Abstract
Non-amyloid, ubiquitinated cytoplasmic inclusions containing TDP-43 and its C-terminal fragments are pathological hallmarks of amyotrophic lateral sclerosis (ALS), a fatal motor neuron disorder, and frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U). Importantly, TDP-43 mutations are linked to sporadic and non-SOD1 familial ALS. However, TDP-43 is not the only protein in disease-associated inclusions, and whether TDP-43 misfolds or is merely sequestered by other aggregated components is unclear. Here, we report that, in the absence of other components, TDP-43 spontaneously forms aggregates bearing remarkable ultrastructural similarities to TDP-43 deposits in degenerating neurons of ALS FTLD-U patients. The C-terminal domain of TDP-43 is critical for spontaneous aggregation. Several ALS-linked TDP-43 mutations within this domain (Q331K, M337V, Q343R, N345K, R361S, and N390D) increase the number of TDP-43 aggregates and promote toxicity in vivo. Importantly, mutations that promote toxicity in vivo accelerate aggregation of pure TDP-43 in vitro. Thus, TDP-43 is intrinsically aggregation-prone, and its propensity for toxic misfolding trajectories is accentuated by specific ALS-linked mutations.
TDP-43 is a ubiquitously expressed and highly conserved metazoan nuclear protein (1), which contains two RNA recognition motifs (RRMs)3 and a glycine-rich region in its C-terminal domain (see Fig. 1A). TDP-43 function is uncertain, but it likely plays important roles in pre-mRNA splicing and transcriptional repression (2, 3). In ALS and FTLD-U, TDP-43 is depleted from the nucleus and accumulates in ubiquitinated cytoplasmic inclusions (4). These and other situations of TDP-43 pathology, including some forms of Alzheimer and Parkinson diseases, are now known as TDP-43 proteinopathies (5). Importantly, mutations in the TDP-43 gene (TARDBP) are linked to sporadic and non-SOD1 familial ALS, implying that TDP-43 abnormalities are likely one cause of disease (6–11). However, despite this synthesis of pathology and genetics, the mechanisms by which TDP-43 might contribute to disease remain unknown and controversial (12, 13).
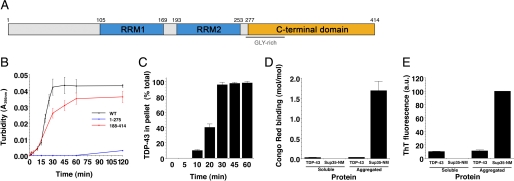
FIGURE 1.
Aggregation of TDP-43 and TDP-43 fragments in vitro. A, a diagram of the domain structure of TDP-43 indicating both RNA recognition motifs (RRM1 and RRM2) and the glycine-rich C-terminal domain. B, TDP-43 or the indicated TDP-43 fragment (1–275 or 188–414) (3 μm) were incubated at 25 °C with agitation for 0–120 min. The extent of aggregation was determined by turbidity. Values represent means ± S.D. (n = 3). C, aggregation of WT TDP-43 as in B assessed by sedimentation analysis. Values represent means ± S.D. (n = 3). D, His-TDP-43 or Sup35-NM (5 μm) was incubated for either 0 min (soluble protein) or 2 h (aggregated protein) at 25 °C with agitation. Reactions were then processed for Congo Red Binding. Values represent means ± S.D. (n = 3). E, TDP-43 or Sup35-NM (5 μm) was incubated for either 0 min (soluble protein) or 2 h (aggregated protein) at 25 °C with agitation. Reactions were then processed for Thioflavin-T fluorescence. Values represent means ± S.D. (n = 3).
A key unresolved question is whether TDP-43 is inherently aggregation-prone or whether TDP-43 is sequestered by other aggregated components and is merely a marker of disease (13–16). Indeed, multiple proteins aside from TDP-43 are found in Sarkosyl-insoluble fractions from FTLD-U patients (14). Moreover, deconvolution imaging reveals that TDP-43 appears to be excluded from some regions of the ubiquitinated inclusions in ALS (15).
Here, we assess TDP-43 aggregation in the absence of other components. We then define which domains of TDP-43 are important for this process and determine the direct effects of several ALS-linked TDP-43 mutations on TDP-43 misfolding and toxicity. Our findings bring to light several intrinsic properties of TDP-43 and ALS-linked TDP-43 mutants that likely play important roles in the aberrant TDP-43 proteostasis (17) that contributes to the pathogenesis of ALS, FTLD-U, and other TDP-43 proteinopathies.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Plasmids
Yeast cells were grown in rich medium (YPD; yeast/peptone/dextrose) or in synthetic media lacking uracil and containing 2% glucose (SD/-Ura), raffinose (SRaf/-Ura), or galactose (SGal/-Ura).
A TDP-43 Gateway entry clone was obtained from Invitrogen, containing full-length human TDP-43 in the vector pDONR221. To generate C-terminally YFP-tagged TDP-43 constructs, we used PCR to amplify TDP-43 without a stop codon and incorporate SpeI and HindIII restriction sites along with a Kozak consensus sequence. The resulting PCR product was cloned into SpeI/HindIII-digested pRS416GAL-YFP to generate the CEN TDP-43YFP fusion construct. Each ALS-linked TDP-43 mutant construct was generated by using the QuikChange® site-directed mutagenesis system (Stratagene) with pRS416GAL-TDP-43-YFP as template. All constructs were verified by DNA sequencing. CEN plasmid constructs (e.g. pRS416GAL-TDP-43-YFP) were transformed into BY4741 (MATa his3 leu2 met15 ura3).
Yeast Transformation and Spotting Assays
Yeast procedures were performed according to standard protocols. We used the polyethylene glycol/lithium acetate method to transform yeast with plasmid DNA. For spotting assays, yeast cells were grown overnight at 30 °C in liquid media containing SRaf/-Ura until they reached log or mid-long phase. Cultures were then normalized for A600 nm, serially diluted and spotted with a Frogger (V&P Scientific) onto synthetic solid media containing glucose (SD/-Ura) or galactose (SGal/-Ura) lacking uracil and were grown at 30 °C for 2–3 days.
Yeast Survivorship Assay
We performed survivorship assays as described previously (18). Briefly, after induction of empty vector, wild-type (WT) or mutant TDP-43 in 2% galactose, survivorship was determined at the indicated time points by harvesting cells at an A600 nm of 1, diluting them 1:1000, and plating 300 μl of these cells on synthetic media containing 2% glucose (represses TDP-43 expression). Plates were incubated at 30 °C for 2 days. Colony forming units were then determined.
Immunoblotting
Yeast lysates were subjected to SDS-PAGE (4–12% gradient, Invitrogen) and transferred to a polyvinylidene difluoride membrane (Invitrogen). Membranes were blocked with 5% nonfat dry milk in PBS for 1 h at room temperature or overnight at 4 °C. Primary antibody incubations were performed at room temperature for 1 h. After washing with PBS, membranes were incubated with a horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature, followed by washing in PBS plus 0.1% Tween 20 (PBST). Proteins were detected with Immobilon Western HRP Chemiluminescent Substrate (Millipore). The anti-GFP monoclonal antibody (Roche Applied Science) was used at 1:10,000, and phosphoglycerate kinase 1 antibody (Invitrogen) at 1:500. The HRP-conjugated anti-mouse secondary antibody was used at 1:5000.
Immunocytochemistry
For immunocytochemistry experiments, yeast cells expressing untagged TDP-43 constructs were grown to a final A600 of 0.2–0.8 and then fixed with 3.7% formaldehyde for 1 h. Cells were collected by centrifugation at 2000 rpm for 5 min and washed in PBS. The cells were diluted and washed once before resuspending in 1 ml of Solution A (0.5 mm MgCl2, 1.2 m Sorbitol, 40 mm K3PO4, pH 6.5). Cells were incubated with 10 μl of 2-mercaptoethanol and 25 μl of 10 mg/ml lyticase for 15 min at 37 °C. Spheroplasted cells were collected at 4000 rpm for 5 min, washed twice with Solution A and once with PBS, and resuspended in PBS+BSA (1× PBS, 1 mg/ml bovine serum albumin). Spheroplasts were diluted 1:5 and incubated on Teflon-covered slides treated with 1 mg/ml polylysine. Wells were blocked with PBS+BSA for 30 min at room temperature. Primary antibody incubations using 1:200 anti-TDP-43 mouse polyclonal antibody (Novus) were performed for 1.5 h at room temperature. After washing with PBS+BSA, wells were incubated with 1:2000 Alexa 488 nm donkey anti-mouse polyclonal antibody (Invitrogen) for 1.5 h at room temperature. After washing with PBS+BSA, wells were incubated with Vectashield mounting medium containing 1.5 μg/ml 4′,6-diamidino-2-phenylindole (Vector Labs) for 5 min before visualization using fluorescence microscopy.
Sedimentation Analysis
We performed sedimentation analysis as described in a previous study (19). Cells expressing either YFP-tagged TDP-43 constructs or CFP-tagged polyglutamine-expanded huntingtin constructs for 4 or 6 h were lysed in 1× native yeast lysis buffer (30 mm HEPES, pH 8.0, 150 mm NaCl, 1% glycerol, 1 mm dithiothreitol, 0.5% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 50 mm N-ethylmaleimide, 1× protease inhibitor mixture (Roche Applied Science)). Cells were disrupted in a bead beater for 3 min at 4 °C. Cellular debris was removed by centrifugation at 6,000 × g for 5 min at 4 °C. The yeast lysate was separated into a total fraction and a pellet fraction. After the pellet fraction was spun at either 16,000 × g or 85,000 rpm in a TLA 100.1 rotor for 30 min at 4 °C the supernatant was recovered and designated the soluble fraction. The pellet fraction was resolubilized by boiling in 50 μl of 1× SDS sample buffer. The total and soluble fractions were boiled in equal volumes of 4× SDS sample buffer. 20% of the pellet fraction and 10% of soluble and total fractions were resolved by SDS-PAGE followed by immunoblotting with anti-GFP antibody.
Fluorescence Microscopy
For fluorescence microscopy experiments, single colony isolates of the yeast strains were grown to mid-log phase in SRaf/-Ura media at 30 °C. Cultures were spun down and resuspended in the same volume of SGal/-Ura to induce expression of the TDP-43-YFP constructs. Cultures were induced with galactose for 6 h before being fixed with 70% ethanol and stained with 4′,6-diamidino-2-phenylindole in Vectashield mounting medium (Vector Laboratories) to visualize nuclei. Images were obtained using a Zeiss Axioplan Upright Microscope and a Zeiss AxioCam HRm high resolution monochrome charge-coupled device camera. The images were deblurred using a nearest neighbor algorithm in the AxioVision 4.5 software, and representative cells were chosen for figures.
Quantification of TDP-43 Aggregation
To assess differences in aggregation between wild-type and mutant TDP-43, yeast cultures were grown, induced, and processed as described above after having normalized all yeast cultures to A600 nm = 0.2 prior to galactose induction. After 6 h of induction, the identities of the samples were blinded to the observer before being examined. Several fields of cells were randomly chosen using the 4′,6-diamidino-2-phenylindole filter to prevent any bias toward populations of cells with increased amounts of aggregation in addition to obtaining the total number of cells in any given field. At least 200 cells per sample were counted for each replicate. Only cells with >3 foci under the YFP channel were considered as cells with aggregating TDP-43.
TDP-43 Purification
TDP-43 and various missense (G294A, M337V, or Q331K) or deletion mutants (1–275 or 188–414) were expressed and purified from E. coli as either His-tagged or GST-tagged proteins. For His-tagged preparations, TDP-43 was cloned into pCOLD I (Takara) and overexpressed in E. coli BL21(RIL). Cells were lysed by sonication on ice in 40 mm Hepes-KOH, pH 7.4, 500 mm KCl, 20 mm MgCl2, 10% glycerol, 20 mm imidazole, 2 mm β-mercaptoethanol, and protease inhibitors (Complete, EDTA-free, Roche Applied Science). The proteins were purified over a nickel-nitrilotriacetic acid column (Qiagen). For GST-tagged preparations, TDP-43 was cloned into GV13 to yield a tobacco etch virus (TEV) cleavable GST-TDP-43 fusion protein, GST-TEV-TDP-43, and overexpressed in E. coli BL21(DE3)RIL or Rosetta2 (Novagen). Protein was purified over a glutathione-Sepharose column (Amersham Biosciences) according to the manufacturer's instructions. GST was then removed by cleavage with TEV protease (Invitrogen), and protease and GST were removed using nickel-nitrilotriacetic acid and glutathione-Sepharose. His-tagged and untagged TDP-43 proteins were ∼95% pure as assessed by SDS-PAGE. His-tagged and untagged proteins aggregated with identical kinetics and formed aggregates with very similar morphologies.
After purification, proteins were buffer exchanged into assembly buffer (AB): 40 mm HEPES-KOH pH 7.4, 150 mm KCl, 20 mm MgCl2, 1 mm dithiothreitol. Proteins were filtered through a 0.22-μm filter. After filtration, the protein concentration was determined by Bradford assay (Bio-Rad), and the proteins were used immediately for aggregation reactions.
For size-exclusion chromatography, a Superdex-200 10/300 GL analytical gel-filtration column (Amersham Biosciences) was calibrated with thyroglobulin (669 kDa), ferritin (440 kDa), bovine serum albumin (67 kDa), β-lactoglobulin (35 kDa), ribonuclease A (13.7 kDa), and aprotinin (6.5 kDa). TDP-43 was incubated in AB at 25 °C with agitation for 5 min, and any insoluble material was removed by centrifugation at 16,100 × g for 5 min. Protein was loaded onto the calibrated Superdex-200 10/300 GL column equilibrated in AB and eluted at 0.4 ml/min. Oligomeric fractions were pooled and processed for electron microscopy (see below).
TDP-43 Aggregation in Vitro
Filtered, purified TDP-43 was used immediately for aggregation assays. TDP-43 or missense mutant TDP-43 or deletion mutants (3 μm) were incubated at 25 °C in AB for 0–120 min with agitation at 1400 rpm in an Eppendorf Thermomixer. Turbidity was used to assess aggregation by measuring absorbance at 395 nm. For sedimentation analysis, reactions were centrifuged at 16,100 × g for 30 min at 25 °C. Supernatant and pellet fractions were then resolved by SDS-PAGE and stained with Coomassie Brilliant Blue, and the amount in either fraction was determined by densitometry in comparison to known quantities of TDP-43. Alternatively, reactions were processed for Congo Red binding or Thioflavin-T fluorescence as described before (20).
For electron microscopy (EM) of in vitro aggregation reactions, TDP-43 protein samples (10 μl of a 3 μm solution) were adsorbed onto glow-discharged 300-mesh Formvar/carbon-coated copper grid (Electron Microscopy Sciences) and stained with 2% (w/v) aqueous uranyl acetate. Excess liquid was removed, and grids were allowed to air dry. Samples were viewed using a JEOL 1010 transmission electron microscope. Images were captured with a Hamamatsu digital camera using AMT acquisition software.
Electron Microscopy of Yeast Cells
Conventional EM was performed as previously described (21). Briefly, the cells were fixed in 3% glutaraldehyde contained in 0.1 m sodium cacodylate, pH 7.4, 5 mm CaCl2, 5 mm MgCl2, and 2.5% sucrose for 1 h at 25 °C with gentle agitation; spheroplasted; embedded in 2% ultra low temperature agarose (prepared in water); cooled; and subsequently cut into small pieces (∼1 mm3). The cells are then post-fixed in 1% OsO4/1% potassium ferrocyanide contained in 0.1 m cacodylate/5 mm CaCl2, pH 7.4, for 30 min at room temperature. The blocks are washed thoroughly 4× with ddH2O, 10 min total; transferred to 1% thiocarbohydrazide at room temperature for 3 min; washed in ddH2O (4×, 1 min each); and transferred to 1% OsO4/1% potassium ferrocyanide in cacodylate buffer, pH 7.4, for an additional 3 min at room temperature. The cells are then washed 4× with ddH2O (15 min total); en bloc stained in Kellenberger's uranyl acetate for 2 h to overnight; dehydrated through a graded series of ethanol; and subsequently embedded in Spurr resin. Sections were cut on a Reichert Ultracut T ultramicrotome; post stained with Kellenberger's uranyl acetate and lead citrate; and observed on a Philips TEM 420 at 80 kV. Images were recorded with a Soft Imaging System Megaview III digital camera, and figures were assembled in Adobe Photoshop 10.0.
RESULTS
TDP-43 Is Inherently Aggregation Prone
To test whether TDP-43 is inherently aggregation-prone, bacterially expressed recombinant TDP-43 was purified as a soluble protein under native conditions. Upon incubation at 25 °C with agitation, TDP-43 rapidly aggregated after a lag phase of ∼5–10 min, as determined by an increase in turbidity (Fig. 1B) and by the amount that entered the pellet fraction after centrifugation (Fig. 1C). Several control proteins, including BSA, soybean trypsin inhibitor, creatine kinase, and GFP, did not aggregate under identical conditions. After 30 min, no further TDP-43 aggregation occurred (Fig. 1, B and C). This timeframe for TDP-43 aggregation is extended to several hours if we omit agitation during incubation (data not shown). Thus, TDP-43 is an inherently aggregation-prone protein.
It is likely that sophisticated cellular proteostasis mechanisms (17, 22), not reconstituted here, prevent such rapid TDP-43 aggregation in vivo. However, age-associated decline in proteostatic control in concert with environmental factors might enable TDP-43 to aggregate in disease. Regardless of the triggers of TDP-43 aggregation in disease, in vitro assays similar to the one we report here have been tremendously powerful tools in exploring basic mechanisms underpinning the aggregation events in Parkinson disease (PD) and Alzheimer disease (23–25).
Aggregates formed by pure TDP-43 did not react with the amyloid-diagnostic dyes Congo Red and Thioflavin-T, in contrast to those formed by Sup35-NM, the prion domain of the yeast prion protein Sup35 (26) (Fig. 1, D and E). Thus, pure TDP-43 aggregates are likely to be non-amyloid just like aggregated species of TDP-43 in ALS and FTLD-U patients (27). In ALS and FTLD-U, TDP-43 is ubiquitinated, phosphorylated, and proteolytically cleaved (4). The relative extent and contribution of these modifications to the pathogenicity of TDP-43 remain to be defined. Our in vitro aggregation assays will provide the foundation for future studies aimed at determining the effects of TDP-43 post-translational modification and processing on aggregation.
C-terminal Domain of TDP-43 Is Important for Aggregation
Next, we determined which regions of TDP-43 are critical for aggregation in vitro. We purified TDP-43 fragments: 1–275, which comprises the N-terminal domain, RRM1 and RRM2, and 188–414, which comprises RRM2 and the C-terminal domain (Fig. 1A). 1–275 is soluble, whereas 188–414 is the minimal fragment able to confer toxicity and aggregation in a yeast model of TDP-43 proteinopathies (28). Importantly, pure 1–275 did not aggregate, whereas 188–414 aggregated with similar kinetics to full-length TDP-43 (Fig. 1B). Thus, the C-terminal domain plays an important role in TDP-43 aggregation, which is striking because of the >25 recently reported ALS-linked TDP-43 mutations, all but one are within this domain (Fig. 2A) (12). Furthermore, similar aggregated C-terminal fragments accumulate in ALS and FTLD-U (4). Therapeutic strategies aimed at targeting this region, which we have defined as responsible for driving aggregation, may be efficacious.
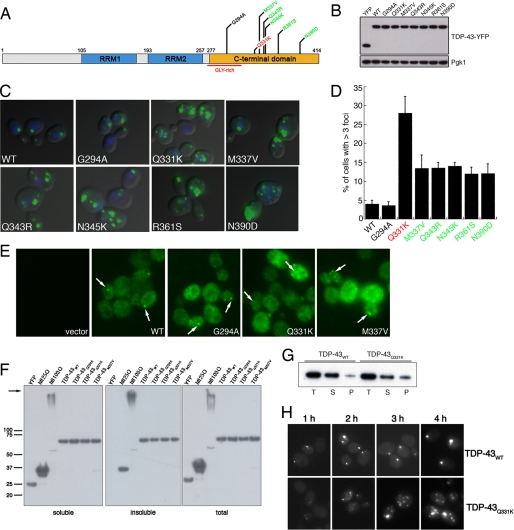
FIGURE 2.
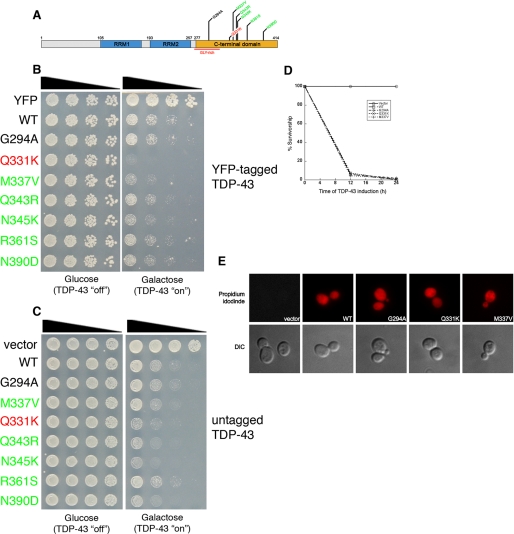
Aggregation of TDP-43 and ALS-linked mutants in vivo. A, schematic indicating disease-associated TDP-43 mutations. Color code of mutations indicates aggregation compared with WT (red = considerably more aggregates than WT, green = more aggregates than WT, and black = as many aggregates as WT). B, YFP alone, WT, and mutant TDP-43-YFP expression levels were determined by immunoblotting with an anti-GFP antibody. Phosphoglycerate kinase 1 (Pgk1) was used as a loading control. C, representative fluorescent microscopy images of 4′,6-diamidino-2-phenylindole-stained (blue to denote the position of the nucleus) WT or mutant TDP-43-YFP (green). D, the effect of TDP-43 mutations on aggregation in vivo was quantified by counting the number of cells containing >3 foci. Values represent means ± S.E. (n ≥ 3, at least 200 cells per sample). Cells expressing YFP alone did not contain foci (data not shown). E, untagged TDP-43 also forms aggregates in yeast cells. Immunocytochemistry with an anti-TDP-43 antibody was used to visualize TDP-43 expression. Arrows point to cytoplasmic TDP-43 inclusions. Cells transformed with an empty vector were used as a control for antibody specificity. F, sedimentation assay to demonstrate TDP-43 forms insoluble aggregates in yeast cells. Yeast cells expressing YFP, htt25QCFP, htt103Q-CFP, or WT and mutant TDP-43-YFP for 6 h were lysed and processed for sedimentation assay as in a previous study (19). Soluble and insoluble fractions were separated by centrifugation. 20% of the pellet fraction and 10% of soluble and total fractions were resolved by SDS-PAGE followed by immunoblotting with anti-GFP antibody. Whereas YFP was completely soluble, WT and TDP-43 partitioned into soluble and insoluble fractions, similar to the aggregation-prone fragments of the huntingtin protein (htt25Q (mostly soluble) or htt103Q (mostly insoluble)). The arrow indicates the top of gel. G, sedimentation assay performed as in F except that WT and Q331K TDP-43 constructs were induced for 4 h. At this time point, a greater relative amount of Q331K is present in the insoluble pellet fraction than WT TDP-43. T, total; S, soluble; P, pellet. Representative image from three separate experiments is shown. H, time-course analysis of WT and Q331K TDP-43 aggregation demonstrates Q331K forms more discrete aggregates than WT at early time points.
ALS-linked TDP-43 Mutants Form Multiple Aggregates in Yeast
Having established that TDP-43 is inherently aggregation-prone, we next asked if ALS-linked TDP-43 mutations affect aggregation in vivo. We have developed a yeast TDP-43 proteinopathy model to investigate mechanisms of TDP-43 aggregation and toxicity (28). This model recapitulates several important features seen in human disease. In yeast, TDP-43 is initially localized to the nucleus but eventually forms cytoplasmic inclusions (28), mimicking the pathobiology of TDP-43 in human neurons (4). Importantly, expressing high levels of TDP-43 is toxic to yeast (28), thus possibly modeling, in a simple cell, features of neurodegeneration. We tested the effects of seven recently reported ALS-linked mutations (6–9, 11) (Fig. 2A) on TDP-43 aggregation in this system. Wild-type (WT) and mutant TDP-43-YFP were expressed from a low copy (CEN) plasmid, under control of a galactose-inducible promoter, and cells were visualized by fluorescence microscopy. We confirmed that the TDP-43 proteins were expressed at comparable levels (Fig. 2B).
We compared aggregation in cells expressing WT TDP-43 to those expressing each of the seven mutants. YFP alone was diffusely distributed between the cytoplasm and nucleus (data not shown). Strikingly, with the exception of G294A, ALS-linked mutants formed more numerous aggregates than WT TDP-43, which formed more than three cytoplasmic foci in ∼4% of cells (Fig. 2, C and D). Of the mutants, Q331K formed more than three cytoplasmic inclusions in >25% of cells, compared with >10% of cells for the other TDP-43 mutants (Fig. 2, C and D). GFP-tagged protein fusions can occasionally produce artifactual aggregation (29). Hence, we confirmed these results by performing immunocytochemistry, with a TDP-43-specific antibody, on cells expressing untagged WT and a representative selection of mutant TDP-43 constructs (Fig. 2E). Untagged Q331K and M337V typically formed more discrete aggregates per cell than WT or G294A (Fig. 2E).
To confirm that TDP-43 foci visualized by microscopy represented insoluble aggregates, we performed a sedimentation analysis (19). As a negative control, we used YFP-expressing cells. YFP was entirely soluble (Fig. 2F). As positive controls, we used 25Q- and 103Q-containing fragments from exon 1 of huntingtin. In our yeast sedimentation assay, these proteins also partitioned between soluble and insoluble, with 25Q being mostly soluble and 103Q being mostly insoluble (Fig. 2F). WT and mutant TDP-43 partitioned between soluble and insoluble fractions (Fig. 2F). Thus, TDP-43 forms insoluble aggregates in yeast. In contrast to our fluorescence microscopy data (Fig. 2, C and D), after 6 h of expression similar quantities of TDP-43 formed insoluble aggregates in cells expressing WT, G294A, M337V, and Q331K TDP-43 (Fig. 2F). This total amount of insoluble protein appears to be distributed among more numerous discrete aggregates for M337V and Q331K (Fig. 2, C and D) and is concentrated in fewer foci for G294A and WT (Fig. 2, C and D). This could reflect a saturable end point for aggregation. Therefore, we focused on Q331K, which aggregated the most extensively of the mutants tested (Fig. 2C) and performed sedimentation analysis at earlier time-points. At 4 h, there was a greater relative amount of Q331K in the pellet fraction than WT TDP-43 (Fig. 2G), consistent with this mutation accelerating aggregation. However, the sedimentation assay may not be sensitive enough to detect subtle differences in TDP-43 aggregation between WT and the other ALS-linked mutants in yeast, like those we observe by fluorescence microscopy. Therefore, to resolve these issues, additional in vitro assays are required to define quantitatively the contribution of ALS-linked mutations to the biochemical properties of TDP-43 (i.e. aggregation; for example, see Fig. 5 below).
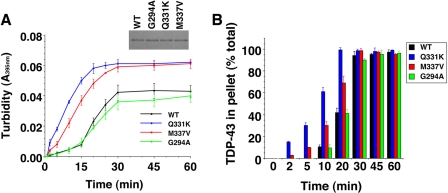
FIGURE 5.
Q331K and M337V accelerate spontaneous TDP-43 aggregation. A and B, TDP-43 or the indicated ALS-linked TDP-43 mutant (Q331K, M337V, or G294A) (3 μm) were incubated at 25 °C with agitation for 0–60 min. The extent of aggregation was determined by turbidity (A) or sedimentation analysis (B). Values represent means ± S.D. (n = 3). Inset (A) the equal input of TDP-43 proteins is shown on a Coomassie-stained gel.
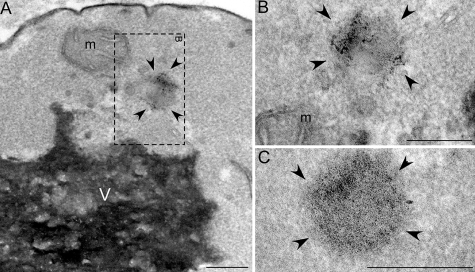
Next, we used electron microscopy to visualize aggregates formed by TDP-43 in vivo. In the yeast cytoplasm, Q331K TDP-43 formed discrete aggregated foci of ∼0.2 μm in diameter (Fig. 3) that were not observed in control cells (data not shown). TDP-43 aggregates formed in yeast cells possessed a granular morphology (Fig. 3) similar to TDP-43 aggregates in ALS and FTLD-U (30–32) and to those formed in vitro using pure protein (see below).
FIGURE 3.
EM of TDP-43 aggregates in yeast. A–C, low magnification TEM shows an example of a granular aggregate in yeast cells expressing the Q331K TDP-43 mutant (boxed region (A), high magnification +25° tilt view of boxed region in A (B), and additional high magnification view of TDP-43 aggregate (C)). m = mitochondria; V = vacuole; arrowheads delineate TDP-43 aggregate; bars = 0.25 μm.
Finally, we performed a time-course experiment to compare aggregation between WT TDP-43 and one of the mutants (Q331K). Even at very early time points (e.g. 2 h), we observed many more aggregates per cell with Q331K TDP-43 than with WT (Fig. 2H). Thus, ALS-linked mutations in TDP-43 can increase the number of discrete aggregates in the yeast cytoplasm.
ALS-linked TDP-43 Mutants Are More Toxic in Yeast
Next, we investigated the effects of ALS-linked TDP-43 mutations on toxicity. In yeast, expression of WT TDP-43 at high levels (2μ plasmid) is extremely toxic (28). Because we wanted to compare toxicity between WT and mutant TDP-43, we used a low copy CEN plasmid to express TDP-43-YFP at a level that was only slightly toxic. We performed spotting assays to compare growth defects elicited by WT and by ALS-linked TDP-43 mutants (Fig. 4). The six ALS-linked mutants that formed more discrete aggregates than WT (Fig. 2) were also more toxic (Fig. 4, B and C). The Q331K mutant that consistently formed the most numerous aggregated foci in yeast (Fig. 2D) was considerably more toxic than WT or the other mutants (Fig. 4, B and C), despite being expressed at the same level as the other proteins (Fig. 2B). G294A was similar to WT in both number of aggregated foci per cell (Fig. 2D) and toxicity (Fig. 4B). We obtained similar results using either YFP-tagged (Fig. 4B) or untagged constructs (Fig. 4C).
FIGURE 4.
The effect of ALS-linked TDP-43 mutations on toxicity. A, schematic indicating disease-associated TDP-43 mutations shown above. Color code of mutations indicates toxicity compared with WT (red = considerably more toxic than WT; green = slightly more toxic than WT; black = as toxic as WT). B, spotting assay to compare the toxicity of WT and mutant TDP-43. Serial dilutions of yeast cells transformed with galactose-inducible YFP, WT, or mutant TDP-43-YFP constructs. Transformants were spotted on glucose- (non-inducing) or galactose- (inducing) containing agar plates, and growth assessed after 48–72 h. C, spotting assay to compare the toxicity of WT and mutant TDP-43. Serial dilutions of yeast cells transformed with galactose-inducible untagged WT or mutant TDP-43 constructs. Transformants were spotted on glucose- (non-inducing) or galactose- (inducing) containing agar plates, and growth was assessed after 48–72 h. D, survivorship curve during TDP-43 induction, using the high copy 2μ vector. After induction of empty vector, TDP-43 WT, G294A, Q331K, or M337V, survivorship was determined at the indicated time points by harvesting cells at A600 nm = 1, diluting 1:1000, and plating 300 μl of these cells onto synthetic media containing 2% glucose (represses TDP-43 expression). Plates were incubated at 30 °C, and colony forming units were determined after 2 days. E, TDP-43 expression causes cell death. TDP-43 expression was induced for 6 h, and cells were stained with propidium iodide (PI) to assess viability. WT and mutant TDP-43-expressing cells were positive for PI staining (indicating cell death), whereas empty vector-containing cells were negative for PI staining (indicating viability).
The growth defect elicited by TDP-43 in our spotting assays could reflect either cell death or simply a growth arrest. To distinguish between these two possibilities, we performed survivorship assays, in which we determined the ability of cells to form a colony upon cessation of TDP-43 expression. When we used the high copy 2μ plasmid to express WT TDP-43 and a subset of the mutants, fewer than 10% of cells were able to form colonies following 12 h of expression, and by 24 h fewer than 2% were still alive (Fig. 4D). We were able to detect differences in survivorship between WT and the mutants by using the low copy CEN plasmid. Whereas 100% of cells containing an empty vector were able to form a colony, following 12 h of galactose induction, only ∼30% of cells that had expressed WT, M337V, or G294A TDP-43 were able to form a colony and only ∼10% of Q331K-expressing cells formed colonies. This survivorship assay is not as sensitive as the spotting assays. Thus, with the survivorship assay, we were unable to detect the subtle differences in growth between WT and M337V that we saw by spotting (Fig. 4, B and C).
To further confirm cell death as opposed to simple growth arrest, we employed propidium iodide (PI), a fluorescent dye that binds DNA. PI is membrane-impermeant and is generally excluded from viable cells. TDP-43-expressing cells stained positively for PI (Fig. 4E), which indicates a loss of cell viability. Thus, in yeast TDP-43 aggregation leads to cell death and not simply a growth arrest. Moreover, several ALS-linked TDP-43 mutations can increase the number of TDP-43 aggregates and enhance toxicity.
ALS-linked TDP-43 Mutations Can Accelerate TDP-43 Aggregation
We then used pure proteins to determine whether three of the ALS-linked TDP-43 mutations affected the aggregation process directly. We selected Q331K and M337V, which formed more aggregated foci than WT, and G294A, which formed similar numbers of aggregated foci to WT in yeast (Fig. 2D). The formation of multiple aggregated foci by Q331K and M337V in vivo might reflect more rapid nucleation of aggregation, which can result in more numerous aggregates (33). Consistent with our in vivo data, the Q331K and M337V mutants aggregated much more rapidly than WT (Fig. 5, A and B), whereas G294A did not accelerate TDP-43 aggregation and resembled WT (Fig. 5, A and B). Specifically, the lag phase of aggregation was greatly reduced for both Q331K and M337V compared with WT and G294A (Fig. 5, A and B). Q331K aggregated even more rapidly than M337V (Fig. 5, A and B), consistent with Q331K forming more numerous aggregates in yeast. Moreover, more Q331K protein was insoluble than WT protein after 4 h of expression in yeast, consistent with more rapid assembly kinetics (Fig. 2G). These data suggest that the Q331K and M337V mutations likely alter the TDP-43 folding landscape such that aggregation is more energetically favorable. However, even though Q331K and M337V aggregated more rapidly than WT or G294A in vitro, the final amount of aggregated protein after 30–60 min was similar for all TDP-43 variants (Fig. 5, A and B). Similarly, after 6 h of expression in vivo these TDP-43 variants formed approximately equal quantities of insoluble protein (Fig. 2F).
Pure TDP-43 Aggregates Resemble TDP-43 Aggregates in Degenerating Neurons in ALS and FTLD-U
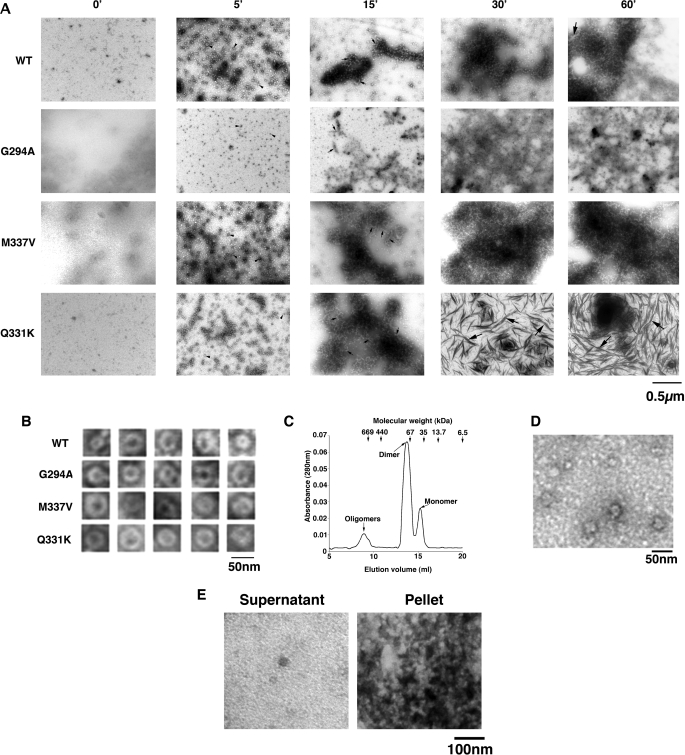
EM revealed that WT TDP-43 and G294A rapidly formed small oligomeric complexes after 5 min (Fig. 6A, examples denoted by arrowheads), which would sometimes adopt a pore-like conformation (Fig. 6B), strikingly akin to the pathological oligomers formed by Aβ42 and α-synuclein (34). To confirm that these conformers were TDP-43 oligomers, we performed size-exclusion chromatography. Consistent with previous reports (35), WT TDP-43 eluted from a Superdex 200 gel-filtration column as predominantly dimeric and monomeric species (Fig. 6C). However, a small fraction eluted as higher order oligomers (Fig. 6C). Inspection of the pooled oligomeric fraction by EM revealed oligomeric profiles similar to those observed at early times during aggregation reactions (Fig. 6D).
FIGURE 6.
Pure TDP-43 and ALS-linked mutant aggregates resemble TDP-43 aggregates in degenerating neurons of ALS patients. A, TDP-43 or the indicated ALS-linked TDP-43 mutant (Q331K, M337V, or G294A) (3 μm) were incubated at 25 °C with agitation for 0–60 min. At various times, reactions were processed for EM. Arrowheads denote small oligomers, small arrows denote filament-like structures, and large arrows denote thread-like structures. Bar, 0.5 μm. B, gallery of TDP-43 and ALS-linked TDP-43 mutant oligomers formed after 5 min. Bar, 50 nm. C, TDP-43 was incubated at 25 °C with agitation for 5 min and processed for Superdex 200 size-exclusion chromatography. TDP-43 eluted as a predominantly dimeric and monomeric species, but a small fraction eluted as oligomeric species. Arrows at the top indicate elution volumes of the molecular weight standards. D, EM of oligomeric pool eluted from the Superdex 200 column. Bar, 50 nm. E, TDP-43 (3 μm) was incubated at 25 °C with agitation for 30 min. Supernatant and pellet fractions were separated and processed for EM. Bar, 100 nm.
Incubation of TDP-43 for longer times (15–60 min) with agitation induced the formation of short filament-like structures (Fig. 6A, examples denoted by small arrows), which clustered together with oligomers to form large masses by 30–60 min (Fig. 6A). EM of supernatant and pellet fractions demonstrated that some small oligomers remain in the supernatant fraction, whereas clusters of oligomers and larger species enter the pellet fraction (Fig. 6E).
G294A formed aggregates that tended to be more amorphous and less well resolved than WT (Fig. 6A). The large masses of oligomeric forms populated by pure TDP-43 are remarkably similar to those observed in ALS and FTLD-U patients, which can displace cytoplasmic organelles (30–32). For example, WT TDP-43 aggregates in Fig. 6A (30 or 60 min) appear morphologically similar to the TDP-43 inclusions in the neurons and glial cells of ALS patients (see Fig. 3 (c–f) of Ref. 31 and Fig. 2 of Ref. 32).
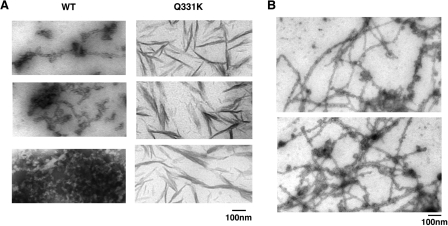
M337V aggregated in a similar manner (Fig. 6, A and B), except that oligomers and filament-like structures clustered together much earlier and proceeded to form larger masses than WT or G294A (Fig. 6A). By contrast, Q331K passed through similar oligomeric forms as WT, G294A, and M337V (Fig. 6, A and B), but eventually accessed morphologically distinct aggregated forms, with a thread- or skein-like appearance (Fig. 6A, examples denoted by large arrows). Similar thread-like structures that immunolabel for TDP-43 are observed regularly in the degenerating neurons of ALS and FTLD-U patients (30–32). For example, compare Q331K TDP-43 aggregates formed with pure protein in Fig. 7A to those in neurons of FTLD-U patients (see Fig. 2 (b and d) of Ref. 30) or in neurons of ALS patients (see Fig. 6 (c and d) of Ref. 30). The pure Q331K structures differ subtly from those observed in patients, in that they are slightly less regular and are typically more filamentous and not granulo-filamentous. Nonetheless, their overall similarity is striking.
FIGURE 7.
Thread-like aggregates formed by TDP-43. A, gallery of thread-like aggregates of TDP-43 and Q331K formed after 60 min of agitation at 25 °C. Bar, 100 nm. B, longer and more regular granulo-filamentous forms populated by WT TDP-43 after 2 h of agitation at 25 °C. Bar, 100 nm.
Although less abundant, similar threadlike structures were also formed by WT TDP-43 (Fig. 7A), as well as G294A and M337V (data not shown). Typically, these forms are shorter and more ragged than those observed in ALS or FTLD-U patients. However, after extended incubations, occasionally longer, smoother granulo-filamentous WT TDP-43 aggregates were observed (Fig. 7B). These structures are strikingly reminiscent of aggregated species populated by TDP-43 in the degenerating neurons of ALS and FTLD-U patients (30–32).
DISCUSSION
In sum, we have faithfully reconstructed several aspects of the pathological TDP-43 aggregation process using solely pure TDP-43. Our data establish that pure TDP-43 forms aggregated species in vitro that are remarkably similar to the TDP-43 aggregates in FTLD-U and ALS patients. Further, we show that pathogenic mutations in the TDP-43 gene can accelerate aggregation in vitro and elicit the formation of more numerous aggregates in vivo. Thus, it seems likely that TDP-43 is a key aggregated protein in ALS and FTLD-U, just as α-synuclein is in PD and tau and amyloid-β are in Alzheimer disease (36).
Six of seven ALS-linked TDP-43 mutants that we tested, especially Q331K, induced the formation of multiple TDP-43 aggregates per cell. Although, at 4 h we observed an increase in aggregation of Q331K compared with WT (Fig. 2G), by sedimentation analysis we could not detect differences in the total amount of insoluble TDP-43 between WT and the mutants expressed for 6 h in yeast (Fig. 2F). This may simply reflect the end point of aggregation in vivo, just as similar amounts of TDP-43 eventually form aggregates in vitro, regardless of the TDP-43 variant. Furthermore, the sedimentation assay might not be sensitive enough to allow us to discern the significant differences in aggregation patterns observed by microscopy. The 25Q versus 103Q huntingtin constructs clearly showed a difference in insolubility in the sedimentation assay. However, huntingtin fragments with polyglutamine tracts of ∼25Q do not typically aggregate in vitro or in vivo (37, 38). Thus, a large difference was expected. Moreover, our TDP-43 mutants only differ from WT by a single residue.
Rather, we suggest that the ALS-linked mutations might affect the morphology and/or localization of the aggregates in vivo rather than the total amount of insoluble TDP-43 (as detected by sedimentation assay). For example, with WT TDP-43, we frequently observe one large juxtanuclear focus (Fig. 2C), reminiscent of the recently described JUNQ (“juxtanuclear quality control”) compartment where misfolded proteins accumulate (19). The observation that ALS-linked TDP-43 mutants tend to form multiple foci in vivo (Fig. 2C) might be attributable to the accelerated kinetics of aggregation, which we observe in vitro (Fig. 5), enabling them to escape this quality control mechanism and thus be more toxic than WT. Nevertheless, the two mutants that consistently formed multiple aggregated foci in yeast, Q331K and M337V, also aggregated more rapidly in vitro, in our pure protein assays. Thus, a single amino acid change is sufficient to accelerate TDP-43 misfolding, supporting the notion that some ALS-linked mutations can cause disease by a toxic “gain-of-function mechanism” at the protein level. The mutations that accelerate spontaneous aggregation in vitro and form multiple aggregated foci in vivo are also more toxic (Fig. 4), suggesting that a subset of ALS-linked TDP-43 mutations may cause disease by accelerating TDP-43 aggregation and contingent toxicity. ALS-linked TDP-43 mutants with accelerated aggregation and toxicity are reminiscent of PD-linked α-synuclein mutants (A30P, A53T, and E46K) that oligomerize or fibrillize more rapidly and are connected with early-onset PD (23, 24, 39).
So far, it does not appear that TDP-43 mutations are a common cause of ALS (40). There are likely other pathogenic mechanisms, some that converge on TDP-43 and others that do not. Nonetheless, it is apparent that TDP-43 pathology is very commonly associated with sporadic and non-SOD1 familial ALS (41). Thus, studying TDP-43 aggregation and toxicity in vitro and in vivo, as reported here, will likely help to elucidate the role of TDP-43 in pathogenesis. Furthermore, given the dose-dependent toxicity (28) and aggregation-prone nature of TDP-43, increased TDP-43 levels might accelerate ALS just as increased wild-type α-synuclein levels accelerate PD (42). Therefore, TDP-43 copy number, promoter, and regulatory regions should also be analyzed in the context of ALS, FTLD-U, and other TDP-43 proteinopathies.
Why is Q331K considerably more toxic than the other mutants we tested in our yeast model? We note that pure Q331K more readily accessed thread-like aggregated forms, and grossly similar inclusions are consistently found in motor neurons undergoing neuronophagia, the final step in motor neuron death in sporadic ALS (43). Thus far, there has been one reported patient with this mutation (7), and it has not been found in thousands of control samples. Future studies using animal models (e.g. mouse, Drosophila, Caenorhabditis elegans, and zebrafish) will provide additional insights into the effects of WT versus mutant TDP-43 in neurodegeneration.
Other TDP-43 mutations, e.g. M337V, have been identified in multiple individuals and segregate with disease in familial ALS (7). Another mutation, Q343R, is also associated with familial ALS and patients present with more widespread TDP-43 pathology than most non-mutant TDP-43 cases (11). Remarkably, Q343R also aggregates more extensively and is more toxic in our yeast model (Figs. 2 and 4). However, because exhaustive genetic studies of TDP-43 in ALS and FTLD-U have not yet been performed, it is probably too soon to judge the potential pathogenicity of every single TDP-43 mutation that has been found in patients. One recent report found that TDP-43 pathology in a patient harboring the G294A mutation was similar to that of patients without this lesion (44). Interestingly, G294A behaved like WT TDP-43 in our yeast and pure protein assays, further corroborating these approaches.
Our yeast model will facilitate assessment of the effects of newly identified mutations on aggregation and toxicity. Given the previous success defining disease mechanisms and mammalian genetic suppressors of PD using a yeast synucleinopathy model (18, 45, 46), genome-wide approaches using the yeast TDP-43 model are likely to provide insight into disease mechanism, identify potential biomarkers, and suggest avenues for therapeutic strategies. Finally, the pure TDP-43 aggregation assay described here will empower us to identify small molecules and cellular factors able to inhibit or even reverse TDP-43 aggregation.
Acknowledgments
We thank Greg Van Duyne for pGV13 and Mark Lemmon, Jon Epstein, and Nancy Bonini for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 1DP2OD004417-01 (Director's New Innovator Award to A. D. G.) and 1DP2OD002177-01 (Director's New Innovator Award to J. S.). This work was also supported by a pilot grant from the University of Pennsylvania Institute on Aging (to A. D. G.).
- RRM
- RNA recognition motif
- ALS
- amyotrophic lateral sclerosis
- FTLD-U
- frontotemporal lobar degeneration with ubiquitin-positive inclusions
- YFP
- yellow fluorescent protein
- PBS
- phosphate-buffered saline
- HRP
- horseradish peroxidase
- GFP
- green fluorescent protein
- BSA
- bovine serum albumin
- CFP
- cyan fluorescent protein
- GST
- glutathione S-transferase
- EM
- electron microscopy
- PD
- Parkinson disease
- WT
- wild type
- PI
- propidium iodide
- TEV
- tobacco etch virus.
REFERENCES
- 1.Ayala Y. M., Pantano S., D'Ambrogio A., Buratti E., Brindisi A., Marchetti C., Romano M., Baralle F. E. (2005) J. Mol. Biol. 348, 575–588 [DOI] [PubMed] [Google Scholar]
- 2.Ayala Y. M., Misteli T., Baralle F. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buratti E., Baralle F. E. (2001) J. Biol. Chem. 276, 36337–36343 [DOI] [PubMed] [Google Scholar]
- 4.Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 5.Forman M. S., Trojanowski J. Q., Lee V. M. (2007) Curr. Opin. Neurobiol. 17, 548–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P. F., Camu W., Meininger V., Dupre N., Rouleau G. A. (2008) Nat. Genet. 40, 572–574 [DOI] [PubMed] [Google Scholar]
- 7.Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., Miller C. C., Nicholson G., Shaw C. E. (2008) Science 319, 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Deerlin V. M., Leverenz J. B., Bekris L. M., Bird T. D., Yuan W., Elman L. B., Clay D., Wood E. M., Chen-Plotkin A. S., Martinez-Lage M., Steinbart E., McCluskey L., Grossman M., Neumann M., Wu I. L., Yang W. S., Kalb R., Galasko D. R., Montine T. J., Trojanowski J. Q., Lee V. M., Schellenberg G. D., Yu C. E. (2008) Lancet Neurol. 7, 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutherford N. J., Zhang Y. J., Baker M., Gass J. M., Finch N. A., Xu Y. F., Stewart H., Kelley B. J., Kuntz K., Crook R. J., Sreedharan J., Vance C., Sorenson E., Lippa C., Bigio E. H., Geschwind D. H., Knopman D. S., Mitsumoto H., Petersen R. C., Cashman N. R., Hutton M., Shaw C. E., Boylan K. B., Boeve B., Graff-Radford N. R., Wszolek Z. K., Caselli R. J., Dickson D. W., Mackenzie I. R., Petrucelli L., Rademakers R. (2008) PLoS Genet. 4, e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gitcho M. A., Baloh R. H., Chakraverty S., Mayo K., Norton J. B., Levitch D., Hatanpaa K. J., White C. L., 3rd, Bigio E. H., Caselli R., Baker M., Al-Lozi M. T., Morris J. C., Pestronk A., Rademakers R., Goate A. M., Cairns N. J. (2008) Ann. Neurol. 63, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokoseki A., Shiga A., Tan C. F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A., Okamoto K., Nishizawa M., Takahashi H., Onodera O. (2008) Ann. Neurol. 63, 538–542 [DOI] [PubMed] [Google Scholar]
- 12.Banks G. T., Kuta A., Isaacs A. M., Fisher E. M. (2008) Mamm. Genome 19, 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothstein J. D. (2007) Ann. Neurol. 61, 382–384 [DOI] [PubMed] [Google Scholar]
- 14.Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. (2006) Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 15.Sanelli T., Xiao S., Horne P., Bilbao J., Zinman L., Robertson J. (2007) J. Neuropathol. Exp. Neurol. 66, 1147–1153 [DOI] [PubMed] [Google Scholar]
- 16.Xiao S., Tjostheim S., Sanelli T., McLean J. R., Horne P., Fan Y., Ravits J., Strong M. J., Robertson J. (2008) J. Neurosci. 28, 1833–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 18.Cooper A. A., Gitler A. D., Cashikar A., Haynes C. M., Hill K. J., Bhullar B., Liu K., Xu K., Strathearn K. E., Liu F., Cao S., Caldwell K. A., Caldwell G. A., Marsischky G., Kolodner R. D., Labaer J., Rochet J. C., Bonini N. M., Lindquist S. (2006) Science 313, 324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaganovich D., Kopito R., Frydman J. (2008) Nature 454, 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chernoff Y. O., Uptain S. M., Lindquist S. L. (2002) Methods Enzymol. 351, 499–538 [DOI] [PubMed] [Google Scholar]
- 21.Rieder S. E., Banta L. M., Köhrer K., McCaffery J. M., Emr S. D. (1996) Mol. Biol. Cell 7, 985–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimoto R. I. (2008) Genes Dev. 22, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conway K. A., Harper J. D., Lansbury P. T. (1998) Nat. Med. 4, 1318–1320 [DOI] [PubMed] [Google Scholar]
- 24.Conway K. A., Lee S. J., Rochet J. C., Ding T. T., Williamson R. E., Lansbury P. T., Jr. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lashuel H. A., Hartley D. M., Petre B. M., Wall J. S., Simon M. N., Walz T., Lansbury P. T., Jr. (2003) J. Mol. Biol. 332, 795–808 [DOI] [PubMed] [Google Scholar]
- 26.Shorter J., Lindquist S. (2005) Nat. Rev. Genet. 6, 435–450 [DOI] [PubMed] [Google Scholar]
- 27.Kwong L. K., Uryu K., Trojanowski J. Q., Lee V. M. (2008) Neurosignals 16, 41–51 [DOI] [PubMed] [Google Scholar]
- 28.Johnson B. S., McCaffery J. M., Lindquist S., Gitler A. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6439–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean P. J., Kawamata H., Hyman B. T. (2001) Neuroscience 104, 901–912 [DOI] [PubMed] [Google Scholar]
- 30.Lin W. L., Dickson D. W. (2008) Acta Neuropathol. 116, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori F., Tanji K., Zhang H. X., Nishihira Y., Tan C. F., Takahashi H., Wakabayashi K. (2008) Acta Neuropathol. 116, 193–203 [DOI] [PubMed] [Google Scholar]
- 32.Nishihira Y., Tan C. F., Onodera O., Toyoshima Y., Yamada M., Morita T., Nishizawa M., Kakita A., Takahashi H. (2008) Acta Neuropathol. 116, 169–182 [DOI] [PubMed] [Google Scholar]
- 33.Scheibel T., Parthasarathy R., Sawicki G., Lin X. M., Jaeger H., Lindquist S. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4527–4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lashuel H. A., Hartley D., Petre B. M., Walz T., Lansbury P. T., Jr. (2002) Nature 418, 291. [DOI] [PubMed] [Google Scholar]
- 35.Kuo P. H., Doudeva L. G., Wang Y. T., Shen C. K., Yuan H. S. (2009) Nucleic Acids Res. 37, 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skovronsky D. M., Lee V. M., Trojanowski J. Q. (2006) Annu. Rev. Pathol. 1, 151–170 [DOI] [PubMed] [Google Scholar]
- 37.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. (1997) Cell 90, 549–558 [DOI] [PubMed] [Google Scholar]
- 38.Krobitsch S., Lindquist S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi W., Zibaee S., Jakes R., Serpell L. C., Davletov B., Crowther R. A., Goedert M. (2004) FEBS Lett. 576, 363–368 [DOI] [PubMed] [Google Scholar]
- 40.Daoud H., Valdmanis P. N., Kabashi E., Dion P., Dupré N., Camu W., Meininger V., Rouleau G. A. (2008) J. Med. Genet. 46, 112–114 [DOI] [PubMed] [Google Scholar]
- 41.Mackenzie I. R., Bigio E. H., Ince P. G., Geser F., Neumann M., Cairns N. J., Kwong L. K., Forman M. S., Ravits J., Stewart H., Eisen A., McClusky L., Kretzschmar H. A., Monoranu C. M., Highley J. R., Kirby J., Siddique T., Shaw P. J., Lee V. M., Trojanowski J. Q. (2007) Ann. Neurol. 61, 427–434 [DOI] [PubMed] [Google Scholar]
- 42.Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M. R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K. (2003) Science 302, 841. [DOI] [PubMed] [Google Scholar]
- 43.Pamphlett R., Kum Jew S. (2008) Acta Neuropathol. 116, 221–222 [DOI] [PubMed] [Google Scholar]
- 44.Pamphlett R., Luquin N., McLean C., Jew S. K., Adams L. (2009) Neuropathol. Appl. Neurobiol. 35, 222–225 [DOI] [PubMed] [Google Scholar]
- 45.Gitler A. D., Chesi A., Geddie M. L., Strathearn K. E., Hamamichi S., Hill K. J., Caldwell K. A., Caldwell G. A., Cooper A. A., Rochet J. C., Lindquist S. (2009) Nat. Genet. 41, 308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeger-Lotem E., Riva L., Su L. J., Gitler A. D., Cashikar A. G., King O. D., Auluck P. K., Geddie M. L., Valastyan J. S., Karger D. R., Lindquist S., Fraenkel E. (2009) Nat. Genet. 41, 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]