Abstract
Focal Adhesion Kinase (FAK) activity is controlled by growth factors and adhesion signals in tumor cells. The scaffolding protein RACK1 (receptor for activated C kinases) integrates insulin-like growth factor I (IGF-I) and integrin signaling, but whether RACK1 is required for FAK function is unknown. Here we show that association of FAK with RACK1 is required for both FAK phos pho ryl a tion and dephos pho ryl a tion in response to IGF-I. Suppression of RACK1 by small interfering RNA ablates FAK phos pho ryl a tion and reduces cell adhesion, cell spreading, and clonogenic growth. Peptide array and mutagenesis studies localize the FAK binding interface to blades I-III of the RACK1 β-propeller and specifically identify a set of basic and hydrophobic amino acids (Arg-47, Tyr-52, Arg-57, Arg-60, Phe-65, Lys-127, and Lys-130) as key determinants for association with FAK. Mutation of tyrosine 52 alone is sufficient to disrupt interaction of RACK1 with FAK in cells where endogenous RACK1 is suppressed by small interfering RNA. Cells expressing a Y52F mutant RACK1 are impaired in adhesion, growth, and foci formation. Comparative analyses of homology models and crystal structures for RACK1 orthologues suggest a role for Tyr-52 as a site for phos pho ryl a tion that induces conformational change in RACK1, switching the protein into a FAK binding state. Tyrosine 52 is further shown to be phos pho ryl a ted by c-Abl kinase, and the c-Abl inhibitor STI571 disrupts FAK interaction with RACK1. We conclude that FAK association with RACK1 is regulated by phos pho ryl a tion of Tyr-52. Our data reveal a novel mechanism whereby IGF-I and c-Abl control RACK1 association with FAK to facilitate adhesion signaling.
RACK12 is a tryptophan-aspartate (WD) repeat containing protein that acts as a scaffolding protein in a wide array of signaling events (1, 2). It has been reported to both regulate and promote cell migration in different cell types (3–5). RACK1 scaffolds proteins at focal adhesions and is capable of mediating both focal adhesion assembly and disassembly (4, 6, 7). RACK1 also scaffolds core kinases of the ERK pathway in response to adhesion signals and modulates the phosphorylation of focal adhesion proteins including focal adhesion kinase (FAK) and paxillin (8, 9). In transformed cells RACK1 integrates signaling from the IGF-I receptor (IGF-IR) and β1 integrin by forming a scaffolding complex that includes these receptors as well as signaling molecules that promote cell migration (5, 10, 11). Cooperation between IGF-IR and β1 integrin signaling is essential for growth of certain tumors (12), and we propose that RACK1 has an important role in this.
The interaction of RACK1 with the IGF-IR requires integrins to be ligated and also requires a domain in the C terminus of the IGF-IR that is essential for IGF-IR function in anchorage-independent growth, cell survival, and cell migration (13, 14). Ligand-mediated activation of the IGF-IR leads to recruitment of certain proteins to RACK1 such as IRS-1, β1 integrin, and dissociation of other proteins from RACK1 such as PP2A and Src. Competitive binding to RACK1 occurs for some of these proteins. For example, IGF-I-mediated dissociation of PP2A from RACK1 is required for recruitment of β1 integrin, and both PP2A and β1 integrin compete for binding to tyrosine 302 in RACK1 (5, 15).
RACK1 is located in areas of cell protrusion that are rich in paxillin (4, 7) and can increase the phosphorylation of FAK (7). FAK is a well characterized kinase in mediating integrin signaling and is associated with the enhanced migratory potential of several cancer cell types (16–18). FAK is phosphorylated on tyrosine 397 in response to the clustering of integrins (for review, see Ref. 19) or by activation of the EGF and platelet-derived growth factor receptors (20–23). This results in recruitment of Src and subsequent phosphorylation of target proteins that are associated with focal adhesion formation and activation of mitogen-activated protein kinase pathways. FAK becomes rapidly dephosphorylated when cells are detached, and this is thought to be essential for focal adhesion dissolution and cell migration. FAK dephosphorylation can be stimulated by IGF-I (5, 24–27). Interestingly, we have observed that IGF-I-mediated dephosphorylation of FAK is enhanced in cells overexpressing RACK1, which also have enhanced migratory potential and increased activation of mitogen-activated protein kinase pathways (28). However, it is not known how the phosphorylation and subsequent dephosphorylation of FAK are coordinated. In particular, the role of RACK1 in regulation of FAK phosphorylation remains undefined. Here we investigated this in the context of IGF-I and adhesion signaling by determining the role of RACK1 in FAK function.
MATERIALS AND METHODS
Reagents and Antibodies
Recombinant IGF-I and EGF were purchased from Pepro Tech. Inc. (Rocky Hill, NJ). The anti-RACK1 and anti-Abl (554148) were from BD Transduction Laboratories. The anti-IGF-IR polyclonal antibody and anti c-Abl antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-phospho-Akt, anti-Akt, and anti-phospho FAK (Tyr-397) polyclonal antibodies were from Cell Signaling Technology (Beverly, MA). The anti-Shc polyclonal antibodies, the anti-phosphotyrosine monoclonal antibody 4G10, anti-ERK-2, anti-FAK antibodies, and recombinant glutathione S-transferase (GST)-Crk was purchased from Upstate Biotechnology Inc. (Lake Placid, NY). The anti-actin monoclonal antibody was from Sigma. Recombinant GST-FAK and anti-c-Abl (OP14) were purchased from Calbiochem. STI571 was a kind gift from Novartis Pharma AG, (Basel, Switzerland).
Cell Culture, Transfection, and IGF-I-mediated Stimulation of MCF-7 and R− Cells
The MCF-7 breast carcinoma cell line and R− cell line (the mouse embryonic fibroblast cell line derived from the IGF-IR knock out mouse (29) was maintained in Dulbecco's modified Eagle's medium (BioWhittaker, Verviers, Belgium) were supplemented with 10% (v/v) fetal bovine serum, 10 mm l-Glu, and 5 mg/ml penicillin/streptomycin. R− cells were transiently transfected with pcDNA3/IGF-IR WT or IGF-IR Y1250F/Y1251F or empty pcDNA3 vectors (8 μg of DNA) using Lipofectamine transfection reagent (Invitrogen). After 24 h in culture the transfected cells were seeded into 10-cm plates and cultured for an additional 18 h. For analysis of signaling responses in adherent cells, the cells were then washed with PBS and starved from serum for 4 h before stimulation with IGF-I or EGF for the indicated times. For analysis of signaling responses in non-adherent cells (Fig. 1A) confluent R− cells were detached with trypsin/EDTA and then washed with PBS. Cells were resuspended in serum-free medium and maintained in suspension for 4 h before stimulation with IGF-I for the indicated times.
FIGURE 1.
RACK1 is required for FAK phosphorylation and dephosphorylation. A, MCF-7 cells were transfected with siRNA1, siRNA2, and siRNA3 targeting RACK1 or control siRNA and serum-starved for 4 h before stimulating with IGF-I for the indicated times. Cell lysates were prepared and separated on a 4–20% gradient PAGE and transferred to nitrocellulose membrane. The membrane was probed with antibodies directed against phospho-FAK (Tyr-397) and FAK. B, R− cells were transiently transfected with pcDNA3 plasmids encoding full-length WT IGF-IR or the Y1250F/Y1251F mutant IGF-IR. The cells were serum-starved under adherent conditions or while maintained in suspension culture for 4 h before stimulation with IGF-I for the indicated times. Cell lysates were prepared followed by Western blotting using antibodies directed against phospho-FAK (Tyr-397) and FAK to determine protein loading. C, MCF-7 cells, R+ cells, and R− cells were serum-starved for 4 h and stimulated with IGF-I before cell lysates were prepared. RACK1 was immunoprecipitated (IP) from the lysates and analyzed for associated FAK by Western blotting with anti-phospho-FAK (Tyr-397) and anti-FAK antibodies. RACK1 was also assessed to show that equal amounts of the protein were immunoprecipitated from each sample.
RNA Interference
siRNAs targeted to human RACK1 were purchased from Ambion. The sequences are: siRNA ID 135615 (siRNA 1) 5′-ccaucaagcuauggaauactt-3′ (sense), 5′-guauuccauagcuugauggtt-3′ (antisense); siRNA ID 135616 (siRNA 2) 5′-gcuauggaauacccugggutt-3′ (sense), 5′-acccaggguauuccauagctt-3′ (antisense); siRNA ID135617 (siRNA 3) 5′-ccuuuacacgcuagauggutt-3′ (sense), 5′-accaucuagcguguaaaggtg-3′ (antisense). MCF-7 cells were seeded at 50% confluency and transfected with 100 μm siRNA using the OligofectAMINE transfection reagent (Invitrogen) as described previously (5). After 24 h the cells were starved and stimulated as described above or lifted using trypsin/EDTA and replated for cell assays.
Preparation of Cellular Protein Extracts and Immunoprecipitation
Cellular protein extracts were prepared by washing cells with PBS and then scraping into lysis buffer consisting of 10 mm Tris HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40 plus the tyrosine phosphatase inhibitor Na3VO4 (1 mm), and the protease inhibitors phenylmethylsulfonyl fluoride (1 mm), pepstatin (1 μm), and aprotinin (1.5 μg/ml). After incubation at 4 °C for 20 min nuclear and cellular debris were removed by microcentrifugation at 14,000 rpm for 15 min at 4 °C. For immunoprecipitation of endogenous proteins from stimulated or unstimulated cells, cells were initially precleared using bovine serum albumin-coated protein G-agarose beads (15 μl of beads per 400 μg of total protein in 700 μl of lysis buffer) by incubation at 4 °C for 1 h with gentle rocking. The lysates were recovered from the beads by centrifugation at 3000 rpm for 3 min and transferred to fresh tubes for incubation with primary antibody (3 μg of each antibody) overnight at 4 °C with gentle rocking. Immune complexes were obtained by adding 20 μl of protein G-agarose beads for 3 h at 4 °C, washing (×3) with ice-cold lysis buffer, and removal from the beads by boiling for 5 min in 20 μl of 2× SDS-PAGE sample buffer before electrophoresis and Western blot analysis. For RACK1 immunoprecipitations, 700 μg of protein was incubated with 1 μg of anti-RACK1 IgM antibodies, 5 μg of goat anti-mouse IgM Fab fragment, 30 μl of protein G-agarose beads, 500 μl of immunoprecipitation buffer (1 mm EGTA, 1 mm EDTA, 10 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 1% deoxycholate together with the tyrosine phosphatase inhibitor Na3VO4 (1 mm) and protease inhibitors as described previously (Kiely et al. (25)).
Western Blot Analysis
Protein samples were resolved by SDS-PAGE on 4–20% gradient gels and then transferred to nitrocellulose membranes, which were blocked for 1 h at room temperature in Tris-buffered saline containing 0.05% Tween 20 and 5% milk (w/v). Primary antibody incubations were overnight at 4 °C. Secondary antibody incubations were at room temperature for 1 h. Where indicated, membranes were stripped in 62.5 mm Tris-Cl, 1% SDS, and 0.7% 2-mercaptoethanol for 30 min at 50 °C followed by extensive washing in 0.2 and 0.05% Tris-buffered saline containing 0.05% Tween 20. Secondary antibodies conjugated with horseradish peroxidase were used for detection of Shc with enhanced chemiluminescence (Super Signal from Pierce). In all other cases we used Alexa Fluor 680- and 800-coupled anti-rabbit and anti-mouse secondary antibodies (LI-COR Biosciences) for detection with the Odyssey® infrared imaging system (LI-COR Biosciences).
Cell Adhesion and Cell Spreading Assay
Plates (96 wells) were coated with collagen I (10 μg/ml) overnight at 4 °C and washed twice with 2× PBS and then blocked with 2.5% bovine serum albumin for 2 h at 37 °C. MCF-7 cells transfected with siRNA targeting RACK1 or a control siRNA for 24 h were-serum starved for 4 h before harvesting with trypsin/EDTA, washing with serum-free media (SFM), and resuspending in SFM to give a final density of 2.0 × 105 cells/ml. This cell suspension (100 μl/2.0 × 104 cells) was plated onto collagen-coated plates and allowed to attach for the indicated times at 37 °C. Unbound cells were removed by inverting and gentle washing in PBS before the cells were fixed in methanol at −20 °C for 5 min. Cells were stained with 0.1% crystal violet and measured by reading the absorbance at 595 nm. To assess cell spreading, MCF-7 cells (transfected with siRNA targeting RACK1 or a control siRNA) were harvested with trypsin/EDTA and replated onto multiple wells of a collagen-coated 24-well plate. After 8 h the cells were inspected and photographed.
Cell Proliferation and Plating Efficiency Assays
MCF-7 cells transfected with siRNA targeting RACK1 or a control siRNA were harvested after 24 h with trypsin/EDTA, washed in serum-free medium, and resuspended in Dulbecco's modified Eagle's medium/10% fetal bovine serum at a final density of 3.0 × 104 cells/well in multiple wells of a 24-well plate. At regular intervals (48, 72, and 96 h post-transfection) the cells were removed from triplicate wells and counted using a hemocytometer and trypan blue exclusion. To assess plating efficiency cells were seeded in triplicate wells of a 6-well plate at 500 cells/well in 3 ml of Dulbecco's modified Eagle's medium, 10% fetal bovine serum and cultured for 14 days. Cells were fixed in methanol at −20 °C for 5 min and stained with Giemsa. Colonies were examined, counted, and photographed.
Spot Synthesis of Peptides and Overlay Analysis
Peptides arrays of RACK1 in nitrocellulose were generated as previously described (30–32). Essentially, scanning libraries of overlapping 25-mer peptides covering the entire sequence of RACK1 were produced by automatic SPOT synthesis and synthesized on continuous cellulose membrane supports on Whatman 50 cellulose using Fmoc (9-fluorenylmethyloxycarbonyl) chemistry with the AutoSpot-Rosbot ASS 222 (Intavis Bioanalytical Instruments). The interaction of GST, GST-FAK, and c-Abl with the RACK1 array was investigated by overlaying the cellulose membranes with 10 μg/ml concentrations of each recombinant protein. Bound protein was detected with specific rabbit antisera for each protein and a secondary anti-rabbit antibody coupled with horseradish peroxidase. Once the binding site of FAK on the full-length RACK1 array was determined, specific alanine scanning substitution arrays were generated for selected peptides using the same synthesis procedure.
In Vitro Kinase Assays
c-Abl kinase was first immunoprecipitated from 500 μg of cell lysate by incubating 40 μl of protein G-Sepharose beads and 3 μg of anti-c-Abl (K-12) overnight. Beads were washed twice in lysis buffer and once with the kinase buffer (20 mm HEPES, pH 7.1, 150 mm NaCl, 1% Triton X-100, 10% glycerol, 1 mm MgCl2, 2 mm MnCl2, 1 mm phenylmethylsulfonyl fluoride, 20 μg/ml aprotinin, and 100 μm sodium vanadate). The in vitro kinase reaction was performed using 10 μg of GST-Crk as an exogenous substrate in the presence of 10 μm ATP and 10 μCi of [γ-32P]ATP at 30 °C for 15 min. The reaction mixture was then subjected to SDS-PAGE, Western blotting, and autoradiography phosphorimaging analysis. To assess the phosphorylation of peptide arrays, the membrane was placed in membrane phosphorylation buffer (20 mm HEPES, pH 7.4, 100 mm NaCl, 5 mm MgCl2, 1 mm dithiothreitol, 0.2 mg/ml bovine serum albumin for 1 h at room temperature. The membrane was blocked overnight at 4 °C in membrane phosphorylation buffer containing 1 mg/ml bovine serum albumin, 100 μm ATP. Each membrane was phosphorylated in 20 ml of membrane phosphorylation buffer containing 50 μm ATP and 10 μCi of [γ-32P]ATP with and without kinase at 30 °C for 30 min. After extensive washing with 1 m NaCl (10 × 10 min), H2O (3 × 5 min), 5% H3PO4 (3 × 15 min), and H2O (3 × 5 min), the membrane was exposed by autoradiography phosphorimaging analysis.
RESULTS
Association with RACK1 Is Required for FAK Phosphorylation and Dephosphorylation
We have shown previously that FAK phosphorylation on Tyr-397 is increased in RACK1-overexpressing cells and that IGF-I induced FAK dephosphorylation is accelerated (25). We, therefore, investigated whether RACK1 is required for FAK phosphorylation and dephosphorylation by cell adhesion and IGF-I stimulation, respectively. To do this we suppressed RACK1 expression with siRNA in MCF-7 cells. In cells transfected with control siRNA, FAK phosphorylation on Tyr-397 was evident in adherent serum-starved cell cultures and was decreased after IGF-I stimulation. However, in cells transfected with three different siRNAs targeting RACK1, FAK phosphorylation on Tyr-397 was not detectable in either the presence or absence of IGF-I (Fig. 1A). This indicates that FAK phosphorylation on Tyr-397 requires RACK1.
We next investigated whether FAK phosphorylation was dependent on an interaction between RACK1 and the IGF-IR. To test this, R− cells were transfected with either wild type IGF-IR (IGF-IR WT) or an IGF-IR mutant (Y1250F/Y1251F) that does not interact with RACK1 and does not promote cell migration (25). As can be seen in Fig. 1B, FAK phosphorylation was decreased in response to IGF-I in cells expressing wild type IGF-IR but was unresponsive to IGF-I in cells expressing the Y1250F/Y1251F mutant. Moreover, when R− cells expressing WT IGF-IR were maintained in suspension culture, FAK phosphorylation was not detected (Fig. 1B). These data confirm that FAK phosphorylation requires cell adhesion and demonstrate that IGF-I-mediated dephosphorylation of FAK requires association of RACK1 with the IGF-IR.
These results led us to hypothesize that FAK association with RACK1 may be regulated by IGF-I. To test this we investigated the co-immunoprecipitation of FAK with RACK1 in MCF-7 cells, R+ cells, and R− cells. FAK was found to be constitutively associated with RACK1 in immunoprecipitates from all three cell lines (Fig. 1C). This suggests that the association is not dependent on the presence of the IGF-IR. However, although RACK1-associated FAK was phosphorylated in serum-starved cultures with all cell lines, IGF-I-mediated dephosphorylation of FAK was observed in both MCF-7 and R+ cells but not in R− cells. This indicates that phosphorylation of FAK in response to integrin ligation and dephosphorylation of FAK in response to IGF-I, both, occur when RACK1 is associated with the IGF-IR.
RACK1 Is Required for Cell Spreading, Proliferation, and the Transformed Phenotype of MCF-7 Cells
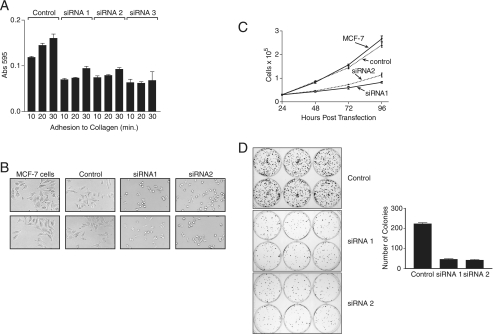
We next investigated whether suppression of RACK1 would affect the ability of MCF-7 cells to attach and spread on collagen and proliferate under different conditions. In cells with RACK1 suppressed, attachment to collagen was decreased by almost 50% in comparison to control siRNA- transfected MCF-7 cells (Fig. 2A). Furthermore, as can be seen in Fig. 2B, cell spreading after 8 h of attachment was greatly impaired compared with untransfected or control cells. Although cells in which RACK1 was suppressed were attached and viable, they clearly occupied a smaller surface area than control cells (Fig. 2B).
FIGURE 2.
RACK1 is required for cell adhesion, spreading, and foci formation. A, MCF-7 cells were transfected with RACK1 siRNA. After 20 h cells were serum-starved for 4 h, removed with trypsin EDTA, and resuspended in serum-free medium at 2 × 105 cells/ml for analysis of adhesion to collagen-coated plates. At the indicated times cells were stained with crystal violet, and absorbance was determined at 595 nm. Data are presented for quadruplicate samples. B, MCF-7 cells were transfected with siRNA, cultured for 20 h, removed with trypsin EDTA, seeded into collagen-coated wells of a 24-well plate, and allowed to adhere for 8 h before being examined and photographed with an inverted microscope (20× magnification). C, MCF-7 cells transfected with siRNA were seeded in multiple wells of a 24-well plate at a density of 3 × 104 cells/well 24 h post-transfection. At the indicated time points cells were collected, and the number and viability were determined using a hemocytometer and trypan blue exclusion. Data are presented as the mean and S.D. of live cell numbers in triplicate wells. D, MCF-7 cells transfected with siRNA were seeded in multiple wells of a six-well plate. After 24 h cells were seeded at a density of 500 cells per well in medium supplemented with 10% fetal calf serum and incubated for 14 days at 37 °C. After 14 days the cells were stained with Giemsa. The plates were photographed, and the colonies were counted. Data are presented as the mean and S.D. of colonies per well for six wells and are representative of three independent experiments with similar results.
Proliferation was assessed in normal monolayer cultures and in plating efficiency assays, which measure the ability to form foci at low density, a feature of transformed cells. In monolayer cultures the proliferation rate of MCF-7 cells with RACK1 suppressed was reduced by 60% compared with controls (Fig. 2C). In plating efficiency assays these cells also yielded fewer and smaller foci than control cells (Fig. 2D). Taken together, these data indicate that RACK1 is required for FAK phosphorylation, cell attachment, initiation of cell spreading, proliferation, and foci formation. The data suggest that RACK1 may be required for the initial stages of cell attachment necessary for both proliferation and migration.
The Binding Site for FAK in RACK 1 Resides within WD Repeats 1–3
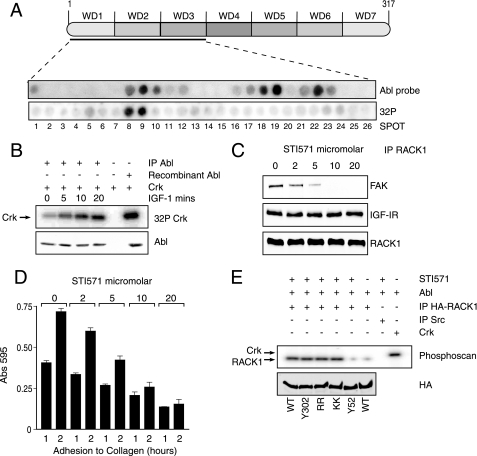
Our co-immunoprecipitation results suggest that FAK is constitutively associated with RACK1 (Fig. 1C). To determine whether this is a direct interaction and to define the site of interaction, we employed peptide arrays, which we have recently used to identify the binding site for PP2A and β1 integrin on RACK1 (15) and sites for RACK1 and β arrestin2 on PDE4D5 (33, 34). A library of overlapping peptides (25-mers) each shifted by 5 amino acids encompassing the entire sequence of RACK1 (60 in all) was spot-synthesized on nitrocellulose membranes to generate RACK1 peptide arrays. These arrays were then probed with recombinant GST or GST-FAK. GST did not bind to any peptide spot within the RACK1 array. GST-FAK bound to a number of peptides with the positive reactions indicated by dark spots (8, 9, 12, 22, and 23) (Fig. 3A). All of these peptides are derived from the RACK1 sequence spanning WD repeats 1–3.
FIGURE 3.
Identification of FAK interaction sites on RACK1 peptide arrays and alanine scanning peptide arrays. A, the amino acid sequence of the entire RACK1 protein is shown schematically (1–317) with the 7 WD repeat sequences indicated. Peptides arrays of immobilized overlapping 25-mer peptides, each shifted to the right by 5 amino acids encompassing the entire RACK1 sequence, were generated. Arrays were probed with GST-FAK, which was detected by immunoblotting with anti-GST antibody. The array shown is a representative of three arrays which gave a similar pattern of FAK binding. Positively interacting peptides generated dark spots, and non-interacting peptides left blank spots. The spots were blank in all sections of the array except for spots representing peptides in WD2 (8, 9, and 10) and WD3 (22 and 23) of RACK1. Purified GST did not interact with any peptides (not shown). B, arrays in which the 25 amino acids in peptide 10 and peptide 22 of RACK1 were sequentially substituted with alanine along the entire sequence of the 25-mer parent peptide. In cases where the parent amino acid was alanine, it was substituted for aspartate. The top panel shows the alanine substitution array for peptide 10 (Thr-46 to Val-70), and the bottom panel shows the alanine substitution array for peptide 22 (Lys-106 to Lys-130). Double alanine substitutions were also included for peptide 10 (Val-69 and Val-70, Arg-57 and Arg-60, His-62 and His-64) and for peptide 22 (Lys-106 and Asp-107, Arg-125 and Asp-126, Ser-114 and Ser-115, and Lys-127 and Lys-130). The array was probed with GST-FAK, which was detected by immunoblotting with anti-FAK antibody. A decrease in intensity in binding to the peptides after alanine substitution is indicative of decreased binding of FAK to RACK1. The binding of FAK to each alanine-substituted RACK1 peptide was quantified by densitometry and presented as a percentage of the control “parent” sequence.
To identify the specific amino acids within these peptides required for FAK binding we generated an array of peptides derived from the 25-mer parent peptides corresponding to spot 10 (Thr-46 to Val-70) and spot 22 (Lys-106 to Lys-130). For each parent peptide, 25 progeny were generated where each new peptide in the array had a single alanine substitution in successive amino acids in the sequence. These two alanine-scanning peptide arrays were then probed with recombinant GST-FAK. Results showed that FAK binding to these peptides was either severely attenuated or ablated by alanine substitution of Arg-47, Tyr-52, Arg-57, Arg-60, and Phe-65 in peptide 10 and by alanine substitution of Lys-127 and Lys-130 in peptide 22 (Fig. 3B). Dual substitution of the positively charged Arg-57 and Arg-60 in peptide 10 and the positively charged Lys-127 and Lys-130 in peptide 22 completely ablated binding of FAK. Dual substitutions of either Val-69 and Val-70 or His-62 and His-64 in peptide 10 and Lys-106 and Asp-107 or Ser-114 and Ser-115 in peptide 22 had no effect on the binding of FAK to RACK1 and, thus, provide internal controls. The interaction of FAK with the peptides was quantified by densitometry and is presented in Fig. 3B as a percentage of binding of FAK to the control parent peptide. Together these analyses confirm that the binding of FAK to RACK1 is direct and demonstrates that key amino acids required for binding are located in the RACK1 sequence immediately after WD1 and running into WD2 as well as into WD3.
Mutagenesis Substantiates the Involvement of RACK1 Blades I-III in FAK Binding
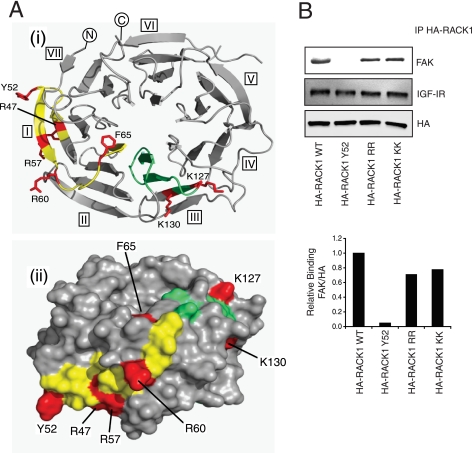
During the course of this work no crystal structure was available for RACK1, although we had previously developed a seven-bladed β-propeller RACK1 model based on homologous proteins (35). To direct subsequent mutagenesis studies, this model was used to assess the likely surface exposure of array peptide residues implicated in FAK binding. The clustered residues from peptide 10 (Arg-47, Tyr-52, Arg-57, and Arg-60) were predicted to occupy surface-exposed positions on a single β-hairpin motif (Fig. 4Ai), and peptides encompassing this region might reasonably preserve the native structure of the intact protein. In contrast, Phe-65 fell within a loop region in WD2, and its structural organization in the array peptides and mimicry of the intact protein was considered to be less assured. Nevertheless, the model predicted surface exposure of Phe-65 proximal both to the preceding peptide 10 residues and those implicated in FAK binding from array peptide 22. Lys-127 and Lys-130, residues present in peptide 22, were also predicted to occupy surface locations on RACK1 (Fig. 4Ai), consistent with their possible involvement in a protein docking interface. Other residues in peptides 10/22 (or indeed neighboring regions of RACK1) may potentially engage FAK, albeit insufficiently strong to be identified from the scanning peptide array.
FIGURE 4.
Predicted locations of key residues implicated in FAK binding. A, the sequences corresponding to array peptides 10 and 22 (respectively, yellow and green) are mapped onto a homology model of RACK1 (1); amino acids whose individual mutation to alanine significantly compromises FAK binding are highlighted in red: (i) top view of β-propeller; (ii) edge view with solvent accessible surface rendition. B, MCF-7 cells were transfected with plasmids encoding HA-RACK1, WT, Y52F, R57A/R60A, or K127A/K130A. Cell lysates were prepared and immunoprecipitated (IP) with anti-HA antibody followed by Western blotting to detect associated FAK and IGF-IR.
We focused our attention on the RACK1 residues (Tyr-52, Arg-57, Arg-60, Lys-127, and Lys-130) that had the most prominent predicted surface exposure and that were, therefore, considered strong candidates for direct interaction with FAK. These residues were mutated (Y52F, R57A/R60A, and K127A/K130A) in the context of full-length, HA-tagged RACK1, and the mutants were expressed in MCF-7 cells for comparison with HA-RACK1-WT. Each HA-tagged RACK1 protein was then immunoprecipitated with anti-HA antibody and assessed for associated FAK. The R57A/R60A and K127A/K130A mutants had decreased binding of FAK, but the Y52F mutant completely lost binding of FAK (Fig. 4B). These data clearly indicate a critical role for Tyr-52 in RACK1 for either directly or indirectly binding to FAK. Given the conservative nature of Y52F substitution (in terms of steric demand and aromatic character of the side chain), this suggests that phosphorylation of Tyr-52 in the intact RACK1 protein may be required for binding of FAK.
Tyr-52 in Propeller Blade I Is Required for RACK1 Function in Cell Adhesion
Our data suggest that RACK1 is essential for cell adhesion in MCF-7 cells and FAK phosphorylation by growth factor and adhesion signals. FAK is important for focal adhesion formation, and given our observation that Y52F mutation in RACK1 disrupts FAK binding, we predicted that cells expressing HA-RACK1 (Y52F) would have defective adhesion. To test this, we generated MCF-7 cells stably overexpressing HA-RACK1 (Y52F) and tested their ability to attach to collagen. Cells overexpressing HA-RACK1 (WT) displayed increased attachment at 1 and 2 h, but MCF-7 cells expressing HA-RACK1 (Y52F) showed a dramatic decrease in attachment to collagen compared with untransfected cells and WT cells (Fig. 5A). We then investigated whether the Y52F mutation affects the ability of RACK1 to support the transformed phenotype of MCF-7 cells in foci-forming assays. WT cells displayed increased numbers of foci (Fig. 5B). In marked contrast to this, Y52F cells exhibited decreased numbers of foci (Fig. 5B), indicating that interaction of FAK with RACK1 is essential for foci formation. These data also suggest that Y52F RACK1 may have dominant negative function in MCF-7 cells. Together, these results indicate that the interaction between RACK1 and FAK at Tyr-52 is essential for cell adhesion and foci formation.
FIGURE 5.
The RACK1 Tyr-52 mutant suppresses cell adhesion and foci formation. A, MCF-7 cells stably overexpressing HA-RACK1 Y52F were serum-starved for 4 h, removed from plates, resuspended in serum-free medium, and allowed to adhere to collagen for the indicated times. Cells were washed and stained with crystal violet, and absorbance was determined at 595 nm. Data are presented for quadruplicate samples for three individual clones and are representative of that obtained for five clones. B, MCF-7 cells stably overexpressing HA-RACK1 Y52F were seeded in multiple wells of a 6-well plate 24 h after transfection at a density of 500 cells per well and incubated for 14 days at 37 °C. After 14 days cells were stained with Giemsa, the plate was photographed, and the colonies were counted. Data are presented as the mean and S.D. of colonies per well for six wells and are representative of data obtained with five independent clones.
RACK1 Can Be Phosphorylated by c-Abl, and Inhibition of c-Abl Activity Suppresses FAK Binding to RACK1
Because Tyr-52 in RACK1 is required for interaction with FAK, we considered it likely that phosphorylation of this tyrosine may regulate FAK association with RACK1. We also hypothesized that c-Abl may be a candidate to act as the kinase to phosphorylate Tyr-52 because c-Abl associates with RACK1 in transformed cells (36) and phosphorylates focal adhesion-associated proteins (37–39). We first investigated the interaction of c-Abl with RACK1 by probing a RACK1 peptide array with recombinant c-Abl. This revealed positive reactions for peptides derived from WD repeats 1–3 (Fig. 6A) and a pattern of interaction that was very similar to that of FAK (Fig. 3A). This included the WD1/2-connecting sequence and WD repeats 2 and 3 represented by peptides 8 and 9 and peptides 21, 22, and 23. c-Abl also interacted strongly with peptides 18 and 19, which were not implicated in the FAK interaction, suggesting some differences in the way in which FAK and c-Abl interact with RACK1.
FIGURE 6.
c-Abl phosphorylates RACK1 at the FAK interaction site and c-Abl kinase activity is required for interaction of FAK with RACK1. A, A RACK1 peptide array was probed with recombinant active c-Abl, which was detected by immunoblotting with anti-c-Abl antibody. Blank spots indicate no binding and were evident in all sections of the array except for spots in WD2 (corresponding to peptides 8, 9, and 10) and WD3 (corresponding to peptides 18, 19, 22, and 23) (top). To measure phosphorylation of the RACK1 array by c-Abl kinase, the membrane was incubated in 20 ml of membrane phosphate buffer together with 1 μg of c-Abl and 32P at 30 °C for 30 min. After extensive washing, radioactivity was detected with phosphorimaging. The spots were blank in all sections of the array except for spots corresponding to peptides 8 and 9 in WD2. B, c-Abl was immunoprecipitated (IP) from MCF-7 cells which had been serum-starved for 4 h and stimulated with IGF-I for the indicated times. The immunoprecipitated proteins were subjected to an in vitro kinase assay using GST-Crk as a substrate. The total amount of c-Abl used in the kinase assay was detected by Western blotting. C, MCF-7 cells maintained in serum were pretreated with increasing concentrations of STI571. RACK1 was immunoprecipitated (IP) from the lysates and investigated for associated FAK before probing with antibodies against the IGF-IR and RACK1 to show that equal amounts of the protein were immunoprecipitated from each sample. D, MCF-7 cells maintained in serum were pretreated with increasing concentrations of STI571. The cells were then serum-starved for 4 h and resuspended in serum-free medium and allowed to adhere to collagen coated plates for the indicated times before staining with crystal violet and analysis of absorbance at 595 nm. Data are presented for quadruplicate samples. E, MCF-7 cells were transfected with plasmids encoding HA-RACK1 WT, Y302F, R57A/R60A, K127A/K130A, or Y52F. Cell lysates were prepared and immunoprecipitated with anti-HA antibody and used as a substrate in an in vitro kinase assay with c-Abl kinase. Recombinant Crk and immunoprecipitated Src were used as positive and negative controls, respectively. Incorporated radioactivity was detected by phosphorimaging.
We next asked if c-Abl could phosphorylate a target peptide within the RACK1 peptide array. To do this we incubated the RACK1 peptide array with active recombinant c-Abl and [32P]ATP in a kinase assay. A radioactive signal was detected only for peptides 8 and 9, both of which include Tyr-52 (Fig. 6A, lower panel). All peptides containing other tyrosines (Tyr-140, -194, -228, -248, and -302) in RACK1 were not phosphorylated by c-Abl (not shown). Furthermore, all other peptides where c-Abl bound (17, 18, 19, 21, 22, and 23) were negative in the kinase assay, indicating that the radioactive signal on the array did not come from RACK1-bound c-Abl but rather from the phosphorylated RACK1 peptide itself. Src did not phosphorylate the RACK1 peptide array incorporating this site (not shown). Together these data indicate that c-Abl and FAK bind to a shared region in RACK1 and suggest that phosphorylation of RACK1 on Tyr-52 is important for the interaction with FAK.
IGF-I has recently been shown to activate c-Abl kinase in MCF-7 cells (40). Furthermore, either pharmacological inhibition of c-Abl activity or siRNA-mediated suppression of c-Abl reduced cell proliferation and anchorage-independent growth (40). In agreement with Srinivasan et al. (40), we found that IGF-I induces an increase in c-Abl activity as measured by its ability to phosphorylate Crk in MCF-7 cells (Fig. 6B). We also reasoned that if c-Abl associates with and phosphorylates RACK1, then inhibition of c-Abl activity should disrupt the RACK1-FAK interaction and cell adhesion. To test this, MCF-7 cells were preincubated with increasing concentrations of the Abl kinase inhibitor ST1571 (41, 42) before immunoprecipitating RACK1 and measuring associated FAK. In untreated cells FAK was associated with RACK1, but strikingly, the amount of RACK1-associated FAK decreased with increasing concentrations of STI571. No FAK was detected in the RACK1 immunoprecipitation (Fig. 6C) in the presence of 10 μm STI571, which has been shown in previous reports to inhibit phosphorylation of c-Abl by 75% activity and switch off kinase activity in MDA-MB-435 and BT-549 cells (43). STI571 also reduced the adhesion of MCF-7 cells to collagen in a dose-dependent manner with almost complete loss of adhesion in cells cultured in the presence of 10 μm STI571 (Fig. 6D).
We next asked if phosphorylation of Tyr-52 by c-Abl occurs in vivo. We have consistently observed that tyrosine phosphorylation of RACK1 in cell lysates is barely detectable. We also predict that phosphorylation of Tyr-52 in RACK1 is likely to be a dynamic event during cell adhesion and de-adhesion. Therefore, we designed experiments that measure the ability of c-Abl to phosphorylate RACK1 that was transiently expressed in cells and then immunoprecipitated for analysis in in vitro kinase assays. If c-Abl is the correct kinase for Tyr-52, then under normal culture conditions this tyrosine should be phosphorylated by endogenous c-Abl and would not be available for further phosphorylation by c-Abl when immunoprecipitated RACK1 is subjected to in vitro kinase assays. However, in cells pretreated with 10 μm STI571, where c-Abl activity is suppressed, RACK1 Tyr-52 should not be phosphorylated and should be available for phosphorylation by Abl in in vitro kinase assays.
To test this, MCF-7 cells were transfected with WT RACK1 or a series of point mutants of RACK1 (Tyr-52, Tyr-302, Arg-57/Arg-60, Lys-127/Lys-130). Cells were pretreated with STI571 or not, and each RACK1 was then immunoprecipitated and subjected to in vitro kinase assays with recombinant c-Abl. Crk was used as a positive control for c-Abl activity (Fig. 6E). Strong phosphorylation of RACK1 WT, Y302F, R57A/R60A, and K127A/K130A was observed from cells that were pretreated with STI571 (first four lanes). This indicates that c-Abl can phosphorylate tyrosines in these immunoprecipitated RACK1 proteins. No phosphorylation of the RACK1 Y52F mutant was observed (fifth lane), indicating that this tyrosine is required for c-Abl phosphorylation of RACK1. No phosphorylation of WT RACK1 from cells that were not pretreated with ST1571 was observed (sixth lane), suggesting that the cAbl target tyrosine is already phosphorylated in these cells. Taken together these data indicate that Abl may phosphorylate RACK1 on Tyr-52 in vitro and in vivo and that c-Abl activity is required for interaction of FAK with RACK1 as well as cell adhesion.
DISCUSSION
FAK is widely regarded as being the convergence point of growth factor and integrin signaling (16, 44), yet how FAK phosphorylation is regulated within the growth factor and integrin network is not fully understood. The aim of this study was to investigate whether RACK1 is important for regulating FAK activity and the early events of cell adhesion and cell proliferation promoted by IGF-I in transformed cells. IGF-I and insulin have both been shown to induce FAK dephosphorylation, which is in contrast to both platelet-derived growth factor and EGF, which both induce FAK phosphorylation (45). These different responses may relate to the capacity of these receptors to associate with RACK1. Indeed, in contrast to the IGF-IR, there are no reports of RACK1 interacting with either the EGF or platelet-derived growth factor receptors.
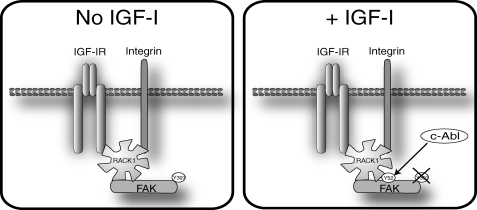
Our findings are summarized in Fig. 7. We propose that RACK1 is an essential scaffolding protein for the phosphorylation of FAK on Tyr-397 by adhesion signals and for dephosphorylation of FAK in response to IGF-I stimulation. The interaction of RACK1 and FAK occurs on an exposed surface of RACK1 involving propeller blades I-III but is disrupted by mutation of Tyr-52. This tyrosine can be phosphorylated by c-Abl, which interacts with RACK1 on the same face, and whose kinase activity is enhanced by IGF-I. Thus, we conclude that phosphorylation of Tyr-52 by c-Abl stabilizes the interaction of FAK and RACK1, which facilitates FAK activation and function (phosphorylation and dephosphorylation on Tyr-397). Thus, RACK1 is a scaffold for cross-talk between the IGF-IR and integrins in regulating FAK activity.
FIGURE 7.
Model illustrating our findings on how RACK1 association with FAK is regulated. In adherent serum-starved cells (left panel) RACK1 associates with the IGF-IR. FAK associates with RACK1 and is phosphorylated on Tyr-397, presumably because of integrin ligation. Upon stimulation with IGF-I (right panel), c-Abl activity is enhanced, and RACK1 is phosphorylated on Tyr-52, which regulates and stabilizes the interaction with FAK. FAK then becomes dephosphorylated at Tyr-397, which promotes cell migration. Suppression of RACK1 expression, inhibition of c-Abl kinase activity, or mutation of Tyr-52 are all sufficient to ablate FAK phosphorylation. Thus, the association of FAK with RACK1 at Tyr-52 is essential for activity of FAK.
IGF-I-mediated dephosphorylation of FAK has previously been associated with cell migration (24). Cell migration is enhanced when RACK1 is overexpressed and is decreased when RACK1 expression is suppressed (5). In this study we show that RACK1 expression is required for cell adhesion and spreading, which are key initiating steps in cell migration (3, 4, 6). FAK activity is essential for cell spreading (46, 47). Enhanced cell spreading is also associated with enhanced cell growth (48, 49). When RACK1 expression is suppressed, FAK is not phosphorylated on Tyr-397 and remains unresponsive to IGF-I stimulation. Our data indicate that RACK1 scaffolding is necessary for FAK to receive signals from both the IGF-IR and β1 integrin to initiate the early events of cell spreading and migration. This is supported by our observations in R− cells expressing the Y1250F/Y1251F IGF-IR mutant. In these cells RACK1 does not associate with the IGF-IR, FAK dephosphorylation is not induced by IGF-1, and cells do not migrate. The role of RACK1 in regulating FAK function is likely to be important for cooperation between IGF-IR and integrin signaling in transformed cells. Suppression of RACK1 expression dramatically reduced the proliferation and plating efficiency of MCF-7 cells (Fig. 2). Consistent with this, in prostate carcinoma cells suppression of RACK1 inhibits androgen-mediated cell growth (50). Other studies have shown that lack of Integrin-IGF-IR interaction leads to suppressed tumor growth (12).
The interaction of FAK with RACK1 appears to occur independently of Tyr-397 phosphorylation in FAK as recombinant FAK interacts with RACK1 peptide arrays and FAK can be co-immunoprecipitated constitutively with RACK1. FAK interacts with the N terminus of RACK1. To date, the N terminus encompassing WD repeats 1–4 is known to contain binding sites for protein kinase Cs, STAT3, and the IGF-IR (10, 51, 52), whereas β1 integrin, PP2A, and Src interact within sites in the C terminus (WD repeats 4–7) (4–6, 15)) The interaction of FAK with the N terminus of RACK1 is likely to facilitate recruitment of FAK to integrins and, at the same time, provide a mechanism for integration of integrin signaling with the IGF-IR signaling pathway. Clustering of integrins facilitates FAK phosphorylation on Tyr-397, which in turn facilitates the recruitment of Src to FAK (for review, see Ref. 53). Subsequent interactions with downstream signaling proteins, many of which are shared by the IGF-IR including the adaptor proteins Shc, Grb2, and the p85 subunit of phosphatidylinositol 3-kinase, Cas, and paxillin, would facilitate FAK signaling in focal adhesion formation (19, 45, 54).
RACK1 residues contributing to a putative FAK binding interface were initially identified through scanning peptide array analysis. This identified a group (Arg-47, Tyr-52, Arg-57, Arg-60, and Phe-65) in the sequence immediately after WD1 and running into WD2 and two amino acids (Lys-127 and Lys-130) in the WD3 repeat. With no crystal structure available, we used a RACK1 homology model (Fig. 4Ai) to assess the likely contribution of the identified amino acids to a FAK binding surface and to select residues for further evaluation through mutagenesis. Recent emergence (55) of a crystal structure for Arabidopsis thaliana RACK1A, a protein that shares 66% sequence identity with human RACK1, has subsequently confirmed the validity of our homology model and the predicted surface exposure of Arg-47, Tyr-52, Arg-57, Arg-60, Phe-65, Lys-127, and Lys-130. Although alanine mutation of the Arg-57/Arg-60 and Lys-127/Lys-130 pairings reduced the binding of FAK to RACK1, the single Y52F mutation was sufficient to completely abolish the interaction. Thus, Tyr-52 is another critical tyrosine in RACK1 important for protein-protein interaction. Tyr-246 in WD6 of RACK1 is important for Src binding (4, 6, 56), and we have recently shown that Tyr-302 in WD7 of RACK1 controls the binding of PP2A and β1 integrin (15).
The fact that semi-conservative mutation of Tyr-52 (to Phe) was sufficient to ablate FAK binding suggests that it is phosphorylation of this tyrosine in vivo that is the critical determinant for FAK binding. However, the absence of known phosphotyrosine binding motifs in FAK also suggests that this phosphorylation may control FAK binding indirectly by mediating conformational change in RACK1. Indeed, comparative analysis of A. thaliana RACK1A and a second RACK1 structure, recently acquired for a yeast orthologue (Asc1) with 52% sequence identity to human RACK1 (57), strongly supports this contention. As predicted by our homology model, the cognate tyrosine to Tyr-52 in RACK1A is located on a loop connecting adjacent antiparallel β-strands (Fig. 8). The tyrosine phenolic group is hydrogen-bonded to Lys-8 (conserved as Arg-8 in human RACK1). Significantly the Tyr-52 cognate residue in the yeast protein is phenylalanine 54 (Fig. 8B), and yet the presentation of this latter residue is essentially identical to Tyr-52 in RACK1A. Thus, it is unlikely that removal of the phenolic OH afforded by Y52F mutation would ablate FAK binding by inducing conformational change in RACK1. Moreover, in the RACK1 basal state the Tyr-52 phenolic group is predicted to be sequestered by interaction with Arg-8 and is unlikely to be able to engage FAK directly by hydrogen bonding. The ablation of FAK binding seen with Y52F mutation in our studies is, therefore, likely because of the loss of a required phosphorylation site rather than direct or indirect disruption of an association with RACK1 in a nonphosphorylated state.
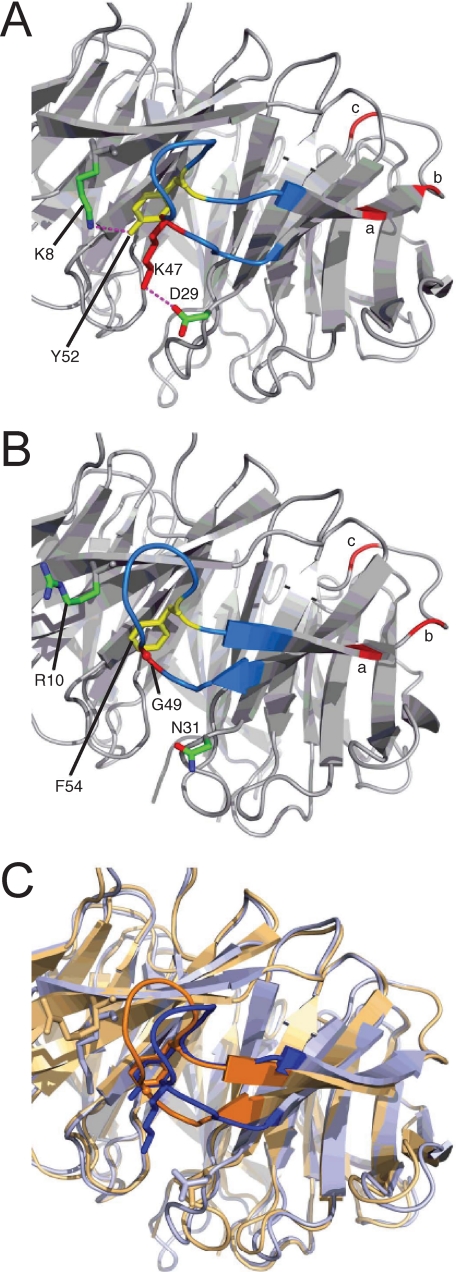
FIGURE 8.
Structural detail for the cognate Tyr-52-containing loops in RACK1 orthologues. A, crystal structure (PDB code 3DM0) of A. thaliana RACK1A (loop sequence, KLTKDDKAYGV) (55). B, crystal structure (3FRX) of Saccharomyces cerevisiae RACK1 orthologue, Asc1 (loop sequence KLTGDDQKFGV) (57). C, superimposition of A. thaliana and S. cerevisiae protein structures (respectively, blue and orange), highlighting the conserved presentation of Tyr-52/Phe-54 side chains and the local conformational difference arising from the absence (in the yeast protein) of the Asp-29–Lys-47 ion-pairing present in the A. thaliana protein.
In principle, a direct electrostatic interaction between the putative RACK1 Tyr(P)-52 tyrosine phosphate and FAK might contribute to the association of these proteins notwithstanding the absence of established phosphotyrosine binding motifs in FAK. However, an indirect role for the phosphate in driving conformational change required for FAK binding is consistent with our observation that FAK binds to the unphosphorylated RACK1 array peptides, where greater flexibility in these peptides, abstracted from the full-length RACK1 protein, could allow their adoption of a FAK binding conformation without the need for activation by a phosphate switch. Compromised FAK binding capacity with Y52A mutation in array peptide 10 may, however, signify partial involvement of the tyrosine benzylic side chain for interaction with FAK. The structural impact of Tyr-52 phosphorylation is unclear at present, although the predicted orientation of Arg-47 conserved as (Lys-47 in RACK1A, Fig. 8A) on the opposing limb of the Tyr-52-containing loop may be significant. Although the tyrosine phenol and terminal ammonium functionalities of these residues are proximal (4.8 Å), they lack a tight hydrogen-bonded pairing in the RACK1A protein. Instead, the side chain of Lys-47 forms a salt bridge to Asp-29 (also conserved in human RACK1). Given the strong sequence conservation in this region of the protein, it is probable that the unphosphorylated Tyr-52-containing loop in human RACK1 adopts a similar conformation to that seen in the RACK1A crystal structure. Tyrosine phosphorylation would reasonably lead to a significant conformational change in this loop region, and the predicted proximity of Tyr-52 and Arg-47 suggests the likely establishment of a strong Arg-47–Tyr(P)-52 salt bridge pairing. This could serve to disengage the putative Arg-47–Asp-29 interaction in the unphosphorylated protein, whereas the predicted contact with Arg-8 may or may not be retained post-phosphorylation. Notably the cognate residues to Arg-47 and Asp-29 are, respectively, Gly and Asn in the yeast protein (Gly-49 and Asn-31, Fig. 8B), contrasting with the Arabidopsis and Drosophila orthologues that conserve Asp, Arg/Lys, and Tyr residues at the positions corresponding to Asp-29, Arg-47, and Tyr-52. It may be, therefore, that a basic residue corresponding to Arg-47 is redundant in the absence of a neighboring tyrosine phosphorylation site. Moreover, analysis of the yeast RACK1 protein structure suggests that it is the absence of the Arabidopsis Lys-47–Asp-29 ion-pairing that gives rise to the local conformational differences between the two crystal structures (Fig. 8C). Thus, phosphorylation-driven reorganization of this loop region of human RACK1 may plausibly involve Arg-47–Asp-29 disengagement resulting from the establishment of a Tyr(P)-52–Arg-47 ion pairing, and this may contribute to local conformational changes that facilitate FAK binding across propeller blades I-III.
Given the evidence suggesting a role for Tyr-52 as a site for phosphorylation that induces conformational change in RACK1, switching the protein into a FAK binding state, we next sought to identify a candidate kinase that might mediate this phosphorylation. Our attention focused on c-Abl kinase, which has previously been shown to bind to RACK1 (36). We determined that c-Abl interacts with RACK1 within WD repeats 2 and 3 and demonstrated that c-Abl kinase phosphorylates RACK1 peptides containing Tyr-52 in RACK1 peptide arrays. Furthermore, Y52F mutation is sufficient to remove c-Abl-mediated phosphorylation of RACK1. Our finding that inhibition of c-Abl activity with STI571 disrupts the RACK1-FAK interaction and cell adhesion in a dose-dependent manner confirms that c-Abl activity may modulate FAK binding to RACK1 and strongly suggests that c-Abl phosphorylates RACK1 at Tyr-52 in cells under physiological conditions. FAK activity, cell adhesion, and foci formation in MCF-7 cells were all significantly reduced in cells expressing the RACK1 Y52F mutant. The mutant exhibits dominant negative function, which further highlights the importance of Tyr-52 for interaction with FAK. In summary we have identified FAK as a novel RACK1-interacting protein. We conclude that the association of FAK with RACK1 is dependent on phosphorylation at Tyr-52 by c-Abl and that this is essential for IGF-I-mediated cell migration.
Acknowledgments
We are grateful to Kurt Tidmore for assistance with the illustrations and to our colleagues in the Cell Biology Laboratory for helpful discussions and critical review.
This work was supported by the Health Research Board of Ireland, Cancer Research Ireland, and Science Foundation Ireland as well as Medical Research Council (UK) Grants G0600765 and G0400053, European Union Grant 037189, and Fondation Leducq (Paris) Grant 06VD02 (to M. D. H. and G. S. B.).
- RACK1
- receptor for activated C kinase
- siRNA
- small interfering RNA
- ERK
- extracellular signal-regulated kinase
- GST
- glutathione S-transferase
- FAK
- focal adhesion kinase
- IGF-IR
- insulin-like growth factor I receptor
- EGF
- epidermal growth factor
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin.
REFERENCES
- 1.McCahill A., Warwicker J., Bolger G. B., Houslay M. D., Yarwood S. J. (2002) Mol. Pharmacol. 62, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 2.Sklan E. H., Podoly E., Soreq H. (2006) Prog. Neurobiol. 78, 117–134 [DOI] [PubMed] [Google Scholar]
- 3.Buensuceso C. S., Woodside D., Huff J. L., Plopper G. E., O'Toole T. E. (2001) J. Cell Sci. 114, 1691–1698 [DOI] [PubMed] [Google Scholar]
- 4.Cox E. A., Bennin D., Doan A. T., O'Toole T., Huttenlocher A. (2003) Mol. Biol. Cell 14, 658–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiely P. A., O'Gorman D., Luong K., Ron D., O'Connor R. (2006) Mol. Cell. Biol. 26, 4041–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doan A. T., Huttenlocher A. (2007) Exp. Cell Res. 313, 2667–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onishi I., Lin P. J., Diering G. H., Williams W. P., Numata M. (2007) Cell. Signal. 19, 194–203 [DOI] [PubMed] [Google Scholar]
- 8.Mamidipudi V., Chang B. Y., Harte R. A., Lee K. C., Cartwright C. A. (2004) FEBS Lett. 567, 321–326 [DOI] [PubMed] [Google Scholar]
- 9.Vomastek T., Iwanicki M. P., Schaeffer H. J., Tarcsafalvi A., Parsons J. T., Weber M. J. (2007) Mol. Cell. Biol. 27, 8296–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermanto U., Zong C. S., Li W., Wang L. H. (2002) Mol. Cell. Biol. 22, 2345–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynch L., Vodyanik P. I., Boettiger D., Guvakova M. A. (2005) Mol. Biol. Cell 16, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goel H. L., Breen M., Zhang J., Das I., Aznavoorian-Cheshire S., Greenberg N. M., Elgavish A., Languino L. R. (2005) Cancer Res. 65, 6692–6700 [DOI] [PubMed] [Google Scholar]
- 13.Leahy M., Lyons A., Krause D., O'Connor R. (2004) J. Biol. Chem. 279, 18306–18313 [DOI] [PubMed] [Google Scholar]
- 14.O'Connor R., Kauffmann-Zeh A., Liu Y., Lehar S., Evan G. I., Baserga R., Blättler W. A. (1997) Mol. Cell. Biol. 17, 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiely P. A., Baillie G. S., Lynch M. J., Houslay M. D., O'Connor R. (2008) J. Biol. Chem. 283, 22952–22961 [DOI] [PubMed] [Google Scholar]
- 16.Mitra S. K., Schlaepfer D. D. (2006) Curr. Opin. Cell Biol. 18, 516–523 [DOI] [PubMed] [Google Scholar]
- 17.Mon N. N., Ito S., Senga T., Hamaguchi M. (2006) Ann. N. Y. Acad. Sci. 1086, 199–212 [DOI] [PubMed] [Google Scholar]
- 18.Schlaepfer D. D., Hou S., Lim S. T., Tomar A., Yu H., Lim Y., Hanson D. A., Uryu S. A., Molina J., Mitra S. K. (2007) J. Biol. Chem. 282, 17450–17459 [DOI] [PubMed] [Google Scholar]
- 19.Schlaepfer D. D., Mitra S. K. (2004) Curr. Opin. Genet. Dev. 14, 92–101 [DOI] [PubMed] [Google Scholar]
- 20.Hsia D. A., Mitra S. K., Hauck C. R., Streblow D. N., Nelson J. A., Ilic D., Huang S., Li E., Nemerow G. R., Leng J., Spencer K. S., Cheresh D. A., Schlaepfer D. D. (2003) J. Cell Biol. 160, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W., Duzgun A., Sumpio B. E., Basson M. D. (2001) Am. J. Physiol. Gastrointest. Liver Physiol. 280, G75–G87 [DOI] [PubMed] [Google Scholar]
- 22.Wang J. G., Miyazu M., Xiang P., Li S. N., Sokabe M., Naruse K. (2005) Life Sci. 76, 2817–2825 [DOI] [PubMed] [Google Scholar]
- 23.Webb D. J., Donais K., Whitmore L. A., Thomas S. M., Turner C. E., Parsons J. T., Horwitz A. F. (2004) Nat. Cell Biol. 6, 154–161 [DOI] [PubMed] [Google Scholar]
- 24.Guvakova M. A., Surmacz E. (1999) Exp. Cell Res. 251, 244–255 [DOI] [PubMed] [Google Scholar]
- 25.Kiely P. A., Leahy M., O'Gorman D., O'Connor R. (2005) J. Biol. Chem. 280, 7624–7633 [DOI] [PubMed] [Google Scholar]
- 26.Mañes S., Mira E., Gómez-Mouton C., Zhao Z. J., Lacalle R. A., Martínez-A C. (1999) Mol. Cell. Biol. 19, 3125–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mauro L., Sisci D., Bartucci M., Salerno M., Kim J., Tam T., Guvakova M. A., Ando S., Surmacz E. (1999) Exp. Cell Res. 252, 439–448 [DOI] [PubMed] [Google Scholar]
- 28.Kiely P. A., Sant A., O'Connor R. (2002) J. Biol. Chem. 277, 22581–22589 [DOI] [PubMed] [Google Scholar]
- 29.Sell C., Dumenil G., Deveaud C., Miura M., Coppola D., DeAngelis T., Rubin R., Efstratiadis A., Baserga R. (1994) Mol. Cell. Biol. 14, 3604–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank R. (2002) J. Immunol. Methods 267, 13–26 [DOI] [PubMed] [Google Scholar]
- 31.Frank R., Overwin H. (1996) Methods Mol. Biol. 66, 149–169 [DOI] [PubMed] [Google Scholar]
- 32.Kramer A., Schneider-Mergener J. (1998) Methods Mol. Biol. 87, 25–39 [DOI] [PubMed] [Google Scholar]
- 33.Bolger G. B., Baillie G. S., Li X., Lynch M. J., Herzyk P., Mohamed A., Mitchell L. H., McCahill A., Hundsrucker C., Klussmann E., Adams D. R., Houslay M. D. (2006) Biochem. J. 398, 23–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baillie G. S., Adams D. R., Bhari N., Houslay T. M., Vadrevu S., Meng D., Li X., Dunlop A., Milligan G., Bolger G. B., Klussmann E., Houslay M. D. (2007) Biochem. J. 404, 71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steele M. R., McCahill A., Thompson D. S., MacKenzie C., Isaacs N. W., Houslay M. D., Bolger G. B. (2001) Cell. Signal. 13, 507–513 [DOI] [PubMed] [Google Scholar]
- 36.Huang C. C., Liu C. H., Chuang N. N. (2008) Int. J. Biochem. Cell Biol. 40, 423–431 [DOI] [PubMed] [Google Scholar]
- 37.Mitsushima M., Takahashi H., Shishido T., Ueda K., Kioka N. (2006) FEBS Lett. 580, 4288–4295 [DOI] [PubMed] [Google Scholar]
- 38.Renshaw M. W., Lewis J. M., Schwartz M. A. (2000) Oncogene 19, 3216–3219 [DOI] [PubMed] [Google Scholar]
- 39.Zvara A., Fajardo J. E., Escalante M., Cotton G., Muir T., Kirsch K. H., Birge R. B. (2001) Oncogene 20, 951–961 [DOI] [PubMed] [Google Scholar]
- 40.Srinivasan D., Sims J. T., Plattner R. (2008) Oncogene 27, 1095–1105 [DOI] [PubMed] [Google Scholar]
- 41.Druker B. J., Sawyers C. L., Kantarjian H., Resta D. J., Reese S. F., Ford J. M., Capdeville R., Talpaz M. (2001) N. Engl. J. Med. 344, 1038–1042 [DOI] [PubMed] [Google Scholar]
- 42.Druker B. J., Talpaz M., Resta D. J., Peng B., Buchdunger E., Ford J. M., Lydon N. B., Kantarjian H., Capdeville R., Ohno-Jones S., Sawyers C. L. (2001) N. Engl. J. Med. 344, 1031–1037 [DOI] [PubMed] [Google Scholar]
- 43.Srinivasan D., Plattner R. (2006) Cancer Res. 66, 5648–5655 [DOI] [PubMed] [Google Scholar]
- 44.Guan J. L. (1997) Matrix Biol. 16, 195–200 [DOI] [PubMed] [Google Scholar]
- 45.Sieg D. J., Hauck C. R., Ilic D., Klingbeil C. K., Schaefer E., Damsky C. H., Schlaepfer D. D. (2000) Nat. Cell Biol. 2, 249–256 [DOI] [PubMed] [Google Scholar]
- 46.Owen J. D., Ruest P. J., Fry D. W., Hanks S. K. (1999) Mol. Cell. Biol. 19, 4806–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlaepfer D. D., Hauck C. R., Sieg D. J. (1999) Prog. Biophys. Mol. Biol. 71, 435–478 [DOI] [PubMed] [Google Scholar]
- 48.Sechler J. L., Corbett S. A., Wenk M. B., Schwarzbauer J. E. (1998) Ann. N.Y. Acad. Sci. 857, 143–154 [DOI] [PubMed] [Google Scholar]
- 49.Sottile J., Hocking D. C., Swiatek P. J. (1998) J. Cell Sci. 111, 2933–2943 [DOI] [PubMed] [Google Scholar]
- 50.Kraus S., Gioeli D., Vomastek T., Gordon V., Weber M. J. (2006) Cancer Res. 66, 11047–11054 [DOI] [PubMed] [Google Scholar]
- 51.Ron D., Jiang Z., Yao L., Vagts A., Diamond I., Gordon A. (1999) J. Biol. Chem. 274, 27039–27046 [DOI] [PubMed] [Google Scholar]
- 52.Zhang W., Zong C. S., Hermanto U., Lopez-Bergami P., Ronai Z., Wang L. H. (2006) Mol. Cell. Biol. 26, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sieg D. J., Hauck C. R., Schlaepfer D. D. (1999) J. Cell Sci. 112, 2677–2691 [DOI] [PubMed] [Google Scholar]
- 54.Schlaepfer D. D., Mitra S. K., Ilic D. (2004) Biochim. Biophys. Acta 1692, 77–102 [DOI] [PubMed] [Google Scholar]
- 55.Ullah H., Scappini E. L., Moon A. F., Williams L. V., Armstrong D. L., Pedersen L. C. (2008) Protein Sci. 17, 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mamidipudi V., Dhillon N. K., Parman T., Miller L. D., Lee K. C., Cartwright C. A. (2007) Oncogene 26, 2914–2924 [DOI] [PubMed] [Google Scholar]
- 57.Coyle S. M., Gilbert W. V., Doudna J. A. (2009) Mol. Cell. Biol. 29, 1626–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]