Abstract
Sphingolipids are important signaling molecules in many biological processes, but little is known regarding their physiological roles in the mitochondrion. We focused on the biochemical characters of a novel sphingomyelinase (SMase) and its function in mitochondrial cer a mide generation in zebrafish embryonic cells. The cloned SMase cDNA encoded a polypeptide of 545 amino acid residues (putative molecular weight, 61,300) containing a mitochondrial localization signal (MLS) and a predicted transmembrane domain. The mature endogenous enzyme was predicted to have a molecular weight of 57,000, and matrix-assisted laser de sorp tion ionization time-of-flight mass spectrometry analysis indicated that the N-terminal amino acid residue of the mature enzyme was Ala-36. The purified enzyme optimally hydrolyzed [14C]sphingomyelin in the presence of 10 mm Mg2+ at pH 7.5. In HEK293 cells that overexpressed SMase cDNA, the enzyme was localized to the mitochondrial fraction, whereas mutant proteins lacking MLS or both the MLS and the transmembrane domain were absent from the mitochondrial fraction. Endogenous SMase protein co-localized with a mitochondrial cytostaining marker. Using a protease protection assay, we found that SMase was distributed throughout the intermembrane space and/or the inner membrane of the mitochondrion. Furthermore, the overexpression of SMase in HEK293 cells induced cer a mide generation and sphingomyelin hydrolysis in the mitochondrial fraction. Antisense phosphorothioate oligonucleotide-induced knockdown repressed cer a mide generation and sphingomyelin hydrolysis in the mitochondrial fraction in zebrafish embryonic cells. These observations indicate that SMase catalyzes the hydrolysis of sphingomyelin and generates cer a mide in mitochondria in fish cells.
Sphingomyelinase (SMase,2 sphingomyelin phosphodiesterase, EC 3.1.4.12) hydrolyzes sphingomyelin and produces ceramide and phosphocholine. Ceramide plays an important role as a signaling molecule in cell proliferation, apoptosis, cell cycle arrest, differentiation, and the stress response in animal cells (1–5). To date, three distinct classes of acid, neutral, and alkaline SMases have been identified according to optimum pH, cation dependence, amino acid sequence, and subcellular localization (3).
The Mg2+-dependent neutral SMases have emerged as major candidates in the mediation of ceramide-induced cell signaling (6). Recent research has identified at least three distinct neutral SMases in human and mouse, designated as neutral SMase 1, SMase 2, and SMase 3 (7–9). Neutral SMase 1 was the first SMase identified in human and mouse. Although mammalian enzymes exhibited Mg2+-dependent neutral SMase activity in vitro (9), no significant biological functions in sphingomyelin and ceramide metabolism were identified in SMase 1-overexpressing cells (10) or neutral SMase 1 knock-out mice (11). In zebrafish embryos, Mg2+-dependent neutral SMase 1 produced ceramide and caused thalidomide-induced vascular defects (12). In addition, SMase 1 was found to mediate heat-induced ceramide generation and apoptosis (13).
The neutral SMase 2 gene SMPD3, has also been identified based on its similarity to Bacillus cereus SMase DNA sequences (7). This gene encodes a membrane-bound protein expressed in the brain and liver that has two highly hydrophobic segments near the N-terminal region, both of which are thought to function as transmembrane domains. Unlike neutral SMase 1, neutral SMase 2 possesses Mg2+-dependent neutral SMase activity in vivo in MCF-7 cells (14). When overexpressed in the confluent phase of MCF-7 cells, mouse neutral SMase 2 was palmitoylated via thioester bonds and localized in the inner leaflet of the plasma membrane (15). In MCF-7 cells stably expressing neutral SMase 2, the enzyme inhibited cell growth and was required for cells to undergo confluence-induced cell cycle arrest (16). Interestingly, neutral SMase 2 was isolated as the confluent 3Y1 cell-associated 1 gene (cca1) in rat 3Y1 cells (17). Neutral SMase 2 has been implicated in signal transduction events in cell growth and the cellular response to cytokines (18, 19), oxidative stress (20), and amyloid β-peptide (21).
Stoffel et al. (22) demonstrated that gene-targeted mice deficient for neutral SMase 2 developed a novel form of dwarfism and had delayed puberty as part of a hypothalamus-induced pituitary hormone deficiency. Strikingly, positional cloning of the recessive mutation fragilitas ossium in mice identified a deletion in the gene that encodes neutral SMase 2, leading to the complete loss of neutral SMase activity (23). The mutant fragilitas ossium mice develop severe osteogenesis and dentinogenesis imperfecta, with no collagen defect. Thus, mouse neutral SMase 2 is essential for late embryonic and postnatal development.
Mitochondria contain small amounts of a variety of sphingolipids, including ceramide and sphingomyelin (24–26), which may be derived from the endoplasmic reticulum via intimate membrane contacts or produced in response to apoptosis. For mitochondria isolated from HL-60 cells, treatment with ceramide inhibited the mitochondrial respiratory chain complex III (27). Birbes et al. (28) found that the selective hydrolysis of a mitochondrial pool of sphingomyelin induced apoptosis. They transfected MCF-7 cells with B. cereus SMase targeted to various subcellular organelles, but they observed cytochrome c release and apoptosis induction only when the enzyme was targeted to the mitochondria. Ceramide activated the mitochondrial protein phosphatase 2A, which dephosphorylated Bcl-2 and led to apoptosis (29). In MCF-7 cells, mitochondrial ceramide generation in response to tumor necrosis factor-α induced Bax translocation to mitochondria and subsequent cytochrome c release and apoptosis (30). The permeability of the mitochondrial outer membrane correlates directly with the level of ceramide in the membrane (31). The concentration of ceramide at which significant channel formation occurs is consistent with the level of mitochondrial ceramide that occurs during the induction phase of apoptosis (31). In isolated mitochondria, ceramide can also form membrane channels large enough to release cytochrome c and other small proteins (32). Ceramide-metabolizing enzymes, such as a bovine liver ceramide synthase (33) and human ceramidase (34), are localized to the mitochondrion. These observations suggest the existence of a mitochondrial pool of sphingomyelin and the function of a sphingomyelin-specific metabolic pathway in mitochondria. However, no SMase has been identified in mitochondria.
We identified and examined the biochemical properties of a novel SMase localized to the zebrafish mitochondrion. The enzyme was cloned from a cDNA library of embryonic zebrafish cells. It was found to regulate mitochondrial ceramide levels.
EXPERIMENTAL PROCEDURES
Reagents
The C6–7-nitro-2–1,3-benzoxadiazol-4-yl (C6-NBD) sphingomyelin was purchased from Matreya (Pleasant Gap, PA). The [N-methyl-14C]sphingomyelin (2 GBq/mmol), l-3-phosphatidyl [N-methyl-14C]choline, 1,2-dipalmitoyl (2.11 GBq/mmol), [1-O-octadecyl-3H]lyso-platelet activating factor (5.99 TBq/mmol), protein G-Sepharose, polyvinylidine fluoride membrane, and ECLTM Western blotting detection kit were purchased from GE Healthcare. Anti-actin monoclonal antibody, anti-FLAG M2 monoclonal antibody, anti-HSP60 monoclonal antibody, anti-catalase monoclonal antibody, anti-cytochrome c polyclonal antibody, and p3XFLAG-CMVTM-14 expression vector were purchased from Sigma. Rabbit anti-cadherin polyclonal antibody and goat anti-aldolase polyclonal antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-KDEL mouse monoclonal antibody and anti-58-kDa Golgi protein mouse monoclonal antibody were purchased from Abcam (Cambridge, MA). Cell culture reagents, the ThermoScript RT-PCR system, XpressTM system synthetic oligonucleotides, goat anti-rabbit and anti-mouse IgG Alexa Fluor 488-labeled antibody, goat anti-mouse IgG Alexa Fluor 594-labeled antibody, and MitoTracker Red were purchased from Invitrogen. The Premix Taq (ExTaqTM version 2) and PrimeSTARTMHS DNA polymerase were purchased from Takara Biomedical (Shiga, Japan).
Cell Culture
A zebrafish embryonic (ZE) cell line was cultured in Leibovitz's L-15 medium (Invitrogen) supplemented with 2% fetal calf serum (FCS; JRH Biosciences, Lenexa, KS) at 28.5 °C (13). Human embryonic kidney (HEK) 293 cells were obtained from the Health Science Research Resources Bank (Osaka, Japan). HEK293 cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% FCS at 37 °C in a humidified incubator in 5% CO2.
Cloning of Mitochondrial Neutral SMase and Neutral SMase 2
The amino acid sequence of human neutral SMase 2 (GenBankTM accession number AJ250460) was used to search the zebrafish EST data base of GenBankTM. Zebrafish EST clones for mitochondrial neutral SMase (GenBankTM accession numbers EH455427, EH443470, and DT055903) and zebrafish genomic sequences (GenBankTM accession numbers EH455427, EH443470, and DT055903) for neutral SMase 2 (GenBankTM accession number NW_001512806) homologous to human neutral SMase 2 were obtained from the National Center for Biotechnology Information (NCBI) EST data base. Total RNA from zebrafish ZE cells was extracted using TRIzol® reagent (Invitrogen), according to the manufacturer's protocol. For first strand cDNA synthesis, 5 μg of RNA was reverse-transcribed in a 20-μl reaction volume using the ThermoScript RT-PCR system (Invitrogen), according to the manufacturer's protocol. The RT products were diluted by 10 times with distilled water and stored at −20 °C until use. PCR primers for mitochondrial SMase and neutral SMase 2 were synthesized. The following primers were used to amplify mitochondrial SMase: upstream, 5′-GAGTA ACTCA GTAGG GTGTT GAGGA CACGG-3′; downstream, 5′-AGCTG ATCAG AGGTG GGGTT GTATT GATCT-3′ (PCR product, 1820 bp). The following primers were used to amplify neutral SMase 2: upstream, 5′-AAGGT GAGCC AGAAA TGGTC TTGCA CACC-3′; downstream, 5′-AAGGT GAGCC AGAAA TGGTC TTGCA CACC-3′ (PCR product, 2083 bp). PCR was carried out in a total reaction volume of 50 μl using PrimeSTARTMHS DNA polymerase. The reaction conditions were as follows: 96 °C for 2 min; 30 cycles of denaturation at 98 °C for 10 s, annealing at 60 °C for 5 s, and polymerization at 72 °C for 2 min; and a final extension at 72 °C for 5 min. The PCR products were subcloned into the pGEM-T Easy vector, and the cloned nucleotide sequences were determined using a DNA sequencer (ABI 3100; Applied Biosystems, Foster City, CA) with a BigDye Terminator cycle sequencing kit (Applied Biosystems).
Sequence Alignment
The proteins, including zebrafish mitochondrial neutral SMase (AB361066), zebrafish neutral SMase 2 (AB361067), human neutral SMase 2 (AJ250460), and mouse neutral SMase 2 (AJ250461), were subjected to sequence alignment using the DNASTAR program (DNASTAR, Madison, WI).
Preparation of Rabbit Polyclonal Antibody against Mitochondrial SMase
To generate a polyclonal antibody for zebrafish mitochondrial SMase, the recombinant protein was purified as an antigen. The cDNA sequence of mitochondrial neutral SMase was amplified by PCR using the sense primer 5′-CATAT GCCAC TGCAA GCAAT ACGCC GACCG-3′ and the antisense primer 5′-GGATC CTCAG TTCTG CTCTG AATCC AGAGA-3′, each of which contained a BamHI and an NdeI site; the amplified DNA was subcloned into the pGEM-T Easy plasmid vector using the TA cloning method (Promega, Madison, WI). The cloned nucleotide sequence was confirmed by sequencing and then subcloned into both the NdeI and BamHI sites in the multiple cloning site of the pET-16b vector (Novagen, Madison, WI) to fuse to a His tag sequence at the N terminus of the mitochondrial SMase open reading frame. The construct was designated pETZMTSMase. Mitochondrial SMase was expressed in Escherichia coli BL21(DE3)pLysE cells (Novagen) transformed with pETZMTSMase. The cells were inoculated to 200 ml of Luria-Bertani (LB) broth and grown overnight at 30 °C in a shaker at 200 rpm. The culture was transferred to 2000 ml of fresh LB broth supplemented with 100 μg/ml ampicillin in a 5-liter flask, and the above incubation conditions were continued until turbidity at 600 nm reached 0.8. Isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.5 mm, and the culture was grown for a further 4 h to induce the expression of the transgene products. Bacterial cells were collected by centrifugation at 4000 × g for 15 min. The N-terminal His-tagged mitochondrial SMase was purified from the bacterial extract by affinity chromatography using a His Trap HP column (GE Healthcare), according to the manufacturer's protocol. The bacterial cells were suspended in lysis buffer (50 mm Tris-HCl (pH 7.5), 10% glycerol, 1× proteinase inhibitor mixture, 5 mm MgCl2, 60 mm imidazole, 0.1% Triton X-100, 1 mm EDTA, and 1 mg/ml lysozyme) by passing through a 22-gauge needle. All subsequent procedures were carried out at 4 °C. The supernatant was collected by centrifuging the lysate at 10,000 × g for 30 min and was then dialyzed with wash buffer (50 mm Tris-HCl (pH 7.5), 300 mm NaCl, 60 mm imidazole, 0.1% Triton X-100, and 1 mm EDTA). The dialyzed sample was loaded onto a 5-ml His Trap HP column (GE Healthcare) that had been equilibrated in wash buffer. After sample loading, the column was washed with 100 ml of wash buffer, followed by a 0–100% linear gradient of elution buffer (50 mm Tris-HCl (pH 7.5), 300 mm NaCl, 800 mm imidazole, 0.1% Triton X-100, and 1 mm EDTA). The flow rate was 1 ml/min, and 2-ml fractions were collected. The His Trap HP fractions with neutral SMase activity against C6-NBD-sphingomyelin were pooled and dialyzed with gel filtration buffer (25 mm Tris-HCl (pH 7.5), 150 mm NaCl, 5 mm MgCl2, 0.1% Triton X-100, and 1 mm EDTA) and then loaded onto a Sephacryl S-100 column (HR 16/60, GE Healthcare) equilibrated in gel filtration buffer. The column was eluted at 1 ml/min with 150 ml of gel filtration buffer. The fractions were collected into test tubes and analyzed by SDS-PAGE. The recombinant protein was used to immunize rabbits, and the obtained antiserum was affinity-purified using a protein G-Sepharose column (GE Healthcare) coupled to the same antigen.
Purification of Mitochondrial SMase from ZE Cells
To purify endogenous mitochondrial SMase, protein G-Sepharose coupled to rabbit anti-mitochondrial SMase antibody was used for affinity chromatography. ZE cells (5 × 107) were collected and centrifuged at 1000 × g for 15 min. The cells were suspended in lysis buffer (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Triton X-100, 1% Nonidet P-40, 1 mm dithiothreitol (DTT), 1 mm EDTA, 1 mm EGTA, and 1× protease inhibitor mixture) using a Dounce homogenizer. All subsequent procedures were carried out at 4 °C. The supernatant was collected and centrifuged at 10,000 × g for 30 min and loaded onto a protein G-Sepharose column coupled to an anti-mitochondrial neutral SMase antibody column equilibrated with lysis buffer at a flow rate of 10 ml/h. The column was washed with 2 volumes of lysis buffer, followed by wash buffer (20 mm Tris-HCl (pH 7.5), 300 mm NaCl, 0.1% Triton X-100, 0.1% Nonidet P-40, 1 mm EDTA, and 1 mm EGTA). The enzyme was eluted with 5 ml of elution buffer (20 mm Tris-HCl (pH 7.5), 800 mm NaCl, 0.1% Triton X-100, 1 mm EDTA, and 1 mm EGTA). Fractions (1.5 ml) were collected in test tubes and dialyzed in 20 mm Tris-HCl (pH 7.5), 0.1% Triton X-100, and 150 mm NaCl and then subjected to SDS-PAGE and assay of sphingomyelin hydrolyzing assay.
Protein Identification by Mass Spectrometry
N-terminal identification of the mature enzyme was performed using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS). The protein derived from Coomassie-stained polyacrylamide gels was digested using an in-gel digestion procedure with Staphylococcus aureus V8 protease (Wako Pure Chemical, Osaka, Japan) according to methods described previously (35). For TOF-MS, mass measurements were made after peak smoothing and internal calibration (Autoflex III, Bruker Daltonics, Bremen, Germany). The N terminus of the mature enzyme was identified based on the deduced amino acid sequence of the cDNA.
SMase Assay
For characterization of purified enzyme, sphingomyelin hydrolyzing activity was determined using a radiolabeled substrate in a mixed micelle assay system, as described previously (36). In the SMase assay, the reaction mixture contained an enzyme preparation that was pH-adjusted by the addition of the following buffers at a final concentration of 100 mm (reaction volume, 100 μl): sodium acetate (pH 4 and 5), PIPES (pH 6 and 7), Tris (pH 7.5, 8, 8.5, and 9), 5 nmol of [14C]sphingomyelin (100,000 dpm), 5 mm DTT, 0.1% Triton X-100, and 5 mm MgCl2. Typically, [14C]sphingomyelin and bovine brain sphingomyelin were placed in a glass tube and dried under nitrogen. A mixed micelle solution was prepared by sonicating the tube for 5 min in a bath sonicator and vortexing for 5 min at room temperature. The mixture was incubated for 30 min at 37 °C, and the enzymatic reactions were quenched by the addition of 0.2 ml of water and 1.5 ml of chloroform/methanol (2:1, by volume). After vortexing and two-phase separation by centrifugation, 0.2 ml of the upper aqueous phase was removed and added to 2 ml of scintillation solution for radioactivity counting. The reaction was linear with incubation times of up to 3 h. The amount of enzyme added to the reaction mixtures was chosen such that <10% of the substrate was hydrolyzed. An appropriate blank, containing denatured enzyme, was run with each reaction and subtracted from the experimental samples. To examine the effects of metal ions on the activity of the purified enzyme, aliquots of the enzyme were incubated in SMase buffer with or without EDTA in the presence or absence of magnesium.
The phosphatidylcholine hydrolyzing activity of the purified enzyme was measured similarly in a mixed micelle solution, with the [14C]sphingomyelin replaced by 10 nmol of [14C]phosphatidylcholine (l-3-phosphatidyl[N-methyl-14C]choline,1,2-dipalmitoyl) (100,000 dpm).
Hydrolyzing activity against lyso-platelet activating factor was determined similarly, as described previously (13). The enzyme was added to 100 μl of a mixed micelle system containing 10 nmol of [1-O-octadecyl-3H]lyso-platelet activating factor (200,000 dpm) in 100 mm Tris-HCl (pH 7.5), 5 mm DTT, and 5 mm MgCl2, and the mixture was incubated at 37 °C for 30 min. The lipids were extracted (36) and separated by TLC in a solvent system consisting of chloroform, methanol, 15 mm CaCl2(aq) (60:35:8, v/v). To determine the monoalkylglycerol content, we exposed the TLC plate to imaging film, and the radioactivity of the positive spots scraped from the TLC plate was established using liquid scintillation counting.
For the assay of neutral SMase activity in the cellular lysates, synthesized fluorescent substrate, i.e. C6-NBD-sphingomyelin, was used as a substrate. The cells were lysed by passing them through a 27-gauge needle in a lysis buffer (10 mm Tris-HCl (pH 7.5), 1 mm EDTA, 0.1% Triton X-100, and 1× protease inhibitor mixture). The lysate was centrifuged at 10,000 × g for 15 min at 4 °C. The supernatant was used as an enzyme source. Supernatant protein (60 μg) or the mitochondrial fraction (20 μg) was mixed in reaction buffer (100 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 5 mm DTT, 50 μm C6-NBD-sphingomyelin, and 0.1% Triton X-100) and incubated at 37 °C for 1 h. The reaction was quenched by the addition of 900 μl of water and 2 ml of chloroform/methanol (2:1, v/v), mixed well, and then centrifuged. The lower phase was collected, and the solvent was evaporated. Aliquots were applied to TLC plates. The solvent system used to separate C6-NBD-ceramide and C6-NBD-sphingomyelin was chloroform, methanol, 12 mm MgCl2 in water (65:25:4, v/v). C6-NBD-ceramide contents were measured with a fluorescent spectrophotometer (475 nm excitation, 525 nm emission).
Immunofluorescence and Confocal Microscopy
ZE cells were cultured on a coverslip and then fixed with 4% paraformaldehyde in PBS for 15 min. After rinsing in PBS, the cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. After treatment with PBS containing 3% FCS for 15 min, the fixed cells were incubated with anti-zebrafish mitochondrial SMase polyclonal antibody (1:1000) and anti-HSP60 monoclonal antibody (1:100) or anti-KDEL monoclonal antibody (1:250) in blocking buffer at room temperature for 1 h. The cells were washed three times with PBS for 10 min and incubated with goat anti-mouse IgG Alexa Fluor 488-labeled antibody (1:250) and goat anti-rabbit IgG Alexa Fluor 594-labeled antibody (1:250) for 30 min. Finally, cells were washed three times with PBS for 10 min, and the nuclei were counterstained with To-Pro3. Confocal images were obtained with a laser-scanning confocal microscope (LSM 510, Carl Zeiss, Wetzlar, Germany). For mitochondrial staining with MitoTracker Red, the cells were incubated with a mitochondrial fluorescent probe, MitoTracker Red CMXRos (Molecular Probes, Eugene, OR) at a concentration of 100 nm for 15 min to stain the mitochondria before fixation. The cells were stained with anti-mitochondrial SMase antibody and To-Pro3. Fluorescence signals were collected by single-line excitation at 648 nm (blue), 594 nm (red) and 488 nm (green). For immunocytochemical observation of the zebrafish SMase with FLAG tag in HEK293 cells, the transfected cells were incubated with 25 nm MitoTracker Red and fixed with 4% paraformaldehyde in PBS. After treatment with PBS containing 3% FCS for 15 min, the fixed cells were incubated with anti-FLAG monoclonal antibody (1:250) in blocking buffer at room temperature for 1 h. The cells were washed three times with PBS for 10 min and incubated with goat anti-mouse IgG Alexa Fluor 488-labeled antibody (1:250) for 30 min. Finally, cells were washed three times with PBS for 10 min, and then nuclei were counterstained with To-Pro3. Signals for zebrafish SMase with FLAG tag (green color image) and signals for mitochondrial marker (red color image) were observed with a fluorescence microscope (Edge Scientific Instruments, R400). Raw data images were cropped in Adobe Photoshop CS3 (Adobe Systems, San Jose, CA) for publication.
Preparation of the Mitochondrial Fraction
Adherent cells were scraped into a subcellular fractionation lysis buffer, 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 250 mm sucrose, and 1× protease inhibitor mixture, and then homogenized for 50 strokes using a Dounce homogenizer. The lysates were centrifuged at 1300 × g for 15 min to remove nuclei and unbroken cells. The supernatant was collected and centrifuged again at 5600 × g for 30 min to obtain the secondary pellet (the mitochondrial fraction). The supernatant fraction was centrifuged at 100,000 × g for 1 h at 4 °C, and the supernatant was used as the cytosolic fraction. The tertiary pellet was used as the microsomal fraction. After determining the protein concentration, the mitochondrial, cytosolic, and microsomal fractions were stored at −80 °C until use for Western blotting to measure ceramide and sphingomyelin content and to perform the SMase assay against C6-NBD-sphingomyelin.
The mitochondrion of ZE cells was isolated using a density gradient with sucrose via ultracentrifugation according to methods described previously (37). To separate the mitochondrion from the lysosomes and peroxisomes by density gradient, the mitochondrial fraction prepared according to the methods described above was washed with subcellular fractionation lysis buffer. The resulting pellet was suspended in 1 ml of 20 mm Tris-HCl (pH 7.4), containing 250 mm sucrose, 25 mm KCl, and 5 mm MgCl2. This fraction was layered above a continuous sucrose gradient (0.8–1.6 m sucrose containing 20 mm Tris-HCl (pH 7.4)) and centrifuged in the 28SA1 rotor of the HIMAC ultracentrifuge (Hitachi, Ibaragi, Japan) at 82,500 × g for 200 min at 4 °C. After ultracentrifugation, the contents of the tube were collected from the top in 1-ml aliquots using a Pasteur pipette. The aliquot fractions were stored at −80 °C until use.
Marker Enzymes Assay
Subcellular fractions were characterized by measuring organelle-specific marker enzyme activities. Cytochrome c oxidase activity (mitochondrial marker) was assayed with a cytochrome c reductase assay kit (Sigma) as described previously (38). A cytochrome c oxidase activity (endoplasmic reticulum marker) was measured with the cytochrome c oxidase assay kit (Sigma) according to the method reported previously (38). Acid phosphatase activity (lysosomal marker) was measured with 15 μg of protein for each fraction as described previously (39).
Protease Protection Assay
Freshly prepared mitochondrial fraction was incubated at 0 °C for 30 min in a reaction buffer (20 mm HEPES-NaOH (pH 7.4), 150 mm NaCl, and 250 mm sucrose with or without 1 mg/ml proteinase K in the presence or absence of 0.1% Triton X-100) according to methods described previously (40). The residual proteins were precipitated with trichloroacetic acid and used for Western blotting.
Western Blotting
The proteins extracted from whole cells and the mitochondrial and cytosolic fractions were separated by SDS-PAGE and electroblotted onto a polyvinylidine fluoride membrane according to Yabu et al. (41). Anti-mitochondrial SMase polyclonal, anti-neutral SMase 1 polyclonal (13), anti-cytochrome c polyclonal, anti-cadherin polyclonal, anti-KDEL monoclonal, anti-58,000 Golgi protein monoclonal, anti-cathepsin L polyclonal (42), anti-catalase monoclonal, anti-HSP60 monoclonal, and anti-FLAG monoclonal antibodies were used as the primary antibody. Following the addition of the secondary antibody, signals were detected using an ECLTM Western blotting detection kit according to the manufacturer's protocol.
Construction of Mitochondrial SMase Variants
SMase fusion constructs, alanine substitution mutants, and FLAG tag-containing mutants were created by PCR using zebrafish mitochondrial sphingomyelinase cDNA, p3 X FLAG-CMVTM-14 expression vector, pET-16b vector for the recombinant protein, or their derivatives as the templates, and appropriate combinations of the forward and reverse oligonucleotide primers. All constructs were confirmed by nucleotide sequencing.
Ceramide Measurement
Lipids in the whole cell or the mitochondrial fraction were extracted according to Bligh and Dyer (43), and ceramide contents were measured with E. coli diacylglycerol kinase according to methods described previously (5). The solvent system used to separate ceramide 1-phosphate was chloroform/acetone/methanol/acetic acid/water (10:4:3:2:1, v/v). Ceramide contents were measured using a STORM 860 analyzer system (GE Healthcare).
Sphingomyelin Measurement
Lipids in whole cells and the mitochondrial fraction were extracted according to Bligh and Dyer (43) and developed using a solvent system consisting of chloroform/methanol/water (60:35:8, v/v) on a plastic TLC plate with sphingomyelin as a standard. Spots corresponding to sphingomyelin were scraped, and the lipids were extracted according to Bligh and Dyer (43). Inorganic phosphate in the extract was measured using the ammonium molybdate/ascorbic acid method to determine sphingomyelin content (44).
Transfection of Mitochondrial SMase Gene into HEK293 Cells
HEK293 cells were cultured at a density of 5 × 105 cells per 60-mm dish in 4 ml of RPMI 1640 medium supplemented with 10% FCS. At ∼85% confluence, each dish of cells was transiently transfected with 4 μg of SMase fusion constructs, alanine substitution mutants, or FLAG tag-containing mutants using FuGENE 6 transfection reagent (Roche Applied Science), according to the manufacturer's protocol. At 48 h following transfection, the cells were washed twice in PBS and homogenized in the lysis buffer, and neutral SMase activity was measured.
Generation of Stable HEK293 Transfectants
HEK293 cells were cultured in RPMI 1640 medium containing 10% FCS, 5 units/ml penicillin, and 50 μg/ml streptomycin. To obtain stable transfectants, 5 × 106 HEK293 cells were transfected with 4 μg of SMase fusion constructs, alanine substitution mutants, or FLAG tag-containing mutant vectors using FuGENE 6 transfection reagent (Roche Applied Science), according to the manufacturer's protocol, and then the cells were selected in the presence of 0.8 mg/ml geneticin (Invitrogen). The overexpression of wild-type mitochondrial SMase and its alanine substitution mutant was established in three independent cell lines for each construct.
Gene Knockdown of Mitochondrial SMase
The following phosphorothioate oligonucleotides were synthesized to block the translation of mitochondrial SMase: antisense mitochondrial SMase, 5′-GAAAG GAGAC TCTCT TAAAG ACATA-3′; sense mitochondrial SMase, 5′-TATGT CTTTA AGAGA GTCTC CTTTC-3′. The cells were incubated with sense or antisense mitochondrial SMase oligonucleotide (0–20 μm) at a concentration of 5 × 105 cells/ml in Leibovitz's L-15 medium supplemented with 5% FCS for 48 h.
Statistical Analysis
The results are expressed as the mean ± S.D.. Differences among groups were analyzed using one-way analysis of variance followed by Bonferroni's post-hoc t test. Comparisons between two experimental groups were based on two-tailed t-tests. p < 0.01 was deemed statistically significant.
RESULTS
Cloning of Novel SMase
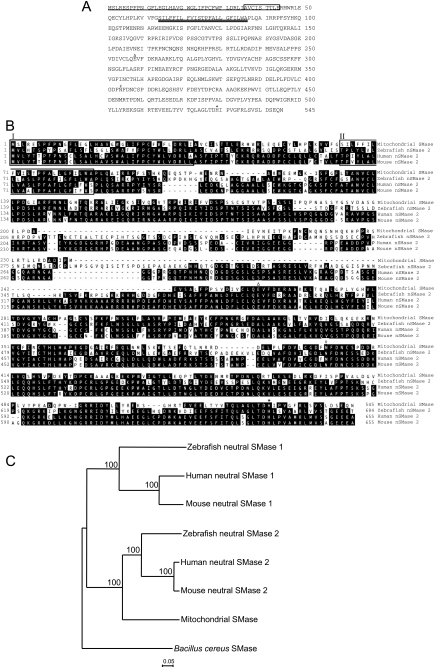
A zebrafish cDNA homologous to human neutral SMase 2 was isolated. This SMase cDNA had a 1635-bp open reading frame that encoded a protein of 545 amino acids with a predicted molecular weight of 61,300 (Fig. 1A). The deduced amino acid sequence of the zebrafish SMase showed 51% identity to that of the known human and mouse neutral SMase 2 (Fig. 1B). The three amino acid residues of the putative magnesium-binding site, the substrate-binding asparagine site, and the histidine residue of the active site on the zebrafish SMase (Gln-258, Asp-404, and His-529, respectively; see Refs. 43–47), which are critical for the enzymatic regulation of mammalian neutral SMase 2, were completely conserved (Fig. 1B). The mammalian neutral SMase 2 had a single collagenous domain between the membrane-anchoring domain and the catalytic domain (7), whereas the zebrafish SMase had no collagenous domain. Additionally, mouse neutral SMase 2 was palmitoylated on four Cys residues, i.e. Cys-53, Cys-54, Cys-395, and Cys-396, creating two Cys clusters that bound to the inner leaflet of the plasma membrane (15). The zebrafish SMase had no Cys cluster structure or palmitoylation, but the Cys-53 and Cys-294 residues were conserved. Based on a phylogenetic analysis, the zebrafish SMase was classified into a cluster with neutral SMase 2 (Fig. 1C).
FIGURE 1.
Amino acid sequence of zebrafish mitochondrial SMase and neutral SMase 2 and the phylogenetic relationship among vertebrate neutral SMases. A, deduced amino acid sequence of zebrafish mitochondrial SMase. Amino acid positions are shown on the right. The putative MLS identified by the SMART program is underlined. The N-terminal amino acid sequence from the purified mature enzyme, as determined by MALDI-TOF-MS, is boxed. The putative transmembrane domain identified by the SMART program is double underlined. The putative Mg2+-complexing glutamine residue (Δ), the asparagine residue involved in substrate binding (#), and the catalytic base histidine residue (*) are shown. B, amino acid sequence alignment for zebrafish mitochondrial SMase, zebrafish neutral SMase 2, human neutral SMase 2, and mouse neutral SMase 2. Proteins with significant amino acid sequence homology were identified using a FASTA search of the GenBankTM data base. The sequences of neutral SMases were aligned using the deduced amino acid sequences of homologous proteins from zebrafish mitochondrial SMase, zebrafish neutral SMase 2, human neutral SMase 2, and mouse neural SMase 2. The putative MLS is boxed (I). The putative transmembrane domain is also boxed (II). C, phylogenetic tree based on the amino acid sequences of the vertebrate neutral SMases. The phylogenetic analysis was performed using the neighbor-joining method in ClustalX. Numbers on the internal branches denote the bootstrap percentages of 1000 replicates. The scale indicates the evolutionary distance of one amino acid substitution per site. The amino acid sequences used in the analysis were obtained from the NCBI protein data base with the following accession numbers: B. cereus SMase (X12854); human neutral SMase 1 and SMase 2 (NM_009213 and AJ250460, respectively); mouse neutral SMase 1 and SMase 2 (NM_009213 and AJ250461, respectively); zebrafish neutral SMase 1 and SMase 2 (AB196165 and AB361067, respectively); and zebrafish mitochondrial SMase (AB361066).
The isolated SMase gene (mtSMase) was located on the zebrafish chromosome 16 (Table 1). Based on an analysis of gene synteny between the zebrafish and human genomes, an ortholog of the zebrafish mtSMase was found on chromosome 8 of the human genome. In contrast, the nSMase 2 was located on chromosome 25 of the zebrafish genome and was orthologous to the human SMPD3 located on chromosome 16. Therefore, the isolated zebrafish SMase is a novel SMase distinct from the mammalian neutral SMase 2 and zebrafish neutral SMase 2 (Table 1).
TABLE 1.
Zebrafish genes neighboring the mtSMase and the nSMase 2
Chr. indicates chromosome.
| Zebrafish genes in the chromosome containing the mtSMase and the nSMase 2 (localization) | Human homolog (chromosomal localization) |
|---|---|
| Chromosome 16 | |
| Myomesin family, member 3 | MYOM3, NM_152372 |
| (61.68 Mb) | (Chr. 1, 24.25 Mb) |
| LIM homeobox 3 | LHX3, NM_178138 |
| (62.69 Mb) | (Chr. 3, 138.22 Mb) |
| SAC1 suppressor of actin mutations 1-like | SACM1L, NM_014016 |
| (62.73 Mb) | (Chr. 3, 45.7 Mb) |
| Mitochondrial sphingomyelinase | ENSG00000204791 |
| (62.38 Mb) | (Chr. 8, 145.177 Mb) |
| N-Methyl-d-aspartate-associated protein 1 | GRINA, NM_001009184 |
| (62.85 Mb) | (Chr. 8, 145.136 Mb) |
| Phosphatidylserine synthase 1 | PTDSS1, NM_014754 |
| (63.07 Mb) | (Chr. 8, 97.343 Mb) |
| Chromosome 25 | |
| 1-O-Acylceramide synthase precursor | LYPLA3, NM_012320 |
| (16.85 Mb) | (Chr. 16, 66.83 Mb) |
| DNA-directed RNA polymerase II 33-kDa polypeptide | POLR2C, NM_032940 |
| (18.56 Mb) | (Chr. 16, 56.05 Mb) |
| Ubiquinone biosynthesis protein COQ9,mitochondrial precursor | COQ9, NM_020312 |
| (18.57 Mb) | (Chr. 16, 56.03 Mb) |
| Cytokine-induced apoptosis inhibitor 1 | CIAPIN1, NM_020313 |
| (18.59 Mb) | (Chr. 16, 56.02 Mb) |
| Protein arginine N-methyltransferase 7 | PRMT7, NM_019023 |
| (18.60 Mb) | (Chr. 16, 66.90 Mb) |
| Neutral sphingomyelinase 2 | SMPD3, NM_018667 |
| (19.61 Mb) | (Chr. 16, 66.94 Mb) |
| Olfactomedin 4 | OLFM4, NM_006418 |
| (19.73 Mb) | (Chr. 13, 52.5 Mb) |
| NDRG family member 4 | NDRG4, NM_022910 |
| (19.99 Mb) | (Chr. 16, 57.05 Mb) |
| Mps one binder kinase activator-like 2 | MOB2, VSP_012298 |
| (20.04 Mb) | (Chr. 11, 1.44 Mb) |
| Potassium inwardly rectifying channel,subfamily J, member 11 | KCNJ11, NM_000525 |
| (20.07 Mb) | (Chr. 11, 17.36 Mb) |
| ATP-binding cassette, subfamily C (CFTR/MRP), member 8 | ABCC8, NM_000352 |
| (20.09 Mb) | (Chr. 11, 17.37 Mb) |
| Usher syndrome 1C | USH1C, NM_005709 |
| (20.23 Mb) | (Chr. 11, 17.47 Mb) |
We also isolated another cDNA clone that encoded neutral zebrafish SMase 2 and possessed significant homology with the human neutral SMase 2, SMPD3 (7). The zebrafish neutral SMase 2 cDNA contained a predicted open reading frame encoding a 684-amino acid protein (predicted molecular weight, 76,000; Fig. 1B). The deduced amino acid sequence showed 55% identity to human and mouse neutral SMase 2, belonged to the neutral SMase 2 cluster in the phylogenetic tree (Fig. 1C), and showed partial conservation of the putative palmitoylation sites (Cys-53, Cys-421, and Cys-422).
When the secondary structure of SMase was predicted using the SMART program (48), a single signal peptide sequence and a hydrophobic transmembrane domain were identified in the N-terminal region (Fig. 1, A and B), suggesting that the SMase protein was membrane-bound. Based on the presence of a long signal peptide sequence in the first 35 amino acid residues and similarity to the mitochondrial targeting sequence, the SMase appeared to be a mitochondrial protein.
Characterization of the Purified Novel SMase from ZE Cells
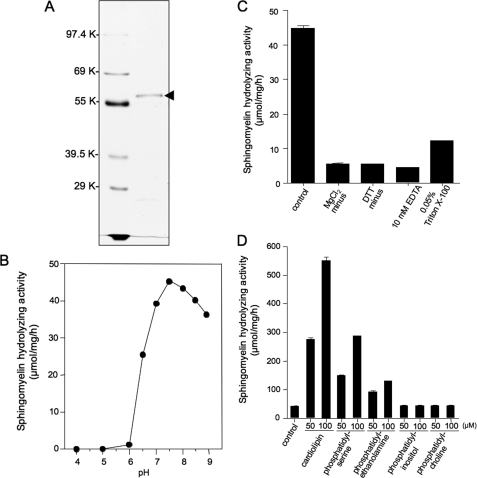
To determine the enzymatic activity and substrate specificity of the SMase, the active enzyme was purified from ZE cells using antibody-affinity chromatography. SDS-PAGE of the purified enzyme revealed a single band with an estimated molecular weight of 57,000 (Fig. 2A). The protein of excised gel sections was reduced and S-carboxymethylated and then digested with Staphylococcus V8 protease. MALDI-TOF-MS analysis of the mixture of peptides detected an ion that corresponds to the amino acid sequence AVCISTTLE (M + H) m/z 995.4 (observed), 995.14 (calculated), containing the S-carboxymethylated Cys residue. Thus, the Ala-36 residue was identified as the N-terminal amino acid of the mature enzyme (Fig. 1A). The protein showed high activity toward the substrate [14C]sphingomyelin, although it showed no activity against the phospholipid [14C]phosphatidylcholine and [1-O-octadecyl-3H]lyso-platelet activating factor (Table 2). The sphingomyelin hydrolyzing activity was optimal at pH 7.5, with half-maximal activity at pH 6.5 (Fig. 2B); the activity was absolutely dependent upon the presence of magnesium ions (Fig. 2C). Finally, several kinds of lipids derived from the mitochondrial and other membranes were tested for modulation of the activity of the purified SMase. The activity against sphingomyelin was induced 12-fold in the presence of 100 μm cardiolipin, 6-fold in the presence of 100 μm phosphatidylserine, and 3-fold in the presence of 100 μm phosphatidylethanolamine, although the activity was not influenced by phosphatidylcholine or phosphatidylinositol (Fig. 2D).
FIGURE 2.
Purification and characterization of the mitochondrial SMase from a zebrafish embryonic cell line. A, PAGE of purified enzyme. The arrowhead indicates the molecular mass of a 57-kDa protein. SDS-PAGE (10% gel) was performed after reduction of the sample. The gel was stained with Coomassie Brilliant Blue R-250. B, pH dependence of neutral SMase activity. The sphingomyelin hydrolyzing activity of the purified enzyme was measured at 37 °C for 30 min at various pH values, with an estimated optimum pH of 7.5. The pH was adjusted by the addition of the following buffers at a final concentration of 100 mm: acetate (pH 4 and 5), PIPES (pH 6, 6.5, and 7), and Tris (pH 7.5, 8, 8.5, and 9). C, effect of Mg2+ ions on neutral sphingomyelin hydrolyzing activity. The basal assay mixture contained 100 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 0.1% Triton X-100, and 5 mm DTT. Effects of Mg2+ ion on the activity were measured in the presence of 10 mm EDTA. D, effect of lipids on mitochondrial SMase activity. The sphingomyelin hydrolyzing activity of the purified enzyme was determined under standard conditions in the presence of [14C]sphingomyelin under standard conditions and the indicated phospholipids (50 or 100 μm). The basal enzyme activity (control) was 45 ± 3 μmol/mg/h.
TABLE 2.
Substrate specificity of the cloned purified enzyme
| Substrate | Activitya |
|---|---|
| μmol/mg/h | |
| [14C]Sphingomyelin | 45 ± 2.76b |
| [14C]Phosphatidylcholine | 0.003 ± 0.0003b |
| [3H]Lyso-PAF | 0.024 ± 0.0012c |
a Values are means ± S.D. (n = 3).
b Activity was determined in the presence of 0.1% Triton X-100.
c Activity was determined in the absence of Triton X-100.
Subcellular Localization of Novel SMase
The subcellular localization of the isolated SMase in ZE cells was examined by centrifugal fractionation (Fig. 3, A and B). According to Western blotting, the SMase was mainly detected in the mitochondrial fraction (Fig. 3A, lane 2) when using HSP60 as a mitochondrion marker, aldolase as a cytosol marker, and cadherin and neutral SMase 1 (13) as cell membrane markers, although SMase was not detected in the cytosolic (Fig. 3A, lane 3) or microsomal fractions (Fig. 3A, lane 4).
FIGURE 3.
Subcellular localization and distribution of SMase. A, whole lysate of ZE cells (lane 1) was fractionated into the mitochondrial fraction (lane 2), cytosolic fraction (lane 3), and microsomal fraction (lane 4) via ultracentrifugation. These fractions were analyzed by Western blotting using antibodies against zebrafish mitochondrial SMase, HSP60 (a mitochondrial marker), aldolase (a cytosolic marker), and neutral SMase 1 and cadherin (cell membrane markers). B, The mitochondrial fraction was separated into eight fractions using the sucrose gradient ultracentrifugation. The separated proteins in each fraction were subjected to Western blotting using antibodies against zebrafish mitochondrial SMase, HSP60, and cytochrome c (a mitochondrial marker), KDEL protein (an endoplasmic reticulum marker), 58-kDa protein (a Golgi marker), cathepsin L (a lysosomal marker), and catalase (peroxisomal marker), and neutral SMase activities in each fraction were determined. The lysosomal marker, endoplasmic reticulum marker, and Golgi marker, mitochondrial marker, or peroxisomal marker were detected in the fraction numbers 1 and 2, fraction numbers 4–6, and fraction numbers 7 and 8, respectively. C, specific activities of marker enzymes in subcellular fractionation of ZE cells. The C6-NBD-sphingomyelin hydrolyzing activities of the SMase, mitochondrial cytochrome c oxidase, endoplasmic reticulum cytochrome c reductase, and lysosomal acid phosphatase were measured in each fraction by subcellular fractionation. Values and bars indicate the mean ± S.D. of three independent experiments. D, ZE cells were fixed and permeabilized with 0.1% Triton X-100. The cells were incubated with 100 nm MitoTracker Red and then fixed with 4% paraformaldehyde in PBS. The cells were co-stained with anti-zebrafish SMase antibody and an antibody against either HSP60 protein (a mitochondria marker) or KDEL protein (an endoplasmic reticulum marker) and stained with fluorescent secondary antibodies. Signals for SMase (green color image) and signals for subcellular markers such as mitochondria and endoplasmic reticulum (red color image) were observed. The overlay images indicate the SMase, and the subcellular markers were co-localized either in the sample place or adjacent to one another as described under “Experimental Procedures.” Scale bar, 10 μm. E, distribution of SMase in the mitochondrion. Mitochondrial fractions were obtained from a zebrafish embryonic cell line and incubated at 0 °C for 30 min in the absence (lane 1) or presence (lanes 2–4) of proteinase K, and under swelling condition (20 mm HEPES-NaOH (pH 7.4), 10 mm sodium orthovanadate, 20 mm NaF, 250 mm sucrose, 2 mm CaCl2) (lanes 3 and 4) or in the presence (lane 4) of Triton X-100. The samples were subjected to Western blotting with anti-mitochondrial SMase, anti-cytochrome c, and anti-HSP60 antibodies.
To confirm the subcellular localization of the SMase in the mitochondrion, the cells were fractionated using sucrose density gradient ultracentrifugation (37). The separated proteins in each fraction in the sucrose density gradient were applied to Western blotting, and the neutral SMase was detected with specific polyclonal antibody. When we used HSP60 and cytochrome c as mitochondrial markers, KDEL protein as an endoplasmic reticulum marker, 58K protein as a Golgi marker, cathepsin L as a lysosomal marker, and catalase as a peroxisomal marker, the neutral SMase was mainly detected in the fractions 4–6 rich in the mitochondrion (Fig. 3B) but not in the lysosomal fractions (Fig. 3B, fraction numbers 1 and 2), endoplasmic reticulum fractions (Fig. 3B, fraction numbers 1 and 2), Golgi fractions (Fig. 3B, fraction numbers 1 and 2), and peroxisomal fractions (Fig. 3B, fraction numbers 7 and 8). In addition, higher neutral SMase activity was also detected in the fraction numbers 4–6 rich in the mitochondrial fractions containing cytochrome c oxidase activity, but not in the endoplasmic reticulum showing cytochrome c reductase activity and the lysosomal fractions showing acid phosphatase activity (Fig. 3C).
The ZE cells were co-stained with the anti-SMase antibody together with the specific mitochondrial probe MitoTracker Red, anti-HSP60 antibody (mitochondrial marker), or anti-KDEL antibody (endoplasmic reticulum marker) by confocal microscopy. Immunocytochemical localization of the SMase with that of MitoTracker Red and that of HSP60 showed that the SMase exhibited major signals in the mitochondria (Fig. 3D). Consequently, staining with the antibody against endoplasmic reticulum marker, i.e. KDEL protein, the SMase was not localized in the endoplasmic reticulum (Fig. 3D). Therefore, the SMase was located in the mitochondria.
To establish the topology of enzyme localization, we performed a protease protection assay. Mitochondrial fractions prepared from ZE cells were treated with proteinase K to digest peripheral proteins and then subjected to Western blotting with antibodies against mitochondrial SMase and cytochrome c, as an intermembrane space marker, or against HSP60, as a matrix marker (40). The expressed proteins, as well as the endogenous SMase, were recovered from mitochondria (Fig. 3E, lane 1) but not from the post-mitochondrial supernatant (data not shown). The endogenous SMase recovered from mitochondria was unaffected by the presence of proteinase K (Fig. 3E, lane 2). Upon disruption of the outer membrane, the endogenous SMase was completely degraded by proteinase K, whereas HSP60 protein remained undigested (Fig. 3E, lane 3). As a control, all proteins were digested by proteinase K when mitochondrial membranes were solubilized in Triton X-100 (Fig. 3E, lane 4). These results indicate that this SMase is bound to both the intermembrane space and/or the inner membrane in the mitochondrion.
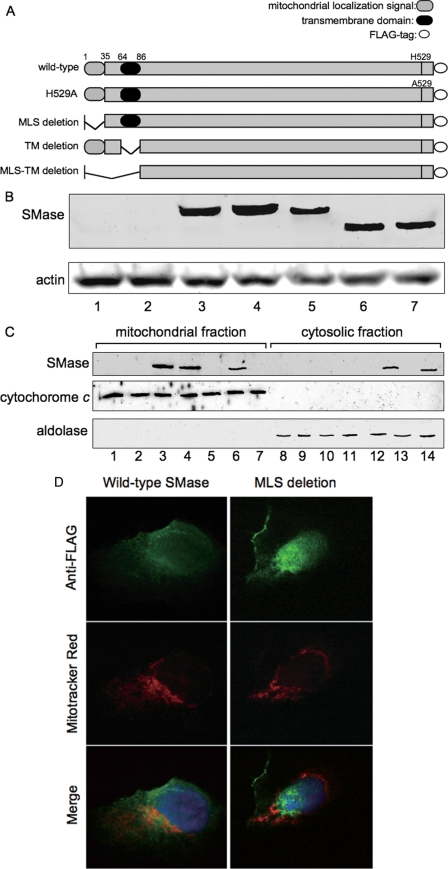
Identification of the MLS with the First 35 Residues
Based on the structural features of zebrafish SMase, a putative MLS was identified within the first 35 residues of the N terminus; a putative hydrophobic transmembrane domain was predicted within Ser-64 to Ala-86, and a putative catalytic center site was identified at His-529 (Fig. 1A). To confirm the presence of these features, we constructed various SMase-FLAG fusion proteins as follows: a histidine to alanine substitution at the catalytic site (H529A mutant); a deletion mutant lacking the N-terminal MLS (MLS deletion mutant); a deletion mutant lacking the transmembrane domain (TM deletion mutant); and a deletion mutant lacking both the MLS and the transmembrane domain (MLS-TM deletion mutant, Fig. 4A). These constructs were transiently expressed in HEK293 cells, and the FLAG tag of the expressed proteins was detected via Western blotting (Fig. 4B). We examined the intracellular distribution of these mutants and the wild-type protein in HEK293 cells via subcellular fractionation. When expressed in HEK293 cells, the wild type, H529A mutant, and TM mutant proteins localized correctly to the mitochondrial fraction (Fig. 4C and Table 3), whereas the MLS deletion and the MLS-TM deletion mutants were not detected in the mitochondrial fraction but were instead found in the cytosolic fraction. To further clarify the protein localization, HEK293 cells transfected with the wild type and the MLS deletion were stained with both anti-FLAG antibody and MitoTracker Red. The SMase with FLAG tag was co-localized with the mitochondrial probe, whereas the MLS deletion mutant was not co-localized with the mitochondrial probe (Fig. 4D). These findings indicate that first 35 residues of the N-terminal sequence represent an MLS essential for localization in mitochondria. These findings indicate that first 35 residues of the N-terminal sequence represent an MLS essential for localization in mitochondria.
FIGURE 4.
Mitochondrial localization of SMase variants. A, schematic representation of SMase constructs. B, HEK293 cells were transiently transfected with their constructs alone. Lane 1, control; lane 2, mock; lane 3, wild type; lane 4, H529A mutant; lane 5, MLS deletion mutant; lane 6, TM deletion mutant; lane 7, MLS-TM deletion mutant. At 48 h after transfection, the expressed proteins were detected using anti-FLAG antibody or anti-actin via Western blotting as described under “Experimental Procedures.” C, HEK293 cells expressing the indicated constructs were fractionated into mitochondrial (lanes 1–7) and cytosolic (lanes 8–14) fractions, and each fraction was analyzed by Western blotting using antibodies against FLAG, SMase, cytochrome c, and aldolase. Lanes 1 and 8, control; lanes 2 and 9, mock; lanes 3 and 10, wild type; lanes 4 and 11, H529A mutant; lanes 5 and 12, MLS deletion mutant; lanes 6 and 13, TM deletion mutant; lanes 7 and 14, MLS-TM deletion mutant. D, mitochondrial localization of the wild-type construct. The HEK293 cells transfected with wild type of SMase and MLS deletion mutant were incubated with 25 nm MitoTracker Red and fixed with 4% paraformaldehyde in PBS. The cells were co-stained with anti-FLAG antibody. Signals for zebrafish SMase with FLAG tag (green color image) and signals for mitochondrial marker (red color image) and the overlay images (merge) were observed as described under “Experimental Procedures.”
TABLE 3.
Subcellular distribution of various SMase constructs in HEK293-transfected cells
Various SMase constructs (Fig. 4) were transiently expressed in HEK293 cells.
| Construct | Distribution |
|---|---|
| Wild type | Mitochondrial fraction |
| H529A mutant | Mitochondrial fraction |
| MLS deletion mutant | Cytosolic fraction |
| TM deletion mutant | Mitochondrial fraction |
| MLS-TM deletion mutant | Cytosolic fraction |
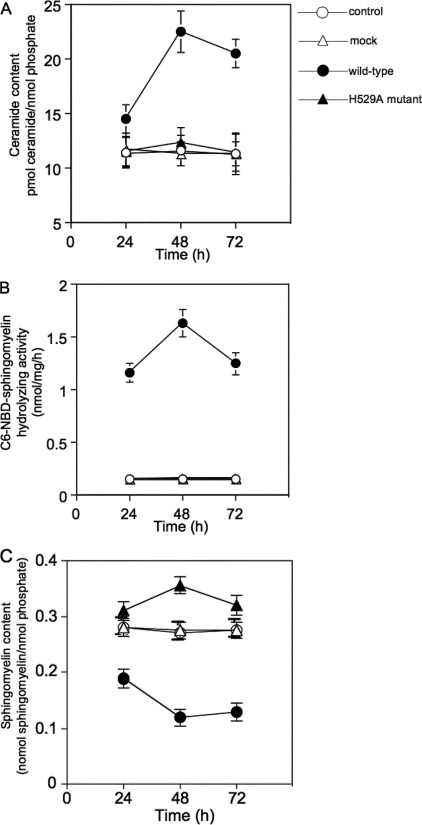
Ceramide and Sphingomyelin Levels and Neutral SMase Activity in Transient Transfectants
We used HEK293 cells transiently transfected with the wild-type or H529A mutant constructs to examine whether SMase is involved in ceramide generation. When the intercellular ceramide content was measured using a diacylglycerol kinase assay, the wild-type transfectants showed significantly higher ceramide levels than cells transfected with the H529A mutant or mock vector; in H529A mutants, peak ceramide levels (2-fold) occurred 48 h after transfection (Fig. 5A). At 24, 48, and 72 h post-transfection, cells harboring wild-type SMase showed higher Mg2+-dependent neutral SMase activity than cells harboring the H529A mutant or mock vector (Fig. 5B). In wild-type transfectants, a decrease in cellular sphingomyelin levels was accompanied by an increase in cellular ceramide levels at 48 h post-transfection (Fig. 5C). These changes in ceramide and sphingomyelin levels in association with neutral SMase activity in SMase-overexpressing cells indicate a significant role for this enzyme in ceramide metabolism.
FIGURE 5.
Ceramide and sphingomyelin levels in association with neutral SMase activity in SMase transfectants. Cellular lipids were extracted at the indicated times after transfection. The levels of ceramide (A) and sphingomyelin (C) were quantified using the diacylglycerol kinase assay and phosphate measurement after TLC separation, respectively. The cells were lysed, and C6-NBD-sphingomyelin hydrolyzing activity (B) was determined as described under “Experimental Procedures.” Values and bars indicate the mean ± S.D. of three independent experiments.
Ceramide and Sphingomyelin Levels and Neutral SMase Activity in Mitochondria
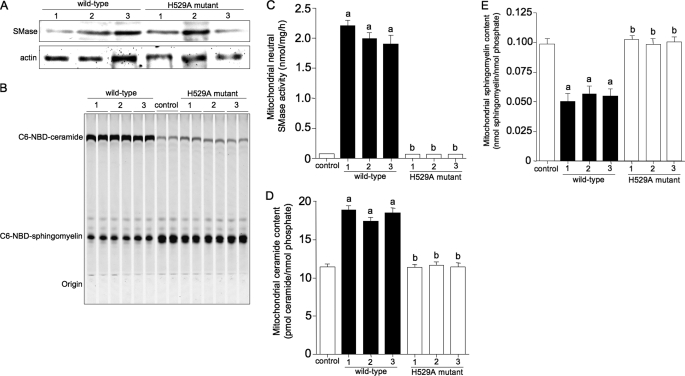
To examine SMase-mediated ceramide metabolism in mitochondria, we established three wild-type and three H529A mutant cell lines and confirmed that the transfected HEK293 cells overexpressed the transgene products (Fig. 6A). When Mg2+-dependent neutral SMase activity was measured in the mitochondrial fraction using C6–7-NBD-4-yl-sphingomyelin as a substrate, neutral SMase activity was 25 times higher in the wild-type transfectants than in the H529A mutants (Fig. 6, B and C). In addition, the mitochondrial ceramide content increased by 1.5–1.8 times in the wild-type transfectants compared with the H529A mutants (Fig. 6D). This increase in mitochondrial ceramide was accompanied by a decrease in mitochondrial sphingomyelin (Fig. 6E). These changes in mitochondrial ceramide and sphingomyelin levels in association with increased enzyme activity indicate a significant role for SMase in sphingomyelin metabolism.
FIGURE 6.
Increase in neutral SMase activity with increased ceramide and decreased sphingomyelin levels in the mitochondrial fraction of SMase transfectants. HEK293 cells were stably transfected with the wild-type or H529A mutant construct. Three wild-type and three H529A mutant lines were established as described under “Experimental Procedures.” The expressed proteins in each group of three lines (A) were detected via Western blotting using anti-FLAG antibody as described under “Experimental Procedures.” B, neutral SMase activity against C6-NBD-sphingomyelin in the mitochondrial fraction was demonstrated via TLC separation and visualized by UV irradiation at 254 nm. C, increase in neutral SMase activity in wild-type cell lines. The cells were lysed, and C6-NBD-sphingomyelin hydrolyzing activity was determined as described under “Experimental Procedures.” D, increasing ceramide content in the mitochondrial fraction isolated from three wild-type lines. E, decreasing sphingomyelin content in the mitochondrial fraction isolated from three wild-type lines. Cellular lipids in the mitochondrial fraction were extracted, and the levels of ceramide and sphingomyelin were quantified using the diacylglycerol kinase assay and phosphate measurement, respectively. Values and bars indicate the mean ± S.D. of three independent experiments. Different letters denote a statistical difference between wild-type and H529A mutant cells (p < 0.01).
Effects of SMase Knockdown on ZE Cells
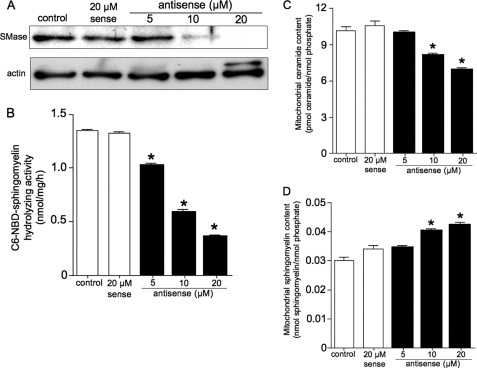
To confirm whether SMase regulates ceramide generation, a phosphorothioate oligonucleotide was used to repress mitochondrial SMase levels. Anti-mitochondrial SMase antibodies detected a 57-kDa protein band (Fig. 7A). At 48 h after antisense oligonucleotide treatment, both the protein band and the activity level of Mg2+-dependent neutral SMase in the mitochondrial fraction decreased in a dose-dependent manner (Fig. 7B). Treatment with 20 μm of the antisense oligonucleotide reduced the basal activity of Mg2+-dependent neutral SMase from 1.32 to 0.34 nmol/mg/h (Fig. 7B). The antisense oligonucleotide treatment repressed basal endogenous ceramide content in mitochondrial fractions (Fig. 7C), although it induced an increase in the sphingomyelin content (Fig. 7D). In contrast, treatment with control sense oligonucleotides had no effect. Thus, antisense oligonucleotide-induced enzyme deficiency affected ceramide metabolism.
FIGURE 7.
Antisense oligonucleotides against mitochondrial SMase protein inhibit ceramide generation. ZE cells were pretreated with 0–20 μm antisense or 20 μm sense oligonucleotides against mitochondrial SMase for 48 h. A, expressed proteins in oligonucleotide-treated cells were detected with anti-mitochondrial SMase or anti-actin antibodies by Western blotting, as described under “Experimental Procedures.” B, C6-NBD-sphingomyelin hydrolyzing activity; C, ceramide content; and D, sphingomyelin content in the mitochondrial fractions. Values and bars indicate the mean ± S.D. of three independent experiments. *, p < 0.01 versus sense oligonucleotide-treated cells.
DISCUSSION
A novel mitochondrial SMase was cloned, and the active endogenous enzyme was purified from ZE cells. The purified enzyme showed specific activity against [14C]sphingomyelin in the presence of 10 mm Mg2+ at pH 7.5. Cytoimmunostaining revealed that the enzyme was localized to the mitochondrion, and subcellular fractionation indicated that the enzyme was distributed in the mitochondrial fraction. A protease protection assay revealed that the enzyme was distributed in the intermembrane space and/or the inner membrane of the mitochondrion. The overexpression of wild-type and mutant proteins showed that an MLS was required for translocation of the enzyme to the mitochondrion. Mutants lacking this MLS were instead directed to the cytoplasm. Cytostaining for zebrafish SMase in ZE cells indicated that the enzyme was co-localized with MitoTracker Red and HSP60 in the mitochondrion. The transient and stable overexpression of mitochondrial SMase cDNA in HEK293 cells resulted in enhanced neutral SMase activity and increased ceramide levels paralleled by reduced sphingomyelin levels. Antisense oligonucleotide-induced SMase deficiency confirmed that SMase was required for mitochondrial ceramide generation. Therefore, the identified zebrafish SMase is a novel mitochondrial SMase that regulates mitochondrial ceramide production.
The activity and substrate specificity of this novel enzyme are similar to those of the known mammalian neutral SMase 2s (7, 16). The purified zebrafish SMase showed specific hydrolyzing activity against sphingomyelin, producing ceramide, although it showed no activity against phosphatidylcholine. In addition, the enzyme regulated the metabolism of sphingomyelin and ceramide in cells. When SMase was overexpressed in cultured cells, we observed decreases in sphingomyelin and corresponding increases in ceramide levels in the mitochondrial fraction. Thus, the zebrafish enzyme is a typical Mg2+-dependent neutral SMase that accelerates the catabolism of sphingomyelin in the mitochondrion.
The previously purified bovine brain neutral SMase, the partially purified rat brain neutral SMase, yeast Isc1p, and mouse neutral SMase 2 showed dependence upon anionic phospholipids such as phosphatidylserine for in vitro activity (14, 49–51). Interestingly, site-directed mutagenesis in Isc1p indicated the importance of positively charged C-terminal amino acid residues in protein-anionic phospholipid interaction (52). The zebrafish SMase described here was also activated by cardiolipin and phosphatidylserine, similar to mammalian SMase 2 (7). Cardiolipin and phosphatidylserine are highly enriched in the outer and inner membranes of the mitochondrion. Therefore, cardiolipin and phosphatidylserine may be a candidate for the activator of mitochondrial SMase.
Molecular phylogenetic analysis based on amino acid sequences showed that the zebrafish mitochondrial SMase falls into a cluster consisting of human, mouse, and zebrafish neutral SMase 2s. Both zebrafish mitochondrial SMase and SMase 2 share common structural features, such as a magnesium-binding site, a substrate-binding site, and an active center His residue (Fig. 1B). We found that the mitochondrial SMase also possesses an MLS and a transmembrane domain as a stop-transfer sequence in the N-terminal region. This MLS was also conserved in other SMase 2 enzymes, suggesting that all known SMase 2s and zebrafish mitochondrial SMase may be mitochondrial enzymes.
In eukaryotes, the MLS has been identified in the majority of pre-sequences cleaved by mitochondrial processing peptidases. This signal sequence is characterized by an overall positive charge, a predicted ability to form an amphiphilic α-helix, and the presence of an Arg residue at the −2 or −3 position from the cleavage site (53). Four cleavage site motifs have been identified (54) as follows: XRX↓X(S/X) (R-2 motif); XRXl(Y/X)↓(S/A/X)X (R-3 motif); XRX↓(F/L/I)XX(S/T/G)XXXX (R-10 motif); and XX↓X(S/X) (R-none motif). The identified zebrafish mitochondrial SMase was consistent with the R-3 motif (i.e. DRLI↓AV).
Hoffman et al. (7) first identified that mammalian neutral SMase 2 was localized in the Golgi in a number of cell lines by immunofluorescent staining. Tani and Hannun (15) reported that mouse neutral SMase 2 was palmitoylated at four Cys residues (Cys-53, Cys-54, Cys-395, and Cys-396) that were localized to the inner leaflet of the cell membrane. Zebrafish neutral SMase 2 also showed three conserved Cys residues (Cys-53, Cys-421, and Cys-422), but zebrafish mitochondrial SMase showed no palmitoylated Cys cluster. This work suggests that in contrast to zebrafish mitochondrial SMase, neutral SMase 2 might be localized to the cell membrane as a result of palmitoylation. Further studies of the intracellular localization of SMase 2 are required to examine whether SMase 2 and mitochondrial SMase are co-localized in the mitochondrion.
Based on an analysis of gene synteny between zebrafish and human genomes, mtSMase is located on zebrafish chromosome 16. The ortholog of mtSMase is found on chromosome 8 of the human genome and is expressed in human breast tumors (55). In contrast, the nSMase 2 is located on zebrafish chromosome 25 and was orthologous to the neutral SMase 2 gene SMPD3 located on human chromosome 16. Therefore, in terms of gene synteny, the isolated zebrafish mitochondrial SMase is distinct from other neutral SMase 2s.
At least five SMases, i.e. acidic SMase, neutral SMase 1, SMase 2, SMase 3, and mitochondrial SMase, have been characterized in mammalian and other animal cells, and all of these enzymes were expressed in ZE cells.3 Thus, ZE cells represent an important model for characterizing the precise roles of mitochondrial and other types of SMases, as well as the mechanism of ceramide signaling. Our results indicate that mitochondrial SMase has a specific function in mitochondrial ceramide generation.
Previous studies have demonstrated that ceramidase (34), ceramide synthase (33), dihydroceramide synthase (56), glycosyltransferase (57), yeast inositol sphingolipid phospholipase C (58), and sphingosine kinase (59) are localized to the mitochondrion. Therefore, a set of enzymes, including mitochondrial SMase and other ceramide-related enzymes, may regulate sphingolipid metabolism in the mitochondrion and regulate cell growth and apoptosis in response to mitochondrial functions.
This work was supported in part by grants from the Japan Science and Technology Corporation, the Japan Society for Promotion of Science, and the Ministry of Agriculture, Forestry, and Fisheries of Japan.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) AB361066 and AB361067.
T. Yabu and M. Yamashita, unpublished data.
- SMase
- sphingomyelinase
- [14C]sphingomyelin
- [N-methyl-14C]sphingomyelin
- C6-NBD-ceramide
- C6–7-nitro-2–1,3-benzoxadiazol-4-yl-ceramide
- C6-NBD-sphingomyelin
- 6–7-nitro-2–1,3-benzoxadiazol-4-yl-sphingomyelin
- DTT
- dithiothreitol
- FCS
- fetal calf serum
- HEK293 cells
- human embryonic kidney 293 cells
- MLS
- mitochondrial localization signal
- TM
- transmembrane domain
- ZE cells
- zebrafish embryonic cells
- MALDI-TOF-MS
- matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- PBS
- phosphate-buffered saline
- PIPES
- 1,4-piperazinediethanesulfonic acid
- TM
- transmembrane domain
- RT
- reverse transcription
- EST
- expressed sequence tag.
REFERENCES
- 1.Hannun Y. A., Luberto C. (2000) Trends Cell Biol. 10, 73–80 [DOI] [PubMed] [Google Scholar]
- 2.Levade T., Malagarie-Cazenave S., Gouazé V., Ségui B., Tardy C., Betito S., Andrieu-Abadie N., Cuvillier O. (2002) Neurochem. Res. 27, 601–607 [DOI] [PubMed] [Google Scholar]
- 3.Marchesini N., Hannun Y. A. (2004) Biochem. Cell Biol. 82, 27–44 [DOI] [PubMed] [Google Scholar]
- 4.Obeid L. M., Linardic C. M., Karolak L. A., Hannun Y. A. (1993) Science 259, 1769–1771 [DOI] [PubMed] [Google Scholar]
- 5.Okazaki T., Bell R. M., Hannun Y. A. (1989) J. Biol. Chem. 264, 19076–19080 [PubMed] [Google Scholar]
- 6.Clarke C. J., Snook C. F., Tani M., Matmati N., Marchesini N., Hannun Y. A. (2006) Biochemistry 45, 11247–11256 [DOI] [PubMed] [Google Scholar]
- 7.Hofmann K., Tomiuk S., Wolff G., Stoffel W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5895–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krut O., Wiegmann K., Kashkar H., Yazdanpanah B., Krönke M. (2006) J. Biol. Chem. 281, 13784–13793 [DOI] [PubMed] [Google Scholar]
- 9.Tomiuk S., Hofmann K., Nix M., Zumbansen M., Stoffel W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3638–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawai H., Domae N., Nagan N., Hannun Y. A. (1999) J. Biol. Chem. 274, 38131–38139 [DOI] [PubMed] [Google Scholar]
- 11.Zumbansen M., Stoffel W. (2002) Mol. Cell. Biol. 22, 3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yabu T., Tomimoto H., Taguchi Y., Yamaoka S., Igarashi Y., Okazaki T. (2005) Blood 106, 125–134 [DOI] [PubMed] [Google Scholar]
- 13.Yabu T., Imamura S., Yamashita M., Okazaki T. (2008) J. Biol. Chem. 283, 29971–29982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchesini N., Luberto C., Hannun Y. A. (2003) J. Biol. Chem. 278, 13775–13783 [DOI] [PubMed] [Google Scholar]
- 15.Tani M., Hannun Y. A. (2007) J. Biol. Chem. 282, 10047–10056 [DOI] [PubMed] [Google Scholar]
- 16.Marchesini N., Osta W., Bielawski J., Luberto C., Obeid L. M., Hannun Y. A. (2004) J. Biol. Chem. 279, 25101–25111 [DOI] [PubMed] [Google Scholar]
- 17.Hayashi Y., Kiyono T., Fujita M., Ishibashi M. (1997) J. Biol. Chem. 272, 18082–18086 [DOI] [PubMed] [Google Scholar]
- 18.Clarke C. J., Truong T. G., Hannun Y. A. (2007) J. Biol. Chem. 282, 1384–1396 [DOI] [PubMed] [Google Scholar]
- 19.Karakashian A. A., Giltiay N. V., Smith G. M., Nikolova-Karakashian M. N. (2004) FASEB J. 18, 968–970 [DOI] [PubMed] [Google Scholar]
- 20.Levy M., Castillo S. S., Goldkorn T. (2006) Biochem. Biophys. Res. Commun. 344, 900–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jana A., Pahan K. (2004) J. Biol. Chem. 279, 51451–51459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoffel W., Jenke B., Blöck B., Zumbansen M., Koebke J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4554–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aubin I., Adams C. P., Opsahl S., Septier D., Bishop C. E., Auge N., Salvayre R., Negre-Salvayre A., Goldberg M., Guénet J. L., Poirier C. (2005) Nat. Genet. 37, 803–805 [DOI] [PubMed] [Google Scholar]
- 24.Tepper A. D., de Vries E., van Blitterswijk W. J., Borst J. (1999) J. Clin. Invest. 103, 971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ardail D., Popa I., Alcantara K., Pons A., Zanetta J. P., Louisot P., Thomas L., Portoukalian J. (2001) FEBS Lett. 488, 160–164 [DOI] [PubMed] [Google Scholar]
- 26.Colbeau A., Nachbaur J., Vignais P. M. (1971) Biochim. Biophys. Acta 249, 462–492 [DOI] [PubMed] [Google Scholar]
- 27.Gudz T. I., Tserng K. Y., Hoppel C. L. (1997) J. Biol. Chem. 272, 24154–24158 [DOI] [PubMed] [Google Scholar]
- 28.Birbes H., El Bawab S., Hannun Y. A., Obeid L. M. (2001) FASEB J. 15, 2669–2679 [DOI] [PubMed] [Google Scholar]
- 29.Ruvolo P. P., Deng X., Ito T., Carr B. K., May W. S. (1999) J. Biol. Chem. 274, 20296–20300 [DOI] [PubMed] [Google Scholar]
- 30.Birbes H., Luberto C., Hsu Y. T., El Bawab S., Hannun Y. A., Obeid L. M. (2005) Biochem. J. 386, 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siskind L. J., Kolesnick R. N., Colombini M. (2006) Mitochondrion 6, 118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siskind L. J., Kolesnick R. N., Colombini M. (2002) J. Biol. Chem. 277, 26796–26803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimeno H., Soeda S., Sakamoto M., Kouchi T., Kowakame T., Kihara T. (1998) Lipids 33, 601–605 [DOI] [PubMed] [Google Scholar]
- 34.El Bawab S., Roddy P., Qian T., Bielawska A., Lemasters J. J., Hannun Y. A. (2000) J. Biol. Chem. 275, 21508–21513 [DOI] [PubMed] [Google Scholar]
- 35.Thongboonkerd V., Luengpailin J., Cao J., Pierce W. M., Cai J., Klein J. B., Doyle R. J. (2002) J. Biol. Chem. 277, 16599–16605 [DOI] [PubMed] [Google Scholar]
- 36.Okazaki T., Bielawska A., Domae N., Bell R. M., Hannun Y. A. (1994) J. Biol. Chem. 269, 4070–4077 [PubMed] [Google Scholar]
- 37.Kalra J., Brosnan J. T. (1974) J. Biol. Chem. 249, 3255–3260 [PubMed] [Google Scholar]
- 38.Shiao Y. J., Lupo G., Vance J. E. (1995) J. Biol. Chem. 270, 11190–11198 [DOI] [PubMed] [Google Scholar]
- 39.Sugiura Y., Kawabe H., Tanaka H., Fujimoto S., Ohara A. (1981) J. Biol. Chem. 256, 10664–10670 [PubMed] [Google Scholar]
- 40.Otera H., Ohsakaya S., Nagaura Z., Ishihara N., Mihara K. (2005) EMBO J. 24, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yabu T., Kishi S., Okazaki T., Yamashita M. (2001) Biochem. J. 360, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita M., Konagaya S. (1991) J. Agric. Food Chem. 39, 1402–1405 [Google Scholar]
- 43.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 44.Sawai H., Okazaki T., Yamamoto H., Okano H., Takeda Y., Tashima M., Sawada H., Okuma M., Ishikura H., Umehara H., Okazaki T. (1995) J. Biol. Chem. 270, 27326–27331 [DOI] [PubMed] [Google Scholar]
- 45.Matsuo Y., Yamada A., Tsukamoto K., Tamura H., Ikezawa H., Nakamura H., Nishikawa K. (1996) Protein Sci. 5, 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujii S., Ogata K., Inoue B., Inoue S., Murakami M., Iwama S., Katsumura S., Tomita M., Tamura H., Tsukamoto K., Ikezawa H., Ikeda K. (1999) J. Biochem. 126, 90–97 [DOI] [PubMed] [Google Scholar]
- 47.Fujii S., Inoue B., Yamamoto H., Ogata K., Shinki T., Inoue S., Tomita M., Tamura H., Tsukamoto K., Ikezawa H., Ikeda K. (1998) J. Biochem. 124, 1178–1187 [DOI] [PubMed] [Google Scholar]
- 48.Letunic I., Copley R. R., Pils B., Pinkert S., Schultz J., Bork P. (2006) Nucleic Acids Res. 34, D257–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernardo K., Krut O., Wiegmann K., Kreder D., Micheli M., Schäfer R., Sickman A., Schmidt W. E., Schröder J. M., Meyer H. E., Sandhoff K., Krönke M. (2000) J. Biol. Chem. 275, 7641–7647 [DOI] [PubMed] [Google Scholar]
- 50.Liu B., Hassler D. F., Smith G. K., Weaver K., Hannun Y. A. (1998) J. Biol. Chem. 273, 34472–34479 [DOI] [PubMed] [Google Scholar]
- 51.Sawai H., Okamoto Y., Luberto C., Mao C., Bielawska A., Domae N., Hannun Y. A. (2000) J. Biol. Chem. 275, 39793–39798 [DOI] [PubMed] [Google Scholar]
- 52.Okamoto Y., Vaena de Avalos S., Hannun Y. A. (2003) Biochemistry 42, 7855–7862 [DOI] [PubMed] [Google Scholar]
- 53.Gakh O., Cavadini P., Isaya G. (2002) Biochim. Biophys. Acta 1592, 63–77 [DOI] [PubMed] [Google Scholar]
- 54.Gavel Y., von Heijne G. (1990) Protein Eng. 4, 33–37 [DOI] [PubMed] [Google Scholar]
- 55.Dias Neto E., Correa R. G., Verjovski-Almeida S., Briones M. R., Nagai M. A., da Silva W., Jr., Zago M. A., Bordin S., Costa F. F., Goldman G. H., Carvalho A. F., Matsukuma A., Baia G. S., Simpson D. H., Brunstein A., de Oliveira P. S., Bucher P., Jongeneel C. V., O'Hare M. J., Soares F., Brentani R. R., Reis L. F., de Souza S. J., Simpson A. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3491–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bionda C., Portoukalian J., Schmitt D., Rodriguez-Lafrasse C., Ardail D. (2004) Biochem. J. 382, 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ardail D., Popa I., Bodennec J., Louisot P., Schmitt D., Portoukalian J. (2003) Biochem. J. 371, 1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kitagaki H., Cowart L. A., Matmati N., Vaena de Avalos S., Novgorodov S. A., Zeidan Y. H., Bielawski J., Obeid L. M., Hannun Y. A. (2007) Biochim. Biophys. Acta 1768, 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu H., Toman R. E., Goparaju S. K., Maceyka M., Nava V. E., Sankala H., Payne S. G., Bektas M., Ishii I., Chun J., Milstien S., Spiegel S. (2003) J. Biol. Chem. 278, 40330–40336 [DOI] [PubMed] [Google Scholar]