Abstract
Continuous macroautophagic activity is critical for the maintenance of neuronal homeostasis; however, unchecked or dysregulated autophagy can lead to cell death. Cultured Purkinje neurons die by an autophagy-associated cell death mechanism when deprived of trophic support. Here, we report that insulin-like growth factor-I (IGF-I) completely blocked the autophagy-associated cell death of Purkinje neurons. To examine the mechanism by which IGF-I influences autophagy, neurons were infected with adeno-RFP-LC3 and subjected to trophic factor withdrawal, and the size and number of autophagosomes were analyzed by live-cell fluorescence imaging. In control neurons, autophagy occurred at a constitutive low level with most autophagosomes measuring less than 0.75 μm. Trophic factor withdrawal increased the number and size of autophagosomes with most autophagosomes ranging between 0.75 and 1.5 μm and some reaching 1.5–2.25 μm. IGF-I added at the time of trophic factor withdrawal prevented the accumulation of the larger autophagosomes; however, it had no effect on the conversion of LC3, an indicator of autophagy induction. Instead, the rate of autophagosome-to-lysosome fusion measured by colocalization of RFP-LC3 and LysoSensor Green was accelerated by IGF-I. Treating the neurons with bafilomycin A1 in the presence of IGF-I led to the accumulation of autophagosomes even larger than those induced by trophic factor withdrawal alone, indicating that IGF-I regulates autophagic vesicle turnover. Finally, the effect of IGF-I on autophagy was mediated by an Akt/mTOR-de pend ent and an ERK-independent pathway. These data suggest a novel role for IGF-I in protecting Purkinje neurons from autophagy-associated cell death by increasing autophagy efficiency downstream of autophagy induction.
Autophagy is a regulated, catabolic pathway for the turnover of long-lived proteins, macromolecular aggregates, and damaged organelles by lysosomal degradation (1). It also plays a role in clearing the cell of invading bacteria and viruses (2, 3). In mammalian cells, the lysosomal pathway of intracellular degradation is divided into three distinct pathways: macroautophagy, microautophagy, and chaperone-mediated autophagy (4). Macroautophagy (5, 6) begins with the formation of a unique double-membrane vesicle (autophagosome) that engulfs cytoplasmic constituents such as proteins, lipids, and damaged organelles, including mitochondria. The outer membrane of the autophagosome then docks and fuses with the lysosome to deliver the sequestered cargo. The inner membrane of the fused vesicle (autolysosome) along with the interior contents of the autophagosome are degraded by lysosomal hydrolases, a process that generates nucleotides, amino acids, and free fatty acids that are recycled to provide raw materials and energy to the cell. Microautophagy (7) circumvents the autophagosome sequestration step and begins with the direct uptake of cytosolic components by invaginations and pinching off of the lysosomal membrane. As in macroautophagy, the internalized cytosolic components are digested by lysosomal enzymes, and the macromolecules are released when the vacuolar membrane disintegrates. In chaperone-mediated autophagy (8, 9) specific chaperone proteins bind to targeted proteins containing a KFERQ sequence and direct these proteins to the surface of the lysosome. These proteins bind to LAMP-2A on lysosomal membranes and are then transported across the membrane with the assistance of chaperone proteins where they are degraded by the lysosomal proteases.
In eukaryotic cells, macroautophagy (hereon referred to as autophagy) occurs constitutively at low levels in most cells to perform housekeeping functions such as degradation of proteins and destruction of dysfunctional organelles. Dramatic up-regulation of autophagy occurs in the presence of external stressors such as starvation, hormonal imbalance, oxidation, extreme temperature, and infection as well as internal needs such as cellular remodeling and removal of protein aggregates (10).
Dysregulated autophagy is thought to contribute to many human diseases, particularly neurodegeneration. Morphological studies of diseased human postmortem brain consistently reveal signs of enhanced autophagy in degenerating neuronal populations. Increased autophagy and disturbances in the lysosomal degradative system have been reported in many neurodegenerative conditions including Alzheimer disease (11, 12), Parkinson disease (13), Huntington disease (14), prion encephalopathies (15, 16), and diffuse Lewy body disease (17). Many of these neurodegenerative diseases are associated with the accumulation of mutant proteins. Several of the mutant proteins including α-synuclein and huntingtin are degraded by an autophagic-lysosomal pathway, suggesting that autophagy may be a protective mechanism for the clearance of toxic aggregate-prone proteins (18, 19). In mouse and Drosophila models of Huntington disease, early treatment with the mTOR inhibitor and autophagy stimulator, rapamycin, and a rapamycin analogue (CCI-779) decreased neurodegeneration and improved behavioral performance (20).
In other animal models of neurodegeneration, it has been suggested that enhanced autophagy contributes to cell death. Autophagy is induced in vulnerable populations of neurons in transgenic mouse models of Alzheimer disease in which human mutant presenilin-1 and the Swedish variant of amyloid precursor protein are overexpressed (21, 22). In both conditions autophagosomes proliferate in dendrites, and the level of LC3-II, a specific marker for autophagosome formation, is elevated. This process occurs before β-amyloid pathology develops, indicating that autophagy induction is an early response in the disease. Likewise, an ultrastructural study on neuronal degeneration in transgenic mice expressing mutant human tau demonstrated the involvement of autophagic processes (23). In experimental animal models of Creutzfeldt-Jakob disease and scrapie disease, ultrastructural features of autophagy are abundant in cortical neurons (24). Extensive autophagic vacuole formation occurs in degenerating cerebellar Purkinje neurons in models of ischemia (25, 26) and in spinocerebellar ataxia 1 transgenic mice (27). Finally, neuronal death in the Lurcher mouse, which suffers from extensive Purkinje neuron degeneration caused by a point mutation in the δ2 glutamate receptor (28) appears to result from the direct activation of autophagy by Lurcher GRID2 receptors through a signaling pathway consisting of GRID-2, n-PIST, and Beclin 1 (29, 30). It is possible that autophagy is initially up-regulated in the nervous system to provide macromolecular nutrients or clear protein aggregates from neurons and that once a certain level of intracellular damage is reached, accumulation of large autophagic vacuoles exacerbate the damage eventually leading to cell death.
We have established a novel model system utilizing primary cultured cerebellar neurons in which Purkinje neurons undergo up-regulation of autophagy when trophic factors are removed from their medium. The prolonged up-regulated autophagy and the accumulation of autophagic vesicles contribute to the death of these neurons, thus providing a model to examine the relationship between enhanced autophagy and cell death (31). In this report we have utilized live-cell imaging techniques to monitor autophagy in Purkinje neurons. We demonstrate that IGF-I, a potent neurotrophic factor in the cerebellum, prevents the accumulation of autophagic vesicles and cell death of Purkinje neurons by increasing the rate of autophagosome-lysosome fusion and degradation via an Akt/mTOR-dependent pathway, indicating a novel mechanism by which IGF-I maintains neuronal homeostasis.
EXPERIMENTAL PROCEDURES
Materials
The polyclonal rabbit antibody to calbindin D-28K and LC3 were obtained from Chemicon (Temecula, CA) and Abgent (San Diego, CA), respectively. Cy3-conjugated secondary antibody was obtained from Jackson ImmunoResearch (West Grove, PA). Horseradish peroxidase-linked secondary antibodies and reagents for enhanced chemiluminescence detection were obtained from GE Healthcare and Pierce, respectively. LysoSensor Green was obtained from Molecular Probes (Eugene, OR). Rapamycin, SH-5, and PD98059 were purchased from Calbiochem. Hoechst, IGF-I,2 bafilomycin A1, and lithium chloride were purchased from Sigma-Aldrich. The adenoviral RFP-LC3 was a gift from Aviva Tolkovsky (University of Cambridge, Cambridge, England). All other chemicals were purchased from Sigma-Aldrich unless otherwise stated.
Cell Culture
Animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Colorado Denver and were conducted in accordance with policies for the ethical treatment of animals established by the National Institutes of Health. Primary rat cerebellar granule neurons were isolated from 7-day-old Sprague-Dawley rat pups as described previously (32). Briefly, cells were plated at a density of 2.0 × 106 cells/ml on poly-d-lysine/laminin (BD Biosciences)-coated coverslips in basal modified Eagle's medium containing 10% fetal bovine serum, 25 mm KCl, 2 mm l-glutamine, and penicillin (100 units/ml)-streptomycin (100 μg/ml) (Invitrogen), which is referred to as 25K+S medium. Cytosine arabinoside (10 μm) was added to the culture medium 24 h after plating to limit the growth of non-neuronal cells. On day 5, neurons were infected with adenoviral vector expressing MAP1-LC3 labeled with red fluorescent protein (RFP-LC3) at a multiplicity of infection of 100 for 24 h. Autophagic cell death was induced by removing the plating medium and replacing it with trophic factor withdrawal 5K-S medium (serum-free basal modified Eagle's medium containing 5 mm KCl).
Immunocytochemistry
Purkinje neurons in cerebellar cultures were identified by immunostaining with a polyclonal antibody to calbindin D-28K. Neurons were plated and induced to undergo death as indicated above. After the appropriate treatment, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS, pH 7.4) for 30 min at room temperature. The neurons were then permeabilized and blocked in PBS containing 0.2% Triton X-100 and 5% bovine serum albumin for 1 h at room temperature. Cells were incubated overnight at 4 °C with calbindin D-28K (1:250) diluted in PBS containing 0.2% Triton X-100 and 2% bovine serum albumin. The primary antibody was aspirated, and cells were washed six times with PBS at room temperature for at least 1 h. Neurons were then incubated with the appropriate Cy3-conjugated secondary antibody (diluted 1:500) and 4′,6-diamidino-2-phenylindole (1 μg/ml) for 1 h at room temperature. The cells were then washed 6 more times with PBS at room temperature, and coverslips were adhered to glass slides with mounting medium (0.1% p-phenylenediamine in 75% glycerol in PBS). Slides were sealed with clear nail polish and stored at −20 °C before imaging. Fluorescence imaging was performed on a Zeiss Axioplan 2 microscope equipped with a Cooke sensicam deep-cooled CCD camera, and images were analyzed and subjected to digital deconvolution using the Slidebook 4.2 software program (Intelligent Imaging Innovations Inc., Denver, CO).
Western Blotting
Cells were washed twice with PBS and lysed in Laemmli sample buffer (60 mm Tris pH 6.8, 10% glycerol, and 2% SDS), and lysates were briefly sonicated. Lysates were boiled for 5 min, and equal volumes of total protein were separated by SDS-PAGE (NuPAGE Novex system; Invitrogen) and transferred to a polyvinylidene fluoride membrane. Nonspecific binding sites were blocked in TBS containing 0.1% Tween 20 (TBS-T) and 5% bovine serum albumin for 1 h at room temperature. Rabbit anti-LC3 (1:1,000; Abgent) was diluted in blocking solution and incubated with the membranes overnight at 4 °C while on a rotating platform. Excess primary antibody was removed by washing the membranes three times in TBS-T at room temperature for at least 30 min. The blots were then incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (anti-rabbit-horseradish peroxidase (1:5000)) diluted in TBS-T for 1 h at room temperature and were subsequently washed 3 times in TBS-T for 30 min. Immunoreactive bands were detected by use of the enhanced chemiluminescence reagent (GE Healthcare) and exposed to CL-Exposure film (Pierce). In some experiments membranes were reprobed after stripping in Restore Western blot Stripping Buffer (Pierce). The blots were rinsed three times in TBS-T and processed as above with a different primary antibody. Autoluminograms shown are representative of at least three independent experiments.
Quantitation of Purkinje Neuron Survival
Purkinje neuron numbers were measured by counting the number of calbindin-positive cells in 152 fields using a 63× oil objective that were randomly selected by following a fixed grid pattern over the coverslip. The total area counted per coverslip was 14.6 mm2 or ∼13% of the coverslip. In addition to calbindin staining, cells were also judged by their morphology. Cells counted as Purkinje neurons had large rounded cell bodies, elaborate processes, and larger, more oval nuclei in comparison to the smaller, rounder nuclei of granule neurons. At least three coverslips were counted per experimental condition. The numbers were averaged and expressed as a percentage of the average number counted in the appropriate controls. This was repeated for at least three independent experiments.
Quantitation of Autophagy
Vesicle and fusion analysis from live-cell imaging was performed as described previously (33). Briefly, cerebellar neurons were infected with RFP-LC3 as described above. Twenty-four hours later, autophagic cell death was induced by removing the plating medium and replacing it with either control medium (25K+S) or trophic factor withdrawal medium (5K-S) containing the appropriate drug for various time points (0–24 h). After the appropriate treatment times, cells were stained at 37 °C with Hoechst (20 ng/ml) for 15 min to visualize cellular nuclei and LysoSensor Green (2 μm) for 5 min to visualize lysosomes. The cells were then washed 3 times for 5 min with 37 °C phenol red-free 25K+S control medium (Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, 25 mm KCl, 2 mm l-glutamine, and penicillin (100 units/ml)-streptomycin (100 μg/ml; Invitrogen)) or phenol red-free 5K-S medium (serum-free DMEM containing 5 mm KCL) to remove nonspecifically bound dyes. The individually timed coverslips to be imaged were attached to a 35-mm dish using Vaseline petroleum jelly and submerged in 3 ml of the appropriate phenol red-free medium. The dish was then fitted into a pre-warmed 37 °C heated stage which maintains the temperature at 37 °C. Imaging was performed as described above. Images of LysoSensor Green, RFP-LC3, and Hoechst fluorescence were captured on the fluorescein isothiocyanate, Cy3, and 4′,6-diamidino-2-phenylindole channels, respectively, using the 63× water immersion objective. The length of exposure on the 4′,6-diamidino-2-phenylindole channel was calibrated by the Slidebook program (Slidebook 4.2). Collected images were used to measure the diameters of all the visible RFP-and LysoSensor Green-positive vacuoles using Slidebook. Autophagosome-to-lysosome fusion was measured as the degree of co-localization between RFP-LC3 positive vacuoles and LysoSensor Green positive vacuoles, which was quantified using the Pearson correlation analysis tool in Slidebook 4.2. Vesicle diameter and fusion measurements were collected from at least 7–10 Purkinje live cell images per treatment condition for at least three independent experiments.
Data Analysis
Results shown represent the mean ± S.E. from three independent experiments performed in triplicate. Statistical differences between the means of unpaired sets of data were evaluated using one-way analysis of variance followed by post hoc Tukey's test. A p value of <0.05 was considered statistically significant. The degree of co-localization between RFP-LC3-positive vacuoles and LysoSensor Green positive vacuoles was quantified using the Pearson correlation coefficient (r), which calculates the proportion of all red intensities that contain green signals within all red intensities and was assessed using the Pearson correlation analysis tool in Slidebook 4.2.
RESULTS
IGF-I Blocks the Accumulation of Autophagic Vacuoles and Cell Death of Purkinje Neurons
Previous studies by our laboratory have demonstrated that trophic factor withdrawal leads to the death of cultured cerebellar neurons. Granule neurons die by an apoptotic mechanism involving Bax translocation to the membrane, cytochrome c release, activation of caspase 9 and caspase 3, and nuclear condensation (34–36). In contrast, Purkinje neurons die by an autophagy-associated cell death mechanism, characterized by extensive cytoplasmic vacuolation, the presence of autolysosomes detected by electron microscopy, and the absence of caspase activation and nuclear condensation (31). The apoptosis induced in granule neurons deprived of trophic support was completely inhibited with IGF-I, which has been well characterized in inhibiting pathways that promote apoptosis (36–38). However, the ability of IGF-I to inhibit cell death associated with autophagy has not been fully investigated. Using live-cell imagining techniques, we examined the ability of IGF-I to inhibit the autophagy-associated cell death of Purkinje neurons.
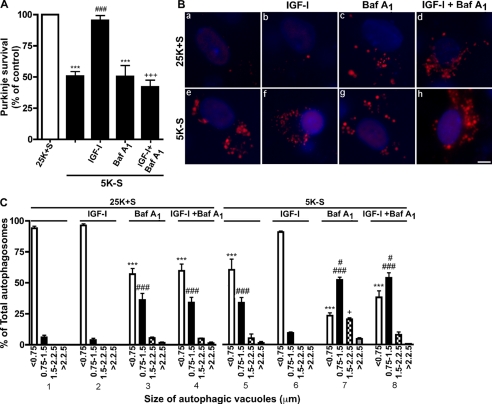
To examine the effect of IGF-I on cell death of Purkinje neurons, we quantified the number of surviving Purkinje neurons after trophic factor withdrawal in the absence or presence of IGF-I (Fig. 1A). Purkinje neurons were identified by staining cultures with the Purkinje marker, calbindin D-28K. After 24 h of trophic factor withdrawal (5K-S), only 50% of Purkinje neurons remained compared with controls, which supports our previously published data (31) (Fig. 1A, ***, p < 0.001 compared with 25K+S). By 72 h all of the neurons have died and were lifted from the dish. The addition of IGF-I (200 ng/ml) to the cultures prevented the loss of Purkinje neurons induced by trophic factor withdrawal and resulted in 95% survival, which was comparable with untreated control cells (Fig. 1A, ###, p < 0.001 compared with 5K-S). The addition of bafilomycin A1, a vacuolar ATPase inhibitor that disrupts lysosomal vesicle acidification and inhibits vesicle degradation (39), did not decrease Purkinje survival more than 5K-S treatment alone, but it completely reversed the neuroprotective effects of IGF-I on Purkinje neuron survival, indicating that lysosomal vesicle turnover is required for IGF-I mediated neuroprotection (Fig. 1A, ***, p < 0.001 compared with 25K+S; +++, p < 0.001 compared with 5K-S + IGF-I). In the presence of trophic support (25K+S), IGF-I and bafilomycin A1 had no additional effect on Purkinje survival compared with 25K+S alone (data not shown).
FIGURE 1.
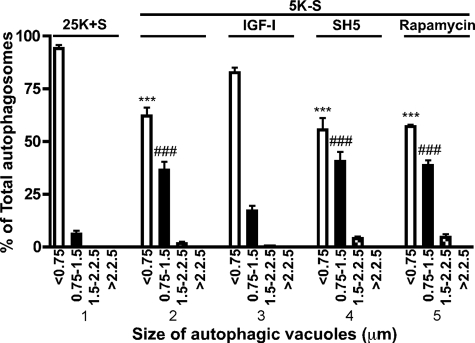
IGF-I blocks the accumulation of autophagic vacuoles and cell death of Purkinje neurons. A, Purkinje neurons were maintained in control medium (25K+S), trophic factor-withdrawal medium (5K-S), 5K-S +IGF-I (200 ng/ml), 5K-S +bafilomycin A1 (Baf A1 100 nm), and 5K-S +IGF-I +Baf A1 for 24 h. The cultures were fixed and stained with the Purkinje neuron marker calbindin D-28K, and Purkinje neurons were counted. Numbers are plotted as percent survival of control. B and C, RFP-LC3-infected Purkinje neurons were maintained in 25K+S, 25K+S +IGF-I, 25K+S +Baf A1, 25K+S +IGF-I+Baf A1, 5K-S, 5K-S +IGF-I, 5K-S +Baf A1, or 5K-S +IGF-I+Baf A1 for 16 h after which neurons were stained with Hoechst, and images were captured via live-cell imaging. Representative live-cell images are shown in B; the scale bar represents 5 μm. C, autophagosome vacuole size was quantified by measuring the diameters of all RFP-LC3-positive vacuoles in 7–12 Purkinje neurons per treatment (n = 3). The size distribution was graphed as the percent of total vacuoles within the indicated size ranges. Data represent the mean ± S.E. of three independent experiments, each performed in triplicate. A, ***, p < 0.001 compared with 25K+S control; ###, p < 0.001 compared with 5K-S; +++, p < 0.001 compared with 5K-S+ IGF-I. C, ***, p < 0.001 compared with 25K+S, <0.75 μm; ###, p < 0.001 compared with 25K+S, 0.75–1.5 μm; #, p < 0.05 compared with 5K-S, 0.75–1.5 μm; +, p < 0.05 compared with 5K-S, 1.5–2.25 μm. One-way analysis of variance was followed by Tukey's post hoc test.
We then analyzed the effects of IGF-I on various steps of the autophagy pathway beginning with autophagosome formation using an adenoviral vector expressing RFP-tagged LC3, the mammalian equivalent of yeast Atg8. Atg8 was the first autophagy-related protein shown to localize to autophagosomes (40, 41). In mammals LC3 is required for the expansion phase of autophagosome formation (42) and, therefore, acts as a cellular marker for autophagosomes.
To determine whether IGF-I could prevent the extensive autophagic vacuolation observed after trophic factor withdrawal, the size of autophagosomes were measured in RFP-LC3-infected Purkinje neurons subjected to 16 h trophic factor withdrawal in the absence or presence of IGF-I. After 16 h, the cells were stained with Hoechst and LysoSensor Green, and live-cell imaging was performed. Quantitative analysis of vacuolation was performed on captured images by measuring the diameters of all RFP-LC3 vacuoles present after 16 h of trophic factor withdrawal, which was used to create a vesicle size distribution profile (33). In the presence of trophic support (25K+S), the majority (>95%) of Purkinje neurons contained small (<0.75 μm) RFP-LC3-positive vacuoles (Fig. 1, Ba and C, section 1). IGF-I (200 ng/ml) had no additional effect on vesicle size under control 25K+S conditions (Fig. 1, Bb and C, section 2). After 16 h of trophic factor withdrawal (5K-S), there was a significant shift in the size distribution profile with a ∼40% decrease in RFP-LC3 positive vacuoles less than 0.75 μm and a ∼30% increase in RFP-LC3 positive vacuoles between 0.75 and 1.5 μm (Fig. 1, Be and C, section 5; p < 0.001 (***) and p < 0.001 (###) compared with 25K+S control). The addition of IGF-I to the trophic factor withdrawal media inhibited the autophagosome accumulation induced by trophic factor withdrawal, resulting in a shift in the vesicle size distribution toward control conditions (Fig. 1, Bf and C, section 6).
To examine whether IGF-I prevented vesicle accumulation by regulating the formation of autophagosomes or vesicle turnover, we added the lysosomal degradation inhibitor, bafilomycin A1 (100 nm), in the absence or presence of IGF-I and analyzed the vesicle size distribution. Under control 25K+S conditions, bafilomycin A1 induced the accumulation of vesicles due to basal autophagy in these cells (Fig. 1, Bc and C, section 3; p < 0.001 (***) and p < 0.001 (###) compared with 25K+S control). Trophic factor withdrawal plus bafilomycin A1 resulted in an even greater increase in autophagosomes larger than 0.75 μm compared with 5K-S conditions alone (Fig. 1C, section 7; #, p < 0.05 compared with 5K-S, 0.75–1.5 μm; +, p < 0.05 compared with 5K-S, 1.5–2.25 μm). When treated in the presence of bafilomycin A1, IGF-I no longer blocked the accumulation of autophagosomes greater than 0.75 μm (Fig. 1, Bh and C, section 8, p < 0.001 (***) and p < 0.001 (###) compared with 25K+S control and p < 0.05 (#) compared with 5K-S). The inhibition of IGF-I effects on vacuole accumulation by bafilomycin A1 demonstrates that IGF-I acts downstream of autophagosome formation to increase vesicle turnover.
IGF-I Does Not Influence the Up-regulation of LC3-II That Is Induced during Autophagy
Another method to quantify vesicle formation is by analyzing LC3 protein levels. LC3 exists in two states, LC3-I and LC3-II. Newly synthesized LC3 is cleaved to form LC3-I, which is localized in the cytosol. When autophagy is induced, LC3-I is lipidated (phosphatidylethanolamine-conjugated) to form LC3-II, which binds to the autophagosomal membrane (42, 43). Thus, expression of LC3-II is a good indicator of autophagy induction.
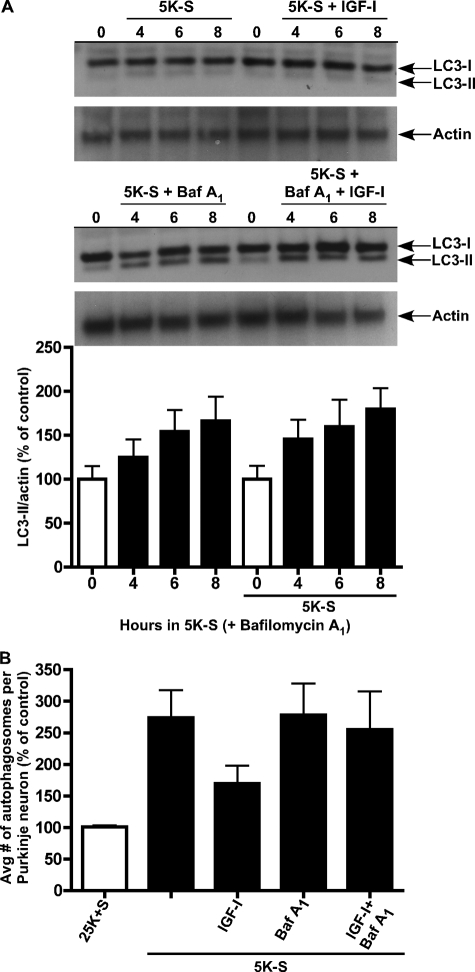
To determine the role of IGF-I on autophagy induction, we examined the protein levels of LC3-II by Western blot (Fig. 2A). Cerebellar cultures were subjected to a 0-, 4-, 6-, and 8-h trophic factor withdrawal time course in the absence or presence of IGF-I (200 ng/ml) and in the absence or presence of bafilomycin A1 (100 nm). Trophic factor withdrawal both in the absence or presence of IGF-I did not induce measurable levels of LC3-II, suggesting that the turnover of vesicles was rapid in these cultures (Fig. 2A, top blot). Therefore, the lysosomal degradation inhibitor, bafilomycin A1, was added to block the turnover of autolysosomes, allowing for the accurate measurement of total LC3-II. At 0 h of trophic factor withdrawal in the absence or presence of IGF-I vehicle (H20), LC3-II expression levels were almost non-existent, in agreement with low basal levels of autophagy under control conditions. Trophic factor withdrawal alone in the presence of bafilomycin A1 induced LC3-II expression over time, indicative of autophagy induction (Fig. 2A, bottom blot). Surprisingly, the addition of IGF-I in the presence of bafilomycin A1 had no effect on the LC3-II induction time-course, as LC3-II expression levels were not statistically different from trophic factor alone conditions. These results suggested that autophagy activation still occurred in the presence of IGF-I but that autophagosomes did not accumulate.
FIGURE 2.
IGF-I does not block LC3-II induction during autophagy. An immunoblot analysis was performed to determine the effects of trophic factor withdrawal and IGF-I on LC3-II expression. Purkinje neurons were maintained in 25K+S and 5K-S in the absence or presence of IGF-I (200 ng/ml) and bafilomycin A1 (Baf A1, 100 nm) for 0, 4, 6, and 8 h. Immunoblot analysis was performed using a polyclonal antibody to LC3. A, LC3-I is 18 kDa, and LC3-II (the lipidated membrane bound form) is 16 kDa. Actin served as a loading control. Data shown are a representative immunoblot of three independent experiments. A, bottom, quantitation of LC3-II induction in the presence of Baf A1. B, Purkinje neurons maintained in 25K+S and 5K-S in the absence and presence of IGF-I and Baf A1 for 6 h were subjected to live-cell imaging. The total number of autophagosomes were counted in 10 Purkinje neurons per treatment condition and averaged for three separate experiments.
Because these cultures represent a heterogeneous population of neurons including granule neurons, which do not exhibit enhanced autophagy upon trophic factor withdrawal, we verified the Western blot data by live-cell imaging of LC3 expression in Purkinje neurons using RFP-LC3. LC3-II expression is localized specifically to the autophagosomal membrane; therefore, counting the total number of RFP-LC3-stained autophagosomes is another method of measuring both autophagy induction and autophagosome formation. Cerebellar cultures were treated with trophic factor withdrawal media in the absence or presence of IGF-I (200 ng/ml) both with and without bafilomycin A1 (100 nm) for 6 h and subjected to live-cell imaging. Bafilomycin A1 was added again to ensure accurate measurements of the total number of autophagosomes. The addition of IGF-I to cerebellar cultures subjected to trophic factor withdrawal in the absence of bafilomycin A1 appeared to reduce the total number of autophagosomes per Purkinje neuron compared with trophic factor withdrawal alone (Fig. 2B). In trophic factor withdrawal media containing bafilomycin A1, the average number of autophagosomes formed per Purkinje neuron in the absence and presence of IGF-I revealed no significant difference, confirming Western blot data that IGF-I does not regulate autophagy at the step of autophagic vesicle formation.
IGF-I Enhances the Fusion Rate between Autophagosomes and Lysosomes
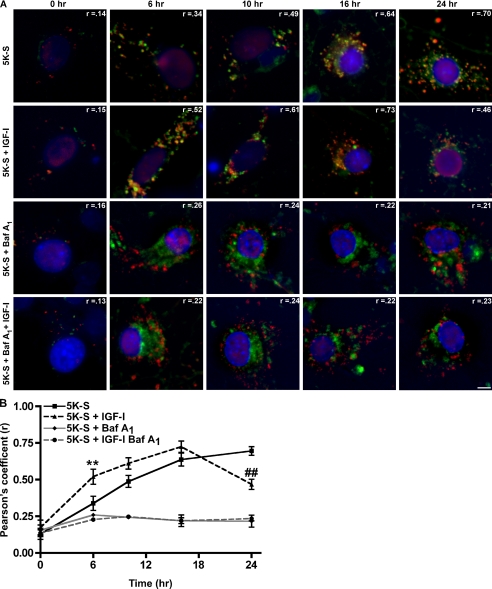
Downstream of autophagosome formation is the fusion of the autophagosome with the lysosome and degradation of both bulk cytoplasm within the autophagosome and the autophagosome itself. Using live-cell imaging techniques, we analyzed whether IGF-I could regulate the fusion of autophagosomes with lysosomes. Cerebellar cultures were infected with RFP-LC3 and subjected to a trophic factor withdrawal time course (0, 6, 10, 16, 24 h) in the absence or presence of IGF-I and repeated in trophic factor withdrawal media containing bafilomycin A1. Neurons were then stained with Hoechst to visualize cellular nuclei and LysoSensor Green to visualize lysosomes, and images were captured via live-cell imaging (Fig. 3A). Co-localization of RFP-LC3 and LysoSensor Green is a representation of fused vesicles or autolysosomes. Therefore, fusion was quantified by measuring the degree of co-localization between the red (autophagosome) and green (lysosome) signal using the Pearson correlation coefficient (Slidebook 4.2). The Pearson correlation calculates the correlation between the intensity distribution of the red and green channel and expresses the correlation as an r-value between −1 and +1, indicating no co-localization to perfect co-localization, respectively.
FIGURE 3.
IGF-I increases the autophagosome-to-lysosome fusion rate in Purkinje neurons. RFP-LC3-infected cerebellar cultures were subjected to a 5K-S time course (0, 6, 10, 16, 24 h) in the absence or presence of IGF-I (200 ng/ml) and bafilomycin A1 (100 nm). Neurons were stained with Hoechst to visualize cellular nuclei and LysoSensor Green to visualize lysosomes, and images were captured via live cell imaging. A, images represent merged images of Hoechst (blue), LysoSensor Green (green), and RFP-LC3 (red). Areas of co-localization (yellow) between RFP-LC3-and LysoSensor Green-positive vacuoles are indicative of autophagosome to lysosome fusion. The scale bar represents 5 μm. B, co-localization between RFP-LC3- and LysoSensor Green-positive vacuoles was quantified using Pearson's correlation coefficient analysis in Slidebook 4.2. Data represent the mean ± S.E. of three independent experiments, each performed in triplicate. **, p < 0.01 compared with 6 h 5K-S; ##, p < 0.01 compared with 24 h 5K-S. One-way analysis of variance was followed by Tukey's post hoc test.
In 5K-S trophic factor withdrawal conditions, the degree of co-localization, expressed as the Pearson correlation coefficient (r) increased with time and at 24 h was 0.70, which indicates a high degree of co-localization and, thus, a high degree of fusion of autophagosomes with lysosomes (Fig. 3, A, top panel, and B). The autophagosomes also appear to increase in size over time, supporting the vesicle size distribution data from Fig. 1. In comparison, the amount of fusion in IGF-I-treated coverslips also increased with time; however, fusion peaked at 16 h with a Pearson correlation of 0.73 (Fig. 3, A, second from top panel, and B). Furthermore, at 6 h the degree of co-localization was significantly higher than 5K-S-alone conditions (Fig. 3B, **, p < 0.01 compared with 6 h 5K-S) and remained higher than 5K-S for up to 16 h. By 24 h the degree of co-localization in the presence of IGF-I was less than 0.50, significantly lower than 5K-S alone (Fig. 3B, ##, p < 0.01 compared with 24 h 5K-S), and the neurons were comparable in morphology to control conditions (smaller vesicles). In comparison, no RFP-LC3 and LysoSensor Green colocalization was observed in treatment conditions containing bafilomycin A1, demonstrating that lysosomal inhibition via bafilomycin A1 also blocks vesicle fusion (Fig. 3, A, third and fourth panel from the top and B). These results suggested that IGF-I regulates autophagic flux by increasing the autophagosome-lysosome fusion rate at earlier time points, resulting in a more rapid turnover of autophagic vacuoles. The results are also consistent with the observed decrease in total autophagosomes in trophic withdrawal conditions treated with IGF-I compared with trophic factor withdrawal alone in Fig. 2B.
IGF-I Effects on Purkinje Neuron Survival and Autophagy-associated Cell Death Are Akt- and mTOR-dependent
IGF-I is known to inhibit apoptotic cell death by activating the serine/threonine kinase Akt (44). We have previously shown that Akt activation is required for the inhibition of granule neuron apoptosis by IGF-I (36) but have yet to investigate its role in the autophagy-associated cell death of Purkinje neurons. Akt has been shown to be a negative regulator of autophagy, as treatments that decrease Akt activity (overexpression of PTEN and ceramide) induce autophagy, whereas those that up-regulate Akt activity (treatment with phosphatidylinositol 3,4-diphosphate and phosphatidylinositol 3,4,5-trisphosphate and overexpression of a constitutively active Akt) inhibit autophagy (45, 46). In addition, Akt has been shown to be required for insulin-dependent inhibition of autophagy in myeloid cells (47). We, therefore, examined the role of Akt in mediating the survival of Purkinje neurons by IGF-I.
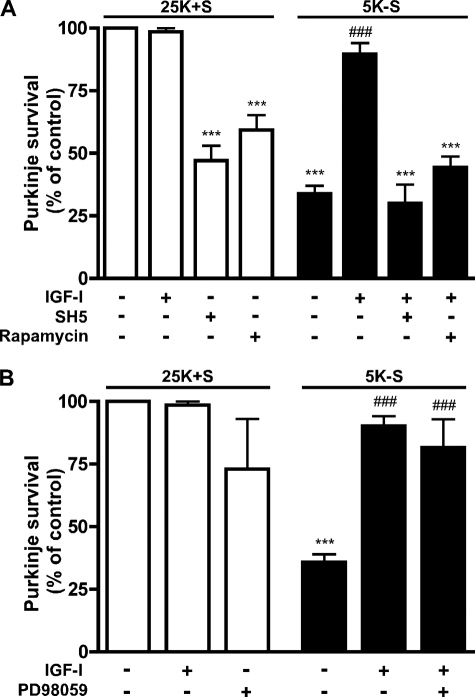
When the specific Akt inhibitor SH-5 (1.0 μm) was included with IGF-I in trophic factor withdrawal conditions (5K-S), it completely blocked the promotion of Purkinje neuron survival by IGF-I as only 30.7 ± 8.2% of Purkinje neurons remained, which is not statistically different from trophic factor withdrawal alone (Fig. 4A, ***, p < 0.001 compared with 25K+S). SH-5 also significantly decreased Purkinje neuron viability under control 25K+S conditions (Fig. 4A, ***, p < 0.001 compared with 25K+S control). The concentration of SH-5 that completely reversed IGF-I neuroprotection corresponds well with the concentrations that are required to completely inhibit Akt activity (47).
FIGURE 4.
IGF-I-mediated neuroprotection of Purkinje neurons against autophagy-associated cell death is Akt- and mTOR-dependent and ERK1/2-independent. Purkinje neurons were maintained in control medium (25K+S) in the absence or presence of IGF-I (200 ng/ml), SH-5 (1.0 μm), rapamycin (100 nm), and PD98059 (50 μm) or trophic factor-withdrawal medium (5K-S) in the absence or presence of IGF-I, SH-5, rapamycin, or PD98059 for 48 h. The cultures were fixed and stained with the Purkinje neuron marker, calbindin D-28K, and Purkinje neurons were counted. Numbers are plotted as the percent survival of control. A, Purkinje survival in the absence and presence of IGF-I and in presence of SH-5 or rapamycin plus IGF-I. B, Purkinje survival in the absence and presence of IGF-I and in the presence of PD98059 plus IGF-I. A and B, data represent mean ± S.E. of five independent experiments, each performed in triplicate. ***, p < 0.001 compared with 25K+S control; ###, p < 0.001 compared with 5K-S. One-way analysis of variance followed by Tukey's post hoc test.
Although mTOR is known to regulate autophagy in diverse organisms and the insulin/Akt signaling pathway contributes to mTOR activation, there have been conflicting reports regarding the requirement of mTOR signaling for the inhibition of autophagy by insulin (47, 48). Additionally, there is a paucity of information regarding the role of mTOR signaling in Purkinje neurons. The addition of rapamycin (100 nm), a potent and selective inhibitor of mTOR signaling, reversed the protection of Purkinje neurons by IGF-I, and the remaining Purkinje neuron numbers were reduced to 44.3 ± 4.4% that of controls (Fig. 4A, ***, p < 0.001 compared with 25K+S control). Rapamycin also decreased Purkinje neuron viability under control conditions (Fig. 4A, ***, p < 0.001 compared with 25K+S control).
ERK1/2 Signaling Pathway Is Not Required for Purkinje Neuron Survival Promoted by IGF-I
IGF-I has also been reported to promote cell survival by activating ERK1/2 signaling pathways (49, 50). To determine a possible role of ERK1/2 signaling in mediating the survival of Purkinje neurons promoted by IGF-I, we utilized the ERK1/2 inhibitor, PD98059. The addition of PD98059 (50 μm) with IGF-I (200 ng/ml) had no effect on the IGF-I-mediated protection of Purkinje neurons as cultures treated with both IGF-I and PD98059 maintained Purkinje neuronal counts at 81.7 ± 11.3% of control (Fig. 4B, ###, p < 0.001 compared with 5K-S), which was not statistically different from either control conditions or treatment with IGF-I alone. IGF-I inhibition of Purkinje neuron death is independent of its ability to activate the ERK signaling pathway.
IGF-I Effects on Autophagic Vesicle Accumulation Are Akt- and mTOR-dependent
We have demonstrated that the degree of autophagic vacuolation of Purkinje neurons is negatively correlated with survival. To determine whether the effect of IGF-I on vesicle accumulation was Akt- and mTOR-dependent, as was Purkinje survival, cerebellar cultures were subjected to a 24-h trophic factor withdrawal in the absence or presence of IGF-I and the Akt and mTOR inhibitors, SH5 and rapamycin, respectively. Quantitative analysis of vesicle size was performed from captured live cell RFP-LC3 images (Fig. 5). As observed previously, the majority of Purkinje neurons contained small (<0.75 μm) autophagic vacuoles in the presence of trophic support. After 24 h of trophic factor withdrawal there was a significant decrease in 0.75–1.5-μm vacuoles and increase in 1.5–2.25-μm vacuoles (Fig. 5, section 2, p < 0.001 (***) and p < 0.001 (###) compared with 25K+S, section 1). The addition of IGF-I (200 ng/ml) to the trophic factor withdrawal media decreased the overall vacuolation of Purkinje neurons, as the vesicle size distribution was not significantly different from 25K+S control (Fig. 5, compare sections 1 and 3). The addition of SH-5 (1.0 μm) and rapamycin (100 nm) to IGF-I-treated cultures completely reversed the inhibition of Purkinje neuron vacuolation by IGF-I as the percentage of small to large autophagosomes was statistically identical to conditions of trophic factor withdrawal alone (Fig. 5, sections 4 and 5, p < 0.001 (***) and p < 0.01 (###) compared with 25K+S, section 1). Control 25K+S conditions treated with SH5 and rapamycin also increased the accumulation of vesicles greater than 0.75 μm (data not shown). These data demonstrate that IGF-I acts through a Akt/mTOR-dependent pathway to prevent both vesicle accumulation and Purkinje cell death.
FIGURE 5.
IGF-I effects on autophagic vesicle accumulation are Akt- and mTOR-dependent. RFP-LC3-infected Purkinje neurons were maintained in 25K+S, 5K-S, 5K-S +IGF-I (200 ng/ml), 5K-S +SH5 (1.0 μm), or 5K-S +rapamycin (100 nm) for 24 h after which neurons were stained with Hoechst, and images were captured via live-cell imaging. Autophagosome vacuole size was quantified by measuring the diameters of all RFP-LC3-positive vacuoles in 7–12 Purkinje neurons per treatment (n = 3). The size distribution was graphed as the percent of total vacuoles within the indicated size ranges. Data represent the mean ± S.E. of three independent experiments, each performed in triplicate. ***, p < 0.001 compared with 25K+S, <0.75 μm; ###, p < 0.001 compared with 25K+S, 0.75–1.5 μm. One-way analysis of variance followed by Tukey's post hoc test.
DISCUSSION
Previous research in our laboratory has demonstrated in cultured cerebellar granule neurons that Purkinje neurons up-regulate autophagy and undergo cell death when trophic factors are removed from their medium. Factors that block the accumulation of large autophagic vesicles, i.e. 3-methyladenine and expression of DN-FADD (dominant negative Fas-associated death domain protein), blocked cell death, suggesting that the accumulation of vesicles per se contributes to abnormal neuronal function and death (31, 34). Here, we report that IGF-I, a potent neurotrophic factor in vivo, also blocks the accumulation of large autophagic vesicles and prevents the subsequent cell death of Purkinje neurons, again supporting the notion that the accumulation of large autophagic vacuoles causes neuronal death. The effect of IGF-I on the accumulation of large vacuoles was not due to changes in autophagosome formation as the induction of LC3-II expression and the total number of RFP-LC3 labeled autophagosomes (in the presence of bafilomycin A1) formed at 6 h was similar in the absence or presence of IGF-I. Treating neurons with the lysosomal degradation inhibitor, bafilomycin A1, in the presence of IGF-I, abolished the IGF-I effect on autophagic vacuole accumulation, suggesting that IGF-I prevents vacuole accumulation by increasing their turnover. Indeed, a time-course of autophagosome-to-lysosome fusion demonstrated that in the presence of IGF-I, fusion rates were significantly higher than control at 6 h and lower than control by 24 h, indicating that IGF-I regulates autophagic flux.
Recent papers have provided additional insight regarding the molecular machinery that drives the autophagy pathway. Of particular importance is the role that Atg8, the yeast homolog of LC3, may play in autophagosome expansion. Xie et al. (51) demonstrate that differential levels of Atg8 expression correlates directly with autophagosome size, such that lower levels of Atg8 form smaller autophagosomes without affecting the total number of autophagosomes formed. In their model of autophagosome formation, Atg8 is specifically recruited to the pre-autophagosomal structure (PAS) to regulate PAS expansion and autophagosome size and is released from the PAS before autophagosome maturation. These data provide strong evidence that changes in autophagosome size are indicators of autophagy initiation. The vesicle size distribution data presented in this paper confirms the observations made by Xie et al. (51) in that removal of trophic support, a classic inducer of autophagy, significantly increased the size distribution of autophagic vesicles.
Measuring the vesicle size distribution and LC3-II levels (via total counts of RFP+ autophagosomes) in the absence and presence of bafilomycin A1 provides additional information on autophagic flux. In the presence of bafilomycin A1, the effect of IGF-I on vesicle size and accumulation is no longer apparent, suggesting that IGF-I may prevent vacuole accumulation by increasing vesicle turnover. The apparent decrease in total autophagosomal vesicles per Purkinje neuron in the presence of IGF-I compared with 5K-S alone when degradation is not blocked also supports the role of IGF-I in preventing vesicle accumulation. Furthermore, when these two conditions (5K-S in the absence or presence of IGF-I) are compared in the presence of bafilomycin A1, there is no significant difference in the total number of autophagosomes per Purkinje neuron, demonstrating that IGF-I does not regulate the formation but, rather, the turnover of vesicles.
Our data describing the ability of IGF-I to regulate vesicular fusion are supported by a recent publication by Yamamoto et al. (52), which demonstrated an IGF-I-regulated increase in the autophagy-mediated degradation and clearance of accumulated mutant exon 1htt, a mutant form of the huntingtin exon 1 protein. This IGF-I-mediated increased autophagic clearance of mutant exon1htt occurred in the presence of a stimulated class I phosphatidylinositol 3-kinase pathway, which included both Akt and mTOR activation, a pathway generally thought to inhibit starvation-induced autophagy (53). Yamamoto et al. (52) along with our data showing that regulation of autophagic vacuole turnover by IGF-I requires a phosphatidylinositol 3-kinase-Akt-mTOR pathway appears inconsistent with growth factor-mediated activation of the phosphatidylinositol 3-kinase-Akt-mTOR pathway as a repressor of autophagy induction (45, 54, 55). One would have predicted that IGF-I would block autophagosome formation. On the other hand, the observed effect of IGF-I on autophagy fusion and turnover would allow for a more rapid degradation and recycling of cellular protein under conditions of continued trophic factor deficiency.
This raises an interesting question on the mechanism by which the Akt/mTOR pathway regulates autophagy, which at large remains unclear. In yeast, TOR has been shown to inhibit autophagy by regulating Apg13 phosphorylation and repressing Atg8 expression, two essential proteins for autophagy induction and autophagosome membrane expansion, respectively (41, 56, 57). To date, the mammalian ortholog of yeast Atg13 or its interacting protein, Atg1, have not been identified. Considering the greater complexity with higher eukaryotes compared with yeast, it remains possible that mTOR may target autophagy-related proteins other than those required for autophagosome formation.
Clearly, the regulation of autophagy is complex, and further work is required to elucidate mechanistically how IGF-I regulates autophagic vacuole turnover by examining downstream targets of the IGF-I/Akt/mTOR pathway. It will also be interesting to determine whether other neurotrophic factors regulate autophagy in a similar manner. These studies may reveal new potential targets for the treatment or prevention of neurodegenerative diseases.
Acknowledgments
We are grateful to Dr. Aviva M Tolkovsky for the generous gift of adeno-RFP-LC3, Dr. Andrew Thorburn for helpful discussions and critically reviewing the manuscript, and Janna Mize-Berge for technical help. Molecular Biology Core Services were supported by National Institutes of Health Diabetes Endocrinology Research Center Grant P30-DK57516.
This work was supported, in whole or in part, by National Institutes of Health Grants NS045560 and 1F32NS062534-01A1 (NINDS). This work was also supported by a Department of Veterans Affairs merit awards and Department of Defense Grant 03281009 (United States Army Medical Research and Material Command).
- IGF-I
- insulin-like growth factor I
- PBS
- phosphate-buffered saline
- TBS-T
- Tris-buffered saline with Tween
- RFP
- red fluorescent protein
- ERK
- extracellular signal-regulated kinase
- Baf A1
- bafilomycin A1.
REFERENCES
- 1.Levine B., Klionsky D. J. (2004) Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 2.Levine B., Deretic V. (2007) Nat. Rev. Immunol. 7, 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee H. K., Iwasaki A. (2008) Curr. Opin. Immunol. 20, 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuervo A. M. (2004) Mol. Cell. Biochem. 263, 55–72 [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N. (2007) Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 6.Xie Z., Klionsky D. J. (2007) Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 7.Klionsky D. J., Cuervo A. M., Dunn W. A., Jr., Levine B., van der Klei I., Seglen P. O. (2007) Autophagy 3, 413–416 [DOI] [PubMed] [Google Scholar]
- 8.Majeski A. E., Dice J. F. (2004) Int. J. Biochem. Cell Biol. 36, 2435–2444 [DOI] [PubMed] [Google Scholar]
- 9.Massey A. C., Zhang C., Cuervo A. M. (2006) Curr. Top. Dev. Biol. 73, 205–235 [DOI] [PubMed] [Google Scholar]
- 10.Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cataldo A. M., Hamilton D. J., Barnett J. L., Paskevich P. A., Nixon R. A. (1996) J. Neurosci. 16, 186–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nixon R. A., Cataldo A. M., Mathews P. M. (2000) Neurochem. Res. 25, 1161–1172 [DOI] [PubMed] [Google Scholar]
- 13.Anglade P., Vyas S., Javoy-Agid F., Herrero M. T., Michel P. P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E. C., Agid Y. (1997) Histol. Histopathol. 12, 25–31 [PubMed] [Google Scholar]
- 14.Kegel K. B., Kim M., Sapp E., McIntyre C., Castaño J. G., Aronin N., DiFiglia M. (2000) J. Neurosci. 20, 7268–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boellaard J. W., Kao M., Schlote W., Diringer H. (1991) Acta Neuropathol. 82, 225–228 [DOI] [PubMed] [Google Scholar]
- 16.Jeffrey M., Scott J. R., Williams A., Fraser H. (1992) Acta Neuropathol. 84, 559–569 [DOI] [PubMed] [Google Scholar]
- 17.Zhu J. H., Guo F., Shelburne J., Watkins S., Chu C. T. (2003) Brain Pathol. 13, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravikumar B., Duden R., Rubinsztein D. C. (2002) Hum. Mol. Genet. 11, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 19.Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. (2003) J. Biol. Chem. 278, 25009–25013 [DOI] [PubMed] [Google Scholar]
- 20.Ravikumar B., Vacher C., Berger Z., Davies J. E., Luo S., Oroz L. G., Scaravilli F., Easton D. F., Duden R., O'Kane C. J., Rubinsztein D. C. (2004) Nat. Genet. 36, 585–595 [DOI] [PubMed] [Google Scholar]
- 21.Nixon R. A., Wegiel J., Kumar A., Yu W. H., Peterhoff C., Cataldo A., Cuervo A. M. (2005) J. Neuropathol. Exp. Neurol. 64, 113–122 [DOI] [PubMed] [Google Scholar]
- 22.Yu W. H., Cuervo A. M., Kumar A., Peterhoff C. M., Schmidt S. D., Lee J. H., Mohan P. S., Mercken M., Farmery M. R., Tjernberg L. O., Jiang Y., Duff K., Uchiyama Y., Näslund J., Mathews P. M., Cataldo A. M., Nixon R. A. (2005) J. Cell Biol. 171, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin W. L., Lewis J., Yen S. H., Hutton M., Dickson D. W. (2003) J. Neurocytol. 32, 1091–1105 [DOI] [PubMed] [Google Scholar]
- 24.Sikorska B., Liberski P. P., Giraud P., Kopp N., Brown P. (2004) Int. J. Biochem. Cell Biol. 36, 2563–2573 [DOI] [PubMed] [Google Scholar]
- 25.Fessatidis I. T., Thomas V. L., Shore D. F., Hunt R. H., Weller R. O., Goodland F., Rowe D., Venetikou M. V., Bloom S. R. (1993) Eur. J. Cardiothorac. Surg. 7, 465–473 [DOI] [PubMed] [Google Scholar]
- 26.Barenberg P., Strahlendorf H., Strahlendorf J. (2001) Neurosci. Res. 40, 245–254 [DOI] [PubMed] [Google Scholar]
- 27.Skinner P. J., Vierra-Green C. A., Clark H. B., Zoghbi H. Y., Orr H. T. (2001) Am. J. Pathol. 159, 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo J., De Jager P. L., Takahashi K. A., Jiang W., Linden D. J., Heintz N. (1997) Nature 388, 769–773 [DOI] [PubMed] [Google Scholar]
- 29.Selimi F., Lohof A. M., Heitz S., Lalouette A., Jarvis C. I., Bailly Y., Mariani J. (2003) Neuron 37, 813–819 [DOI] [PubMed] [Google Scholar]
- 30.Yue Z., Horton A., Bravin M., DeJager P. L., Selimi F., Heintz N. (2002) Neuron 35, 921–933 [DOI] [PubMed] [Google Scholar]
- 31.Florez-McClure M. L., Linseman D. A., Chu C. T., Barker P. A., Bouchard R. J., Le S. S., Laessig T. A., Heidenreich K. A. (2004) J. Neurosci. 24, 4498–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D'Mello S. R., Galli C., Ciotti T., Calissano P. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10989–10993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bains M., Heidenreich K. A. (2009) Methods Enzymol. 453, 145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linseman D. A., McClure M. L., Bouchard R. J., Laessig T. A., Ahmadi F. A., Heidenreich K. A. (2002) J. Biol. Chem. 277, 24546–24553 [DOI] [PubMed] [Google Scholar]
- 35.Precht T. A., Phelps R. A., Linseman D. A., Butts B. D., Le S. S., Laessig T. A., Bouchard R. J., Heidenreich K. A. (2005) Cell Death Differ 12, 255–265 [DOI] [PubMed] [Google Scholar]
- 36.Linseman D. A., Phelps R. A., Bouchard R. J., Le S. S., Laessig T. A., McClure M. L., Heidenreich K. A. (2002) J. Neurosci. 22, 9287–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galli C., Meucci O., Scorziello A., Werge T. M., Calissano P., Schettini G. (1995) J. Neurosci. 15, 1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Mello S. R., Borodezt K., Soltoff S. P. (1997) J. Neurosci. 17, 1548–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fass E., Shvets E., Degani I., Hirschberg K., Elazar Z. (2006) J. Biol. Chem. 281, 36303–36316 [DOI] [PubMed] [Google Scholar]
- 40.Huang W. P., Scott S. V., Kim J., Klionsky D. J. (2000) J. Biol. Chem. 275, 5845–5851 [DOI] [PubMed] [Google Scholar]
- 41.Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. (2004) J. Cell Sci. 117, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 44.Dudek H., Datta S. R., Franke T. F., Birnbaum M. J., Yao R., Cooper G. M., Segal R. A., Kaplan D. R., Greenberg M. E. (1997) Science 275, 661–665 [DOI] [PubMed] [Google Scholar]
- 45.Arico S., Petiot A., Bauvy C., Dubbelhuis P. F., Meijer A. J., Codogno P., Ogier-Denis E. (2001) J. Biol. Chem. 276, 35243–35246 [DOI] [PubMed] [Google Scholar]
- 46.Scarlatti F., Bauvy C., Ventruti A., Sala G., Cluzeaud F., Vandewalle A., Ghidoni R., Codogno P. (2004) J. Biol. Chem. 279, 18384–18391 [DOI] [PubMed] [Google Scholar]
- 47.Saeki K., Hong Z., Nakatsu M., Yoshimori T., Kabeya Y., Yamamoto A., Kaburagi Y., Yuo A. (2003) J. Leukocyte Biol. 74, 1108–1116 [DOI] [PubMed] [Google Scholar]
- 48.Kanazawa T., Taneike I., Akaishi R., Yoshizawa F., Furuya N., Fujimura S., Kadowaki M. (2004) J. Biol. Chem. 279, 8452–8459 [DOI] [PubMed] [Google Scholar]
- 49.di Mari J. F., Davis R., Safirstein R. L. (1999) Am. J. Physiol. 277, F195–203 [DOI] [PubMed] [Google Scholar]
- 50.Remacle-Bonnet M. M., Garrouste F. L., Heller S., André F., Marvaldi J. L., Pommier G. J. (2000) Cancer Res. 60, 2007–2017 [PubMed] [Google Scholar]
- 51.Xie Z., Nair U., Klionsky D. J. (2008) Mol. Biol. Cell 19, 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto A., Cremona M. L., Rothman J. E. (2006) J. Cell Biol. 172, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meijer A. J., Codogno P. (2004) Int. J. Biochem. Cell Biol. 36, 2445–2462 [DOI] [PubMed] [Google Scholar]
- 54.Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P. (2000) J. Biol. Chem. 275, 992–998 [DOI] [PubMed] [Google Scholar]
- 55.Klionsky D. J. (2005) J. Cell Sci. 118, 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000) J. Cell Biol. 150, 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reggiori F., Klionsky D. J. (2002) Eukaryot. Cell 1, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]