Abstract
The binding and structural studies of bovine lactoperoxidase with three aromatic ligands, acetylsalicylic acid (ASA), salicylhydoxamic acid (SHA), and benzylhydroxamic acid (BHA) show that all the three compounds bind to lactoperoxidase at the substrate binding site on the distal heme side. The binding of ASA occurs without perturbing the position of conserved heme water molecule W-1, whereas both SHA and BHA displace it by the hydroxyl group of their hydroxamic acid moieties. The acetyl group carbonyl oxygen atom of ASA forms a hydrogen bond with W-1, which in turn makes three other hydrogen-bonds, one each with heme iron, His-109 Nϵ2, and Gln-105 Nϵ2. In contrast, in the complexes of SHA and BHA, the OH group of hydroxamic acid moiety in both complexes interacts with heme iron directly with Fe-OH distances of 3.0 and 3.2Å respectively. The OH is also hydrogen bonded to His-109 Nϵ2 and Gln-105Nϵ2. The plane of benzene ring of ASA is inclined at 70.7° from the plane of heme moiety, whereas the aromatic planes of SHA and BHA are nearly parallel to the heme plane with inclinations of 15.7 and 6.2°, respectively. The mode of ASA binding provides the information about the mechanism of action of aromatic substrates, whereas the binding characteristics of SHA and BHA indicate the mode of inhibitor binding.
Lactoperoxidase (LPO)4 (EC 1.11.1.7) is a member of the family of glycosylated mammalian heme-containing peroxidase enzymes which also includes myeloperoxidase (MPO), eosinophil peroxidase (EPO), and thyroid peroxidase. These enzymes also show functional similarities to non-homologous plant and fungal peroxidases because they follow a similar scheme of reaction (1–3), but their modes of ligand binding differ considerably. Furthermore, the association of the prosthetic heme group in mammalian peroxidases is through covalent bonds (4–9), whereas covalent linkages are absent in other peroxidases (10–14). Among the four mammalian peroxidases, the prosthetic heme group is linked through three covalent bonds in MPO, whereas in LPO, EPO, and thyroid peroxidase only two covalent linkages are formed. So far the detailed crystal structures of only two mammalian peroxidases, MPO and LPO, are known (15–20). One of the most striking differences between these two mammalian peroxidases is concerned with the basic structural organization in which MPO exists as a covalently linked dimer, whereas LPO is a monomeric protein. At present the fundamental questions pertaining to mammalian heme peroxidases are (i) what distinguishes between the aromatic ligands that one ligand acts as a substrate, whereas the other ligand works as an inhibitor and (ii) how the substrate and inhibitor specificities differ between two enzymes lactoperoxidase and myeloperoxidase.
Lactoperoxidase oxidizes inorganic ions, preferentially thiocyanate (SCN−), and to a lesser extent, bromide (Br−), whereas in the case of myeloperoxidase the chloride (Cl−) ion is a preferred substrate (21, 22). The mammalian peroxidases including LPO are also involved in catalyzing the single electron oxidation of various physiologically important organic aromatic substrates including phenols (23, 24), catecholamines, and catechols (25–27) as well as other experimental model compounds such as aromatic amines (28), polychlorinated biphenyls (29), steroid hormones (30–32), and polycyclic aromatic hydrocarbons (33). However, the mode of binding of aromatic ligands and associated functional implications are not yet clearly understood. Surprisingly, the structural data on the complexes of mammalian peroxidases with aromatic ligands are conspicuously lacking. The only available structural report is on the complex of MPO with salicylhydroxamic acid (SHA) (34). Even in this case, the coordinates of this structure are not available for a detailed analysis. In the case of non-homologous plant peroxidases, a few crystal structures of their complexes with aromatic compounds are available (35–38), but their modes of binding are not very similar to those of mammalian peroxidases because the distal ligand binding sites in mammalian and plant peroxidases differ markedly. In this regard it is pertinent to note that the substrate binding site in peroxidases, in general, is observed at the δ-heme edge of the heme moiety on the distal side; in plant peroxidases an additional ligand binding site has also been observed at γ-heme edge (39–41). Unlike those in mammalian peroxidases where the heme moiety is buried deeply in the protein core, in plant peroxidases it is located close to the surface of the protein. Therefore, to characterize the mode of binding of the aromatic substrates and aromatic inhibitors and also for defining the subsites in the substrate binding site, we have determined the crystal structures of three complexes of bovine lactoperoxidase with aromatic ligands, acetylsalicylic acid (ASA), SHA, and benzylhydroxamic acid (BHA). Acetylsalicylic acid can be oxidized by lactoperoxidase to ASA free radical (42), whereas both salicylhydroxamic acid and benzylhydroxamic acid act as potent inhibitors of mammalian peroxidases (43–47). The determination of binding characteristics of these compounds having different actions has helped in establishing the relationship between the modes of binding and their potential actions as the substrates and inhibitors. To the best of our knowledge, this is the first structural report on the modes of binding of three aromatic ligands, ASA, SHA, and BHA, to LPO as well as the first structural study of the complexes of any mammalian peroxidase with ASA and BHA. These studies have shown that ASA, SHA, and BHA bind to LPO at the substrate binding site on the distal side. The SHA and BHA directly interact with the heme iron, whereas ASA interacts through the heme water molecule, which in turn is hydrogen-bonded to the heme iron. These studies have provided a greater insight into the mechanisms of substrate and inhibitor binding in the two mammalian peroxidases.
EXPERIMENTAL PROCEDURES
Purification of the Protein and Measurement of Its Activity and Inhibition
Fresh bovine mammary gland secretions were collected from Indian Veterinary Research Institute, Izatnagar, India. The protein was isolated and purified using the procedures reported earlier (18). The purified samples were lyophilized. To find out the effects of binding of ASA, SHA, and BHA to LPO in the crystals, the activity tests were carried out using various protein samples. The activity of the protein samples before crystallization was estimated by dissolving lyophilized protein samples in 0.1 m phosphate buffer, pH 6.0, using the procedure described earlier by Kumar et al. (48). 3.0 ml of 1 mm 2,2′-azino-bis(3-ethylbenzthiazoline-sulfonic acid (ABTS) in phosphate buffer (0.1 m, pH 6.0) was mixed with 0.1 ml of 0.5 mg/ml protein solution containing 0.1% gelatin to initialize the spectrophotometer Lambda 25 (PerkinElmer Life Sciences). 0.1 ml of 3.2 mm hydrogen peroxide was added to the above solution. The absorbance was measured at 412 nm as a function of the oxidized product of ABTS. The protein samples were also obtained from the crystals of LPO complexes with ASA, SHA, and BHA. These crystal samples were also used for activity measurements under identical conditions. In the crystal samples the protein and ligand concentrations were assumed to be at 1:1 molar ratio.
Co-crystallizations of LPO with ASA, SHA, and BHA
The freshly purified and lyophilized samples of protein were dissolved in 0.01 m phosphate buffer to a concentration of 25 mg/ml. The ligands (ASA, SHA, and BHA) were dissolved in the above buffer containing 30% (v/v) methanol. Both solutions were mixed in equal volume, giving protein to ligand molar ratios of ∼1:10. A reservoir solution containing 0.2 m ammonium iodide and 20% (w/v) PEG-3350 was prepared. 6 μl of protein-ligand solution was mixed with 6 μl of reservoir solution to prepare 12 μl of drops for the hanging drop vapor diffusion method. This experiment was repeated for all the three set-ups of crystallization for LPO with ASA, SHA, and BHA. The rectangular dark brown-colored crystals measuring up to 0.4 × 0.3 × 0.2 mm3 were obtained after 5 days.
Detection of ASA, SHA, and BHA in Crystals
To confirm the presence of ASA, SHA, and BHA in the crystals of LPO complexes, the crystals of three complexes were used for the identifications of ASA, SHA, and BHA. Initially, the crystals were washed with distilled water and then dissolved separately in the buffer containing 50 mm Tris-HCl, pH 8.0. These crystal solutions were ultrafiltered in the same buffer containing 1 m NaCl using a membrane with a cutoff of 1 kDa.
The presence of ASA in the filtrate was observed by adding Fe3+ ions. When treated with basic solution the acetylsalicylic acid hydrolyzes quickly to salicylic acid. The salicylate ions form an intensely colored complex with the ferric ion in acidic solution, and the absorption of this complex was measured at wavelength of 530 nm using the spectrophotometer Lambda 25 (PerkinElmer Life Sciences).
For the detection of the hydroxamic acid in the crystals of the complexes of LPO with SHA and BHA in respective filtrates, the color reaction with ferric chloride was employed (49). Ferric-hydroxamic acid complex gives a purple color. This was observed in both filtrates.
X-ray Intensity Data Collection
Three x-ray intensity data sets were collected, one each for LPO complexes with ASA, SHA, and BHA at 283 K using a 345-mm diameter MAR Research dtb imaging plate scanner mounted on a Rigaku RU-300 rotating anode x-ray generator operating at 100 mA and 50 kV. The CuKα radiation was produced using Osmic Blue confocal optics. All the three data sets were processed using programs DENZO and SCALEPACK (50). The summary of final data collection statistics is given in Table 1.
TABLE 1.
Data collection and refinement statistics for the three complexes of LPO with ASA, SHA, and BHA
The values in the parentheses correspond to the values in the highest resolution shell. NAG, N-acetylglucosamine.
| LPO-ASA | LPO-SHA | LPO-BHA | |
|---|---|---|---|
| Parameters | |||
| Space group | P21 | P21 | P21 |
| Unit cell dimensions | |||
| a (Å) | 54.6 | 54.6 | 54.5 |
| b (Å) | 80.5 | 80.5 | 80.1 |
| c (Å) | 77.9 | 77.8 | 68.6 |
| β (°) | 102.6 | 102.6 | 94.0 |
| Vm(Å3/Da) | 2.4 | 2.4 | 2.2 |
| Solvent content (%) | 48 | 48 | 45 |
| Resolution range (Å) | 51.8-2.5 | 51.8-2.3 | 51.8-2.9 |
| Highest resolution shell | 2.59-2.5 | 2.40-2.3 | 3.00-2.9 |
| Total number of measured reflections | 159,343 | 155,826 | 136,457 |
| Number of unique reflections | 22,651 | 27,454 | 12,029 |
| Completeness of data (%) | 98.9 (95.1) | 98.5 (95.2) | 91.5 (94.6) |
| Rsym (%) | 14.3 (39.2) | 13.5 (43.2) | 15.3 (48.9) |
| I/σ(I) | 5.3 (1.7) | 6.2 (1.4) | 5.1 (1.4) |
| Refinement | |||
| Rcryst (%) | 20.1 | 18.9 | 19.9 |
| Rfree (%) | 22.2 | 20.6 | 22.4 |
| Protein atoms | 4,774 | 4,774 | 4,774 |
| Hem group (1) atoms | 43 | 43 | 43 |
| Ligand (1) atoms | 13 | 11 | 10 |
| Thiocyanate molecules | 1 | 1 | 1 |
| Iodide ions | 8 | 8 | 8 |
| Calcium ion | 1 | 1 | 1 |
| NAG residues (8) (N-linked) | 112 | 112 | 112 |
| MAN residues (2) (N-linked) | 22 | 22 | 22 |
| Water oxygen atoms | 315 | 351 | 164 |
| r.m.s.d. in bond lengths (Å) | 0.01 | 0.01 | 0.01 |
| r.m.s.d. in bond angles (°) | 2.1 | 1.6 | 1.9 |
| r.m.s.d. in torsion angles (°) | 24.6 | 24.3 | 24.3 |
| Mean B-factor for main-chain atoms | 34.3 | 34.4 | 34.6 |
| Mean B-factor for side-chain atoms and waters | 36.8 | 37.4 | 36.6 |
| Mean B-factor for all atoms | 35.7 | 36.0 | 35.7 |
| Ramachandran plot | |||
| Residues in the most allowed regions (%) | 86.7 | 89.3 | 85.5 |
| Residues in the additionally allowed regions (%) | 11.9 | 9.6 | 13.3 |
| Residues in the generously allowed regions (%) | 1.4 | 1.2 | 1.2 |
| Protein Data Bank codes | 3GCL | 3GCJ | 3GCK |
Structure Determination and Refinement
The structures of the three complexes of LPO with ASA (LPO-ASA), SHA (LPO-SHA), and BHA (LPO-BHA) were refined using the model of native structure of bovine lactoperoxidase (Protein Data Bank code 3GC1). Initially, the coordinates were subjected to 25 cycles of rigid body refinement with REFMAC (51) from the CCP4i, Version 6.1 program package (52). After the first round of refinement, the Rcryst and Rfree factors reduced approximately to values of 0.30 and 0.40, respectively (5% of the reflections were used for the calculations of Rfree; these reflections were not included in the refinement). The initial maps showed good electron densities for the heme groups in all the three structures. The coordinates of heme groups were included in the further rounds of refinement that were carried out with intermittent manual model building of the protein using 2 Fo − Fc Fourier and Fo − Fc difference Fourier maps with graphics programs O (53) and Coot (54) on a Silicon Graphics O2 Work station. At the end of these steps of refinements, the Rcryst and Rfree factors converged to approximate values of 0.25 and 0.30, respectively. The difference Fourier Fo − Fc maps calculated at this stage revealed interpretable electron densities for glycan chains at four sites with two N-acetylglucosamine (NAG) residues and one mannose (MAN) at Asn-95, two NAG residues at Asn-205, two NAG and one MAN residues at Asn-241, and two NAG residues at Asn-332. The additional non-protein electron densities were also observed at the distal heme cavity into which the molecules of ASA, SHA, and BHA were modeled unambiguously (Fig. 1, A, B, and C). The electron density for one calcium ion was also observed in all the three structures. These maps also revealed additional electron densities for eight iodide ions in the structures. The coordinates of all these were included in the subsequent rounds of refinements. The difference Fo − Fc Fourier maps at the final stages of refinement also indicated additional electron densities for the phosphorylation of Ser-198 in all the three structures. The positions of 315 water oxygen atoms in LPO-ASA, 351 water oxygen atoms in LPO-SHA, and 164 water oxygen atoms in LPO-BHA were determined using the usual criteria of correctness. The refinements finally converged with Rcryst (Rfree) values of 0.201 (0.222), 0.189 (0.206), and 0.199 (0.224) for LPO-ASA, LPO-SHA, and LPO-BHA, respectively. The final refinement statistics are included in Table 1. The refined atomic coordinates and structure factors for LPO-ASA, LPO-SHA, and LPO-BHA have been deposited in the Protein Data Bank with accession codes 3GCL, 3GCJ, and 3GCK, respectively.
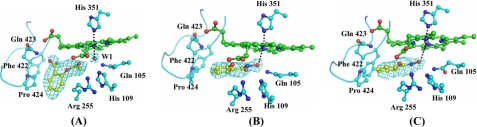
FIGURE 1.
A, results showing ASA fitted into 1.5 σ ( Fo − Fc ) electron density. A few residues in the proximity are also indicated. The W-1 corresponds to W-618 in 3GCL. B, SHA fitted into 1.5 σ ( Fo − Fc ) electron density. C, SHA fitted into 1.5 σ ( Fo − Fc ) electron density. The heme moiety, distal Gln-105, and His-109 are also shown. Figures were drawn using Pymol (55).
RESULTS
Activity/Inhibition of LPO
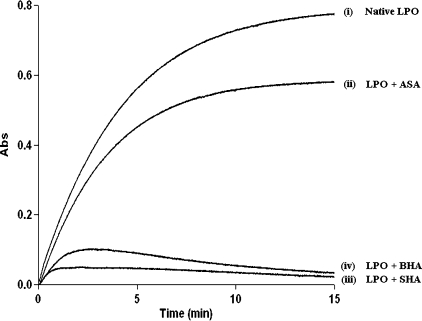
The activity curves obtained for the four samples, protein before crystallization (i) and protein in complex with ASA (ii), SHA (iii), and BHA (iv), are shown in Fig. 2. The activity of LPO was estimated using these curves in the form of units of activity/ml, where 1 unit of activity is defined as that amount of enzyme catalyzing the oxidation of 1 μm of ABTS min−1 at 293 K (molar absorption coefficient 32400 m−1 cm−1). It may be mentioned here that the inhibition of the activity of lactoperoxidase can be caused either by physically blocking the substrate binding site or by preventing interaction of heme iron with the substrate. The protein sample before crystallization showed the peroxidase activity of 6.0 units/ml. The corresponding values for the ASA, SHA, and BHA complexes were estimated to be as 4.5, 0.8, and 1.1 units/ml, respectively. These values indicate that the peroxidase activity was significantly inhibited with SHA and BHA, but with ASA a significant activity was still observed. The fact that all the three ligands were detected in the crystals suggested one mode of binding for ASA and a different mode of binding for SHA and BHA. The structures of the LPO complexes have shown that SHA and BHA bind to heme iron directly. Whereas ASA is locked at the position away from the heme iron. The oxidation of ABTS in the presence of ASA and failure to get oxidized in the presence of SHA and BHA confirms that SHA and BHA block the binding of H2O2 to heme iron, whereas ASA does not block it.
FIGURE 2.
Activity of lactoperoxidase in the native protein (i), in the presence of ASA (ii), in the presence of SHA (iii), and in the presence of BHA (iv) using ABTS as substrate. The protein ligand molar ratios were 1: 1.
Overall Structures
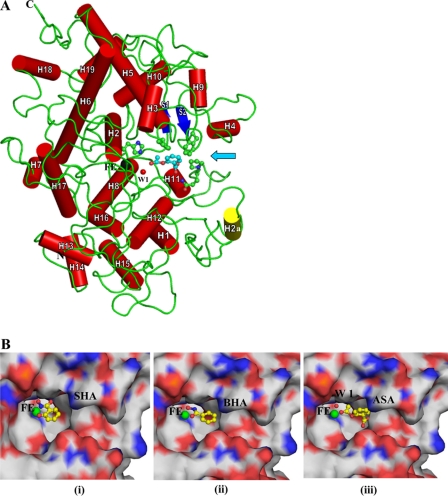
The overall folding of the protein is shown in Fig. 3A. The r.m.s. shifts for the Cα atoms of LPO-ASA, LPO-SHA, and LPO-BHA when superimposed on the native structure of LPO (3GC1) were found to be 0.7, 0.7, and 0.8 Å, respectively. When the side chains of residues, Gln-105, His-109, Phe-113, Arg-255, Glu-258, Phe-380, Phe-381, Phe-422, Gln-423, and Pro-424 of the substrate binding site were superimposed, it showed r.m.s. shifts of 0.6, 0.5, and 0.5 Å, respectively, indicating that the structure of the substrate binding site is not disturbed upon the binding of these ligands. The dimensions of the substrate binding site on the distal heme side are ∼14 × 10 × 8 Å. In all the three structures, the aromatic organic ligands have been observed on the distal heme side. As seen from Fig. 3B (iii), ASA is located at a position that is farther away from the heme iron than the positions of SHA and BHA. The distances of the farthest/nearest ligand atoms from the heme iron for ASA, SHA, and BHA are 10.4/4.6, 8.1/3.0, and 8.8/3.3 Å, respectively. The orientation of ASA in the substrate binding site is also considerably different from those of SHA and BHA as the plane of the aromatic ring of ASA is rotated by 70.7° with respect to the plane of heme moiety, whereas those of SHA and BHA are rotated at 15.7 and 6.2°, respectively.
FIGURE 3.
A, the overall folding of the protein shown as a schematic, where α-helices are indicated as cylinders (red) and numbered. H2a is a unique α-helix (yellow) present only in LPO but absent in MPO. The two anti-parallel β-strands are drawn as arrows (blue). For sake of clarity, only ASA (magenta) is shown in the substrate binding site. Heme iron (Fe) is shown in black, whereas conserved heme water molecule (W-1) is indicated in red. The heme moiety is not shown to avoid crowding. Some of the residues, His-109, Phe-113, Phe-380, Gln-423, and Pro-424, of the substrate binding site are shown in ball and stick representation. An arrow indicates the entrance to the substrate binding site. B, close-up view of ligands bound at the distal site ASA (i), SHA (ii), and BHA (iii). The heme iron atom is indicated in green. The ligands are represented as ball and stick models.
Structure of LPO-Acetylsalicylic Acid
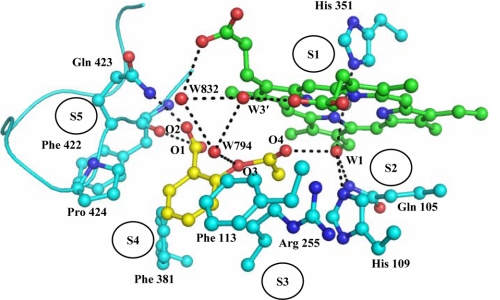
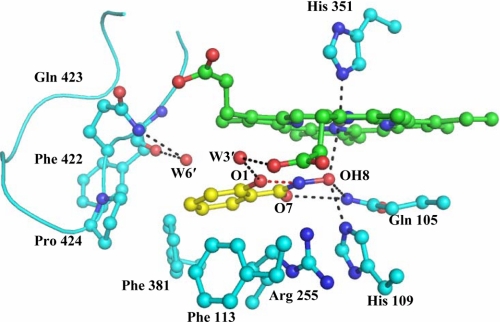
In the native structure of LPO (3GC1) six water molecules, W-1, W-2′, W-3′, W-4′, W-5′, and W-6′ were observed in the substrate binding site on the distal side. As seen from Fig. 4, ASA binds to LPO at the distal cavity by replacing four conserved water molecules, W-2′, W-4′, W-5′, and W-6′. The remaining two conserved water molecules, W-1 and W-3′, and another two water molecules, W-794 and W-832, are part of an extensive hydrogen-bonded network between LPO and ASA. The structure shows that both porphyrin and pyrrole rings of the heme moiety are planar. The aromatic ring of ASA is also planar, and its plane is inclined at an angle of 70.7° with respect to the plane of the heme porphyrin ring. The heme iron is displaced toward the proximal site by 0.1 Å. The conserved water molecule W-1 representing the position of H2O2 is located at a distance of 2.5 Å from the heme iron. It also forms two other hydrogen bonds with His-109 Nϵ2 at a distance of 2.6 Å and Gln-105 Nϵ2 at a distance of 3.2 Å. The acetyl group carboxyl oxygen atom O4 of ASA is also hydrogen-bonded to W-1 at a distance of 3.1 Å. The second oxygen atom of acetyl group O3 is hydrogen-bonded to a water molecule W-794 that is hydrogen-bonded to another water molecule, W-832, which in turn is hydrogen-bonded to the propionic carboxylic oxygen atom of pyrrole ring C. The carboxylic group oxygen atoms O1H and O2 are hydrogen-bonded to Phe-422 O and Gln-423 Nϵ2 at distances of 3.4 and 2.5 Å, respectively (Fig. 4). The latter two interactions are unique in the LPO-ASA complex because of the presence of carboxylic group in ASA, whereas such interactions are not possible in SHA and BHA because a similar moiety does not exist in SHA and BHA to interact with Phe-422 O and Gln-423 Nϵ2. Similarly, such interactions are not possible in MPO as well because the loop Arg-405–Leu-415 in MPO corresponding to the loop Lys-420–Phe-431 of LPO adopts a very different conformation in which the side chain of Phe-407, unlike that of Gln-423 in LPO, protrudes into the substrate binding site. In addition the placement of the aromatic ring of ASA in the hydrophobic pocket formed by residues Phe-113, Arg-255, Glu-258, Phe-380, Phe-381, and Pro-424 is well adjusted, resulting in a large number of hydrophobic interactions (Table 2). The interactions between the residues of the substrate binding site and the ligand can be presented in a better way by dividing the substrate binding site into five subsites (Fig. 4). The subsite S1 is assigned to the front face consisting of the heme moiety including the iron atom, subsite S2 represents one of the side faces containing Gln-105, His-109, and the conserved water molecule W-1, subsite S3 forms the surface on the above having residues Phe-113 and Arg-255 (Cβ, Cγ, and Cδ atoms), subsite S4 makes up the lower surface, containing residues Glu-258 (Cβ and Cγ atoms), Phe-380, and Phe-381, and subsite S5 forms the second side wall with residues Phe-422, Gln-423, and Pro-424 from the characteristic loop Lys-420–Phe-431. The sixth face is an open entrance to the binding site from the surface of the protein (Fig. 4). The subsites S1, S2, and S3 are largely similar in both LPO and MPO. The subsite S4 differs slightly, whereas subsite S5 is formed very differently (20), producing markedly different substrate specificities in the two enzymes. Subsite S5 represents the loop Lys-420–Phe-431, and because S5 is critical in the substrate recognition, the loop Lys-420–Phe-431 is termed as substrate specific loop. The corresponding substrate specific loop in MPO is Arg-405–Leu-415.
FIGURE 4.
Binding of ASA at the distal heme cavity of LPO. The water molecules W-1 and W-3′ correspond to W-618 and W-829, respectively, in Protein Data Bank code 3GCL. The assignment of five subsites has also been shown.
TABLE 2.
Hydrogen-bonded interactions and van der Waals contact distances (<4.0 Å) between ASA/SHA/BHA and atoms of the substrate binding site on the distal side
The numbering of W-1 is W-618 in the PDB file (3GCL), and W-3′ in the LPO-ASA is W-829 (3GCL), W-3′ in LPO-SHA is W-646 (3GCJ), and W-3′ in LPO-BHA is W-772 (3GCK). NA, CAD, and CMD represent the nitrogen atom of heme pyrrole ring A, the α-carbon atom of heme pyrrole ring D, and the methyl carbon atom of heme pyrrole ring D, respectively.
| Interactions/Subsites | LPO/HOH atoms | ASA/HOH atoms | Distance | SHA/HOH atoms | Distance | BHA/HOH atoms | Distance |
|---|---|---|---|---|---|---|---|
| Å | Å | Å | |||||
| Hydrogen bonds | |||||||
| Subsite-S1 | Heme-iron | W-1 | 2.5 | OH8 | 3.0 | OH8 | 3.3 |
| Heme-NA | OH8 | 3.4 | |||||
| Heme O2C | W-3′ | 2.7 | W-3′ | 2.9 | |||
| Heme-ND | OH8 | 3.2 | OH8 | 3.2 | |||
| NH8 | 3.0 | ||||||
| W-3′ | OH6 | 2.8 | O7 | 3.4 | |||
| Subsite-S2 | Gln-105 Nϵ2 | W-1 | 3.2 | OH8 | 3.2 | OH8 | 3.4 |
| O7 | 3.3 | ||||||
| His-109 Nϵ2 | W-1 | 2.6 | OH8 | 2.5 | OH8 | 2.5 | |
| W-1 | O4 | 3.1 | |||||
| Subsite-S5 | Phe-422 O | O1 | 3.4 | ||||
| Gln-423 Nϵ2 | O2 | 2.5 | |||||
| van der Waals contacts <4.0 Å | |||||||
| Subsite S1 | Heme CAD | C1 | 3.6 | C1 | 3.9 | ||
| C2 | 3.7 | ||||||
| Heme CMD | C7 | 3.4 | C5 | 3.9 | C5 | 3.8 | |
| C6 | 3.9 | C6 | 3.8 | ||||
| Heme C1D | C8 | 3.9 | C1 | 3.8 | C7 | 3.5 | |
| C7 | 3.7 | ||||||
| Heme C2D | C8 | 3.7 | C1 | 3.4 | C6 | 3.8 | |
| C6 | 3.7 | C7 | 3.5 | ||||
| C7 | 3.9 | ||||||
| Heme C3D | C8 | 3.6 | C1 | 3.7 | C6 | 3.7 | |
| C6 | 3.5 | C7 | 3.5 | ||||
| C7 | 3.6 | ||||||
| Heme C4D | C8 | 3.7 | C6 | 3.8 | C7 | 3.7 | |
| C9 | 3.6 | C7 | 3.3 | ||||
| Heme CHA | C7 | 3.7 | |||||
| Subsite S2 | His-109 Cϵ1 | C9 | 3.9 | ||||
| Subsite S3 | Arg-255 Cβ | C1 | 3.2 | ||||
| C6 | 3.9 | ||||||
| Arg-255 Cγ | C1 | 3.0 | C1 | 3.8 | C7 | 3.9 | |
| C2 | 3.8 | C7 | 3.8 | ||||
| C6 | 3.5 | ||||||
| C8 | 3.9 | ||||||
| Arg-255 Cδ | C8 | 3.8 | C1 | 3.4 | C7 | 3.6 | |
| C6 | 3.7 | ||||||
| Subsite S4 | Glu-258 Cβ | C9 | 3.5 | C5 | 3.9 | C5 | 3.4 |
| Glu-258 Cγ | C9 | 3.5 | C5 | 3.7 | |||
| Phe-381 Cξ | C5 | 3.8 | C3 | 3.7 | |||
| C4 | 3.1 | ||||||
| C5 | 3.2 | ||||||
| Subsite S5 | Pro-424 Cγ | C4 | 3.0 | C3 | 3.4 | ||
| C5 | 3.3 | ||||||
| Pro-424 Cδ | C4 | 3.3 | C3 | 3.4 | |||
Structure of LPO-Salicylhydroxamic Acid
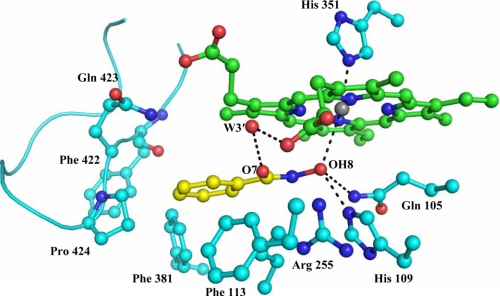
As seen from Fig. 5, SHA binds to LPO in the substrate binding site on the distal side. The reported binding constant for SHA to LPO is 7 × 10−5 m (44). As observed from Table 2, it interacts with four subsites, S1, S2, S3, and S4. Because SHA lacks a moiety corresponding to carboxylic group of ASA, it is unable to interact with residues Gln-423 Nϵ2 and Phe-422 O of subsite S5, although it is fitted well in the substrate binding site. In this complex the heme porphyrin moiety and four pyrrole rings are planar. The planar aromatic ring of SHA is inclined at 15.7° from the plane of the porphyrin ring. The corresponding angle is reported to be 20° in the MPO-SHA complex (34), whereas the aromatic ring of ASA is tilted at 70.7° in the LPO-ASA complex. As a result of the binding of SHA to LPO, four water molecules, W-1, W-2′, W-4′, and W-5′, are replaced by the ligand. The position of conserved heme water molecule W-1 is occupied by hydroxyl group of hydroxamic acid moiety, which makes a direct contact with the heme iron. Because SHA has moved deeper into the hydrophilic center, a notable void is observed in the proximity of loop Lys-420–Phe-431 where an additional water molecule W-6′ is observed which is hydrogen-bonded to both Phe-422 O and Gln-423 Nϵ2 of subsite S5. In this case the heme iron is displaced from the porphyrin plane toward the proximal site by 0.2 Å. The OH group of the hydroxamic acid moiety is hydrogen-bonded to heme iron at a distance of 3.0 Å. The OH group is also hydrogen-bonded to His-109 Nϵ2 and Gln-105 Nϵ2 with distances of 2.5 and 3.2 Å, respectively. It forms yet another hydrogen bond with a water molecule, W-3′. The benzene ring of SHA is located in the proximity of hydrophobic pocket and forms a relatively less number of interactions when compared with those of ASA (Table 2).
FIGURE 5.
Binding of SHA (yellow) at the distal heme cavity of LPO. The H-bonded interactions are shown as dotted lines. The water molecules W-3′ and W-6′ correspond to W-646 and W-774, respectively, in Protein Data Bank code 3GCJ.
Structure of LPO-Benzylhydroxamic Acid
BHA is also observed in the substrate binding site on the distal side (Fig. 6). The binding constant reported for the binding of BHA to LPO is 3 × 10−2 m (44). As observed in the complex of LPO-SHA, the conserved water molecules W-1, W-2′, W-4′, and W-5′ are replaced by BHA molecule. The BHA does not penetrate into the hydrophilic center as deep as SHA; its benzene ring is shifted toward the substrate specific loop Lys-420–Phe-431. As a consequence, the water molecule W-6′ observed in the complex of LPO-SHA is not present in the proximity of Phe-422 O and Gln-423 Nϵ2. This indicates that the position and orientation of BHA are slightly different from those of SHA in the LPO-SHA complex. The heme iron in this case is not shifted appreciably from the plane of heme moiety. The planar benzene ring of BHA is almost parallel to the plane of porphyrin ring with an inclination of 6.2°. The distance between the oxygen atom of the hydroxyl group of BHA and heme iron is 3.3 Å. As observed in the structure of LPO-SHA, it forms three more hydrogen bonds with Gln-105 Nϵ2, His-109 Nϵ2, and a water molecule, W-3′. The overall numbers of interactions observed between BHA and LPO are less than those found between LPO and ASA and SHA.
FIGURE 6.
Binding of BHA (yellow) at the distal heme cavity of LPO. The H-bonded interactions are shown as dotted lines. The water molecule W-3′ corresponds to W-772 in Protein Data Bank code 3GCK.
DISCUSSION
Peroxidases catalyze the H2O2-dependent oxidation of a variety of aromatic electron donor molecules through the formation of intermediate compounds. The structure analyses of three complexes of LPO with ASA, SHA, and BHA indicate a strong correlation between the binding properties of ASA, SHA, and BHA with implications for the ligands to act either as substrates or inhibitors. The structure of the complex of LPO with ASA shows that ASA interacts with heme iron via the conserved heme water molecule W-1. Similarly the complexes of BHA with horseradish peroxidase (35) and Arthromyces ramosus peroxidase (36) and that of SHA with A. ramosus peroxidase (37) also interact with heme iron via an identical water molecule. However, the complexes of SHA with LPO (this work), MPO (34), and ascorbate peroxidase (38) as well as the complex of BHA with LPO (this work) show that the ligands interact directly with the heme iron. The condition of interacting with the heme iron via the heme water molecule mimics the modes of enzyme substrate reaction, whereas a direct interaction with the heme iron results in the inhibition of peroxidases. As seen from Fig. 4, ASA forms one hydrogen bond with conserved heme water molecule W-1, which in turn interacts with heme iron, Gln-105 and His-109. The second oxygen atom O3 of the acetyl group is hydrogen-bonded to another conserved water molecule W-3′, which is hydrogen-bonded to the carboxyl oxygen atom of the propionate of pyrrole rind D. In addition to these two mild interactions from subsites S1 and S2 on one side, it forms two other hydrogen bonds with Phe-422 O and Gln-423 Nϵ2 of subsite S5 from an opposite face. It forms more than a dozen van der Waals contacts with distances less than 4.0 Å (Table 2) primarily involving the atoms of the aromatic ring of ASA and residues of the hydrophobic pocket. Therefore, the position of ASA is balanced by the forces from all sides in the substrate binding site. On the other hand, SHA forms at least nine hydrogen bonds with the heme moiety and hydrophilic center through its hydroxamic acid moiety. However, it lacks a suitable moiety to interact with Phe-422 O and Gln-423 Nϵ2 as a balancing force from the opposite side. Therefore, SHA is shifted toward the hydrophilic center, resulting in the direct interactions with heme iron, Gln-105, and His-109. It also forms a similar number of van der Waals interactions, but unlike those in the complex of LPO-ASA, the majority of the van der Waals interactions are observed with the atoms of the heme moiety. In the complex of LPO-BHA, BHA forms six hydrogen bonds with the heme moiety, Gln-105, and His-109, which indicates a weaker interaction with hydrophilic center when compared with that of SHA (Table 2). In this complex also, a nearly similar number of van der Waals contact distances are observed, but a significant number of them are provided by hydrophobic pocket. As a result of this, BHA is shifted away from the hydrophilic center by more than 1.0 Å with respect to SHA (Fig. 3B (ii)). Overall, it is observed that ASA interacts with all the five subsites in the substrate binding site and does not disturb the position of heme water molecule W-1 and is positioned favorably with respect to the active site of the enzyme, whereas SHA and BHA interact directly with the heme iron, block the peroxide binding site, and prevent the enzymatic action. These binding affinities of 7 × 10−5 and 3 × 10−2 m for SHA and BHA (44) appear to be in agreement with the structural data as indicated by the observed interactions of SHA and BHA with lactoperoxidase.
CONCLUSIONS
The structures of three complexes of LPO with aromatic substrate, ASA, and two aromatic inhibitors SHA and BHA clearly indicate the distinctiveness in their modes of binding as a substrate and as an inhibitor. The aromatic substrate molecule is positioned so as not replace the conserved heme water molecule W-1, whereas an inhibitor replaces the heme water molecule W-1 and interacts directly with the heme iron. For a ligand to function as a substrate of LPO, it ought to have a moiety with the potential hydrogen bond acceptors and donors to interact with W-1 of the hydrophilic center at the distal heme cavity and another moiety such as carboxylic or hydroxyl group with a spacer methyl group at the ortho or meta position of the aromatic ring so that it is able to form hydrogen bonds with Phe-422 O and Gln-423 Nϵ2. The latter interactions are important to induce the right orientation in the aromatic ligands with respect to the heme moiety and hydrophilic center. In the absence of interactions with Phe-422 O and Gln-423 Nϵ2, the aromatic ligands shift toward the hydrophilic center and displace the conserved heme water molecule W-1, resulting in the direct coordination with the heme iron. Consequently a hydrogen peroxide molecule is unable to bind at the active site of LPO. Thus, such ligands act as potent inhibitors. Therefore, ASA as a substrate and SHA and BHA as inhibitors are good examples for demonstrating the modes of binding of substrate and inhibitors to LPO. Similarly, it is important to distinguish the binding requirements of LPO from that of MPO. The hydrophilic centers in the two enzymes both structurally and chemically are almost similar. The real difference in the stereochemistry of the substrate binding sites in two enzymes is caused by the differences in the conformations of the substrate-specific loops Lys-420–Phe-431 in LPO and Arg-405–Leu-415 in MPO. As a consequence of these conformational characteristics the potential interacting partners in LPO are Phe-422 O and Gln-423 Nϵ2, whereas the corresponding player in MPO is the side chain of Phe-407. This clearly indicates the striking differences in the requirements of the interacting moieties in the substrates by the two enzymes. LPO will prefer hydrogen bond acceptors and donors, whereas MPO will recognize hydrophobic moieties. In contrast, there are no differences in the modes of binding of inhibitors, and both enzymes can be inhibited by similar ligands. Thus, it can be stated that LPO and MPO show remarkable substrate specificities, but they can easily exchange inhibitors.
This work was supported by the Department of Science and Technology and Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi.
The atomic coordinates and structure factors (codes 3GC1, 3GCL, 3GCJ, and 3GCK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- LPO
- lactoperoxidase
- MPO
- myeloperoxidase
- EPO
- eosinophil peroxidase
- ASA
- acetylsalicylic acid
- SHA
- salicylhydroxamic acid
- BHA
- benzylhydroxamic acid
- ABTS
- 2,2′-azino-bis (3-ethylbenzthiazoline-sulfonic acid)
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1.Furtmüller P. G., Zederbauer M., Jantschko W., Helm J., Bogner M., Jakopitsch C., Obinger C. (2006) Arch. Biochem. Biophys. 445, 199–213 [DOI] [PubMed] [Google Scholar]
- 2.Poulos T. L., Edwards S. L., Wariishi H., Gold M. H. (1993) J. Biol. Chem. 268, 4429–4440 [DOI] [PubMed] [Google Scholar]
- 3.Gajhede M. (2001) Biochem. Soc. Trans. 29, 91–98 [DOI] [PubMed] [Google Scholar]
- 4.Sievers G. (1981) FEBS Lett. 127, 253–256 [DOI] [PubMed] [Google Scholar]
- 5.Cals M. M., Mailliart P., Brignon G., Anglade P., Dumas B. R. (1991) Eur. J. Biochem. 198, 733–739 [DOI] [PubMed] [Google Scholar]
- 6.Oxvig C., Thomsen A. R., Overgaard M. T., Sorensen E. S., Højrup P., Bjerrum M. J., Gleich G. J., Sottrup-Jensen L. (1999) J. Biol. Chem. 274, 16953–16958 [DOI] [PubMed] [Google Scholar]
- 7.Fayadat L., Niccoli-Sire P., Lanet J., Franc J. L. (1999) J. Biol. Chem. 274, 10533–10538 [DOI] [PubMed] [Google Scholar]
- 8.Kooter I. M., Moguilevsky N., Bollen A., van der Veen L. A., Otto C., Dekker H. L., Wever R. (1999) J. Biol. Chem. 274, 26794–26802 [DOI] [PubMed] [Google Scholar]
- 9.Fenna R., Zeng J., Davey C. (1995) Arch. Biochem. Biophys. 316, 653–656 [DOI] [PubMed] [Google Scholar]
- 10.Wada K., Tada T., Nakamura Y., Ishikawa T., Yabuta Y., Yoshimura K., Shigeoka S., Nishimura K. (2003) J. Biochem. 134, 239–244 [DOI] [PubMed] [Google Scholar]
- 11.Poulos T. L., Freer S. T., Alden R. A., Edwards S. L., Skogland U., Takio K., Eriksson B., Xuong N., Yonetani T., Kraut J. (1980) J. Biol. Chem. 255, 575–580 [PubMed] [Google Scholar]
- 12.Schuller D. J., Ban N., Huystee R. B., McPherson A., Poulos T. L. (1996) Structure 4, 311–321 [DOI] [PubMed] [Google Scholar]
- 13.Gajhede M., Schuller D. J., Henriksen A., Smith A. T., Poulos T. L. (1997) Nat. Struct. Biol. 4, 1032–1038 [DOI] [PubMed] [Google Scholar]
- 14.Kunishima N., Fukuyama K., Matsubara H., Hatanaka H., Shibano Y., Amachi T. (1994) J. Mol. Biol. 235, 331–344 [DOI] [PubMed] [Google Scholar]
- 15.Zeng J., Fenna R. E. (1992) J. Mol. Biol. 226, 185–207 [DOI] [PubMed] [Google Scholar]
- 16.Fiedler T. J., Davey C. A., Fenna R. E. (2000) J. Biol. Chem. 275, 11964–11971 [DOI] [PubMed] [Google Scholar]
- 17.Blair-Johnson M., Fiedler T., Fenna R. (2001) Biochemistry 40, 13990–13997 [DOI] [PubMed] [Google Scholar]
- 18.Singh A. K., Singh N., Sharma S., Singh S. B., Kaur P., Bhushan A., Srinivasan A., Singh T. P. (2008) J. Mol. Biol. 376, 1060–1075 [DOI] [PubMed] [Google Scholar]
- 19.Singh A. K., Singh N., Sharma S., Shin K., Takase M., Kaur P., Srinivasan A., Singh T. P. (2009) Biophys. J. 96, 646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheikh I. A., Singh A. K., Singh N., Sinha M., Singh S. B., Bhushan A., Kaur P., Srinivasan A., Sharma S., Singh T. P. (2009) J. Biol. Chem. 284, 14849–14856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agner K. (1941) Acta Chem. Scand. 2, Suppl. 8, 1–62 [Google Scholar]
- 22.Harrison J. E., Schultz J. (1976) J. Biol. Chem. 251, 1371–1374 [PubMed] [Google Scholar]
- 23.Zhang H., Dunford H. B. (1993) Can. J. Chem. 71, 1990–1994 [Google Scholar]
- 24.Monzani E., Gatti A. L., Profumo A., Casella L., Gullotti M. (1997) Biochemistry 36, 1918–1926 [DOI] [PubMed] [Google Scholar]
- 25.Metodiewa D., Reszka K., Dunford H. B. (1989) Arch. Biochem. Biophys. 274, 601–608 [DOI] [PubMed] [Google Scholar]
- 26.Metodiewa D., Reszka K., Dunford H. B. (1989) Biochem. Biophys. Res. Commun. 160, 1183–1188 [DOI] [PubMed] [Google Scholar]
- 27.Ferrari R. P., Laurenti E., Casella L., Poli S. (1993) Spectrochim. Acta A 49, 1261–1267 [Google Scholar]
- 28.Doerge D. R., Decker C. J. (1994) Chem. Res. Toxicol. 7, 164–169 [DOI] [PubMed] [Google Scholar]
- 29.Oakley G. G., Devanaboyina U., Robertson L. W., Gupta R. C. (1996) Chem. Res. Toxicol. 9, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 30.Sipe H. J., Jr., Jordan S. J., Hanna P. M., Mason R. P. (1994) Carcinogenesis 15, 2637–2643 [DOI] [PubMed] [Google Scholar]
- 31.Cavalieri E. L., Stack D. E., Devanesan P. D., Todorovic R., Dwivedy I., Higginbotham S., Johansson S. L., Patil K. D., Gross M. L., Gooden J. K., Ramanathan R., Cerny R. L., Rogan E. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10937–10942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghibaudi E. M., Laurenti E., Beltramo P., Ferrari R. P. (2000) Redox Rep. 5, 229–235 [DOI] [PubMed] [Google Scholar]
- 33.RamaKrishna N. V., Li K. M., Rogan E. G., Cavalieri E. L., George M., Cerny R. L., Gross M. L. (1993) Chem. Res. Toxicol. 6, 837–845 [DOI] [PubMed] [Google Scholar]
- 34.Davey C. A., Fenna R. E. (1996) Biochemistry 35, 10967–10973 [DOI] [PubMed] [Google Scholar]
- 35.Henriksen A., Schuller D. J., Meno K., Welinder K. G., Smith A. T., Gajhede M. (1998) Biochemistry 37, 8054–8060 [DOI] [PubMed] [Google Scholar]
- 36.Itakura H., Oda Y., Fukuyama K. (1997) FEBS Lett. 412, 107–110 [DOI] [PubMed] [Google Scholar]
- 37.Tsukamoto K., Itakura H., Sato K., Fukuyama K., Miura S., Takahashi S., Ikezawa H., Hosoya T. (1999) Biochemistry 38, 12558–12568 [DOI] [PubMed] [Google Scholar]
- 38.Sharp K. H., Moody P. C., Brown K. A., Raven E. L. (2004) Biochemistry 43, 8644–8651 [DOI] [PubMed] [Google Scholar]
- 39.Sharp K. H., Mewies M., Moody P. C., Raven E. L. (2003) Nat. Struct. Biol. 10, 303–307 [DOI] [PubMed] [Google Scholar]
- 40.Sundaramoorthy M., Kishi K., Gold M. H., Poulos T. L. (1994) J. Biol. Chem. 269, 32759–32767 [PubMed] [Google Scholar]
- 41.Poulos T. L. (2007) Nat. Prod. Rep. 24, 504–510 [DOI] [PubMed] [Google Scholar]
- 42.Chatterjee R., Bandyopadhyay U., Bhattacharyya D., Banerjee R. K. (1993) Biochim. Biophys. Acta 1161, 168–176 [DOI] [PubMed] [Google Scholar]
- 43.Davies B., Edwards S. W. (1989) Biochem. J. 258, 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeda-Saito M., Shelley D. A., Lu L., Booth K. S., Caughey W. S., Kimura S. (1991) J. Biol. Chem. 266, 3611–3616 [PubMed] [Google Scholar]
- 45.Hori H., Fenna R. E., Kimura S., Ikeda-Saito M. (1994) J. Biol. Chem. 269, 8388–8392 [PubMed] [Google Scholar]
- 46.Kimura S., Yamazaki I. (1979) Arch. Biochem. Biophys. 198, 580–588 [DOI] [PubMed] [Google Scholar]
- 47.Kitagawa T., Hashimoto S., Teraoka J., Nakamura S., Yajima H., Hosoya T. (1983) Biochemistry 22, 2788–2792 [DOI] [PubMed] [Google Scholar]
- 48.Kumar R., Bhatia K. L., Dauter Z., Betzel C., Singh T. P. (1995) Acta Crystallogr. D Biol. Crystallogr. 51, 1094–1096 [DOI] [PubMed] [Google Scholar]
- 49.Lipmann F., Tuttle L. C. (1945) J. Biol. Chem. 159, 21–28 [PubMed] [Google Scholar]
- 50.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 51.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 52.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 53.Jones T. A., Zou J. Y, Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 54.Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 55.DeLano W. L. (2002) The PyMol Molecular Graphics System, DeLano Scientific, San Carlos CA [Google Scholar]