treated with calpeptin relative to levels in untreated cells. B, ChIP analysis of USF and Pol II interactions with LCR element HS2 and the β-globin gene promoter in K562 cells grown in the presence (plus calpeptin) or absence (control) of calpeptin. Cells were subjected to ChIP analysis using antibodies specific for USF1 and RNA Pol II as indicated. Precipitated DNA was analyzed by quantitative PCR using primers specific for LCR element HS2 or the β-globin gene promoter. Error bars reflect S.D. from two independent experiments.
We next isolated primary erythroid progenitor cells (c-Kit+/CD71+ cells) from mouse bone marrow using a fluorescence-activated cell sorter. The cells were cultured for 4 days in the presence or absence of 30 μm calpeptin. The data demonstrate that calpeptin induced high level expression of both βmaj-globin and α-globin genes in the primary erythroid progenitor cell culture (Fig. 5A). Incubation with calpeptin did not change expression of the control β-actin (Fig. 5B) or glyceraldehyde-3-phosphate dehydrogenase (data not shown) genes. These experiments were repeated twice, and the results were reproducible. In contrast to MEL cells, the primary erythroid progenitor cells did not increase globin gene expression in response to DMSO only, perhaps rendering the results from the primary cell culture more significant.
FIGURE 5.
Calpeptin increases expression of α- and βmaj-globin genes in primary c-Kit+/CD71+ erythroid progenitor cells. CD71+/c-Kit+ cells were isolated from mouse bone marrow, enriched by fluorescence-activated cell sorting, and incubated in the presence of 0.03% DMSO or in the presence of 30 μm calpeptin, 0.03% DMSO for 4 days. RNA was isolated from the cells, reverse transcribed, and subjected to quantitative PCR using primers against β-actin, α-globin, and βmaj-globin genes. A, α- and βmaj-globin mRNA levels in primary erythroid cells in the absence (Control), in the presence of 0.03% DMSO, or in the presence of 30 μm calpeptin, 0.03% DMSO. Expression was normalized to that of β-actin. B, mRNA levels of β-actin in cells incubated in the absence (DMSO control) or presence of calpeptin. Error bars reflect S.D. from two experiments using different cDNA preparations.
DISCUSSION
In this study we demonstrated that the calpain inhibitor calpeptin stabilizes full-length USF and induces high level β-globin gene expression in erythroid cells. Our work supports and extends previous observations showing that USF is subject to calpain-mediated proteolytic processing and that the differentiation-dependent increase in globin gene expression is associated with a decrease in intracellular calcium concentration (7–9). Our data show for the first time that inhibition of m-calpain, which in vivo may be mediated by decreased availability of calcium, induces expression of the adult α- and β-globin genes.
We showed that USF is subject to proteolytic cleavage by the calcium-sensitive protease m-calpain during differentiation of erythroid cells. Previous work demonstrated that the intracellular calcium concentration of MEL cells changes during DMSO-induced differentiation and that the treatment of MEL cells with calcium ionophores facilitates differentiation but at the same time reduces β-globin gene expression (7). Our results suggest that a decrease in calcium concentration during erythroid cell maturation lowers the activity of m-calpain. The reduction in m-calpain activity increases the level of full-length USF and consequently enhances expression of globin genes.
The protease m-calpain is known to regulate many processes in the cell and cleaves a variety of proteins in the cytoplasm and in the nucleus, including other transcription factors (23). It is therefore likely that stabilization of USF alone does not account for the strong increase in globin gene expression in MEL cells treated with calpeptin. However, we note that overexpression of USF in undifferentiated MEL cells was sufficient to increase expression of the adult βmaj-globin gene, although not to the extent seen in cells treated with calpeptin (13).
The effect of calpeptin on globin gene expression was not restricted to MEL cells but was also found in K562 (Fig. 4) and primary murine c-Kit+/CD71+ erythroid progenitor cells (Fig. 5). The results obtained from the different cell systems are consistent and reveal specific increases in adult α- and β-globin gene expression upon treatment with calpeptin. The mild increase in expression of the ϵ- and γ-globin genes in calpeptin-treated K562 cells (Fig. 4) could be due to the fact that these genes are already expressed at high levels in these cells.
The effect of Ca2+ on erythroid differentiation is likely not limited to regulating the activity of m-calpain. For example, both DMSO- (Fig. 2) and calpeptin (data not shown)-mediated stimulation of globin gene expression in MEL cells was diminished in the presence of a Ca2+ ionophore. This suggests that Ca2+ additionally regulates calpain-independent processes that interfere with erythroid differentiation-induced globin gene expression.
Whether the calpeptin induction of α-globin gene expression is due to a direct effect of USF acting through regulatory elements in the α-globin gene locus remains to be determined. Although USF directly interacts with regulatory elements in the β-globin gene locus (14–16), e.g. LCR HS2 and the adult β-globin gene promoter, such interactions have not been described in the α-globin gene locus. Our unpublished data show that expression of a dominant negative mutant form of USF in erythroid cells of transgenic mice reduces expression of not only the β-type globin genes but also that of the erythroid-specific transcription factors erythroid Krüpple-like factor and NF-E2/p45.4 This suggests that USF functions in concert with erythroid cell-specific transcription factors to enhance expression of both adult α- and β-globin genes.
USF has long been associated with high level gene expression in differentiated cells (22). Previous studies suggest that USF may antagonize the function of c-Myc (24). Expression of c-Myc is associated with the proliferation of cells, whereas expression of USF is associated with cells undergoing differentiation. It will be interesting to examine whether other cellular systems are associated with a differentiation-dependent increase in USF expression. In this respect it is interesting to note that a recent report demonstrates that USF binding activity increases during differentiation of rat Sertoli cells (25).
TFII-I is another helix-loop-helix protein involved in the regulation of the β-globin gene locus (13). It was originally identified as an initiator binding protein that recruits transcription complexes in the absence of the TATA box (26). We previously demonstrated that TFII-I interacts with an initiator sequence located in the β-globin gene promoter and represses its expression in K562 cells (13, 16). More recently it was demonstrated that TFII-I also represses expression of the vascular endothelial growth factor receptor 2 gene by interacting with the initiator (27). When phosphorylated at a specific tyrosine residue, TFII-I is located in the cytoplasm, interacts with phospholipase C-γ, and inhibits cell surface localization of TRPC3 leading to a decrease in calcium influx (28, 29). It is tempting to speculate that TFII-I is relocated to the cytoplasm during differentiation of erythroid cells where it inhibits the influx of calcium and as a consequence stabilizes expression of transcription factors involved in mediating erythroid cell-specific gene expression patterns.
Enzymatic inhibitors have long been shown to be able to induce globin gene expression (30). Well studied examples are inhibitors of histone deacetylases, which increase expression of fetal globin genes in adult erythroid cells. We show here that another such inhibitor, calpeptin, inhibited m-calpain and induced expression of the adult globin genes. These results may impact the use of stem cells and gene targeting for therapies of hemoglobinopathies. For example, genetically modified induced pluripotent stem cells have been successfully used to correct the anemic phenotype in a murine model of sickle cell disease (31). In these experiments, proliferation and differentiation of hematopoietic cells were achieved by transfecting cells with virus encoding transcription factor HoxB4. Perhaps transient exposure of induced pluripotent stem cells to calpeptin could eliminate the requirement for ectopic expression of HoxB4 as transcription of the HoxB4 gene is under the positive control of USF (32).
Acknowledgments
We thank our colleagues in the Bungert and Huang laboratories for support and discussions. We thank Dr. Brandi K. Ormerod (University of Florida) for kindly providing a microscope and camera.
This work was supported, in whole or in part, by National Institutes of Health Grants DK052356 (to J. B.) and HL090589 (to S. H.).
S. Liang, B. Moghimi, V. J. Crusselle-Davis, L. J. Lin, M. Rosenberg, X. Li, J. Strouboulis, S. Huang, J. Bungert, manuscript in preparation.
- TRPC
- transient receptor potential channel
- DMSO
- dimethyl sulfoxide
- MEL
- murine erythroleukemia
- NF-E2
- nuclear factor erythroid 2
- USF
- upstream stimulatory factor
- LCR
- locus control region
- HS2
- hypersensitive site 2
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- ChIP
- chromatin immunoprecipitation
- Pol
- polymerase
- TF
- transcription factor
- βmaj
- βmajor.
REFERENCES
- 1.Oh-hora M., Rao A. (2008) Curr. Opin. Immunol. 20, 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richmond T. D., Chohan M., Barber D. L. (2005) Trends Cell Biol. 15, 146–155 [DOI] [PubMed] [Google Scholar]
- 3.Berridge M. J. (1993) Nature 361, 315–325 [DOI] [PubMed] [Google Scholar]
- 4.Patterson R. L., van Rossum D. B., Ford D. L., Hurt K. J., Bae S. S., Suh P. G., Kurosaki T., Snyder S. H., Gill D. L. (2002) Cell 111, 529–541 [DOI] [PubMed] [Google Scholar]
- 5.Gusella J., Geller R., Clarke B., Weeks V., Housman D. (1976) Cell 9, 221–229 [DOI] [PubMed] [Google Scholar]
- 6.Bridges K., Levenson R., Housman D., Cantley L. (1981) J. Cell Biol. 90, 542–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hensold J. O., Dubyak G., Housman D. E. (1991) Blood 77, 1362–1370 [PubMed] [Google Scholar]
- 8.Faletto D. L., Macara I. G. (1985) J. Biol. Chem. 260, 4884–4889 [PubMed] [Google Scholar]
- 9.Wang J., Ka W., Sun D., Yao W., Wen Z., Chien S. (2006) Cell Biochem. Biophys. 45, 147–156 [DOI] [PubMed] [Google Scholar]
- 10.Goll D. E., Thompson V. F., Li H., Wei W., Cong J. (2003) Physiol. Rev. 83, 731–801 [DOI] [PubMed] [Google Scholar]
- 11.Inomata M., Nakamura M., Imajoh-Ohmi S., Kawashima S. (1993) Biochim. Biophys. Acta 1178, 207–214 [DOI] [PubMed] [Google Scholar]
- 12.Watt F., Molloy P. L. (1993) Nucleic Acids Res. 21, 5092–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crusselle-Davis V. J., Vieira K. F., Zhou Z., Anantharaman A., Bungert J. (2006) Mol. Cell. Biol. 26, 6832–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bresnick E. H., Felsenfeld G. (1993) J. Biol. Chem. 268, 18824–18834 [PubMed] [Google Scholar]
- 15.Elnitski L., Miller W., Hardison R. (1997) J. Biol. Chem. 272, 369–378 [DOI] [PubMed] [Google Scholar]
- 16.Leach K. M., Vieira K. F., Kang S. H., Aslanian A., Teichmann M., Roeder R. G., Bungert J. (2003) Nucleic Acids Res. 31, 1292–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 18.Crusselle-Davis V. J., Zhou Z., Anantharaman A., Moghimi B., Dodev T., Huang S., Bungert J. (2007) FEBS J. 274, 6065–6073 [DOI] [PubMed] [Google Scholar]
- 19.Vieira K. F., Levings P. P., Hill M. A., Crusselle V. J., Kang S. H., Engel J. D., Bungert J. (2004) J. Biol. Chem. 279, 50350–50357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahl J. A., Collas P. (2007) Stem Cells 25, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 21.Song S. H., Hou C., Dean A. (2007) Mol. Cell 28, 810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corre S., Galibert M. D. (2005) Pigment Cell Res. 18, 337–348 [DOI] [PubMed] [Google Scholar]
- 23.Yajima Y., Kawashima S. (2002) Biol. Chem. 383, 757–764 [DOI] [PubMed] [Google Scholar]
- 24.Choe C., Chen N., Sawadogo M. (2005) Exp. Cell Res. 302, 1–10 [DOI] [PubMed] [Google Scholar]
- 25.Wood M. A., Walker W. H. (2009) Biol. Reprod. 80, 24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy A. L., Malik S., Meisterernst M., Roeder R. G. (1993) Nature 365, 355–359 [DOI] [PubMed] [Google Scholar]
- 27.Mammoto A., Connor K. M., Mammoto T., Yung C. W., Huh D., Aderman C. M., Mostoslavsky G., Smith L. E., Ingber D. E. (2009) Nature 457, 1103–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caraveo G., van Rossum D. B., Patterson R. L., Snyder S. H., Desiderio S. (2006) Science 314, 122–125 [DOI] [PubMed] [Google Scholar]
- 29.Kim D. W., Cochran B. H. (2000) Mol. Cell Biol. 20, 1140–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mabaera R., West R. J., Conine S. J., Macari E. R., Boyd C. D., Engman C. A., Lowrey C. H. (2008) Exp. Hematol. 36, 1057–1072 [DOI] [PubMed] [Google Scholar]
- 31.Hanna J., Wernig M., Markoulaki S., Sun C. W., Meissner A., Cassady J. P., Beard C., Brambrink T., Wu L. C., Townes T. M., Jaenisch R. (2007) Science 318, 1920–1923 [DOI] [PubMed] [Google Scholar]
- 32.Giannola D. M., Shlomchik W. D., Jegathesan M., Liebowitz D., Abrams C. S., Kadesch T., Dancis A., Emerson S. G. (2000) J. Exp. Med. 192, 1479–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]