Abstract
The tumor suppressor p53 induces apo pto sis by altering the transcription of pro-apo pto tic targets in the nucleus and by a direct, nontranscriptional role at the mitochondria. Although the post-translational modifications regulating nuclear apo pto tic functions of p53 have been thoroughly characterized, little is known of how transcription-independent functions are controlled. We and others identified acetylation of the p53 DNA binding domain at lysine 120 as a critical event in apo pto sis induction. Although initial studies showed that Lys-120 acetylation plays a role in p53 function in the nucleus, we report here a role for Lys-120 acetylation in transcription-independent apo pto sis. We demonstrate that the Lys-120-acetylated isoform of p53 is enriched at mitochondria. The acetylation of Lys-120 does not appear to regulate the ability of p53 to interact with the pro-apo pto tic proteins BCL-XL and BAK. However, displacement of the inhibitory MCL-1 protein from BAK is compromised when Lys-120 acetylation is blocked. Functional studies show that mutation of Lys-120 to a nonacetylated residue, as occurs in human cancer, inhibits transcription-independent apo pto sis, and enforced acetylation of Lys-120 enhances transcription-independent apo pto sis. These data support a model whereby Lys-120 acetylation contributes to both the transcription-dependent and -independent apo pto tic pathways induced by p53.
The tumor suppressor p53 protects mammalian cells from malignant transformation by preventing the propagation of genetic aberrations required for tumorigenicity (1). Genotoxic insults that cause DNA damage are among the major stimuli that threaten genomic integrity. In cells that are exposed to such insults, p53 can induce either a cell cycle arrest or apoptosis, depending in part on the severity of cellular damage incurred (1).
To promote cell cycle arrest, p53 acts as a transcription factor to directly up-regulate the expression of the cyclin-dependent kinase inhibitor, p21WAF1/CDKN1 (2), and other genes. In contrast, the maximal apoptotic response requires both transcription-dependent and -independent functions of p53 (3). For example, in response to excessive DNA damage, p53 directly activates the transcription of genes that promote apoptosis such as BAX, PUMA, NOXA, BAD, FAS, PERP, p53AIP1, and DRAM (reviewed in Ref. 4). In addition, following an apoptotic stimulus, a fraction of total cellular p53 localizes to mitochondria and directly induces cytochrome c release (5–8). Part of the cytochrome c release is mediated by the ability of p53 to interact with and inhibit anti-apoptotic BCL-XL and BCL-2. These interactions promote the oligomerization of the pro-apoptotic protein BAX (7, 9, 10). Additionally, mitochondrial p53 can interact with the pro-apoptotic BAK protein. Binding of p53 liberates BAK from the anti-apoptotic protein MCL-1, leading to BAK oligomerization (6). The culmination of these events promotes mitochondrial outer membrane permeabilization and subsequent cytochrome c release.
The localization of p53 to the mitochondria is regulated by at least two different pathways. Recent evidence suggests that there is a population of p53 in the cytoplasm that is stabilized following DNA damage and subsequently shuttles to the mitochondria (11). These studies indicate that MDM2-mediated mono-ubiquitylation of cytoplasmic p53 is essential for p53 to re-locate to the mitochondria. Once at the mitochondria p53 must be de-ubiquitylated by the ubiquitin hydrolase USP7/HAUSP to function in a pro-apoptotic manner. The mitochondrial localization of p53 is also influenced by a common polymorphism in p53 that influences MDM2 binding. Specifically the p53-R72 isoform displays both increased levels of mitochondrial p53 and increased apoptotic potential, compared with the p53-P72 isoform (12).
The relative contributions of the mitochondrial and nuclear functions of p53 to apoptosis remain unresolved. However, intriguing data suggest the transcription-dependent and -independent apoptotic functions of p53 are coupled to maximize apoptosis. Specifically, it has been found that the p53 target gene PUMA influences the fraction of anti-apoptotic BCL2 family members that interact with pro-apoptotic members, thereby controlling the threshold of efficacy of mitochondrial p53 (13). In support of a model whereby the transcription-dependent and -independent apoptotic activities of p53 are coordinated, we report here that the form of p53 that is acetylated on Lys-120, and which has enhanced ability to activate pro-apoptotic target gene transcription, is also enriched at mitochondria. Loss of Lys-120 acetylation, such as occurs when this residue is mutated in human cancer, prevents transcription-independent apoptosis by p53. Mechanistically, acetylation of Lys-120 does not affect the localization or stability of mitochondrial or cytoplasmic p53. Furthermore, acetylation at Lys-120 is not required for p53 to interact with the apoptotic mediators BCL-XL and BAK. However, acetylation of Lys-120 is essential for p53 to effectively liberate the pro-apoptotic protein BAK from the oncoprotein MCL-1. These data elucidate a mechanism whereby the transcription-dependent and -independent apoptotic functions of p53 are coordinated.
EXPERIMENTAL PROCEDURES
Plasmids
pcDNA3-FLAG-hMOF was generated by PCR amplification and ligation into the pcDNA3.1-TOPO-TA cloning vector (Invitrogen). pcDNA3-FLAG-p53 was a kind gift of K. Adler-Storthz. All p53-Lys-120 mutants were generated by site-directed mutagenesis (Stratagene). Primer sequences are available upon request. The retroviral pBABE-puro-p53/ER construct was a kind gift of M. Schuler.
Cell Culture
H1299 cells, 293T, LNCaP, and MCF-7 cells (ATCC) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Hyclone). All transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Retroviral packaging plasmid (SVΨ-A-MLV) was described previously (14).
Tetracycline-responsive Cell Lines
The VirapowerTM T-RexTM lentiviral expression system was used to generate a p53 lentivirus expression vector (Invitrogen). For lentiviral packaging, 3 μg of each pLenti4-TO-p53 or pLenti6-TetR vector were co-transfected into 293T cells with the lentiviral packaging plasmids (2 μg of pCMV-ΔR8.2 and 1 μg of pCMV-VSV-G), and viral supernatants were collected, supplemented with 8 μg/ml Polybrene, and added to target cells. Target H1299 cells were first engineered to express the tetracycline repressor protein, and H1299-TetR-expressing cells were subsequently infected with pLenti4-TO-p53 (or p53 mutants) recombinant lentiviruses and selected with 100 μg/ml Zeocin.
Antibodies, Western Blotting, and Immunoprecipitation
To generate whole cell extracts, cells were lysed with a Nonidet P-40-based lysis buffer supplemented with 30 mm sodium butyrate and protease inhibitors. The following antibodies were used to detect protein expression by Western blot, p53 (FL-393G), actin (C-2), and p21 (C-19) (Santa Cruz Biotechnology). The PUMA antibody used was a kind gift of L. Zhang (University of Pittsburgh). For cell fractionation experiments, mitochondrial fractions were blotted with anti-p53 (FL-393G) and either anti-BAK-NT (Upstate) or anti-voltage-dependent anion-selective channel (Abcam). Cytosol was blotted with anti-p53 (FL-393G) and anti-tubulin (Sigma). Nuclear fractions were blotted with p53 (FL-393G), anti-hMOF antibody (where hMOF is the human ortholog of the Drosophila protein males absent of first, the kind gift of E. Smith and J. Lucchessi), and anti-ORC2 (Pharmingen). For immunoprecipitation of acetylated protein substrates, cells were lysed with a Nonidet P-40-based lysis buffer (described above). Following lysis, 300–500 μg of protein lysate was aliquoted and incubated with 0.25 μg of acetyl-Lys-120 antibody or 1.0 μg of pan-acetyl-lysine antibody (Upstate). Antibody-bound protein complexes were then precipitated with protein A/G-agarose (Santa Cruz Biotechnology). Co-immunoprecipitation of p53 was performed using 0.5–1.0 μg of anti-BAK-NT (Upstate) or 0.5–1.0 μg of anti-BCL-XL (S-18, Santa Cruz Biotechnology). Precipitates were then blotted with anti-p53 (FL-393G) or anti-MCL-1 (Abcam) to detect co-precipitation with BAK.
Biochemical Fractionation
The ApoAlertTM cell fractionation kit (BD Biosciences) was used to separate nuclear, cytoplasmic, and mitochondrial cell fractions, according to manufacturer's instructions, with the modifications described in the supplemental material.
Apoptosis Assays
For quantitation of apoptosis, cells were stained with annexin V-FITC2 (Pharmingen). A FACSCalibur flow cytometer was used to collect and analyze cells. 10,000 total events were collected and then subsequently analyzed for the percentage of annexin V-FITC (FL1-H)-positive, propidium iodide (FL2-H)-negative cells.
RESULTS
The p53 tumor suppressor has the ability to induce either cell cycle arrest or apoptosis (1). Although both of these processes require the transcriptional activities of p53, emerging evidence suggests that p53 also has transcription-independent functions that specifically contribute to its ability to induce apoptosis (7, 10). Furthermore, the transcription-dependent and -independent functions of p53 appear to be coupled to maximize apoptotic potential (13). The mechanism by which p53 coordinates transcription-dependent and -independent apoptotic pathways remains unresolved.
Several post-translational modifications on p53 have been specifically associated with p53-mediated apoptosis. These include phosphorylation of serines 15, 20, and 46 as well as acetylation of lysines 373 and 382 (15–17). Although these modifications influence the transcriptional activities of p53, evidence suggests that these modifications are dispensable for p53 transcription-independent apoptosis (18). Recently, mono-ubiquitylation of p53 at an unknown residue was recently shown to be important for transcription-independent apoptosis (11).
As mentioned above, acetylation of Lys-120 within the p53 DNA binding domain is also specifically required for p53-induced apoptosis (14, 19). Mechanistically, Lys-120 acetylation is necessary for p53 to activate the transcription of a subset of pro-apoptotic target genes, such as PUMA and BAX. Specifically, p53 acetylated at Lys-120 (AcLys-120-p53) specifically localizes to certain pro-apoptotic promoters (14). These observations prompted an examination of whether Lys-120 acetylation might also contribute to transcription-independent apoptosis by p53.
AcLys-120-p53 Is Enriched at the Mitochondria
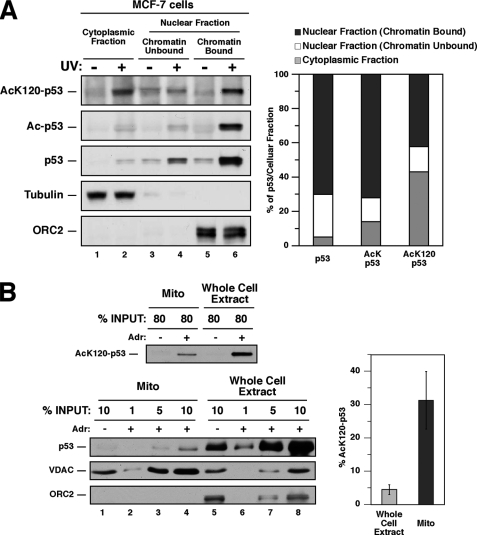
To induce transcription-independent apoptosis, p53 localizes to the outer membrane of mitochondria (7, 20). Therefore, we examined the mitochondrial localization of AcLys-120-p53. A major regulatory step in the localization of p53 to mitochondria occurs at the level of nuclear export (12). Therefore, cytoplasmic levels of p53 are a surrogate for the amount of p53 at the mitochondria (12). As an initial test, both the human breast cancer cell line MCF-7 and human prostate cancer cell line LNCaP were biochemically separated into cytoplasmic and nuclear fractions. Additionally, the nuclear fractions of each condition were biochemically separated into chromatin-bound and unbound fractions (Fig. 1A and supplemental Fig. 1). Following fractionation, extracts from each cellular compartment were divided into aliquots assessed by immunoprecipitation for either lysine 120-acetylated p53 (AcLys-120-p53) or the total cohort of proteins containing acetylated lysines. To quantify p53 acetylation, these precipitates were then Western-blotted with antibodies that recognize all isoforms of p53. In both cell lines, a large portion (∼40% in MCF-7 and ∼30% in LNCaP) of AcLys-120-p53 was found in the chromatin-bound fraction. This observation is consistent with AcLys-120-p53 localizing to select pro-apoptotic promoters (14). In comparison, both total Ac-p53 and overall p53 levels (∼70% each in MCF-7 cells) were present in the chromatin-bound fraction to a far greater extent than AcLys-120-p53 in both cell lines (Fig. 1A and supplemental Fig. 1). Analysis of cytoplasmic fractions from MCF-7 cells revealed that ∼45% of AcLys-120-p53 is localized in this compartment, whereas only ∼15% of Ac-p53 and ∼5% of total p53 are found in the cytoplasm (Fig. 1A). Similar observations were made in LNCaP cells (supplemental Fig. 1).
FIGURE 1.
Acetylated Lys-120-p53 is enriched at chromatin, cytoplasmic, and mitochondrial fractions. A, 18 h following 7.5 J/m2 UV exposure, MCF-7 cells were harvested and biochemically fractionated into nuclear fraction (chromatin-bound and -unbound) and cytoplasmic fractions. Subsequently, each fraction was divided into equal aliquots and subjected to immunoprecipitation with antibodies that recognize either any acetylated protein or specifically the p53 acetylated at lysine 120 (AcK120-p53). Precipitates were then analyzed by Western blot with p53-specific antibodies. Input lysates were also blotted with p53, tubulin (cytoplasmic marker), and ORC2 (nuclear marker) antibodies. B, U2OS cells treated with 0.5 μm adriamycin (Adr) for 18 h and untreated U2OS cells were harvested and separated into 2 equal aliquots. One aliquot from each condition was lysed with a CHAPS-based lysis buffer (described under “Experimental Procedures”) to generate whole cell extracts. Cells from the 2nd aliquot were biochemically fractionated to isolate mitochondria (Mito) and subsequently lysed with a CHAPS-based lysis buffer. Following lysis, 80% of mitochondrial and whole cell extracts were subjected to immunoprecipitation with the AcLys-120 antibody. Precipitates were subjected to Western blot with p53 antibodies to quantify p53 acetylated at lysine 120 (top panel). The amount of p53 localized to the mitochondria is relatively small compared with the amount of total p53 in a stressed cell. Therefore, to compare p53 protein levels between the mitochondria and total p53 lysates in the linear range, 1, 5, and 10% of input lysate from each adriamycin-treated mitochondrial extract (bottom 3 panels, lanes 2–4, respectively) and whole cell extracts (bottom 3 panels, lanes 6–8, respectively) were blotted with p53, voltage-dependent anion-selective channel (mitochondrial marker), and ORC2 (nuclear marker) antibodies. Furthermore, 10% of each untreated fraction was also blotted with the antibodies described (lanes 1 and 5). The acetyl-Lys-120 signals (top panel) from each sample were compared with the titrations probed for total p53 (lower panel). The bar graph to the right represents the percentage of p53 in whole cell extracts acetylated at lysine 120 (light gray, 5 ± 1%) versus the percentage of mitochondrial p53 acetylated at lysine 120 (dark gray, 32 ± 8%).
To extend these observations, mitochondria were biochemically purified from cytoplasmic extracts of the osteosarcoma cell line U2OS, and assessed for the presence AcLys-120-p53 as described above. Although between 4 and 6% of the total p53 population in the cell is acetylated at Lys-120, 24–40% of mitochondrial p53 is acetylated at Lys-120 (Fig. 1B). Also, consistent with the results observed in Fig. 1A, ∼20% of the total AcLys-120-p53 population in the cell is localized at the mitochondria, whereas ∼30 and 50% of AcLys-120-p53 are found in the cytoplasmic and nuclear compartments, respectively (data not shown). Together these data show that AcLys-120-p53 is enriched at the mitochondria and that a significant fraction of mitochondrial p53 is acetylated at Lys-120.
Lysine 120 Acetylation Is Not Required for p53 Mitochondrial Localization
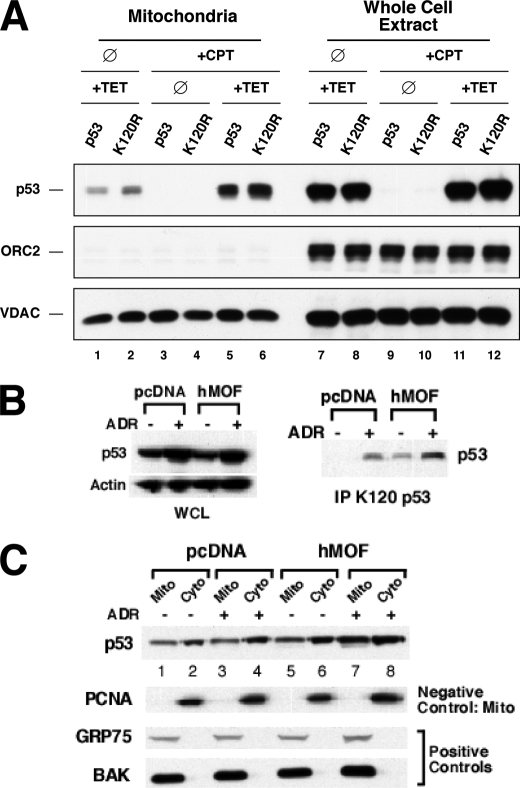
The mitochondrial function of p53 is partly controlled by the regulation of p53 transport to the mitochondria. Therefore, the tetracycline-regulated p53 expression system (TET-p53) was utilized to evaluate whether the Lys-120 acetylation influences the localization of p53 to the mitochondria. In this system apoptosis is induced by the combination of tetracycline to up-regulate p53 and a DNA-damaging agent, such as camptothecin (supplemental Fig. 2). H1299 cells expressing tetracycline-regulated p53 or p53(K120R) were either treated with tetracycline alone, camptothecin alone, or both. Mitochondria recovered from cells of each condition were subjected to Western blot to assess the levels of p53. Although tetracycline treatment alone leads to a small increase in p53 at the mitochondria, the combination of tetracycline and camptothecin strongly increases p53 at the mitochondria (Fig. 2A). It is important to note that eliminating Lys-120 acetylation did not affect the ability of p53 to localize to the mitochondria, indicating that Lys-120 acetylation is not required for p53 mitochondrial localization (Fig. 2A).
FIGURE 2.
Lys-120 acetylation is dispensable for p53 mitochondrial localization. A, H1299 cells expressing either p53 or the K120R mutant under the regulation of a tetracycline (TET)-response element were treated with 500 ng/ml tetracycline or vehicle for 6 h. Following tetracycline treatment, cells were administered 5 μm camptothecin (CPT) or vehicle for an additional 18 h. After treatment, cells were harvested and biochemically fractionated into mitochondria or whole cell extracts. Subsequently, each fraction was subjected to Western blot analysis with p53, voltage-dependent anion-selective channel (mitochondrial marker), and ORC2 (nuclear marker) antibodies. ∅, vehicle-treated. B and C, H1299 cells expressing p53 under the regulation of a tetracycline-response element were transfected with either hMOF or empty vector. Following transfection, cells were treated with 500 ng/ml tetracycline 6 h and then cells were administered 0.5 μm adriamycin (ADR) or vehicle for an additional 18 h. After treatment, cells were harvested and biochemically fractionated into mitochondria (Mito), cytoplasmic (Cyto), and whole cell extracts (WCE). Subsequently, each fraction was subjected to Western blot analysis with the indicated antibodies. PCNA, proliferating cell nuclear antigen.
As mutation of Lys-120 to arginine may mask the requirement for acetylation of this residue in mitochondrial localization of p53, we examined the ability of hMOF, which acetylates p53 on Lys-120, to influence mitochondrial localization of p53. In these studies, the acetyltransferase hMOF was transiently transfected into H1299 cells carrying the tetracycline-regulated p53 transgene. Cells transfected with hMOF or empty vector were treated with either tetracycline alone or in combination with the DNA-damaging agent adriamycin. Overexpression of hMOF in these cells substantially increased Lys-120 acetylation both in the presence and absence of adriamycin (Fig. 2B). However, ectopic expression of hMOF was unable to reproducibly increase steady-state levels of p53 protein at the mitochondria (Fig. 2C).
Lysine 120 Acetylation Is Not Required for p53 to Interact with BCL-XL or BAK
Once at mitochondria, p53 can interact with BCL-2 family proteins. For example, p53 binds to the anti-apoptotic proteins BCL-2 and BCL-XL, leading to the liberation and oligomerization of the pro-apoptotic protein BAX (7, 9, 10). Furthermore, p53 directly interacts with and promotes oligomerization of the pro-apoptotic protein BAK (6). All of these p53-BCL-2 family member interactions utilize the DNA binding domain of p53. Moreover, BAK directly binds to p53 within residues 92–160 of the p53 DNA binding domain. Therefore, the possibility that Lys-120 acetylation influences the binding to BAK or BCL-XL was tested.
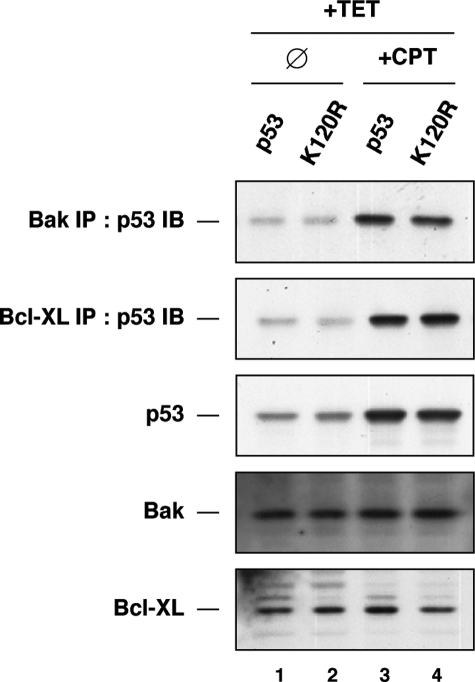
H1299 cells expressing tetracycline-regulated p53 or p53(K120R) were treated with either tetracycline alone, camptothecin alone, or both. Following treatment, cells were biochemically fractionated into crude cytoplasmic and nuclear fractions. Crude cytoplasmic fractions were then separated into 2 aliquots. From each condition, 1 aliquot was subjected to IP with antisera raised against BAK, whereas the other aliquot was immunoprecipitated with BCL-XL antibodies. Following IP, anti-BAK and anti-BCL-XL precipitates were analyzed for the presence of p53 by Western blot. In cells treated with tetracycline alone, some p53 co-precipitated with both BAK and BCL-XL (Fig. 3). However, the amount of p53 bound to each protein was dramatically higher in cells treated with a combination of tetracycline and camptothecin (Fig. 3). Furthermore, both BAK and BCL-XL were able to associate quite well with p53 and p53(K120R), suggesting that Lys-120 acetylation is dispensable for the initial interaction of p53 with BAK or BCL-XL.
FIGURE 3.
Lys-120 acetylation is dispensable for p53 to interact with BAK or BCL-XL. H1299 cells expressing either p53 or the K120R mutant under the regulation of a tetracycline (TET)-response element were treated with 500 ng/ml tetracycline or vehicle for 6 h. Following tetracycline treatment, cells were administered 5 μm camptothecin (CPT) or vehicle for an additional 18 h. After treatment, cells were collected and fractionated in nuclear and cytoplasmic fractions. Cytoplasmic fractions were subjected to immunoprecipitation with either BAK or BCL-XL antibodies. Precipitates were then analyzed by Western blot with p53 antibodies. Input lysates were also blotted with p53, BAK, and BCL-XL antibodies. IB, immunoblot; ∅, vehicle-treated.
Lysine 120 Acetylation Is Necessary for p53 to Displace MCL-1 from BAK
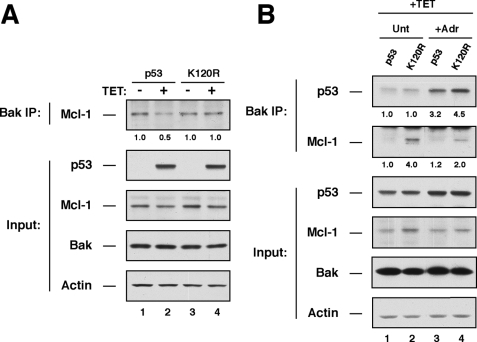
At the mitochondria, p53 promotes BAK oligomerization by directly binding to BAK and displacing the anti-apoptotic protein MCL-1, which normally binds and inhibits BAK (6). Although it is clear that Lys-120 acetylation is dispensable for p53 to bind BAK, the requirement of Lys-120 acetylation for p53 to displace MCL-1 from BAK remained untested. H1299 cells expressing tetracycline-regulated p53 or p53(K120R) were treated with and without tetracycline. Following treatment cells were lysed and subjected to IP with antisera raised against BAK. IPs were subsequently blotted with antibodies that specifically recognize p53 or MCL-1. As reported previously, increased expression of p53 decreases the interaction between BAK and MCL-1 (Fig. 4A). However, when Lys-120 acetylation was abrogated, p53 showed decreased ability to dissociate MCL-1 from BAK (Fig. 4A). Furthermore, the combination of tetracycline and adriamycin, which dramatically enhances p53-BAK interactions, was also insufficient for K120R to fully displace MCL-1 from BAK (Fig. 4B). In these anti-BAK precipitates, both p53 and MCL-1 were detected by Western blotting. This observation is consistent with the possibility that BAK, p53, and MCL-1 associate in a trimeric complex. Together, these data suggest that the acetylation of p53 at Lys-120 is important for transcription-independent apoptosis because of the role of this modification in regulating the disruption of the BAK-MCL-1 complex.
FIGURE 4.
Lys-120 acetylation is required for p53 to displace the anti-apoptotic protein MCL-1 from BAK. A, H1299 cells expressing either p53 or the K120R mutant under the regulation of a tetracycline (TET)-response element were treated with 500 ng/ml tetracycline or vehicle for 24 h. After treatment, cells were collected and lysed to generate whole cell extracts. Whole cell extracts were the subjected to immunoprecipitation with BAK antibodies. BAK precipitates were then analyzed by Western blot with antibodies that recognize p53 or MCL-1. Input lysates were also blotted with p53, BAK, MCL-1, and actin antibodies. B, H1299 cells expressing either p53 or the K120R mutant under the regulation of a tetracycline-response element were treated with 500 ng/ml tetracycline or vehicle for 6 h. Following tetracycline treatment, cells were administered 0.5 μm adriamycin (Adr) or vehicle for an additional 18 h. After treatment, cells were collected and analyzed as in A. Unt, untreated.
Acetylation of Lys-120 Is Required for p53 Transcription-independent Apoptosis
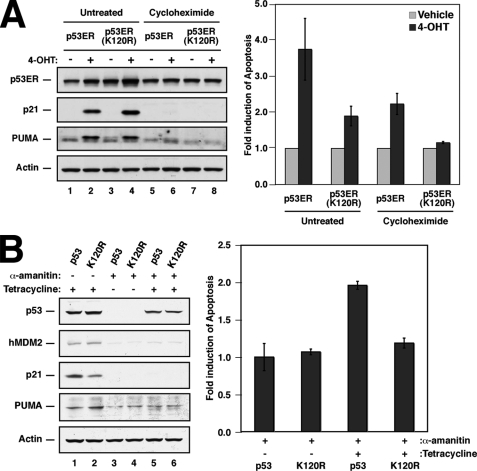
A defect in MCL-1 displacement should lead to decreased levels of transcription-independent apoptosis. To formally evaluate whether Lys-120 acetylation contributes to induction of transcription-independent apoptosis by p53, the p53ER system was employed (10, 14). Here full-length human p53 is fused to a modified version of the ligand binding domain of the human estrogen receptor. When stably expressed in the p53 null human lung cancer line H1299, the p53ER protein is produced but remains inactive. Upon exposure to 4-hydroxytamoxifen (4-OHT), p53ER becomes active for gene transcription and apoptosis (Fig. 5A).
FIGURE 5.
Annexin V staining demonstrates that lysine 120 acetylation is required for p53-mediated transcription-independent apoptosis. A, H1299 cells expressing either p53ER or the p53ER(K120R) mutant were treated with 100 μg/ml cycloheximide or vehicle for 1 h and subsequently treated with 100 nm 4-OHT or vehicle for an additional 48 h. Following treatment, cells were collected and either subjected to Western blot analysis with the indicated antibodies (left panel) or stained with annexin V-FITC and propidium iodide and then analyzed by fluorescence-activated cell sorter (right panel). Right panel is a graphic representation of annexin V-positive (propidium iodide-negative) cells collected after the treatment indicated. Apoptosis levels were normalized to “fold induction” as discussed in the text, and basal levels of apoptosis are ∼2% in the absence of CHX treatment and 15% in the presence of CHX. Error bars represent standard deviation of three independent reactions. B, H1299 cells expressing either p53 or the K120R mutant under the regulation of a tetracycline-response element were treated with 100 μg/ml α-amanitin or vehicle for 2 h and subsequently administered 500 ng/ml tetracycline or vehicle for an additional 24 h. Following treatment, cells were collected and either subjected to Western blot analysis with the indicated antibodies (left panel) or stained with annexin V-FITC and propidium iodide and then analyzed by fluorescence-activated cell sorter (right panel). Right panel is a graphical representation of annexin V-positive (propidium iodide-negative) cells collected after the treatment indicated. Levels of apoptosis are expressed as fold induction. Basal levels of apoptosis were ∼5% in the absence of tetracycline, which is similar to the level observed in response to α-amanitin treatment of parental H1299 cells. Upon activation of p53 by tetracycline treatment, the number of apoptotic cells reached ∼10% of the total population. Error bars represent standard deviation of three independent reactions.
To evaluate transcription-independent apoptosis, p53ER-expressing cells were pretreated with cycloheximide (CHX) to block de novo protein translation (10). Because p53ER is activated post-translationally, CHX treatment does not block p53ER expression or activity. However, p53 target genes transcribed are not translated, and any apoptotic effects of p53 here are thus independent of its transcriptional targets. As CHX is partially toxic to H1299 cells, it is difficult to directly compare the actual percentage of apoptosis between the CHX-treated and -untreated conditions. (Basal levels of apoptosis are ∼2% in the absence of CHX treatment but rise to 15% in the presence of CHX.) Therefore, we plotted p53-mediated apoptosis as the fold induction of apoptosis in response to p53 activation by 4-OHT treatment. Although the presence of CHX inhibits the production of p53 transcriptional targets (Fig. 5A, left panel), p53ER is still partially competent for inducing apoptosis (Fig. 5A, right panel) albeit to a lesser extent than transcriptionally competent p53ER. Although increased levels of PUMA are required for optimal p53-mediated apoptosis, there are sufficient basal levels of PUMA present in the cell that even when transcription of this p53 target is blocked, some apoptosis can proceed. This basal level of PUMA expression was confirmed by Western blotting (Fig. 5, A and B). Our findings are consistent with those of other groups who demonstrated that transcription-independent apoptosis proceeds in the presence of basal PUMA levels, whereas a p53-mediated increase in the levels of PUMA is required for maximum apoptosis (7, 9, 10). As a test of the role of Lys-120 acetylation in this process, cells stably expressing mutant p53ER protein where Lys-120 has been converted to a nonacetylatable arginine (p53ER(K120R)) were evaluated for both p53 transcription-dependent and -independent apoptosis. As reported, loss of Lys-120 acetylation diminishes p53-mediated apoptosis in the absence of CHX (14, 19). However, p53ER(K120R) was also incapable of promoting transcription-independent apoptosis, consistent with acetylation at this site playing a role in this process (Fig. 5A, right panel).
To ensure that this observation was not specific to the p53ER fusion protein, effects of Lys-120 acetylation on p53 transcription-independent apoptosis were also evaluated in a tetracycline-regulated system. H1299 expressing the tetracycline repressor protein and a transgene that expresses p53 under the regulation of a tetracycline-responsive element were generated. Here, levels of p53 are undetectable in the absence of tetracycline. Upon the application of tetracycline, p53 protein levels begin to increase by 2 h, and p53 target gene expression begins to increase at 4 h (data not shown). To assess p53 transcription-independent apoptosis, cells capable of expressing p53 or the p53(K120R) mutant were treated with tetracycline. Two hours following tetracycline or mock treatment, cells were treated with α-amanitin, which inhibits RNA polymerase II-mediated transcription and simultaneously promotes p53 transcription-independent apoptosis by providing a cellular stress signal (21). As expected, α-amanitin treatment suppressed the ability of p53 to up-regulate the expression of its target genes p21WAF1/CDKN1, MDM2, and PUMA (Fig. 5B, left panel). However, even in the absence of transcription, p53 still induced a substantial apoptotic response. As shown, activation of the transcription-independent response resulted in a 2-fold increase in apoptosis, with the total fraction of apoptotic cells reaching ∼10% (Fig. 5B, right panel). Consistent with results obtained using the p53ER system, p53(K120R) was unable to induce transcription-independent apoptosis in the presence of α-amanitin. The total fraction of apoptotic cells remained at ∼5% under these conditions, which is equivalent to the level observed for non-p53 expressing H1299 cells treated with α-amanitin.
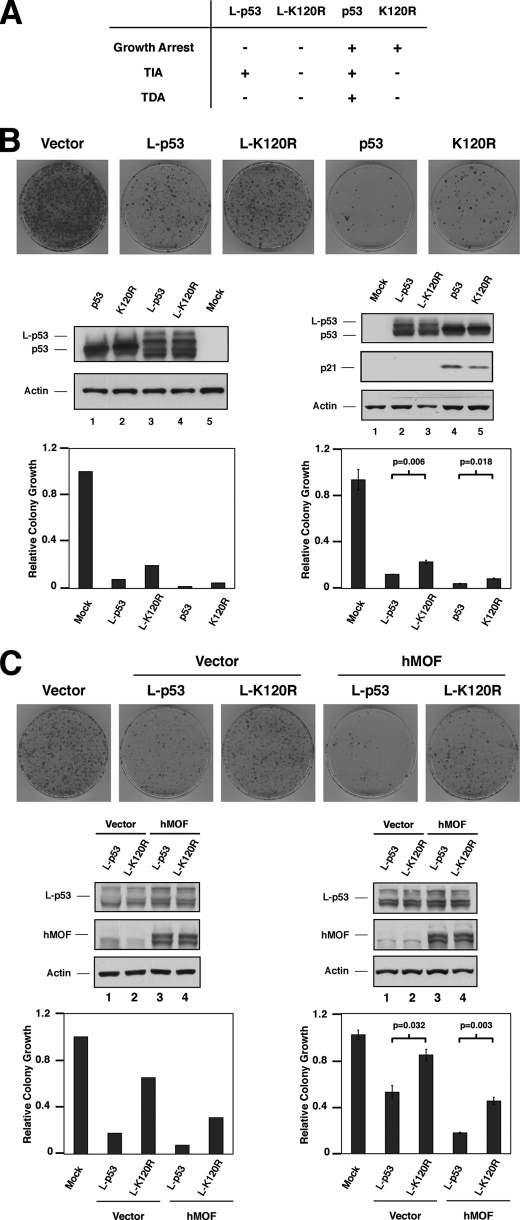
We and others have recently reported that loss of Lys-120 acetylation specifically diminishes p53-induced apoptosis but does not affect the ability of p53 to promote cell cycle arrest (14, 19). Consistent with this, H1299 cells lacking endogenous p53 and transiently transfected with either wild-type p53 or the K120R mutant display severe viability defects as assessed by colony formation assays (Fig. 6B). As the K120R mutant is defective for apoptosis but not growth arrest, the suppression of colony formation by this mutant reflects the growth arrest function of p53, whereas wild-type p53 induces both arrest and apoptosis. Fusion of the mitochondrial import signal from ornithine transcarbamylase to the amino terminus of the p53 protein (L-p53) leads to constitutive localization of p53 at mitochondria (7). Furthermore, because L-p53 is excluded from the nucleus, it cannot induce either cell cycle arrest or transcription of pro-apoptotic target genes. Instead, the L-p53 allele can only suppress colony formation via transcription-independent apoptosis. Similar to previously published data (7), we found that expression of L-p53 effectively suppresses colony formation (Fig. 6B). Remarkably, the L-K120R mutant exhibited reduced ability to suppress colony formation (Fig. 6B). This result was not because of differences in protein expression between experimental conditions, as confirmed by Western blotting. Quantitation of these data confirmed the defect in transcription-dependent apoptosis exhibited by the K120R mutant, a result that was consistently observed over the course of independent experiments (Fig. 6B, experiments 1 and 2).
FIGURE 6.
Colony growth assays demonstrates that lysine 120 acetylation is required for the transcription-independent functions of p53. A, H1299 cells were transiently transfected with plasmids expressing either p53, K120R, or each fused with a mitochondrial import signal (L-p53 and L-K120R). Nontagged versions of p53 are capable of inducing cell cycle arrest, and transcription-dependent (TDA) and -independent apoptosis (TIA). In contrast, the L-p53 isoforms are competent exclusively for transcription-independent apoptosis, as indicated. B, following transfection of the wild-type and mitochondria-targeted p53 isoforms, cells were replated at low density and cultured in media supplemented with G418 for 12 days, then fixed, and stained. Plates of stained cells from experiment 1 are shown (top panel) and colony numbers quantified (left graph). This analysis was repeated, and the quantitation of colony numbers for experiment 2 is displayed (right graph). Western blotting documented appropriate expression levels of p53 isoforms in experiments 1 and 2, as indicated. Experiment 2 was performed as biological triplicates, and results are displayed with standard errors indicated. In addition, p values were generated (Student's t test) by comparing the values for the wild type and K120R isoforms of p53, in the nontagged (p ≤ 0.018) and mitochondria-tagged isoforms (p ≤ 0.006). C, H1299 cells were transfected with the mitochondria-targeted L-p53 constructs described in A, along with an hMOF expression vector where indicated. Colony formation was assayed 12 days post-transfection as described above. Stained colonies from experiment 1 are displayed (top panel) and quantitated (left graph). In experiment 2, biological triplicates were performed, and colony number was quantified (right graph). Western blotting documented appropriate expression levels of p53 isoforms and hMOF in experiments 1 and 2, as indicated. For experiment 2, standard error bars are displayed. In addition, p values were generated (Student's t test) by comparing the values for the wild type and K120R isoforms of p53, in the presence (p ≤ 0.003) and absence of hMOF (p ≤ 0.032).
As the K120R mutant displayed only moderate defects in transcription-independent apoptosis, we reasoned that enforced Lys-120 acetylation might enhance transcription-independent apoptosis. To test this prediction, Lys-120 acetylation was increased by co-transfection of the acetyltransferase hMOF into H1299 cells along with mitochondria-targeted wild-type and K120R p53 expression vectors (Fig. 6C). As demonstrated by increased colony growth suppression when L-p53 was expressed with hMOF, transcription-independent apoptosis was enhanced by increased Lys-120 acetylation (Fig. 6C). This effect of hMOF was in large part related to its ability to acetylate Lys-120, because co-expression of hMOF was substantially less efficient at enhancing transcription-independent apoptosis by the K120R mutant. Quantitation of these results over the course of independent experiments confirmed these observations (Fig. 6C). Combined, both loss-of-function and gain-of-function studies support a model in which Lys-120 acetylation is important for p53 transcription-independent apoptosis.
DISCUSSION
The induction of apoptosis is a central tumor suppressor function of the p53 protein. It has become clear in recent years that maximal apoptosis requires both nuclear and mitochondrial functions of p53 (13). Although the ability of p53 to regulate the transcription of pro-apoptotic targets within the nucleus has been intensely studied, comparatively little is known about how the mitochondrial function of p53 is regulated. We and others have recently reported that acetylation of Lys-120 within the DNA binding domain of p53 is critical for its pro-apoptotic function in the nucleus (14, 19). As shown in this study, blocking Lys-120 acetylation by conservative mutation to arginine also blocks transcription-independent apoptosis by p53. Although somewhat rare, lysine to arginine mutations at Lys-120 have been identified in human cancer (22, 23). This study also reports our efforts at defining the precise biochemical event regulated by Lys-120 acetylation during transcription-independent apoptosis. Although the Lys-120-acetylated isoform of p53 is highly enriched at the mitochondria after DNA damage, our data suggest that this modification is not required for directing p53 to this organelle. Similarly, acetylation of Lys-120 is not required for the interaction of p53 with the BCL-2 family members BCL-XL or BAK. However, subsequent analysis has demonstrated that Lys-120 acetylation is required for efficient displacement of the MCL-1 protein from BAK by p53. This displacement event is critical for the induction of transcription-independent apoptosis by p53 (6), presumably because it facilitates BAK oligomerization and permeabilization of the outer mitochondrial membrane.
The results reported here raise several interesting questions about how p53 acetylation at Lys-120 might enhance displacement of MCL-1 from BAK. For example, it is possible that acetylation of Lys-120 causes a conformational change in p53 that disrupts the otherwise stable trimeric complex of p53, BAK, and MCL-1. Most structural information available for p53 suggests that Lys-120 resides in a flexible domain termed the L1 loop (24, 25). Upon acetylation, this loop may become more rigid or acquire new steric attributes that allow it to block the MCL-1 interface on BAK. The fact that we observe no acetylation-induced change in the ability of p53 to bind BAK itself suggests that it is the displacement event that is directly affected, rather than the ability of p53 to target the BAK/MCL-1 dimer. Future efforts will include the detailed structural analysis required to fully understand the role of Lys-120 acetylation in disruption of the BAK-MCL-1 interaction.
It has recently been reported that the de-ubiquitylation of an unknown lysine plays an important role in activating p53 function at the mitochondria (11). It is possible that Lys-120 acetylation is important for transcription-independent apoptosis because it directly or indirectly participates in controlling the ubiquitylation status of this unknown residue. In fact, Lys-120 can be ubiquitylated (26), and it is thus tempting to speculate that acetylation at this residue enhances p53 function by blocking ubiquitylation of the same lysine.
An intriguing extension of the results reported here relates to the use of therapeutic strategies aimed at reactivating mutant forms of p53 in human cancer. Many of the common cancer-causing mutations in p53 debilitate its nuclear function by eliminating DNA binding capability (24, 25). Evidence suggests that many tumor-derived mutants of p53 constitutively localize to mitochondria but are somehow impaired for apoptotic function (27). This raises the possibility that relieving this block might be a beneficial therapeutic strategy in some human tumors (28). Results such as those reported here will be important for the rational design of these strategies.
Previous studies by us and others have shown that Lys-120 acetylation is critical for the nuclear, transcription-dependent apoptotic function of p53 (14, 19). We report here that Lys-120 acetylation also plays an important role in activating the mitochondrial, transcription-independent function of p53. Ultimately these data suggest a model in which Lys-120 acetylation coordinates the activation of the two critical pro-apoptotic pathways downstream of p53.
Supplementary Material
Acknowledgments
We thank H. Mellert and A. Norvell for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA090465 and CA098172 (to S. B. M.) and R01 CA102184 (to M. E. M.). This work was also supported by the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- FITC
- fluorescein isothiocyanate
- IP
- immunoprecipitation
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- CHX
- cycloheximide
- 4-OHT
- 4-hydroxytamoxifen.
REFERENCES
- 1.Vousden K. H., Lu X. (2002) Nat. Rev. Cancer 2, 594–604 [DOI] [PubMed] [Google Scholar]
- 2.el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 3.Chipuk J. E., Green D. R. (2006) Cell Death Differ. 13, 994–1002 [DOI] [PubMed] [Google Scholar]
- 4.Kuribayashi K., El-Deiry W. S. (2008) Adv. Exp. Med. Biol. 615, 201–221 [DOI] [PubMed] [Google Scholar]
- 5.Erster S., Mihara M., Kim R. H., Petrenko O., Moll U. M. (2004) Mol. Cell. Biol. 24, 6728–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leu J. I., Dumont P., Hafey M., Murphy M. E., George D. L. (2004) Nat. Cell Biol. 6, 443–450 [DOI] [PubMed] [Google Scholar]
- 7.Mihara M., Erster S., Zaika A., Petrenko O., Chittenden T., Pancoska P., Moll U. M. (2003) Mol. Cell 11, 577–590 [DOI] [PubMed] [Google Scholar]
- 8.Marchenko N. D., Zaika A., Moll U. M. (2000) J. Biol. Chem. 275, 16202–16212 [DOI] [PubMed] [Google Scholar]
- 9.Chipuk J. E., Kuwana T., Bouchier-Hayes L., Droin N. M., Newmeyer D. D., Schuler M., Green D. R. (2004) Science 303, 1010–1014 [DOI] [PubMed] [Google Scholar]
- 10.Chipuk J. E., Maurer U., Green D. R., Schuler M. (2003) Cancer Cell 4, 371–381 [DOI] [PubMed] [Google Scholar]
- 11.Marchenko N. D., Wolff S., Erster S., Becker K., Moll U. M. (2007) EMBO J. 26, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumont P., Leu J. I., Della Pietra A. C., 3rd, George D. L., Murphy M. (2003) Nat. Genet. 33, 357–365 [DOI] [PubMed] [Google Scholar]
- 13.Chipuk J. E., Bouchier-Hayes L., Kuwana T., Newmeyer D. D., Green D. R. (2005) Science 309, 1732–1735 [DOI] [PubMed] [Google Scholar]
- 14.Sykes S. M., Mellert H. S., Holbert M. A., Li K., Marmorstein R., Lane W. S., McMahon S. B. (2006) Mol. Cell 24, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao C., Herr D., Chun J., Xu Y. (2006) EMBO J. 25, 2615–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng L., Hollstein M., Xu Y. (2006) Cell Cycle 5, 2812–2819 [DOI] [PubMed] [Google Scholar]
- 17.Knights C. D., Catania J., Di Giovanni S., Muratoglu S., Perez R., Swartzbeck A., Quong A. A., Zhang X., Beerman T., Pestell R. G., Avantaggiati M. L. (2006) J. Cell Biol. 173, 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nemajerova A., Erster S., Moll U. M. (2005) Cell Death Differ. 12, 197–200 [DOI] [PubMed] [Google Scholar]
- 19.Tang Y., Luo J., Zhang W., Gu W. (2006) Mol. Cell 24, 827–839 [DOI] [PubMed] [Google Scholar]
- 20.Chipuk J. E., Green D. R. (2003) J. Clin. Immunol. 23, 355–361 [DOI] [PubMed] [Google Scholar]
- 21.Arima Y., Nitta M., Kuninaka S., Zhang D., Fujiwara T., Taya Y., Nakao M., Saya H. (2005) J. Biol. Chem. 280, 19166–19176 [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto T., Tokuchi Y., Hayashi M., Kobayashi Y., Nishida K., Hayashi S., Ishikawa Y., Tsuchiya S., Nakagawa K., Hayashi J., Tsuchiya E. (1999) Cancer Res. 59, 5572–5577 [PubMed] [Google Scholar]
- 23.Meyers F. J., Chi S. G., Fishman J. R., deVere White R. W., Gumerlock P. H. (1993) J. Natl. Cancer Inst. 85, 1856–1858 [DOI] [PubMed] [Google Scholar]
- 24.Cho Y., Gorina S., Jeffrey P. D., Pavletich N. P. (1994) Science 265, 346–355 [DOI] [PubMed] [Google Scholar]
- 25.Kitayner M., Rozenberg H., Kessler N., Rabinovich D., Shaulov L., Haran T. E., Shakked Z. (2006) Mol. Cell 22, 741–753 [DOI] [PubMed] [Google Scholar]
- 26.Chan W. M., Mak M. C., Fung T. K., Lau A., Siu W. Y., Poon R. Y. (2006) Mol. Cancer Res. 4, 15–25 [DOI] [PubMed] [Google Scholar]
- 27.Erster S., Moll U. M. (2005) Biochem. Biophys. Res. Commun. 331, 843–850 [DOI] [PubMed] [Google Scholar]
- 28.Galluzzi L., Morselli E., Kepp O., Tajeddine N., Kroemer G. (2008) Cell Cycle 7, 1949–1955 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.