Abstract
Lgt1 is one of the glucosyltransferases produced by the Gram-negative bacterium Legionella pneumophila. This enzyme modifies eukaryotic elongation factor 1A (eEF1A) at serine 53, which leads to inhibition of protein synthesis and death of target cells. Here we studied the region of eEF1A, which is essential for substrate recognition by Lgt1. We report that the decapeptide 50GKGSFKYAWV59 of eEF1A is efficiently modified by Lgt1. This peptide covers the loop of the helix-loop-helix region formed by helices A* and A′ of eEF1A and is part of the first turn of helix A′. Substitution of either serine 53, phenylalanine 54, tyrosine 56, or tryptophan 58 by alanine abolished or severely decreased glucosylation. Lgt1 modified the decapeptide 50GKGSFKYAWV59 with a higher glucosylation rate than full-length eEF1A purified from yeast, suggesting that a specific conformation of eEF1A is the preferred substrate of Lgt1. A GenBankTM search on the basis of the substrate decapeptide for similar peptide sequences retrieved heat shock protein 70 subfamily B suppressor 1 (Hbs1) as a target for glucosylation by Lgt1. Recombinant Hbs1 and the corresponding fragment (303GKASFAYAWV312) were gluco syl a ted by Lgt1. NMR studies with the gluco syl a ted eEF1A-derived decapeptide identified an α-anomeric structure of the glucose-serine 53 bond and characterize Lgt1 as a retaining glucosyltransferase.
Legionella pneumophila is a Gram-negative bacterium, causing pulmonary infectious disease in humans. This microorganism is able to infect various free-living protozoa in natural environment as well as macrophages, monocytes, and lung epithelial cells during human disease (1, 2). A plethora of virulence factors, which are important for intracellular proliferation of the bacteria in target eukaryotic cells, and a type IVB secretion system for intracytoplasmic delivery of these effectors have been identified (3). Among the best studied Legionella products are RalF (4) and DrrA (5), which act as exchange factors for Arf1 and Rab1 small GTPases, respectively. Additionally, DrrA has been shown to possess activity of a guanine nucleotide dissociation inhibitor-displacement factor (6). These two proteins were suggested to participate in recruitment of endosomal vesicles and construction of a replicative phagosome, which is a characteristic intracellular niche of Legionella and prerequisite for subsequent proliferation of the bacteria in host cells (7). However, despite considerable progress, many aspects of intracellular biology of L. pneumophila, in particular those apart from processes associated with alterations in vesicular trafficking, remain poorly understood.
In our previous investigations we identified three proteins in L. pneumophila (Lgt1, Lgt2, and Lgt3), which possess enzymatic activity and modify eukaryotic elongation factor eEF1A3 at serine 53 by mono-O-glucosylation (8–10). This modification inhibits protein synthesis and is eventually lethal to target cells. Expression of Lgt1 is strongly increased during late phase of bacterial growth in broth medium and in Acanthamoeba castellanii (10). Because bacteria taken at the stationary phase of growth are known to possess maximal pathogenic potential (11, 12), up-regulation of the glucosyltransferase has been suggested to be involved in virulence of L. pneumophila. Here we studied the recognition of eEF1A1 by Lgt1 and identified the type of glucosylation catalyzed by the enzyme.
EXPERIMENTAL PROCEDURES
Materials, Bacterial Strains, and Plasmids
DNA-modifying enzymes were from Fermentas (St.Leon-Rot, Germany). UDP-[14C]glucose was from American Radiolabeled Chemicals (St. Louis, MO). PhusionTM High-Fidelity DNA Polymerase was from New England Biolabs (Ipswich, MA). Escherichia coli strain BL21(DE3) and pET28a vector were from Novagen (Madison, WI), expression vectors pGEX-4T2 and pGEX-4T3 were from Amersham Biosciences. Wild-type and mutated decapeptides, derived from eEF1A sequence, were synthesized at JPT Peptide Technologies GmbH (Berlin, Germany). E. coli was maintained in Luria-Bertani (LB) broth or agar supplemented with 200 μg/ml ampicillin or 50 μg/ml kanamycin when necessary. Accession numbers of genes coding for eEF1A1 and Hbs1 proteins used throughout the investigations are NM_001402 and AK292656, respectively.
Cloning of Genes
The gene coding for lgt1 was cloned into EcoRI-SalI restriction endonuclease sites of a vector pGEX-4T3 as described previously (9, 10). For eEF1A subcloning, we used a PCR-based protocol utilizing the plasmid with eEF1A1 gene as a matrix (a generous gift from Dr. C. R. Knudsen, University of Aarhus, Denmark). Initially, the full-length eEF1A gene was amplified with primers termed #233/#232, digested with restriction endonucleases BamHI and EcoRI, and cloned into pET28a vector to create the plasmid p28a-266 (for primer description see supplemental Table S1). To generate COOH-terminal truncations of eEF1A, p28a-266 was used as a target and PCR amplification was done with primers #315/#313, #315/#314, #315/#316, and #315/#317. After digesting of amplified DNA with NdeI/EcoRI, NdeI/SacI, or NdeI/SalI and ligation into pET28a vector, these amplicons resulted in plasmids p28a-328 (NdeI/EcoRI), p28a-329 (NdeI/SacI), p28a-330 (NdeI/SalI), and p28a-331 (NdeI/SacI), coding for truncations of eEF1A with molecular masses of ∼38, ∼29 (i.e. G domain), ∼19, and ∼10 kDa, respectively. For NH2-terminal truncations, p28a-331 plasmid was used as a matrix in PCR, and amplification was performed with the primers #350/#352 or #350/#353. The reaction products were digested with NdeI/SmaI and ligated into pET-28a vector cleaved with the same enzymes. These resulted in the plasmids p28a-356 and p28a-357, coding for ∼7- and ∼5-kDa eEF1A fragments. To subclone the latter fragments into pGEX-4T2 vectors, p28a-356 and p28a-357 were used in PCR amplification with primers #385/#386 and #382/#350. The resulting amplicons were digested with BamHI/EcoRI and BamHI/SmaI and ligated into pGEX-4T2 digested with the same enzymes. These experiments resulted in plasmids p4T2–403 and p4T2–402, coding for GST-tagged eEF1A truncations. Further diminishing the size of eEF1A as well as cloning of a decapeptide derived from the sequence of Hbs1 have been done by annealing of the complementary synthetic oligonucleotides and ligating them into pGEX-4T2, digested with EcoRI/SalI or BamHI/SalI, respectively. Plasmid coding for the full-length Hbs1 protein was a generous gift from Dr. T. G. Kinzy (University of Medicine and Dentistry of New Jersey, Piscataway, NJ).
Purification of Recombinant Proteins
For purification of recombinant proteins, the E. coli BL21(DE3) strains, transformed with the corresponding plasmids, were grown in LB broth supplemented with ampicillin or kanamycin on a shaker at 37 °C until the optical density at 600 nm reached 0.5 absorbance units. Expression of the cloned proteins was then induced by supplementation of the culture with 0.2 mm isopropyl-β-d-thiogalactopyranoside (Roth, Karlsruhe, Germany) overnight at 22 °C for pET28-based plasmids, and with 1 mm isopropyl-β-d-thiogalactopyranoside at 37 °C for 3 h with the pGEX-based constructions. Typically, the bacterial cells from 2 liters of culture were harvested by centrifugation at 6,000 × g for 15 min, resuspended in 15 ml of 20 mm Tris-HCl-buffered saline (pH 7.4), and then lysed by French press or sonication. Following clarification by centrifugation, the supernatants from bacterial extracts were subjected to chromatography on a glutathione-Sepharose Fast Flow (for pGEX-based constructs) or nickel-equilibrated chelating Sepharose Fast Flow (for pET-28a-based constructs) columns according to the manufacturer's instructions (Amersham Biosciences). Elution of bound proteins was achieved by 0.5 m imidazole (pET28 plasmids) or 10 mm reduced glutathione or thrombin treatment (both for pGEX plasmids). Insoluble fractions of bacterial extracts (pET28a-based constructs) when necessary were dissolved in 6 m urea and purified as described above, but with addition of 6 m urea into all solutions. Re-folding was done by quick dilution of purified proteins into Tris-HCl-buffered saline with 20% glycerol and overnight dialysis against Tris-HCl-buffered saline with 10% glycerol.
Purification of Yeast Elongation Factor 1A and Determination of its GTPase Activity
Purification of eEF1A was based on the interaction of eEF1A with eEF1Bα.4 E. coli (BL21) was transformed with the plasmid, expressing His-tagged yeast eEF1Bα (pET11d-TEF3, generous gift from Dr. G. Andersen), and induced with isopropyl-β-d-thiogalactopyranoside. The bacterial pellet was taken up in eEF1Bα lysis buffer (50 mm Tris-HCl (pH 7.8), 50 mm KCl, 25 mm imidazole, 5 mm MgCl2, 0.5 mm β-mercaptoethanol, and 0.1 mm phenylmethylsulfonyl fluoride) and lysed by sonication. The cleared lysate was applied to a HisTrap HP column (Amersham Biosciences) equilibrated in eEF1Bα lysis buffer, and the protein was eluted with a linear gradient of 25 to 250 mm imidazole. The eEF1Bα-containing fractions were subsequently dialyzed against eEF1A lysis buffer (100 mm Tris-HCl (pH 7.6), 200 mm KCl, 5 mm MgCl2, 10% glycerol, 0.5 mm β-mercaptoethanol, and 0.1 mm phenylmethylsulfonyl fluoride) and used for eEF1A purification.
As a source of yeast eEF1A, commercial baker's yeast preparation was resuspended in eEF1A lysis buffer, and the cells were lysed by passing them through a French press. The lysate was cleared and mixed with purified eEF1Bα to allow complex formation. This was then applied onto a HisTrap HP column, washed with eEF1A lysis buffer containing 20 mm imidazole, and eluted with a linear gradient of 20 to 250 mm imidazole. Fractions containing the complex of eEF1A and eEF1Bα were dialyzed against 20 mm Tris-HCl, pH 7.6, 0.5 mm dithiothreitol, 50 mm KCl, and 25% glycerol (buffer QA). After dialysis GDP was added to the complex to a final concentration of 100 μm and left on ice for 20 min. Then the protein was applied onto Mono Q column (Amersham Biosciences) equilibrated in buffer QA and dissociated eEF1A was collected in the flow-through fraction. This fraction was diluted in 25% glycerol and applied onto a Resource S column (Amersham Biosciences) equilibrated in 20 mm HEPES (pH 7.2), 50 mm KCl, 25% glycerol, 5 mm MgCl2, and 0.5 mm dithiothreitol. eEF1A was eluted with a linear gradient of 50 to 500 mm KCl and dialyzed against 20 mm Tris-HCl (pH 7.6), 150 mm KCl, 5 mm MgCl2, 0.5 mm dithiothreitol, 15 μm GDP, and 25% glycerol.
GTPase assays were performed as described before (13) with minor modifications. Briefly, eEF1A was incubated for 5 min at 20 °C in a buffer, containing 25 mm Tris-HCl (pH 7.8), 1 mm dithiothreitol, 10 mm MgCl2, 100 mm KCl, 0.5 mm phosphoenolpyruvate, and 0.06 mg/ml pyruvate kinase and then incubated at 30 °C for 5 min. The reaction was started by adding [γ-32P]GTP to a final concentration of 100 μm. Aliquots were removed over a time range of 60 min, mixed with charcoal solution (5% charcoal in 50 mm NaH2PO4, pH 2), and spun down for 10 min at 13,000 rpm, and the amount of radioactive phosphate in the supernatant was analyzed by liquid scintillation counting.
Glucosyltransferase Assay
Eukaryotic cell extract used as a crude substrate was prepared by sonication of EBL cells (9). Target eEF1A- or Hbs1-derived proteins were purified as described above. Glucosylation was carried out in 20 μl of a mixture consisting of 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm MnCl2, 10 μm UDP-[14C]glucose, and different amounts of recombinant Legionella GST-tagged Lgt1 enzyme and crude cell extract or purified substrate protein (for precise concentrations of enzyme and substrates see the “Results”). The mixture was incubated for up to 30 min at 37 °C. The reaction was stopped by the addition of Laemmli sample buffer and heating at 95 °C for 5 min. The samples were then subjected to SDS-PAGE in 12.5 or 15% gels (14) and scanned on a PhosphorImager (Storm 820, Amersham Biosciences). ImageQuant 5.2 (Amersham Biosciences) was used for subsequent data quantification.
NMR Spectroscopy
Two samples were prepared for NMR studies. One sample contained a mixture of 5 mm UDP-glucose and 1 mm eEF1A decapeptide and a second sample with the reaction product of the glucosylation with 5 mm UDP-glucose, 1 mm eEF1A peptide, and 1.5 μm of the enzyme Lgt1. The samples were prepared in 20 mm phosphate buffer (pH 7.4), containing 50 mm NaCl and 1 mm MgCl2. However, because Lgt1 was dialyzed against 20 mm Tris-HCl (pH 7.4), the second sample contained also 1 mm Tris, causing interfering 1H NMR signals. Both samples were lyophilized and transferred into 99.9% D2O prior to NMR experiments. All NMR experiments were carried out on a Bruker DMX 600 spectrometer equipped with a standard room temperature 1H,13C,15N-triple resonance probehead with gradients optimized for the direct detection of protons at 298 K. In addition to proton one-dimensional spectra (512 scans, 32,768 data points with a spectral width of 10,000 Hz, zero-filling to 65,536 points without apodization), a number of TOCSY spectra for the identification of spin systems, a DOSY spectrum for the unambiguous verification of signals originating from the glucosylated peptide and J-resolved spectra for the distinction of homonuclear and heteronuclear coupling constants were recorded. Two-dimensional-TOCSY spectra (15, 16) were implemented using the MOCCA-XY16 multiple pulse sequence for isotropic mixing (17, 18) with a mixing time of 120 ms and the acquisition of 512 and 8192 data points (sweep widths of 6004 Hz and 9615 Hz) for the indirectly and directly detected dimensions, respectively. In addition, selectively excited TOCSY spectra with mixing times of 10, 30, 70, and 120 ms were obtained using excitation sculpting (19) with selective G3 inversion pulses (20) of 70-ms duration on the corresponding H1 protons of the glucose ring. DOSY spectra were obtained using the BPLED pulse sequence scheme with presaturation and spoil gradients (21, 22) by acquiring a series of 32 one-dimensional experiments with pulsed field gradients of 4-ms duration, a diffusion time of 50-ms and linearly incremented gradient strengths. Standard Bruker parameters were used for the DOSY processing. Two-dimensional-J-resolved spectra (23) were recorded for both samples with 64 and 16384 data points (with sweep widths of 64 and 9615 Hz).
RESULTS
Substrate Properties of Truncated eEF1A Molecules
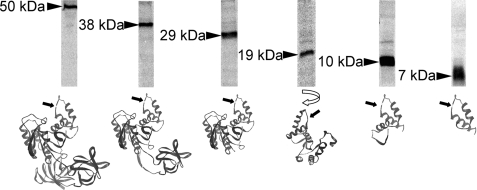
From structural studies it is known that elongation factor eEF1A consists of several domains. Domain 1 is the G domain of ∼240 residues with a Ras-like fold, harboring consensus sequences of typical G proteins. Domains 2 and 3, which consist of 89 and 107 residues, respectively, have both a six-stranded β-barrel structure (24). Glucosylation of eEF1A occurs in domain 1. It was of interest to study whether domains 2 and 3 influence the glucosylation of eEF1A by Lgt1. To this end, we cloned coding sequences of the corresponding eEF1A fragments and expressed them as polyhistidine-fused peptides. As shown in Fig. 1, all truncations tested were glucosylated by Lgt1. In addition, major parts of the G domain of eEF1A were dispensable for modification at serine 53, and even a 7-kDa peptide, covering residues 29–73, was efficiently glucosylated.
FIGURE 1.
Substrate activity of truncated forms of eEF1A. Fragments of eEF1A were produced as described under “Experimental Procedures” and used in 14C-glucosylation assay with Lgt1. Arrowheads on the left of each SDS-PAGE lane indicate approximate molecular mass of the peptide (in kilodaltons). Predicted structural views of each truncated molecule are shown at the bottom. Black arrows indicate the position of serine 53 residue, known to be targeted by glucosylation. The protein with a molecular mass of 29 kDa represents G domain of eEF1A. Structural scheme of the 19-kDa fragment is turned 180° to simplify the presentation of the molecule.
To study whether smaller fragments below 7 kDa could be glucosylated by Lgt1, we generated GST-tagged peptides. As shown in Table 1, a decrease in the size of peptides down to 10 residues did not reduce glucosylation by Lgt1. Further truncations, however, resulted in gradual decrease in the ability to serve as substrates for Lgt1. In particular, elimination of the COOH-terminal valine 59 residue resulted in a roughly 6-fold reduction of glucosylation (Table 1). Subsequent COOH-terminal deletion of tryptophan 58 produced a peptide that was a very poor substrate (1%), whereas further deletions resulted in peptides that were not modified by the glucosyltransferase. The NH2-terminal site was more tolerant toward deletions than the COOH-terminal site. Deletion of glycine 50 only partially decreased glucosylation, and even the hexapeptide SFKYAW was modified by Lgt1.
TABLE 1.
Substrate properties of peptides, derived from the eEF1A sequence
Serine 53, which is glucosylated by Lgt1, is underlined. The glucosylation was performed in reaction buffer with 28 nm GST-tagged Lgt1, 4 μm purified substrate peptide, and 10 μm UDP-[14C]glucose for 5 min and processed thereafter as described under “Experimental Procedures.” Glucosylation of peptides is determined as arbitrary phosphorimaging units. Glucosylation of the decapeptide 50GKGSFKYAWV59 is attributed to 100%. Data from at least three independent experiments are shown as mean ± S.D.
| Constr. ID | Sequence of eEF1A peptide | Glucosylation |
|---|---|---|
| % of maximum | ||
| p403 | 29YKCGGIDKRTIEKFEKEAAEMGKGSFKYAWVLDKLKAERERGITI73 | 21 ± 6.7 |
| p402 | 50GKGSFKYAWVLDKLKAERERGITI73 | 32 ± 11.1 |
| p434 | 50GKGSFKYAWV59 | 100 |
| p485 | 51KGSFKYAWV59 | 55 ± 9.1 |
| p452 | 50GKGSFKYAW58 | 15 ± 7.3 |
| p453 | 50GKGSFKYA57 | 1 ± 0.7 |
| p454 | 50GKGSFKY56 | 1 ± 0.7 |
| p457 | 50GKGSFK55 | <1 |
| p458 | 50GKGSF54 | <1 |
| p455 | 51KGSFKYAW58 | 11 ± 4.9 |
| p456 | 52GSFKYAW58 | 5 ± 0.5 |
| p459 | 53SFKYAW58 | 4 ± 3.6 |
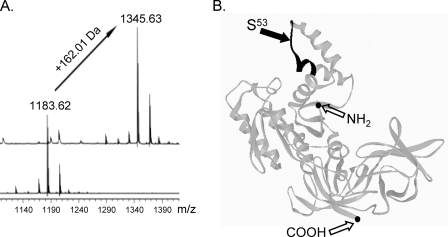
As these data were obtained with GST-fused peptides, we next employed the synthetic peptide acetyl-GKGSFKYAWV-NH2, as well as the peptide with alanine instead of target serine 53. As shown in Fig. 2A, acetyl-GKGSFKYAWV-NH2 was glucosylated by Lgt1, whereas the exchange of serine to alanine blocked the glucosylation (supplemental Fig. S1). Structurally, the glucosylated peptide represents a part of the switch-I region within domain 1 and consists of the NH2-terminal loop and the COOH-terminal α-helix (Fig. 2B).
FIGURE 2.
Analysis of modification in the eEF1A-derived peptide 50GKGSFKYAWV59 by MALDI-TOF mass spectrometry. A, glucosylation reactions with the peptide acetyl-GKGSFKYAWV-NH2 as a substrate were carried out as described under “Experimental Procedures” in the presence of UDP-glucose and with glucosyltransferase Lgt1 (upper spectrum) or without Lgt1 (lower spectrum). The reaction mixtures were subsequently purified using C18 ZipTips (Millipore, Schwalbach, Germany), mixed with a saturated matrix solution of 4-hydroxy-α-cyanocinnamic acid and spotted using the dried droplet method for matrix crystallization. MALDI-TOF mass spectrometry was performed on a Bruker Biflex III mass spectrometer with a nitrogen laser (λ, 337 nm) according to standard methods. The increase in mass of 162.01 Da indicates glucosylation of the peptide by Lgt1. B, predicted structural view of the full-size eEF1A molecule. The fragment corresponding to decapeptide 50GKGSFKYAWV59 is highlighted by a bold line. The black arrow indicates the position of the serine 53 residue, known to be targeted by glucosylation. NH2 and COOH termini are shown by black dots.
Lgt1 Modifies Tryptic Peptides from Yeast eEF1A
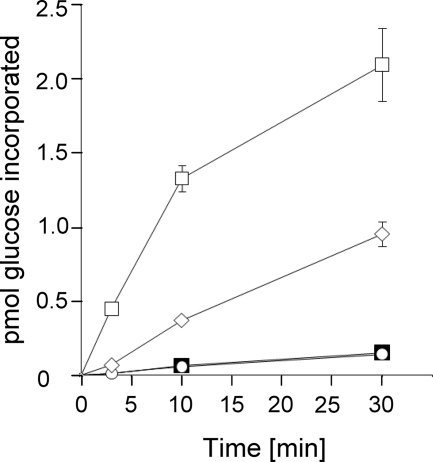
Because glucosylation of small recombinant fragments of eEF1A appeared to be even more efficient than modification of the full-length recombinant eEF1A protein (see Fig. 1, compare 50-kDa with 7-kDa peptide), we asked whether this might be caused by improper folding of the recombinant protein. Therefore, we purified eEF1A from yeast cells, which is >80% identical to mammalian eEF1A. Because yeast eEF1A was purified by specific interaction with eEF1Bα (see purification protocol under “Experimental Procedures” and supplemental Fig. S2A) and exhibited GTPase activity (supplemental Fig. S2B), we concluded proper folding of the purified yeast elongation factor. To study glucosylation of fragments of purified native yeast eEF1A, the elongation factor was partially cleaved by trypsin treatment (Fig. 3A). Thereafter, the fragments were glucosylated by Lgt1 in the presence of UDP-[14C]glucose. As a control, we first [14C]glucosylated full-length yeast eEF1A, and, thereafter, the protein was partially digested by trypsin. As shown in Fig. 3B (lane 3), proteolysis of the full-length elongation factor from yeast produced a small fragment with a molecular mass < 6.5 kDa, which was readily modified by Lgt1. In line with our findings with recombinant proteins and peptides, the small fragment was more efficiently labeled by Lgt1 than the full-length yeast eEF1A (compare signal intensities in lanes 1 and 3, Fig. 3B). Accordingly, trypsin digestion of previously 14C-glucosylated full-length yeast eEF1A resulted in low molecular mass products, which were considerably less labeled as compared with modification of yeast eEF1A-derived peptides (compare lanes 3 and 6 in Fig. 3B). The time courses of the Lgt1-catalyzed glucosylation of the full-length eEF1A protein isolated from yeast, the recombinant G domain of mammalian eEF1A, GST-tagged 25-mer peptide, and the GST-tagged decapeptide (50GKGSFKYAWV59) derived from eEF1A are shown in (Fig. 4) Lgt1 modified the decapeptide ∼20-fold more efficiently than the full-length yeast eEF1A and the G domain of the recombinant mammalian eEF1A.
FIGURE 3.
Coomassie staining (A) and autoradiographic analysis (B) of yeast eEF1A treated with trypsin. Purified eEF1A (1 μm) was incubated without trypsin (lane 1) or with 0.05 μg/ml (lane 2) or with 0.5 μg/ml (lane 3) of trypsin for 30 min at 37 °C. Afterward proteolysis was stopped by the addition of 0.5 mg/ml trypsin soy inhibitor and glucosylation was performed with 70 nm Lgt1 and 10 μm UDP-[14C]glucose for 30 min at 37 °C. In other experiments, after glucosylation of 1 μm yeast eEF1A with 70 nm Lgt1 and 10 μm UDP-[14C]glucose for 30 min at 37 °C the reaction was stopped with 1 mm“cold” UDP-glucose and the samples were treated for 30 min at 37 °C without trypsin (lane 4) or with 0.05 μg/ml (lane 5) or with 0.5 μg/ml (lane 6) of the proteinase. Please note the strong glucosylation of peptides with a molecular mass below 6.5 kDa (panel B, lane 3) obtained by trypsin treatment of full-size yeast eEF1A. Molecular mass markers are shown in lane 7 (A), and their positions are shown on the right in kilodaltons (from top to bottom 66.4, 55.6, 42.7, 34.6, 27.0, 20.0, 14.3, and 6.5 kDa). For simplicity reason only bands of 66.4, 27.0, and 14.3 kDa are indicated on the figure.
FIGURE 4.
Time course of glucosylation of eEF1A-derived substrates by Lgt1. 1 μm of each eEF1A-derived decapeptide (open squares), eEF1A-derived 25-mer peptide (open diamonds), G domain of eEF1A (open circles), and purified yeast eEF1A (filled squares) was incubated with 70 nm Lgt1 and 10 μm UDP-[14C]glucose at 37 °C for 3, 10, and 30 min. Thereafter, autoradiography was performed, and the data were quantified as described under “Experimental Procedures.” Data are given as the mean of at least three independent measurements.
Identification of Hbs1 Protein as a Target of Lgt
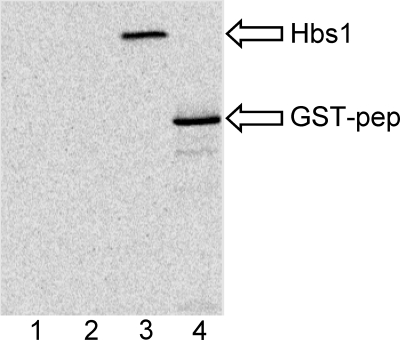
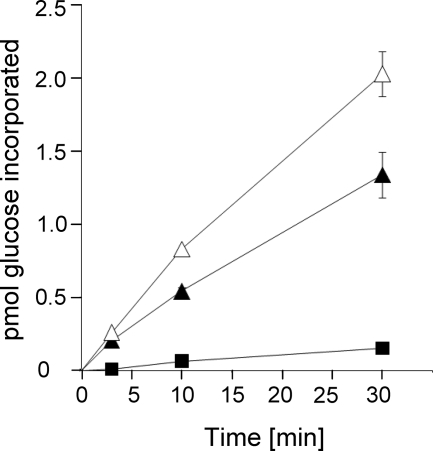
As Lgt1 was able to glucosylate a short peptide of eEF1A, we searched in silico for other proteins containing the decapeptide sequence. A BLAST search retrieved several Hsp70 subfamily B suppressor 1 (Hbs1) proteins of different origin as proteins, containing similar peptides as the decapeptide sequence of eEF1A. In particular, the Hbs1-like protein of Homo sapiens contained the sequence 303GKASFAYAWV312 (locus CAI95161). To test whether this protein is a target for modification by Lgt1, we employed purified recombinant Hbs1 protein as well as the corresponding GST-tagged decapeptide in the 14C-glucosylation assay. In both cases we detected glucosylation by Lgt1 (Fig. 5) (Table 2, construct p407). Furthermore, we compared the rate of glucosylation of full-length yeast eEF1A with recombinant Hbs1 protein and the small peptide derived from Hbs1. As shown in (Fig. 6), the small Hbs1 peptide was glucosylated with a higher rate as full-length yeast eEF1A. Interestingly, full-length Hbs1 was even more efficiently modified than the Hbs1 decapeptide and full-length yeast eEF1A (Fig. 6).
FIGURE 5.
Modification of Hbs1 by 14C-glucosylation. Purified recombinant Hbs1 protein (lane 3) or GST-tagged Hbs1-derived decapeptide (lane 4) (both 1 μm) were incubated with 70 nm Lgt1 in a 14C-glucosylation mixture for 10 min at 37 °C. Thereafter, products formed were analyzed by autoradiography as described under “Experimental Procedures.” Lanes 1 and 2 represent negative controls with Hbs1 and Hbs1-derived decapeptide, respectively, without added Lgt1. Positions of Hbs1 (Hbs1, 76 kDa) and GST-tagged peptide (GST-pep, 30 kDa) are indicated by arrows.
TABLE 2.
Substrate properties of site-mutated GST-tagged peptides derived from the eEF1A sequence
In each sequence the amino-acid changed is underlined. The glucosylation was performed as described in the legend to Table 1. Glucosylation of wild-type decapeptide 50GKGSFKYAWV59 is attributed to 100%. Data from at least three independent experiments are shown as mean ± S.D. Activities of p435- and p480-coded peptides were below the limit of the assay sensitivity (shown as <1%).
| Constr. ID | Sequence of eEF1A peptide | Glucosylation |
|---|---|---|
| % of maximum | ||
| p434 | 50GKGSFKYAWV59 | 100 |
| p478 | AKGSFKYAWV | 84 ± 2.8 |
| p479 | GAGSFKYAWV | 19 ± 1.4 |
| p407 | GKASFAYAWV | 64 ± 4.2 |
| p435 | GKGAFKYAWV | <1 |
| p480 | GKGSAKYAWV | <1 |
| p481 | GKGSFKAAWV | 5 ± 0.7 |
| p484 | GKGSFKYGWV | 12 ± 2.8 |
| p482 | GKGSFKYAAV | 2 ± 0.5 |
| p483 | GKGSFKYAWA | 66 ± 6.4 |
FIGURE 6.
Time course of glucosylation of Hbs1-derived substrates by Lgt1. 1 μm of each full-size Hbs1 (open triangles) and Hbs1-derived decapeptide (filled triangles) was incubated with 70 nm Lgt1 and 10 μm UDP-[14C]glucose at 37 °C for 3, 10, and 30 min. Thereafter autoradiography was performed, and the data were quantified as described under “Experimental Procedures.” A curve with modification of purified yeast eEF1A (filled squares) is shown for comparison. Data are given as mean of at least three independent measurements.
Substrate Properties of Site-mutated eEF1A Decapeptide
The identification of an Hbs1-derived peptide, which contained a sequence around the glucosylated serine residue with two substitutions as compared with eEF1A (303GKASFAYAWV312 in Hbs1 versus 50GKGSFKYAWV59 in eEF1A) suggested that not all amino acid residues of the eEF1A decapeptide are essential for its substrate activity. Therefore, we cloned GST-tagged decapeptides, containing amino acid residues individually substituted by alanine and exchanged alanine 57 of the original peptide by glycine. The peptides were expressed in E. coli, purified, and tested in the glucosylation assay.
As shown in Table 2, change of glycine 50 or valine 59 to alanine resulted in a slight to moderate decrease in modification by Lgt1. Change of lysine 51 or alanine 57 to alanine and glycine, respectively, caused stronger reduction in labeling by Lgt1. Very poor substrates were peptides with exchange of tyrosine 56 and tryptophan 58. No glucosylation was observed with serine 53 and phenylalanine 54 mutants.
NMR Spectroscopy
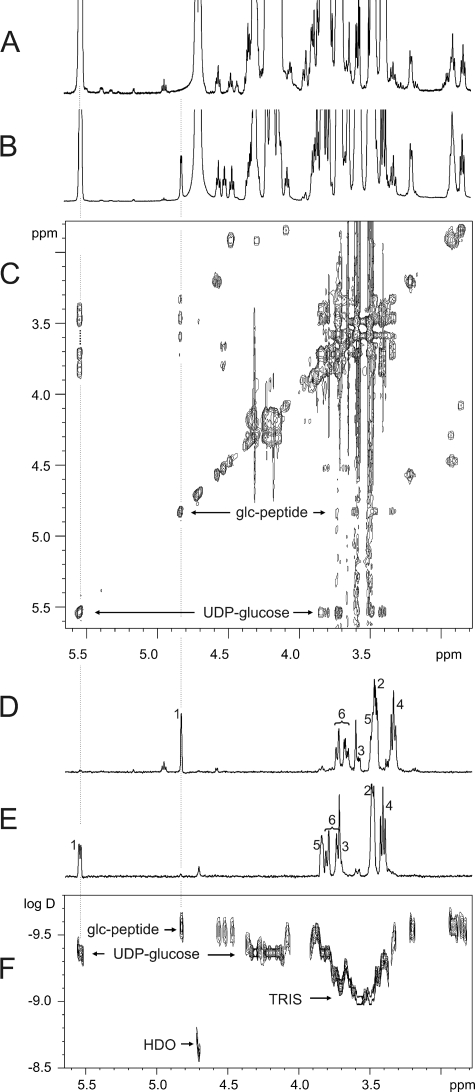
The identification of a small peptide, which is glucosylated by the glucosyltransferase Lgt1, allowed us to study the stereochemical mechanism of its glucosylation by NMR spectroscopy. We addressed the question whether Lgt1 retains or inverts the anomeric center of glucose upon glucosylation. For the NMR investigations (Fig. 7) we compared two samples, one with the educts of the reaction, UDP-glucose and the substrate peptide, and one containing the peptide after glucosylation. The central region of the one-dimensional NMR spectra for the two samples containing all potential sugar signals are shown in Fig. 7, A and B. The region between 3.3 and 4.3 ppm is very crowded with overlap of signals from UDP-glucose, Tris, and the peptide. Visual inspection of the region between 4.4 and 6.0 ppm leads to 4 signals at 4.46, 4.52, 4.56, and 4.83 ppm in the reaction products that are absent or shifted compared with the educts. A two-dimensional-TOCSY spectrum connecting all protons within a spin system reveals that only one of the four signals (at 4.83 ppm) can originate from glucose bound to the eEF1A-derived peptide (Fig. 7C). The identification of the glucose resonances was verified by a two-dimensional DOSY experiment (Fig. 7F).
FIGURE 7.
Identification and assignment of sugar NMR signals of the glucosylated eEF1A-derived decapeptide. A, one-dimensional spectrum of a mixture of UDP-glucose and the decapeptide. The region with potential sugar signals is shown. B, corresponding region of the purified reaction mixture. Signals at 4.46, 4.52, 4.56, and 4.83 ppm do not overlap and are shifted relative to or are not present in A. C, two-dimensional-TOCSY spectrum of the purified reaction mixture. D, selective TOCSY experiment with a 120-ms mixing time starting from the anomeric H1 proton of the glucosylated peptide at 4.83 ppm. The assignment of the sugar protons is annotated. E, corresponding spectrum selectively exciting the H1 proton of UDP-glucose with its assignment. F, two-dimensional BPLED DOSY spectrum of the reaction mixture. At signals without overlap glucosylated peptide, UDP-glucose, Tris, and residual HDO can be distinguished by differences in their diffusion coefficients.
After the spin system of interest was identified, a series of selective TOCSY experiments with increasing mixing times was recorded for UDP-glucose with known assignment and the glucose part of the glucosylation product. A comparison of the spectra led to the assignment shown in Fig. 7D. Using the selective TOCSY experiments and additional two-dimensional J-resolved spectra, most of the 3J(H,H) and nJ(H,P) coupling constants could be determined as shown in Table 3. The central coupling constant for the characterization of the glucosylation is 3J(H1,H2) = 4 Hz, which according to the Karplus relation (25) indicates a dihedral angle between H1–C1 and H2–C2 close to ± 60°. Because all other 3J(H,H) coupling constants are ∼9 Hz, corresponding to a dihedral angle close to 180°, the glucose ring in the reaction product is in the chair conformation and the configuration at the anomeric center can be identified as being α-glucosidic. Because all coupling constants and, therefore, also the configuration is identical to UDP-glucose (Table 3), the activity of the enzyme is unambiguously identified as being retaining.
TABLE 3.
Scalar coupling constants in Hz measured for UDP-glucose and the glucosylated eEF1A-derived decapeptide
Measurement errors are in the order of +/−1.0 Hz for all coupling constants.
| UDP-glucose | Glucosylated peptide | |
|---|---|---|
| 3J(H1,H2) | 4.0 | 4.0 |
| 3J(H2,H3) | 9.9 | 9.3 |
| 3J(H3,H4) | 9.6 | 9.5 |
| 3J(H4,H5) | 9.7 | 8.7 |
| 3J(H1,P) | 8.1 | |
| 4J(H2,P) | 3.4 |
DISCUSSION
Elongation factor eEF1A, with two structurally highly similar isoforms eEF1A1 and eEF1A2, is a large GTP-binding protein, consisting of the N-terminal GTPase domain and two additional domains of β-barrel structure (26). L. pneumophila glucosyltransferase Lgt1 glucosylates eEF1A at serine 53, which is located in the GTPase domain of the elongation factor (9). Here we studied the substrate recognition of eEF1A by Lgt1. Our studies show that domains 2 and 3 of eEF1A are fully dispensable for glucosylation by Lgt1. Moreover, efficient glucosylation by Lgt1 was observed with the decapeptide 50GKGSFKYAWV59 as a substrate. This suggests that the three-dimensional structure of eEF1A is not important for recognition by Lgt1.
Many bacterial protein toxins are characterized by extremely high substrate specificity. These examples include C. difficile toxins A and B, which are at least distantly related to Lgt1 (9), as well as other members of the glucosylating clostridial toxin family. They modify small GTPases of the Rho and/or Ras family in a highly specific manner, depending on the intact folding of the whole substrate protein (27). For example, change of serine 73 to phenylalanine in RhoA turned RhoA into a substrate for lethal toxin of C. sordellii. Similarly, substitution of phenylalanine 85 in RhoD with serine allowed its glucosylation by toxin B of C. difficile (28). In contrast, the cytotoxic necrotizing factors CNF of E. coli needs only a rather small Rho peptide of 20 residues to catalyze deamidation of the target glutamine residue (29). Even smaller is the minimum peptide of eEF1A, which is modified by Lgt1. It contains the sequence 50GKGSFKYAWV59 and forms the loop, which is a part of the helix-loop-helix structure of helices A* and A′ of eEF1A. Whereas glycine 50 is the first residue of this loop, the loop ends with lysine 55. Tyrosine 56 and the other residues of the peptide form the first turn of helix A′. Serine 53 is located in the middle of the loop and apparently is freely accessible at the surface of eEF1A.
Even smaller truncations (e.g. KGSFKYAWV, GKGSFKYAW, and KGSFKYAW) were substrates for Lgt1, although glucosylation was less efficient. As one can deduce from the experiments, the COOH-terminal part of the peptide was more important for its substrate activity than the NH2-terminal part. In line with this notion were results of mutagenesis studies. Substitution of amino acid residues located COOH-terminally (e.g. lysine 55, tyrosine 56, alanine 57, or tryptophan 58) caused major reduction in glucosylation and substitution of phenylalanine 54 completely prevented glucosylation of the neighboring serine 53.
Glucosylation of serine 53 of eEF1A inhibits protein synthesis (9). However, the functional consequences of the modification of serine 53 are not clear on the molecular level. Serine 53, which is glucosylated by Lgt1 in eEF1A, is not present in prokaryotic EF1A (known as EF-Tu) but conserved in many eukaryotic elongation factors 1A. Whereas glucosylation of the small GTPase RhoA by clostridial glucosyltransferases modifies threonine 37, which is essential for nucleotide binding and coordination of magnesium (30), the glucosylation site (serine 53) in eEF1A is not directly involved in nucleotide binding. Glucosylation of RhoA and Ras by clostridial toxins inhibits the active conformation of the protein. The exposed location of serine 53 in eEF1A rather suggests that glucosylation of the elongation factor interferes with binding and/or interaction with other proteins or factors, eventually resulting in inhibition of protein synthesis.
Interestingly, the rate of glucosylation of peptide 50GKGSFKYAWV59 was at least 20-fold higher than the glucosylation of a native full-length eEF1A or its recombinant G domain. This suggests that a specific conformation of eEF1A might be the preferred substrate of Lgt1. However, our attempts to find out conditions that cause a major increase in glucosylation by Lgt1 have been unsuccessful so far.
The short peptide, which was recognized by Lgt1 as a substrate for glucosylation, prompted us to search for other proteins, harboring this sequence. A BLAST search identified Hsp70 subfamily B suppressor 1 (Hbs1) as a putative target for glucosylation. Recombinant Hbs1 was readily modified by Lgt1-induced glucosylation. Initially Hbs1 was identified in yeast screens as a 70-kDa heat shock cognate protein, which suppressed defects caused by a proteasome mutation in Saccharomyces cerevisiae (31). Later it was shown that Hbs1 possesses strong homology to release factor eRF3 (32). Hbs1 and Hbs1-interacting eRF1-related factor Dom34 appear to be important for efficient protein synthesis in and growth of yeast cells under conditions of limiting translation initiation (33). In more recent reports, relationships between Hbs1 function and ribosomal mRNA translation were described (34–36). At present we have no experimental confirmation of the role of glucosylation of Hbs1 in Legionella intracellular biology or disease. However, it should be noted that both identified targets of Lgt1, namely eEF1A and Hbs1, represent important components of protein synthesis machinery in eukaryotic cells. In our previous studies we demonstrated that the glucosylation of eEF1A takes place following intoxication of EBL cells by Lgt1 (9). However we cannot exclude that modification of Hbs1 is also of importance for Lgt1-induced biological effects (e.g. inhibition of protein synthesis).
The identification of a small peptide that is glucosylated by Lgt1 allowed us to study the stereochemical structure of the O-glycosidic bond formed. The anomeric configuration of the product of glucosylation can be either retained or inverted with respect to the donor co-substrate (37). Here we show by NMR spectroscopy that Lgt1 belongs to the retaining type of glucosyltransferases. Therefore, Lgt1 is similar to the clostridial glucosylating toxins, which glucosylate Rho and/or Ras GTPases in a retaining reaction (27, 38–40). Sequence comparison of Lgt1 with clostridial toxins have shown some sequence similarity in the surrounding of the conserved DXD motif, which is involved in coordination of manganese ions and binding of the activated co-substrate UDP-glucose. It remains to be studied whether the structure of the catalytic domain of Lgt1 is related to clostridial glucosyltransferases.
Supplementary Material
Acknowledgments
We thank Dr. C. R. Knudsen for eEF1A-expressing plasmids and helpful hints to establish the GTPase assay, Dr. G. R. Andersen for eEF1Bα-expressing plasmid and for providing protocol of eEF1A purification, and Dr. T. G. Kinzy for generous gift of the Hbs1 gene-containing plasmid.
This work was supported in part by the Bundesministerium für Bildung und Forschung, International Association for the Promotion of Co-operation with Scientists (Project 05-1000004-7756 to Y. B. and K. A.) and the Deutsche Forsch ungs ge mein schaft (DFG, to K. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
G. Andersen, personal communication, University of Aarhus, Denmark.
- eEF1A
- eukaryotic elongation factor 1A
- GST
- glutathione S-transferase
- TOCSY
- total correlation spectroscopy
- Hbs1
- heat shock protein 70 subfamily B suppressor 1
- DOSY
- diffusion-ordered spectroscopy.
REFERENCES
- 1.Bitar D. M., Molmeret M., Abu Kwait Y. (2004) Int. J. Med. Microbiol. 293, 519–527 [DOI] [PubMed] [Google Scholar]
- 2.Fields B. S., Benson R. F., Besser R. E. (2002) Clin. Microbiol. Rev. 15, 506–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juhas M., Crook D. W., Hood D. W. (2008) Cell Microbiol. 10, 2377–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagai H., Kagan J. C., Zhu X., Kahn R. A., Roy C. R. (2002) Science 295, 679–682 [DOI] [PubMed] [Google Scholar]
- 5.Murata T., Delprato A., Ingmundson A., Toomre D. K., Lambright D. G., Roy C. R. (2006) Nat. Cell Biol. 8, 971–977 [DOI] [PubMed] [Google Scholar]
- 6.Ingmundson A., Delprato A., Lambright D. G., Roy C. R. (2007) Nature 450, 365–369 [DOI] [PubMed] [Google Scholar]
- 7.Ninio S., Roy C. R. (2007) Trends Microbiol. 15, 372–380 [DOI] [PubMed] [Google Scholar]
- 8.Belyi I., Popoff M. R., Cianciotto N. P. (2003) Infect. Immun. 71, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belyi Y., Niggeweg R., Opitz B., Vogelsgesang M., Hippenstiel S., Wilm M., Aktories K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16953–16958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belyi Y., Tabakova I., Stahl M., Aktories K. (2008) J. Bacteriol. 190, 3026–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrne B., Swanson M. S. (1998) Infect. Immun. 66, 3029–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer B. K., Swanson M. S. (1999) Mol. Microbiol. 33, 721–731 [DOI] [PubMed] [Google Scholar]
- 13.Kahns S., Lund A., Kristensen P., Knudsen C. R., Clark B. F., Cavallius J., Merrick W. C. (1998) Nucleic Acids Res. 26, 1884–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 15.Braunschweiler L., Ernst R. R. (1983) J. Magn. Reson. 53, 521–528 [Google Scholar]
- 16.Davis D. G., Bax A. (1985) J. Am. Chem. Soc. 107, 2820–2821 [Google Scholar]
- 17.Furrer J., Kramer F., Marino J. P., Glaser S. J., Luy B. (2004) J. Magn. Reson. 166, 39–46 [DOI] [PubMed] [Google Scholar]
- 18.Kramer F., Peti W., Griesinger C., Glaser S. J. (2001) J. Magn. Reson. 149, 58–66 [Google Scholar]
- 19.Stott K., Stonehouse J., Keeler J., Hwang T. L., Shaka A. J. (1995) J. Magn. Reson. 117, 4199–4200 [Google Scholar]
- 20.Emsley L., Bodenhausen G. (1990) Chem. Physiol. Lett. 165, 469–476 [Google Scholar]
- 21.Wu D. H., Chen A. D., Johnson C. S. (1995) J. Magn. Reson. Serie A 115, 260–264 [Google Scholar]
- 22.Cohen Y., Avram L., Frish L. (2005) Angew. Chem. Int. Ed. Engl. 44, 520–554 [DOI] [PubMed] [Google Scholar]
- 23.Freeman R., Hill H. D. (1971) J. Chem. Physiol. 54, 301 [Google Scholar]
- 24.Andersen G. R., Pedersen L., Valente L., Chatterjee I., Kinzy T. G., Kjeldgaard M., Nyborg J. (2000) Mol. Cell 6, 1261–1266 [DOI] [PubMed] [Google Scholar]
- 25.Haasnoot C. A., Deleeuw F. A., Altona C. (1980) Tetrahedron 36, 2783–2792 [Google Scholar]
- 26.Andersen G. R., Nyborg J. (2001) Cold Spring Harb. Symp. Quant. Biol. 66, 425–437 [DOI] [PubMed] [Google Scholar]
- 27.Jank T., Aktories K. (2008) Trends Microbiol. 16, 222–229 [DOI] [PubMed] [Google Scholar]
- 28.Jank T., Pack U., Giesemann T., Schmidt G., Aktories K. (2006) J. Biol. Chem. 281, 19527–19535 [DOI] [PubMed] [Google Scholar]
- 29.Lerm M., Schmidt G., Goehring U. M., Schirmer J., Aktories K. (1999) J. Biol. Chem. 274, 28999–29004 [DOI] [PubMed] [Google Scholar]
- 30.Just I., Selzer J., Wilm M., von Eichel-Streiber C., Mann M., Aktories K. (1995) Nature 375, 500–503 [DOI] [PubMed] [Google Scholar]
- 31.Ohba M. (1994) FEBS Lett. 351, 263–266 [DOI] [PubMed] [Google Scholar]
- 32.Wallrapp C., Verrier S. B., Zhouravleva G., Philippe H., Philippe M., Gress T. M., Jean-Jean O. (1998) FEBS Lett. 440, 387–392 [DOI] [PubMed] [Google Scholar]
- 33.Carr-Schmid A., Pfund C., Craig E. A., Kinzy T. G. (2002) Mol. Cell. Biol. 22, 2564–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doma M. K., Parker R. (2006) Nature 440, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graille M., Chaillet M., van Tilbeurgh H. (2008) J. Biol. Chem. 283, 7145–7154 [DOI] [PubMed] [Google Scholar]
- 36.Lee H. H., Kim Y. S., Kim K. H., Heo I., Kim S. K., Kim O., Kim H. K., Yoon J. Y., Kim H. S., Kim do. J., Lee S. J., Yoon H. J., Kim S. J., Lee B. G., Song H. K., Kim V. N., Park C. M., Suh S. W. (2007) Mol. Cell 27, 938–950 [DOI] [PubMed] [Google Scholar]
- 37.Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 38.Geyer M., Wilde C., Selzer J., Aktories K., Kalbitzer H. R. (2003) Biochemistry 42, 11951–11959 [DOI] [PubMed] [Google Scholar]
- 39.Vetter I. R., Hofmann F., Wohlgemuth S., Herrmann C., Just I. (2000) J. Mol. Biol. 301, 1091–1095 [DOI] [PubMed] [Google Scholar]
- 40.Ziegler M. O., Jank T., Aktories K., Schulz G. E. (2008) J. Mol. Biol. 377, 1346–1356 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.