Abstract
Ubiquitin-like modifications regulate nearly every aspect of cellular functions. A key step in these modifications is the recognition of the carrier enzyme (E2) by the activating enzyme (E1). In this study, we have found that a critical E2-binding surface on the E1 of the small ubiquitin-like modifier has unusually high populations in both ordered and disordered states. Upon binding the E2, the disordered state is converted to the ordered state, which resembles the structure of the bound conformation, providing a mechanism to resolve the “Levinthal Paradox” search problem in a folding-upon-binding process. The significance of the folding-unfolding equilibrium is shown by the loss of functions of the mutations that shift the equilibrium to the folded state. This study highlights the importance of conformational flexibility in the molecular recognition event.
The conjugation of ubiquitin or ubiquitin-like proteins to other cellular proteins is one of the most important mechanisms in regulating cellular functions in eukaryotic systems and is broadly involved in controlling the life spans, trafficking, and functions of a wide range of proteins (1–4). Among the ubiquitin-like modifiers, the small ubiquitin-like modifier (SUMO)5 is the most studied, and its modification regulates many essential functions, such as gene transcription, hormone response, DNA repair, and nuclear import (5–7). Multiple enzymes are required for post-translational modifications by ubiquitin or ubiquitin-like proteins (1, 8). A ubiquitin-like protein (Ublp) is first activated by E1. The E1 required for SUMO modification is a tight heterodimer of two proteins, known as SUMO activation enzymes 1 and 2 (SAE1 and SAE2), which are homologous to the N-terminal and C-terminal portions of the ubiquitin E1, respectively (9). E1 catalyzes the adenylation of the C-terminal carboxyl group of the Ublp, and it then forms a thioester bond between the –SH group of the active site Cys residue and the C-terminal carboxyl group of the Ublp. The Ublp is then transferred to E2 (known as Ubc9 in the SUMO pathway), forming a thioester bond with the –SH group of the active site Cys residue. In the final step, the Ublp is attached to target proteins by forming an isopeptide bond between its C-terminal carboxyl group and the ϵ-amino group of a Lys residue on the target protein. This step generally requires another enzyme, isopeptide ligase.
During the transfer of Ublp from E1 to E2, the E2 enzyme is recruited to the E1-SUMO thioester conjugate by binding to multiple sites on E1, including the Cys domain (the domain containing the active Cys residue) and the ubiquitin-like (Ubl) domain (10, 11). Among these multiple binding sites, the Ubl domain has the highest binding affinity, and thus it is the key E2-binding site. The multivalent interaction produces high affinity binding between E1 and E2, which accounts for the efficiency of E1 at low concentrations. At the same time, the low to medium affinity of each of the individual interactions allows fast turnover of the enzymes upon completion of the reaction.
Recent studies have also indicated that conformational flexibility is important for the E1-E2 recognition. First, the E2-binding surface on the Cys domain of E1 contains an extended and flexible loop (11). Second, the E2-binding surface on the Ubl domains of SUMO and NEDD8 E1s either contains missing segments or has segments with unusually large B-factors in x-ray crystal structures (12, 13), suggesting that conformational flexibility also exists at the E2-binding surface of the Ubl domain. Therefore, conformational flexibility, in addition to the structural features exerted by amino acid side chains, may be an important determinant in the E1-E2 recognition.
In this study, using NMR spectroscopy, we show that the E2-binding site on the Ubl domain of SUMO E1 undergoes exchange between ordered and disordered states. The population of the disordered state is significant and is converted to the ordered state upon binding Ubc9. Using specially designed mutations to shift the conformational equilibrium to the ordered state but not altering the structure at the binding surface, we have shown that the folding-unfolding equilibrium in the E1 Ubl domain is important for the recognition of E2 in SUMO conjugation.
EXPERIMENTAL PROCEDURES
Plasmid Construction
A DNA fragment encoding the SUMO E1 Ubl domain (amino acid residues 441–555 of the SAE2 subunit) was amplified by PCR using primers that generated an NdeI site upstream and an XhoI site downstream. This fragment was cloned into the NdeI-XhoI sites of vector pET-28a to express the E1 Ubl domain protein with a C-terminal His6 tag. Site-directed mutagenesis, protein purification, and preparation of stable isotope-labeled protein samples for NMR experiments were carried out as described previously (11).
Sample Preparation
The plasmids expressing the wild-type and mutant SAE2-(441–555) domains were transfected into Escherichia coli strain BL21(DE3). 13C/15N-Labeled proteins were expressed in cells grown in modified M9 media supplemented with trace minerals and Eagle's basal vitamin mix (Invitrogen) using [13C]glucose and [15N]ammonium chloride as the only carbon and nitrogen sources, respectively, as described previously (14). The proteins were purified with affinity chromatography using nickel-nitrilotriacetic acid columns (Qiagen), and purifications were confirmed by gel electrophoresis. Imidazole was then eliminated by buffer exchange with a 50 mm Tris buffer, pH 7.0 to 7.1, containing 5–10% D2O, 5 mm DTT, and 0.02% sodium azide for NMR analysis. The same protein samples were used for native gel analysis.
Plasmids expressing full-length SAE2 proteins, with either the wild-type Ubl domain or with Ubl domains carrying either of the mutations, were transfected along with the plasmid expressing the SAE1 subunit into the E. coli BL21(DE3) CodonPlus strain. Wild-type and mutant E1 proteins were expressed in Luria-Bertani media and purified as described previously (12). Unlabeled Ubc9 was also expressed and purified as described previously (14).
NMR Measurements
SAE2-(441–555) protein samples for NMR spectroscopy contained 1.8 mm protein in 90% H2O, 10% D2O with 50 mm Tris buffer, pH 7.0, 5 mm DTT, and 0.02% sodium azide. All NMR spectra were collected at 25 °C on a Brucker Avance 600-MHz spectrometer equipped with a cryo-probe or a Brucker Avance 500-MHz spectrometer with a room temperature triple-resonance probe. The following spectra were acquired for backbone resonance assignments: two-dimensional 15N HSQC and three-dimensional HNCA, HNCACB, (H)C(CO)NH-TOCSY, HNCO, and HNCACO (15). All spectra were processed with NMRPipe (16) and analyzed with NMRView (6). Assignments of backbone resonances for proteins carrying the mutant SAE2-(441–555) domains were made by comparison with assignments of the wild-type protein with the assistance of triple-resonance spectra HNCA, HNCACB, CBCACONH, HNCO, and HNCACO (15). 1H chemical shifts were referenced to 2,2-dimethyl-2-silapentanesulfonic acid at 0 ppm, and 13C and 15N shifts were calculated from the 1H spectrometer frequency as described previously (17).
All 15N relaxation data were recorded at 25 °C at 1H frequencies of 600 MHz using a Bruker Avance 600 equipped with a cryo-probe. The spectra were processed using NMRPipe (16) and analyzed using NMRView (6). The 15N relaxation times T1 and T2 and 1H-15N NOE experiments were measured using two-dimensional inversion-recovery, Carr-Purcell-Meiboom-Gill, and steady-state NOE pulse sequences as described (18). 15N T1, T2, and NOE were recorded for each mutant and the wild-type SAE2-(441–555). The relaxation delays for the wild-type protein in T1 experiments were 10, 50, 100, 150, 200, 300, 400, 500, 700, 1000, and 1500 ms with a recycle delay of 1.5 s. The relaxation delays in T2 experiments for the wild-type Ubl domain were 4, 12, 20, 28, 40, 57, 73, 94, 122, 163, and 245 ms with a recycle delay of 1.5 s. The echo repetition time in the Carr-Purcell-Meiboom-Gill sequence was 0.45 ms. 15N T1 relaxation times for the mutant Ubl domains were measured with delays of 50, 210, 370, 530 (repeated), 690, 850, and 1001 ms. 15N transverse relaxation times (T2) for the mutant Ubl domains were measured with delays of 20, 40, 60, 80 (repeated), 100, 120, and 140 ms. For T1 and T2 experiments, the NMRView Rate Analysis module was used to fit the cross-peak intensities (I) as a function of the delay time (t) to a single exponential decay (I = I0e(t/T), where T = T1 or T2) with the peak-picking mode set to jitter. For NOE measurements, a pair of NOE spectra was collected with or without 5 s of proton presaturation. Heteronuclear 1H-15N NOE values were determined by taking the ratio of the peak intensities, as determined by NMRView, with and without 1H saturation. The NOE experiment was duplicated for error estimation.
SUMO Conjugation Assays
Conjugation assays for E1·SUMO and RanGAP1·SUMO complex formation were carried out following procedures described previously (11) unless specified otherwise. The steady-state kinetic assay for RanGAP1·SUMO complex formation under E1-limiting conditions was described previously (11, 19), except that E1 concentration was 1 μm for the GGG mutant and 400 nm for the 484Δ mutant, respectively. The percentages of E1 enzymes that are active are 30 and 37% for GGG and 484D, respectively, judged by the percentage of E1 that can form thioester conjugates with SUMO. The ranges of Ubc9 concentration were 200, 250, 500, and 1000 nm for GGG and 100, 125, 250, and 500 nm for 484Δ, respectively.
Isotope Exchange Assays
Isotope exchange assays were adapted from procedures described previously under initial velocity conditions (20). For ATP:PPi exchange, the reaction mixture included 50 mm Tris-HCl, pH 7.6, 10 mm MgCl2, 1 mm DTT, 1 mm ATP, 1 mm PPi, 10 μm SUMO, 2 μm E1, and tracer [32P]PPi in a final volume of 50 μl. The reaction (20 min, 37 °C) was followed by quenching with 0.5 ml of 5% trichloroacetic acid, and ATP was absorbed onto charcoal, which was washed extensively with 2% trichloroacetic acid; [32P]ATP levels were then counted by Cerenkov radiation. For AMP:ATP exchange, the reaction was carried out in a final volume of 20 μl containing 50 mm Tris-HCl, pH 7.6, 10 mm MgCl2, 0.2 mm DTT, 0.5 mm ATP, 50 μm AMP, 10 μm PPi, 2 μm SUMO, 1 μm E1, and tracer [14C]AMP. The reaction was incubated (10 min, 37 °C) and then stopped by addition of equal volumes of 8 m urea. Reaction samples (4 μl) were loaded onto a polyethyleneimine-cellulose plate, and ATP was separated from AMP by thin layer chromatography with a solution of 0.5 m LiCl, 1 m formic acid. The radiogram was obtained with exposure to a PhosphorImager plate and documented with a Typhoon scanner.
Detection of Ubc9-Ubl Interaction by Gel Electrophoresis
Gel electrophoresis was used to detect the Ubc9·Ubl complex. Approximately 40 μg of Ubc9 was incubated (1 h, room temperature) in a 20-μl volume with ∼6 μg of either the wild-type domain or one of the mutant Ubl domains in 50 mm Tris, 150 mm NaCl, 8% glycerol, and 1 mm DTT. Proteins were separated by nondenaturing 8–16% gel and stained with SimpleBlue.
RESULTS
Two Conformational States at the E2-binding Surface of the Ubl Domain
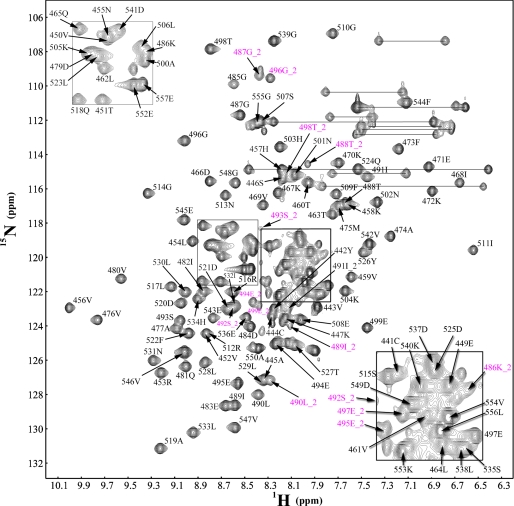
Possible conserved structural disorder in the Ubl domain across E1s of ubiquitin-like modifiers is suggested by missing segments in the x-ray structure of the Ubl domain of NEDD8 E1 (13) and unusually high B-factors in the analogous segments of SUMO E1 (12). To investigate the structural flexibility, we expressed the Ubl domain of SAE2 encompassing residues 441–555. Upon completion of backbone resonance assignments (Fig. 1), we identified two sets of resonances for the SAE2 segment encompassing residues 486 and 499 (Fig. 1, magenta letters). The two sets of resonances suggested that this region (Fig. 2a, purple in the crystal structure) exists in two conformational states that are exchanging at a rate slower than the chemical shift time scale. One set of peaks had intensities at ∼20–30% of the intensities of average cross-peaks (Figs. 1 and 2b). To confirm that the existence of the two sets of resonances was not due to artifacts in protein preparation, three separate purifications of the Ubl domain were made. The 1H-15N HSQC spectra of all three independent samples gave the same two sets of resonances for the residues in this segment. The 1H-15N HSQC spectrum of a longer construct containing the Ubl domain also had the same two sets of resonances for the residues in this segment. Both sets of resonances of residues at the N- and C-terminal ends of the segment (residues 486 and 499) made perfect C-α, C-β, and C-Ο connections with the resonances of residues 485 and 500 in triple-resonance NMR spectra, indicating that the peptide backbone was intact in both conformational states. These through chemical bond connections provide independent evidence of the integrity of the covalent structure.
FIGURE 1.
Identification of the slow conformational conversion at the E2-binding site of the Ubl domain. 1H-15N HSQC spectrum of the Ubl domain of E1 with assignments indicated according to sequence of the full-length SAE2. The assignments corresponding to the minor conformation are indicated with a “_2” in magenta. The boxed areas are enlarged and shown at the corners of the spectra.
FIGURE 2.
Interaction of the Ubl domain with Ubc9. a, crystal structure of the NEDD8 E2 (Ubc12) in complex with the Ubl domain is compared with the crystal structure of the Ubl domain of SUMO E1. Both Ubl domains are shown in a similar orientation to illustrate the conserved binding surface for E2. Residues 484 and 485 of the SAE2 subunit, where the Gly-Gly-Gly sequence was inserted, are highlighted in yellow, and the equivalent residues in the NEDD8 E1 are also highlighted in yellow. The Asp-484 side chain, which was deleted, is directed away from the E2-binding surface. The segment that undergoes folding-unfolding equilibrium is indicated in magenta. b, expanded region of the HSQC spectra showing the titration result of unlabeled Ubc9 with the 15N-labeled SAE2 Ubl domain. Assignments for the major (black letters) and minor (magenta letters) conformations are indicated. The spectra of the complexes of the two proteins with various ratios are overlaid with those of the free Ubl domain (black). Minimal changes were observed when the Ubl:E2 ratio was 3:1 (red). Upon addition of more E2 (2:1 Ubl:E2 ratio, green), there was a significant loss of intensity of the cross-peaks from the major conformer. At a Ubl:E2 ratio of 1:1 (blue), new resonances corresponding to the complex were observed. A further increase in the E2 concentration (magenta) did not alter the spectra but caused greater precipitation.
Structural comparison between the Ubl domains of the SUMO and NEDD8 E1 indicated that the segment encompassing residues 486–499 of SAE2 was equivalent to a segment at the binding surface for the E2 enzyme in NEDD8 E1 (Fig. 2a). To confirm that NEDD8 and SUMO E1s have a conserved mechanism for interactions with their cognate E2, we titrated unlabeled Ubc9 into the labeled SAE2 Ubl domain and examined their interaction using HSQC spectra. The titration results (Fig. 2b) confirmed that the binding interface between Ubc9 and the Ubl domain of SUMO E1 was analogous to that between Ubc12 and the Ubl domain of NEDD8 E1. In addition, the free and Ubc9-bound SAE2 Ubl domains were in slow exchange relative to the NMR time scale because we observed two sets of signals during titration with Ubc9 as follows: one from the free Ubl domain and another from the Ubl domain in the complex with Ubc9 (Fig. 2b). As the ratio of Ubc9 to the Ubl domain increased, signals from the complex appeared, whereas signals from the major conformer of the free SAE2 Ubl domain disappeared gradually. The slow dissociation rate of the Ubl·E2 complex was consistent with observation of the complex by native gel electrophoresis indicating the significant affinity between the Ubl domain and E2 (see below). Titration data also showed that with increasing concentrations of Ubc9, signals from the major conformer (Fig. 2b, black letters) of the Ubl domain disappeared first, whereas signals from the minor conformer (purple letters) disappeared later. This result suggested that the major conformer was responsible for binding E2, whereas the minor conformer was converted to the major conformer upon complex formation. The complex precipitated at concentrations as low as 0.1 mm; thus the sensitivity was not sufficient for resonance assignments and structure determination using triple-resonance three-dimensional NMR experiments.
Structural Insights into the Two Conformers
Secondary structural elements (Fig. 3a) were estimated from the chemical shift index using the TALOS program (21), which predicts backbone dihedral angles empirically from NMR chemical shifts. TALOS analysis indicated that the isolated Ubl domain maintained the structural integrity because the secondary structural elements were very similar to those in the x-ray crystal structure of full-length E1. The minor differences were likely due to differences in the different methods. The major conformer contained short β-strands that encompassed residues 488–491, which corresponded to a β-strand in the x-ray crystal structure of the full-length SUMO E1. The minor conformer did not form any regular secondary structures in this region. We attempted to estimate the exchange rate between the two conformers using exchange spectroscopy (EXSY). Exchange cross-peaks were not observed in either the 1H EXSY or the 1H-detected 15N EXSY experiments (22). This result indicated that the exchange time was slower than the mixing time of the experiment (1.3 s). Increasing the temperature to 35 °C, which was the upper limit before protein precipitation, did not increase the exchange rate sufficiently to produce a visible cross-peak in either the 1H or 1H-detected 15N EXSY spectra.
FIGURE 3.
Structural and dynamic properties of the wild-type and mutant Ubl domains of SAE2. a, comparison of the secondary structures of the SAE2 Ubl domain estimated from NMR chemical shifts of the wild type (WT) (major and minor conformations), GGG insertion mutant, and 484Δ deletion mutant. The secondary structures in the x-ray crystal structure of the full-length protein are also shown. The cylinders and arrows represent helices and β-strands, respectively. The E2-binding regions were estimated through comparison with the NEDD8 structure and confirmed by NMR chemical shift perturbation (Fig. 2). b, 1H-15N NOE of the wild-type and mutant SAE2 Ubl domains versus the residue number. Blue diamonds, major conformation; magenta squares, minor conformation. The secondary structures are shown at the top. The region that undergoes folding-unfolding equilibrium in the wild-type protein is highlighted.
To gain further insights into the structural properties of the two different conformations, 1H-15N NOE and 15N T1 and T2 relaxation measurements were carried out (Fig. 3b and supplemental Fig. S1 and Fig. S2). The 1H-15N NOE values theoretically range from ∼0.83 to −3.6, corresponding to a conformational flexibility that is completely rigid to one that is completely unrestricted. The major conformation of the segment encompassing residues 485–500 had NOE values between 0.6 and 0.83 (Fig. 3b). The residues forming the short β-strand of this segment had average NOE values of ∼0.82. The two flanking loops of the β-strand were more flexible, as indicated by lower NOE values ranging between 0.6 and 0.7. However, the minor conformer had much smaller NOE values that were between 0 and 0.2. Such values indicated that the dynamics of the minor conformer were much more mobile than those of the major conformer and that the minor conformer was similar in mobility to the middle of an unfolded protein (23–26). The high flexibility of the minor conformer was consistent with its lack of regular secondary structures. The flexibility was also indicated by the significantly longer T2 and shorter T1 values (supplemental Figs. S1 and S2). The high mobility of the minor conformer and chemical shift values in the random coil range both indicated that the minor conformer was disordered. Therefore, the segment encompassing residues 486–499 of SAE2 undergoes folding and unfolding equilibrium. In conjunction with previous studies of the NEDD8 E1-E2 interaction, the folded conformation was similar to that of the E2-bound state (27).
Design of Mutations to Investigate the Functional Significance of the Dynamics
To investigate the role of dynamics in the recognition of E2, we evaluated possible ways to alter the dynamics of the SAE2 segment. The sequence from residues 485 to 500 (GKGTILISSEEGETEA) contained mostly charged and polar residues, with only three hydrophobic residues, ILI (Fig. 3a). The lack of hydrophobic and aromatic residues in this segment was consistent with its observed conformational flexibility. The dihedral angle preferences and hydrophobic contacts exerted by the ILI segment likely accounted for the formation of the β-strand in the major conformation. However, these three residues were also at the binding interface with the E2 enzyme (Fig. 2); therefore, a substitution of one hydrophobic residue might not only alter the equilibrium of the folded and unfolded states but could also alter the direct contact with Ubc9. Consequently, we used an alternative strategy.
Sequence variation throughout evolution provides a wealth of information on structure-function relationships. The sequence alignment of the SAE2 Ubl domain indicated that the loop located at the N terminus of the segment, which undergoes folding-unfolding transition, had variable lengths in various species (Fig. 4, boxed region). The variable loop lengths suggested that they were not likely to be important for structural integrity. However, lengthening or shortening of the loop might have a direct effect on the folding-unfolding equilibrium of the segment. We constructed two mutants to evaluate the impact of loop length on the dynamics and functions of the Ubl domain. One mutant contained a deletion of residue Asp-484 (484Δ), which was located at the tip of a turn, with its side chain facing away from the interface with E2; it was therefore not in direct contact with E2 (Fig. 2a). The other mutant had a Gly-Gly-Gly insertion between residues 484 and 485 (Fig. 2a, yellow). This insertion was also located at the tip of the turn and was not at the binding interface. Both mutants were introduced into the Ubl domain of SAE2, and their structural and dynamic effects were investigated.
FIGURE 4.
Sequence alignment of the SUMO Ubl domain of 10 species, including plants and animals. The boxed region is between 484D and 485G, and is the site of both mutations. Conserved residues are shown in red.
Neither mutant altered the structure of the Ubl domain significantly. Upon completion of backbone resonance assignments using 13C/15N-enriched protein samples, the secondary structures of both mutants were estimated using TALOS (Fig. 3a) (21). Both mutants had essentially identical secondary structures as the wild-type protein, suggesting that the mutation did not disrupt the structural integrity. The resonances of both mutants were nearly completely assigned by using triple-resonance experiments, although most resonances had nearly identical chemical shift values as those of the wild-type protein. Although some additional resonances were present in the spectra of both mutants, the intensities of these extra cross-peaks were very low, less than 2% of the intensities of average cross-peaks. This was in contrast to the wild-type Ubl domain, where the cross-peak intensities from the disordered conformation were ∼20–30% of those from the major conformation. This result indicated that the insertion and deletion of the loop shifted the conformation equilibrium to the ordered state, without significantly altering the structural integrity of the major conformer.
To further characterize the dynamic properties of the deletion and insertion mutations, 15N relaxation measurements were carried out for the mutants. 1H-15N NOE values indicated that deletion of residue Asp-484 or insertion of GGG in the loop did not considerably influence the dynamic properties of other regions of the protein (Fig. 3b), which again confirmed that the insertion and deletion mutants maintained overall structural integrity. The only difference appeared to be in the loop where the mutations were introduced; the GGG insertion mutation increased the flexibility in this loop, likely due to the flexibility of the Gly residues. Overall, the structural and dynamic characterization showed that the loop mutations, which were not at the E2-binding interface, did not significantly perturb the major conformation of the Ubl domain but altered the dynamics of the binding site.
Role of Dynamics in Binding E2
To examine the effect of the mutations on E1 activity, we introduced the two site-directed mutations, deletion of Asp-484 (484Δ) and insertion of GGG (GGG), into the full-length E1 for activity assays. Both mutants had lower activities in the overall conjugation reactions of RanGAP1 (Fig. 5a). In particular, the GGG insertion lost most of its activity. A 5-fold increase in its concentration in the assay resulted in slightly raised activity levels. The mutant 484Δ also had much lower activity than wild-type E1, but its activity appeared to be higher than that of the GGG insertion mutant.
FIGURE 5.
E1 mutants do not alter the adenylation and thioester formation activities. a, formation of the RanGAP1-SUMO conjugate catalyzed by the wild-type (WT) and mutant E1. Fifteen μm SUMO was incubated (15 min, 37 °C) with the ATP-regeneration system, 15 μm RanGAP1, 0.5 μm Ubc9, and 0.5 μm E1 or one of the E1 mutants at the indicated concentrations. The assays were resolved by SDS-PAGE and stained with SimpleBlue. Proteins of interest are marked with arrowheads. b and c, ATP:PPi exchange and ATP:AMP exchange rates of the wild-type and mutant E1 enzymes. The rates are normalized to the percentage of the E1 enzyme that is active, judged by the percentage of E1 that can form thioester conjugates with SUMO (see details in text). d, time-dependent formation of SUMO-E1 thioester conjugate (details in text).
E1-dependent reactions were then investigated to determine which step of the conjugation pathway was affected by the mutation. E1 catalyzes two reactions, the adenylation of the C terminus of SUMO and the formation of a thioester bond with SUMO. To investigate whether the mutations altered adenylation activities, we carried out [32P]PPi:ATP exchange assays to examine the rate of radioactive ATP production from radioactive pyrophosphate (Fig. 5b) (20). The results showed that the full-length wild-type and mutant E1 proteins had similar activities, suggesting that the mutations did not affect E1 adenylation activity. The AMP:ATP exchange assay provides information on both steps of the E1-catalyzed reactions (20). Thus, [14C]AMP was used in the AMP:ATP exchange assay, by examining the production of radioactive ATP from radioactive AMP (Fig. 5c). We found that the wild-type and mutant E1 enzymes showed similar activities, although the GGG insertion mutant showed slightly lower activity than the wild-type and 484Δ mutant proteins. Time-dependent formation of the E1-SUMO thioester bond was also examined for the three versions of the enzyme, revealing similarly fast rates (Fig. 5d). Taken together, the data suggested that the altered flexibility of the E1 Ubl domain significantly affected the overall conjugation activity, but it did not affect the catalysis of SUMO adenylation or the formation of the thioester bond between SUMO and the E1 catalytic Cys residue significantly. This was consistent with previous findings that the Ubl domain of SUMO E1 was not important for SUMO adenylation and E1-SUMO thioester formation (12).
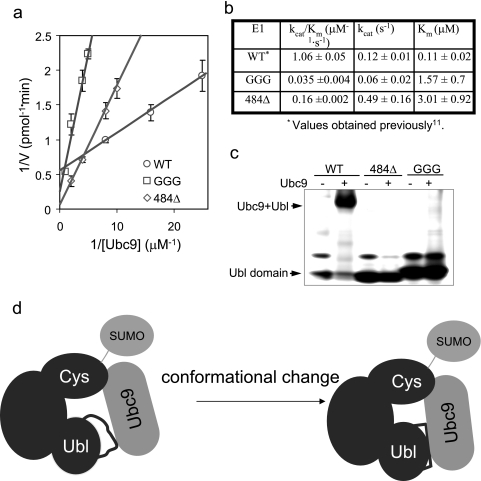
Next, we investigated whether the altered dynamics of the Ubl domain affected the transfer of SUMO from E1 to E2. Quantitative enzyme kinetic analysis was carried out for the various E1 proteins using the approach described in our previous studies (11) and Haas and co-workers (19). The Km values of the mutants are 15- and 30-fold higher than that of the wild-type enzyme, indicating that the mutant E1 proteins have significantly reduced affinity for Ubc9. In addition, the kcat value of GGG is 2-fold less than that of the wild-type protein, but that of 484Δ is 3-fold higher. This result suggests that in addition to the reduced affinity, the mutants may also affect the internal rate constant for trans-thiolation, probably due to slightly altered orientation of Ubc9 in the ternary complex·Ubc9 binding intermediate complex. The altered interaction between E2 and the E1 Ubl domain mutations was further confirmed by native gel mobility shift assays (Fig. 6c), and although the mutations were not located on the binding interface and did not significantly perturb the structure, both mutants were severely deficient in binding Ubc9 (Fig. 6c).
FIGURE 6.
E1 mutants are deficient in recruiting E2. a, double-reciprocal plot of the steady-state kinetics of E1-catalyzed trans-thiolation in overall conjugation reactions. Initial rates for SUMO-RanGAP1 conjugation were determined under E1-limiting conditions (see “Experimental Procedures”). E1 concentration was 1 μm for the GGG mutant and 400 nm for the 484Δ mutant. Ubc9 concentrations are indicated in the figure. Because of its lower activity, higher concentrations of Ubc9 were used for GGG and 484Δ than for the wild-type protein (WT) to reliably measure the initial rates. b, kinetic rate constants extracted from the plots for the mutant E1 proteins along with the values of the wild type reported previously (11). The kinetic constants are normalized to the percentage of the E1 enzyme that is active, judged by the percentage of E1 that can form thioester conjugates with SUMO. c, gel electrophoresis to detect the Ubc9·Ubl complex. Approximately 40 μg of Ubc9 was incubated (1 h, room temperature) with ∼6 μg of either the wild-type or one of the mutant Ubl domains. Proteins were separated on nondenaturing 8–16% gel and stained with SimpleBlue. Because of the charge property, Ubc9 did not run into the gel. d, schematic illustration of the conformational transition during E1 and E2 association.
DISCUSSION
In this study, we have shown that the main E2-binding site of SUMO E1 undergoes conformational conversion that is important for its interaction with E2 for SUMO modification. The conformational flexibility observed here is consistent with previous x-ray crystallographic studies. In the x-ray structure of SUMO E1, this segment has much higher B-factors than other regions (12). The equivalent region in the crystal structure of the homologous NEDD8 E1 is missing, which suggests that the conformational disorder observed in the SUMO E1 also exists in the refined structure of NEDD8 E1 (13).
This study underscores the importance of flexibility in molecular recognition. Mutations in the SUMO E1 that shift the folding-unfolding equilibrium to the folded state, which resembles the bound structure, were impaired in binding E2. The slight difference between the activities of the two mutants may be due to the differences in their dynamics in the loop where the mutations are located. This result indicates that retaining conformational flexibility of the unbound state is necessary for the molecular recognition because the flexibility may be important in lowering the energy barrier of complex formation by allowing adjustment of the interface locally (Fig. 6d) (30). In contrast, a rigid structure would require adjustment of the interface globally, which would encounter a much higher energy barrier. Unlike the NEDD8 and SUMO E1, the corresponding segment in the Ubl domain of the recently reported x-ray structure of ubiquitin E1 does not have unusually elevated B-factor values (28). Consistent with the findings described here, the Ubl domain of the ubiquitin E1 does not bind to its cognate E2s with measurable affinity because an E1·E2 complex could not be detected by gel filtration (29). Such a property may be necessary for the ubiquitin E1 to switch rapidly among its many E2s. In contrast, NEDD8 or SUMO only has a single cognate E2, and both their E1s bind their cognate E2 enzymes with sufficient affinity that is detectable by native gel electrophoresis. Taken together, the conformational flexibility of the Ubl domain is likely to be critical for its binding affinity for E2.
The findings described here provide an example of how flexible segments search the correct bound conformations efficiently. The prevalence of the folding-upon-binding phenomenon has raised the question of how the correct bound conformation is efficiently searched, a problem known as the “Levinthal Paradox” (33). Evidence for residual structure in the denatured states of several proteins characterized to date suggests a way to simplify the Levinthal Paradox search problem by the intrinsic tendency to form certain conformers (32). However, in most of these cases, the residual structures are in very low population or are highly unstable, so it is difficult to characterize them to determine whether they resemble the bound conformations. In this study, a folded conformation is in sufficient population to be shown that it resembles the bound conformation, providing a mechanism to resolve the Levinthal Paradox.
The loop/turn located N-terminal to the segment that displays the folding-unfolding equilibrium is evolutionarily diverse and has variable lengths in different species. Our studies indicate that lengthening or shortening the turn changes the dynamics of the E2-binding site. These data suggest that variation of the loop lengths and composition throughout evolution may be a mechanism to fine-tune the dynamic properties of this E2 recognition site of E1.
In summary, this study has identified an unusual conformational exchange between ordered and disordered states that is important in E1-E2 interaction in SUMO modification. The conformational flexibility is likely to be conserved in some E1s across the various ubiquitin-like modifications. This study is an important addition to a limited number of previous reports showing that the dynamic properties of a protein, in addition to structural properties, are fine-tuned for function (30, 33).
Supplementary Material
Acknowledgments
We thank Dr. Robert London and Dr. Daniel Sem for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM074748 and R01GM086171.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- SUMO
- small ubiquitin-like modifier
- DTT
- dithiothreitol
- E1
- activating enzyme for ubiquitin-like modifications
- E2
- carrier enzyme for ubiquitin-like modifications
- Ubl
- ubiquitin-like
- Ublp
- Ubl protein
- HSQC
- heteronuclear single quantum coherence
- NOE
- nuclear Overhauser effect
- EXSY
- exchange spectroscopy.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. (1997) Trends Biochem. Sci. 22, 383–387 [DOI] [PubMed] [Google Scholar]
- 3.Welchman R. L., Gordon C., Mayer R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 599–609 [DOI] [PubMed] [Google Scholar]
- 4.Kerscher O., Felberbaum R., Hochstrasser M. (2006) Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 5.Guo B., Yang S. H., Witty J., Sharrocks A. D. (2007) Biochem. Soc. Trans. 35, 1414–1418 [DOI] [PubMed] [Google Scholar]
- 6.Johnson E. S. (2004) Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 7.Seeler J. S., Dejean A. (2003) Nat. Rev. Mol. Cell Biol. 4, 690–699 [DOI] [PubMed] [Google Scholar]
- 8.Haas A. L., Siepmann T. J. (1997) FASEB J. 14, 1257–1268 [DOI] [PubMed] [Google Scholar]
- 9.Desterro J. M., Rodriguez M. S., Kemp G. D., Hay R. T. (1999) J. Biol. Chem. 274, 10618–10624 [DOI] [PubMed] [Google Scholar]
- 10.Huang D. T., Hunt H. W., Zhuang M., Ohi M. D., Holton J. M., Schulman B. A. (2007) Nature 445, 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Hu W., Cai S., Lee B., Song J., Chen Y. (2007) Mol. Cell 27, 228–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lois L. M., Lima C. D. (2005) EMBO J. 24, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walden H., Podgorski M. S., Schulman B. A. (2003) Nature 422, 330–334 [DOI] [PubMed] [Google Scholar]
- 14.Liu Q., Jin C., Liao X., Shen Z., Chen D. J., Chen Y. (1999) J. Biol. Chem. 274, 16979–16987 [DOI] [PubMed] [Google Scholar]
- 15.Clore G. M., Gronenborn A. M. (1994) Methods Enzymol. 239, 349–363 [DOI] [PubMed] [Google Scholar]
- 16.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 17.Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 18.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 19.Siepmann T. J., Bohnsack R. N., Tokgöz Z., Baboshina O. V., Haas A. L. (2003) J. Biol. Chem. 278, 9448–9457 [DOI] [PubMed] [Google Scholar]
- 20.Haas A. L., Rose I. A. (1982) J. Biol. Chem. 257, 10329–10337 [PubMed] [Google Scholar]
- 21.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 22.Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E. (1994) J. Biomol. NMR 4, 727–734 [DOI] [PubMed] [Google Scholar]
- 23.Ebert M. O., Bae S. H., Dyson H. J., Wright P. E. (2008) Biochemistry 47, 1299–1308 [DOI] [PubMed] [Google Scholar]
- 24.Schwarzinger S., Wright P. E., Dyson H. J. (2002) Biochemistry 41, 12681–12686 [DOI] [PubMed] [Google Scholar]
- 25.Penkett C. J., Redfield C., Jones J. A., Dodd I., Hubbard J., Smith R. A., Smith L. J., Dobson C. M. (1998) Biochemistry 37, 17054–17067 [DOI] [PubMed] [Google Scholar]
- 26.Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E. (1997) Biochemistry 36, 2390–2402 [DOI] [PubMed] [Google Scholar]
- 27.Huang D. T., Paydar A., Zhuang M., Waddell M. B., Holton J. M., Schulman B. A. (2005) Mol. Cell 17, 341–350 [DOI] [PubMed] [Google Scholar]
- 28.Lee I., Schindelin H. (2008) Cell 134, 268–278 [DOI] [PubMed] [Google Scholar]
- 29.Hershko A., Heller H., Elias S., Ciechanover A. (1983) J. Biol. Chem. 258, 8206–8214 [PubMed] [Google Scholar]
- 30.Peng T., Zintsmaster J. S., Namanja A. T., Peng J. W. (2007) Nat. Struct. Mol. Biol. 14, 325–331 [DOI] [PubMed] [Google Scholar]
- 31.Karplus M. (1997) Fold Des. 2, S69–75 [DOI] [PubMed] [Google Scholar]
- 32.Receveur-Bréchot V., Bourhis J. M., Uversky V. N., Canard B., Longhi S. (2006) Proteins 62, 24–45 [DOI] [PubMed] [Google Scholar]
- 33.Henzler-Wildman K. A., Lei M., Thai V., Kerns S. J., Karplus M., Kern D. (2007) Nature 450, 913–916 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.