Abstract
G protein-coupled receptors mediate cellular responses to a wide variety of stimuli, including taste, light and neurotransmitters. In the yeast Saccharomyces cerevisiae, activation of the pheromone pathway triggers events leading to mating. The view had long been held that the G protein-mediated signal occurs principally at the plasma membrane. Recently, it has been shown that the G protein α subunit Gpa1 can promote signaling at endosomes and requires two components of the sole phosphatidylinositol-3-kinase in yeast, Vps15 and Vps34. Vps15 contains multiple WD repeats and also binds to Gpa1 preferentially in the GDP-bound state; these observations led us to hypothesize that Vps15 may function as a G protein β subunit at the endosome. Here we show an X-ray crystal structure of the Vps15 WD domain that reveals a seven-bladed propeller resembling that of typical Gβ subunits. We show further that the WD domain is sufficient to bind Gpa1 as well as to Atg14, a potential Gγ protein that exists in a complex with Vps15. The Vps15 kinase domain together with the intermediate domain (linking the kinase and WD domains) also contributes to Gpa1 binding, and is necessary for Vps15 to sustain G protein signaling. These findings reveal that the Vps15 Gβ-like domain serves as a scaffold to assemble Gpa1 and Atg14, whereas the kinase and intermediate domains are required for proper signaling at the endosome.
G protein-coupled receptors are highly conserved in organisms from yeast to humans. Upon stimulation, the receptor activates a G protein heterotrimer, causing the Gα subunit to release GDP, bind GTP, and dissociate from the Gβγ heterodimer. Following dissociation, the Gα and Gβγ subunits are able to activate downstream effector proteins (1-3). Examples include adenylyl cyclase, MAP kinases, phosphatidylinositol (PtdIns) 3-kinase, and phospholipase C-β (4).
In simple eukaryotes like yeast, G protein-coupled receptors mediate processes essential for proper cell functioning including mating. MATa and MATα cell types secrete small peptide pheromones, called a-factor and α-factor respectively, that promote cell fusion and the formation of an a/α diploid. Activation of the pathway by pheromone leads to phosphorylation of various proteins that mediate new gene transcription, altered cell morphology, cell division arrest, and eventually cell fusion. Gβγ (Ste4/Ste18) initiates the pathway that originates from the plasma membrane and results in activation of the MAP kinases Fus3 and Kss1 (2). Gα (Gpa1) initiates a parallel pathway that operates at the endosomal membrane and results in activation of Fus3 in preference to Kss1 (5). The pathway originating from the endosome requires Vps15 and Vps34, the regulatory and catalytic subunits of the only PtdIns 3-kinase known in yeast (5-12). Further, Vps15 directly binds the unactivated (GDP-bound) form of Gpa1, while Vps34 directly engages the activated (GTP-bound form) of Gpa1. Additionally, both Vps15 and Vps34 are necessary for recruitment of Gpa1 to endosomes (5). G protein activation of Vps34 results in elevated production of PtdIns 3-P, and leads to the endosomal recruitment of phospholipid binding proteins (5).
Vps15 is an extremely large protein (166 kDa) and contains several distinct domains. These include an N-terminal serine/threonine kinase domain (6, 8, 11, 12), a C-terminal WD-repeat containing domain (5, 13), and an internal domain that bridges the kinase- and WD-domains. It has been shown previously that the kinase activity of Vps15 is necessary for full activation of the G protein signal (5). However, the precise role of the kinase domain including any substrates is unknown. The internal domain contains several HEAT repeats that have been shown to be important for protein-protein interactions (hereafter referred to as the intermediate domain or ID) (13-15). The WD domain was of particular interest to us because of the recognized importance of the WD-containing Gβ protein, Ste4, in the pheromone-mediated pathway that originates from the cell plasma membrane.
Here, we examine the roles of the different domains of Vps15 in binding to Gpa1, as well as in transmitting the G protein signal. We demonstrate that the WD-domain of Vps15 has a seven-bladed propeller structure like that of known Gβ proteins. We show further that the Gβ-like domain acts in the manner of a scaffold to bring together proteins that propagate the pheromone response at the endosome. Finally, we show that it is the kinase domain with the intermediate domain that is necessary for Vps15 to transmit the G protein signal leading to activation of Fus3 as well as for other Vps15 signaling functions in the cell.
EXPERIMENTAL PROCEDURES
Protein expression and purification
The coding sequence of Vps15 WD repeat domain (residues 1027-1454) was amplified by PCR from S. cerevisiae BY4741 genomic DNA and inserted into a modified pFastBac vector through ligation independent cloning (16). Baculovirus coding for production of recombinant His6-Vps15 WD repeat domain was prepared according to the Bac-to-Bac method (Invitrogen). Two liters of High-Five insect cells cultured in Express Five SFM (Gibco) to a density of 2 × 106 cells/ml were infected with the baculovirus for 48 hours at 27 °C.
Cells were harvested by centrifugation at 4,400 x g and resuspended in buffer A [20 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole, 10% (v/v) glycerol, 0.02% NaN3] with Complete EDTA-free protease inhibitor cocktail (Roche) and PMSF. The cells were lysed with three passes through an Emulsiflex C5 homogenizer (Avestin) operating at 15,000 psi, and the lysate was clarified by centrifugation at 105,000 x g. The resulting supernatant was filtered through a 0.45 μm low protein binding filter and applied to a 1 ml HisTrap HP column (GE Healthcare). The column was washed with 30 column volumes (CV) of buffer A followed by 30 CV of 5% buffer B (buffer A containing 1M imidazole). His6-Vps15 WD repeat domain was eluted with a step gradient of 30% buffer B. To prevent aggregation during the elution, fractions of 1 ml were collected into tubes containing 2 ml buffer C [20 mM Tris pH 8.0, 300 mM NaCl, 2 mM EDTA, 10% glycerol (v/v), 2 mM DTT, 0.02% (w/v) NaN3]. The fractions containing the protein were pooled immediately and applied to a Sephacryl S-300 26/60 column (GE Healthcare) equilibrated with buffer D (buffer C without EDTA). Vps15 WD repeat domain was eluted with buffer D and concentrated to 11 mg/ml in a Centriprep YM10 centrifugal concentrator (Millipore). Aliquots of the protein were quick frozen in liquid nitrogen and stored at -80 °C.
Crystallization and data collection
Crystals of the Vps15 WD repeat domain were grown at 18 °C by macroseeding small crystals into microbatch setups. The batch solution was prepared by quickly mixing the protein (6 mg/ml) with an equal volume of precipitant solution composed of 1.4 M ammonium malonate pH 6.5, 100 mM sodium acetate pH 4.6, 4% (v/v) isopropanol. Crystals suitable for x-ray diffraction were obtained within 7–10 days. Prior to data collection, crystals were cryoprotected by stepwise transfer into 20% (v/v) ethylene glycol in synthetic mother liquor (1.2 M ammonium malonate pH 6.5, 100 mM sodium acetate pH 4.6, 2% isopropanol), after which they were suspended in loops (Hampton Research) and quick frozen by immersion into liquid nitrogen. The crystals belong to the space group P3121, with unit cell dimensions of a = b = 86.5 Å, c = 132.8 Å, and one molecule in the asymmetric unit.
Crystals used in derivative screening were stabilized for several hours in synthetic mother liquor prior to soaking in heavy atom solutions. The gold derivative ultimately used to determine the phases was prepared with a 10 min soak in 5.8 mM KAu(CN)2. A single wavelength anomalous dispersion data set (λ = 11.917 keV) was collected from a single crystal at the SER-CAT 22-BM beamline at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). A native data set was subsequently collected from a single crystal at the SER-CAT 22-ID beamline. All data were processed and scaled with HKL2000 (17). Relevant data collection and processing statistics are listed in Table 1.
Table 1.
Data collection parameters and processing statistics. Values in parenthesis refer to the highest resolution shell.
| Native | KAu(CN)2 | |
|---|---|---|
| Unit Cell (□) | a, b = 85.5, c = 132.8 | a, b = 86.0, c = 132.8 |
| Resolution (□) | 50.0Š1.80 | 50.0Š2.20 |
| Observed reflections | 518207 | 637020 |
| Unique reflections | 50964 | 29652 |
| I/σ(I) | 36.0 (1.6) | 39.2 (12.3) |
| Completeness (%) | 96.5 (71.2) | 100 (100) |
| Redundancy | 10.2 (6.8) | 21.5 (22.0) |
| Rsym (%) | 8.1 (52.7) | 9.7 (24.9) |
Phasing, model building and refinement
The positions of the gold atoms in the derivative crystal were found using the program SHELXD (18), and the correct hand for the coordinates was determined with SHELXE. Refinement of the gold atom coordinates and computation of the phases was performed with the CCP4 program MLPHARE, and the phases were subsequently improved with solvent flattening and histogram matching using the method of (19). The initial electron density map was easily interpretable, and 319 of the 437 residues in the protein were automatically fitted with the program ARP/wARP (20). Additional residues were build into the map using COOT (21). The structure of the His6-Vps15 WD repeat domain was completed using native protein data and iterative cycles of model building and refinement with COOT and the CCP4 program REFMAC (22). The final model has a crystallographic R-value of 20.9% (R-free 24.6%) for all data from 50-1.8 Å and includes residues 1031-1040, 1042-1386, 1389-1410, and 1424-1454. No electron density was present for the N-terminal histidine tag or Vps15 residues 1027-1030, 1041, 1387, 1388, and 1411-1423. In the Ramachandran plot, 96.2% of the residues are in the most favored region, 3.0 % are in the generously allowed region, and 0.8% are in the disallowed region. Relevant phasing and refinement statistics are listed in Table 2, and representative electron density is shown in Supporting Figure 1.
Table 2.
Phase calculation and refinement statistics. Values in parenthesis refer to the highest resolution shell.
| Phasing | |
| Resolution (□) | 50Š2.5 |
| Number of sites (SHELX) | 4 |
| FOM after MLPHARE | 0.284 |
| Anomalous Rcullisa | 0.76 (0.82) |
| FOM after DM | 0.819 |
| Refinement | |
| Resolution (□) | 50Š1.80 |
| Rwork/Rfree (%) | 20.9/24.6 |
| Number of atoms | |
| Protein | 3163 |
| Water | 200 |
| Average B-factor (□2) | |
| Protein | 35.4 |
| Water | 41.0 |
| R.m.s. deviations | |
| Bond lengths (□) | 0.014 |
| Bond angles (□) | 1.524 |
Rcullis = lack of closure/anomalous difference
Strains and Plasmids
Standard methods for the growth, maintenance, and transformation of yeast and bacteria and for the manipulation of DNA were used throughout. Yeast Saccharomyces cerevisiae strains used in this study are BY4741 (MATa leu2Δ met15Δ his3Δ ura3Δ) and BY4741-derived gene deletion mutants (Invitrogen). The vps15Δ strain used is described in (5).
Previously described yeast shuttle vectors and expression plasmids are pRS315-ADH-Flag (CEN, ampR, LEU2, ADH1 promoter/terminator and either a Flag epitope that can be added 5′ with a SacI site or 3′ with an XmaI site), pRS316-ADH-Myc (CEN, ampR, URA3, ADH1 promoter/terminator and either a Myc epitope that can be added 5′ with a SacI site or 3′ with an XmaI site) (23), pAD4M-GST, pAD4M-Gpa1-GST, pRS316-ADH-Gpa1-2myc (24), pAD4M-Gpa2-GST (25), pRS423-FUS1-LacZ (26). Truncations of Vps15 were designed based upon the crystal structure of the WD domain of Vps15 (Figure 1) as well as domain predictions from SMART and three different secondary structure predictions (27-32). For naming purposes, Vps15 is considered to possess 3 domains: the kinase domain (KD) from residues 1-294, the WD domain (WD) from residues 1027-1454 and the intermediate domain (ID) spanning the region between the kinase domain and the WD domain. Plasmids pRS315-ADH-Flag-Vps15KD (using primers Vps15fn and Vps15ksrn), pRS315-ADH-Flag-Vps15KD,ID (using primers Vps15fn and Vps15klrn), pRS315-ADH-Flag-Vps15ID,WD (using primers Vps15wdlfn and Vps15rn), pRS315-ADH-Flag-Vps15WD (using primers Vps15wdsfn and Vps15rn), pRS316-ADH-Myc-Vps15ID,WD(using primers Vps15wdlfn and Vps15rn), and pRS316-ADH-Myc-Vps15WD (using primers Vps15wdsfn and Vps15rn) were constructed by PCR amplification of VPS15 from BY4741 genomic DNA, incorporation of the product into a TOPO vector (Invitrogen), digestion by SacI and ligation into pRS315-ADH-Flag or pRS316-ADH-Myc. Plasmids pRS315-ADH-Vps15KD,ID,WD-Flag (using primers Vps15fc and Vps15rc), pRS315-ADH-Vps15ID,WD-Flag (using primers Vps15wdlfc and Vps15rc), pRS315-ADH-Vps15WD-Flag (Vps15wdsfc and Vps15rc) , pRS316-ADH-Vps15WD-Myc (using primers Vps15wdsfc and Vps15rc) were constructed by PCR amplification of VPS15 from BY4741 genomic DNA, incorporation into a TOPO vector (Invitrogen), digestion by XmaI and ligation into pRS315-ADH-Flag or pRS316-ADH-Myc. Plasmids pRS315-ADH-Flag-Atg14 (using primers Atg14fn and Atg14rn) and pRS316-ADH-Myc-Atg14 (using primers Atg14fn and Atg14rn) were generated through PCR amplification of ATG14 from BY4741 genomic DNA, incorporation of the product into TOPO vector, digestion by SacI, and ligation into pRS315-ADH-Flag or pRS316-ADH-Myc. pRS316-ADH-3Myc-Atg14 was constructed by the annealing of two homologous oligonucleotides incorporating the sequence of 2 Myc epitopes as well as overhangs homologous to BamHI and SalI restriction sites (using the primer 2myc and its complement). pRS316-ADH-Myc-Atg14 was then digested with BamHI and SalI and ligated with the annealed oligonucleotides. Myc-Vps15WD R1261A (using primer Vps15R1261A and its complement), Myc-Vps15WD R1261K (Vps15R1261K and its complement), Myc-Vps15ID,WD R1261A (Vps15R1261A and its complement), Myc-Vps15ID,WD R1261K (Vps15R1261K and its complement), Vps15KD,ID,WD R1261A Flag (Vps15R1261A and its complement), and Vps15KD,ID,WD R1261K Flag (Vps15R1261K and its complement) in pRS316-ADH-Myc and pRS315-ADH-Flag respectively were created using QuikChange (Stratagene). Plasmid pRS306 Vps15KD,ID,WD R1261A-Flag was created by digestion of the pRS316 constructs with BamHI and ligation into the corresponding BamHI site of pRS306. Genomic integration was performed using HpaI. All plasmids described were confirmed by sequencing.
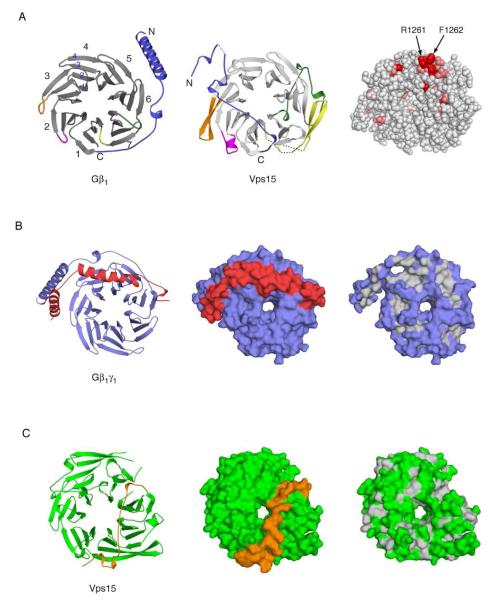
Figure 1. Structure of the Vps15 WD repeat domain and comparison to the beta subunit of transducin (Gβ1).
(A) Left: ribbon diagram of Gβ1. Blades of the propeller are numbered consecutively, as are the strands of one blade. Center: ribbon diagram of Vps15 WD repeat domain, in the same orientation as Gβ1. Breaks within the structure are indicated by dotted lines. The first break occurs at residue 1041; the short stretch of amino acids visible before this break are shown in this part of the figure but deleted in others. Colored regions highlight major differences in the propeller’s structure: blue, N-terminus; magenta, insertion between WD repeats 1 and 2; gold, insertion between WD repeats 2 and 3; green, insertion within WD repeat 6; yellow, insertion within WD repeat 7. The N-terminus and corresponding loop regions of Gβ1 are colored similarly. Right: space filling model of Vps15 WD repeat domain (same orientation) with residues highly conserved across genera colored salmon, and those which are invariant colored red. Arg-1261 and Phe-1262 reside on the surface in the loop between WD repeats 4 and 5. (B) Left: ribbon diagram of Gβγ oriented to highlight the interaction between the beta and gamma (red) subunits. Center: surface representation of Gβγ in the same orientation. Right: surface representation of Gβ1 alone, with hydrophobic residues colored light grey to show the binding cavity for Gγ1. (C) Left: ribbon diagram of Vps15 WD repeat domain oriented as Gβ1 in panel B. The N-terminal loop is colored gold. Center: surface rendering to illustrate partial occlusion of the potential gamma subunit binding surface. Right: surface representation following removal of the N-terminal loop, with hydrophobic residues colored light grey to reveal the absence of a binding cavity for a gamma-like subunit.
Purification of Flag Epitope Tagged Proteins
Plasmids pRS315-ADH-Flag-Vps15KD, pRS315-ADH-Flag-Vps15KD,ID, pRS315-ADH-Vps15KD,ID,WD-Flag, pRS315-ADH-Flag-Vps15ID,WD, pRS315-ADH-Vps15ID,WD-Flag, pRS315-ADH-Flag-Vps15WD, pRS315-ADH-Vps15WD-Flag, and pRS315-ADH-Flag, were transformed into either wildtype or atg14Δ cells expressing pRS316-ADH-Myc, pRS316-ADH-Vps15WD-Myc, pRS316-ADH-Gpa1-2Myc, or pRS316-ADH-Myc-Atg14. A 50 ml culture of cells containing each plasmid was grown in SCD-Ura-Leu until A600 nm∼ 0.8. Cells were disrupted in buffer E (10 mM Tris-Cl pH 8.0, 200 mM NaCl, 0.1% Triton X-100, 25 mM beta-glycerophosphate, 1 mM EDTA, 2 protease inhibitor cocktail pellets per 50 ml (Roche)) for 15 minutes at 4°C using a vortex fitted with a 60-microtube headpiece. Lysates were then combined at 4°C with gentle rotation for 25 minutes (this step omitted in several experiments) to ensure solubilization of membrane bound proteins. The lysates were then centrifuged at 16,000 x g for 1 minute, and then again for 25 minutes. Protein determination was performed for all samples, and each lane contains an equal total concentration and volume of protein. The supernatant was mixed with 40 μl of Flag affinity resin (Sigma-Aldrich) that had been equilibrated with buffer E. The lysates and beads were mixed with gentle rotation for 2 hours at 4°C. The beads were washed five times with 500 μl of buffer E. Bound proteins were then eluted from the beads with two 20 μl washes of 0.25 mg/ml 3X Flag Peptide (Sigma-Aldrich) and collected by centrifugation at 4,900 × g for 1 minute. 6X SDS buffer was added to the eluted proteins to a final concentration of 1X and samples were heated at 95°C for 5 minutes. Samples were resolved by 10% SDS-PAGE or 7.5% SDS-PAGE and immunoblotting with anti-Myc 9E10 cell culture supernatant at a dilution of 1:100 (33), anti-Flag mouse monoclonal M2 antibodies at a dilution of 1:1,000 (Sigma-Aldrich), anti-G6PDH antibodies at a dilution of 1:50,000 (Sigma-Aldrich), and the secondary antibodies goat anti-mouse or goat anti-rabbit at a dilution of 1:10,000 (Biorad). Detection was performed with chemiluminescent ECL substrate (PerkinElmer).
Purification of Glutathione S-transferase (GST) Fusion Proteins
pRS316-ADH-Myc-Vps15WD R1261A, pRS316-ADH-Myc-Vps15WD R1261K, pRS316-ADH-Myc-Vps15ID,WD R1261A, pRS316-ADH-Myc-Vps15ID,WD R1261K, and pRS316-ADH-Myc were co-transformed into wildtype strains with either pRS315-ADH-Flag, pAD4M-Gpa1-GST, or pAD4M-Gpa2-GST. Cells were disrupted in buffer F (200 mM Tris pH 7.6, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100 and 2 protease inhibitor cocktail pellets per 50 ml) and prepared in a manner similar to that described for purification of Flag epitope tagged proteins, except that the lysis time was decreased to 10 minutes. For determination of nucleotide specificity of binding, either 5 mM MgCl2 and 10 μM GDP or 10 mM NaF, 5 mM MgCl2, 10 μM GDP, and 30 μM AlCl3 (GDP-AlF4-) was added to buffer F. Supernatants were mixed with 40 μl of glutathione-Sepharose resin (GE Healthcare) equilibrated with buffer F. The glutathione-Sepharose resin was incubated with the lysate and washed in a manner similar to that described above for the immunoprecipitation of Flag-tagged proteins. Bound proteins were then eluted from the beads with two 20 μl washes of 10 mM reduced glutathione, 50 mM Tris pH 8.0 and were collected by centrifugation at 4,900 x g for 1 minute. 6X SDS buffer was added to the eluted proteins to a final concentration of 1X and samples were heated at 95°C for 5 minutes. Samples were resolved by 10% SDS-PAGE and immunoblotting with anti-GST polyclonal antibody at a dilution of 1:1,500 (a gift of Joan Steitz), anti-Myc 9E10 monoclonal antibodies at a dilution of 1:100, anti-G6PDH antibodies at a dilution of 1:50,000 (Sigma-Aldrich), anti-Ste4 antibodies at a dilution of 1:2000 (a gift of Duane Jenness), and the secondary antibodies goat anti-mouse or goat anti-rabbit at a dilution of 1:10,000 (Biorad). Detection was performed with chemiluminescent ECL substrate (PerkinElmer).
Cell Extract Preparation and Analysis for MAP kinase Phosphorylation and Carboxypeptidase Y Sorting
Plasmids pRS315-ADH-Flag-Vps15KD, pRS315-ADH-Flag-Vps15KD,ID, pRS315-ADH-Vps15KD,ID,WD-Flag, pRS315-ADH-Flag-Vps15ID,WD, pRS315-ADH-Vps15ID,WD-Flag, pRS315-ADH-Flag-Vps15WD, pRS315-ADH-Vps15WD-Flag, and pRS315-ADH-Flag, were transformed into either wildtype or vps15Δ cells. Wildtype, vps15Δ, and vps15R1261A strains were also analyzed for MAP kinase phosphorylation and carboxypeptidase Y sorting without any additional plasmid transformation. Cells were grown to A600 nm ∼ 0.8 in 20 ml SCD-Leu and then split and either treated with 3 μM α-MF pheromone or left untreated at 30°C for 30 minutes. The reaction was stopped by addition of 5% trichloroacetic acid, and the cells were harvested by centrifugation and frozen at -80°C. Cell extracts were prepared as previously described (34), and resolved by a 10% SDS-PAGE and immunoblotted using p44/p42 MAP kinase antibody at 1:500 (Cell Signaling Technology) or anti-CPY antibody at 1:2,000 (a gift of Pat Brennwald), anti-G6PDH antibodies at a dilution of 1:50,000 (Sigma-Aldrich), and the secondary antibody goat anti-rabbit at a dilution of 1:10,000 (Biorad). Detection was performed with chemiluminescent ECL substrate (PerkinElmer).
Pheromone Response Assays
Reporter-transcription (β-galactosidase assay utilizing the pRS423-FUS1-lacZ plasmid) and growth arrest (halo) assays were performed as described previously (26, 35).
RESULTS
The structure of the Vps15 WD domain is presented in Figure 1. The protein folds as a seven-bladed propeller as predicted in a prior homology modeling analysis (5). As described previously, the WD repeat units are not equivalent to the blades of the propeller, but are comprised rather of the fourth strand of one blade and the first three strands of the blade that follows (36). The overall architecture of the Vps15 WD domain is similar to that of the transducin G protein β subunit (Gβ1) (36), and superposition of the structures with the CCP4 topological comparison program TOP yields a r.m.s. deviation of 2.7 Å2 for the Cα carbons (37, 38). There are several key differences in the overall structures of the two proteins. The N-terminus of Gβ folds as an α-helix that extends away from the propeller. In the case of Vps15, the corresponding region is largely unstructured and it packs against the bottom surface of the propeller. There are four insertions in the Vps15 propeller (Figure 1A): an extended loop containing a 310 helix between WD repeats 1 and 2 (residues 1110-1121); an extension of strands 3 and 4 of blade 3 (residues 1156-1172); a large loop with 310 helical twists within the sixth WD repeat (residues 1318-1351), and two additional β-strands within the seventh WD repeat (residues 1385-1426).
Heterotrimeric G protein β subunits form obligate heterodimers with Gγ subunits. The interaction between Gβ and Gγ is largely hydrophobic; the first α-helix of the γ subunit forms a coiled-coil interaction with the Gβ N-terminal α-helix, whereas the second α-helix wraps into a cleft on the bottom surface of the Gβ propeller (36) (Figure 1B). As noted above, the N-terminus of the Vps15 WD repeat domain is folded against the bottom of the propeller (Figure 1C), and the stretch of N-terminal residues immediately before the propeller partially occlude a potential Gγ binding surface. However, inspection of the hydrophobic regions on the bottom surface of the propeller (upon removal of the N-terminus) of Vps15 indicates there is no equivalent binding cleft for interaction with a Gγ subunit. This finding provides an explanation for our ability to express, purify and crystallize the Vps15 WD domain in the absence of an associated Gγ.
The G protein α subunit typically interacts with Gβγ through two interfaces, termed the “switch interface” and the “N-terminal interface” (39). The interaction at the switch interface involves residues in and around switch I and switch II of Gα as well as conserved hydrophobic residues on the top surface of the Gβ propeller (Tyr 59, Trp 99, Met 101 and Leu 117 in Gβ1). The N-terminal interface is formed by interaction of the Gα N-terminal α-helix with four conserved residues in blade 1 of the Gβ propeller (Leu 55, Lys78, Ile 80 and Lys 89 in Gβ1). The Vps15 propeller provides neither of these binding surfaces for interaction with a Gα subunit. The region that corresponds to the switch interface is comprised of polar amino acids (Thr 67, Thr 113, Thr 116 and Lys 131). Further, superposition of the Vps15 structure on the Gαβγ heterotrimer suggests the Vps15 residues corresponding to the N-terminal interface (Glu 62, Val 87, Lys 89 and Ser 104) would not form a tight interaction with the Gα N-terminal α-helix (not shown). The inability of the Vps15 WD domain to form these interactions, or the absence of a corresponding Gγ subunit, may explain our failure to isolate a stable heterodimeric complex of Gpa1 with the WD domain-containing fragment of Vps15 (by size exclusion chromatography, not shown).
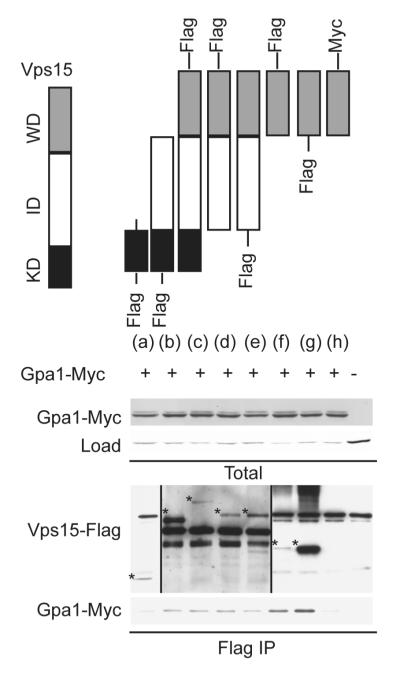
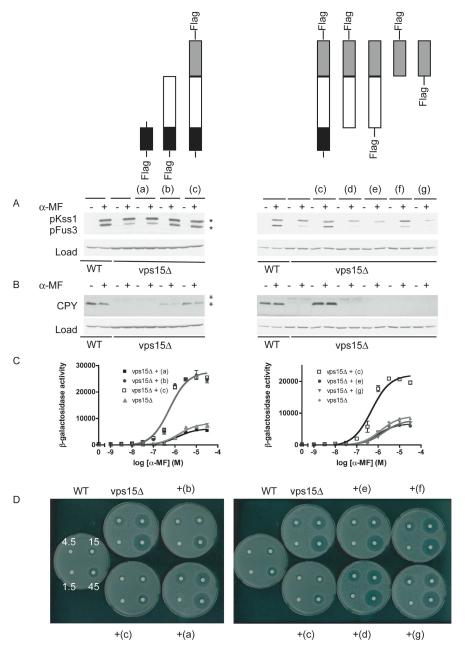
The data presented above reveal the absence of a canonical Gα-binding interface within the Vps15 WD domain. Thus we considered whether other domains of Vps15 might contribute to Gpa1 interactions. In order to determine the possible binding role for these other domains, we co-expressed Gpa1 with several truncations of Vps15 including (a) the kinase domain alone, (b) the kinase domain with the intermediate domain, (c) full length Vps15, (d, e) the intermediate domain with the WD domain, and (f, g) the WD domain alone (Figure 2). Whereas Gpa1 was tagged with the myc epitope (located internally to preserve function), variants of Vps15 were tagged at the N- and C-terminus with the Flag epitope. We eliminated from consideration any Flag-tag fusions that did not express. We then immunoprecipitated Vps15 and monitored the presence of co-precipitating Gpa1 by immunoblotting. Differences in the apparent abundance of Vps15 truncations could reflect differences in actual abundance or more likely differences in the position of the epitope tag (compare lanes f and g, containing Vps15WD-Flag and Flag-Vps15WD respectively) as well as the size of the tagged protein (which could affect the protein transfer and immunoblot detection). As shown in Figure 2, the WD domain of Vps15 is sufficient to bind Gpa1, while the kinase domain alone failed to interact. However the kinase domain with the intermediate domain was capable of binding to Gpa1. These data indicate that the intermediate domain, in addition to the Gβ-like WD domain, of Vps15 contributes to the stable interaction with Gpa1.
Figure 2. The WD domain of Vps15 is sufficient but not necessary to bind Gpa1.
Detergent-solubilized extracts (Total) from cells expressing the indicated Flag fusion proteins and Gpa1 fused to Myc were incubated with Flag resin, eluted with 3X Flag peptide (Flag IP), resolved by 7.5% or 10% SDS-PAGE, and analyzed by immunoblotting with antibodies against Flag, Myc, and G6PDH (Load control). C-terminally tagged forms of (a) and (b) did not express. IP, immunoprecipitation. WD, WD domain. KD, kinase domain. ID, intermediate domain. *, indicates protein of interest.
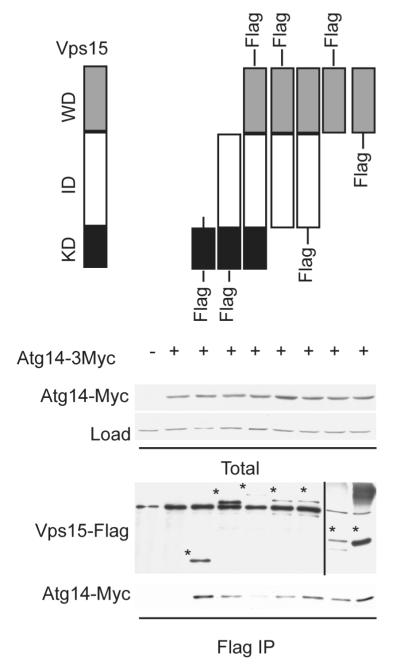
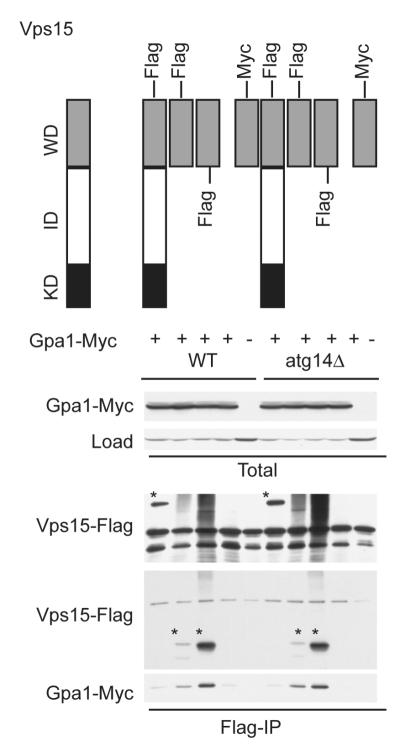
Vps15 is known to exist in a stable multi-protein complex that includes Atg14 or Vps38 (UVRAG in animals), as well as Vps34 and Vps30 (Beclin-1 in animals) (40-45). Atg14 has sequence similarity to canonical Gγ proteins (5). Thus, we investigated if the Vps15 Gβ-like domain binds to the Gγ-like protein Atg14. To this end we immunoprecipitated each of the Vps15 truncation mutants and monitored copurification of Atg14 by immunoblotting, as described above for Gpa1. As shown in Figure 3, the WD domain of Vps15 is indeed sufficient to bind to Atg14. The kinase domain alone was also capable of binding to Atg14 (Figure 3). Typical G protein α subunits bind to the Gγ subunit indirectly, through the common binding partner Gβ. Likewise, Gpa1 bound to the Vps15 WD domain even in the absence of Atg14 expression (Figure 4). Taken together these data indicate that the Vps15 Gβ-like domain exists in a multi-subunit complex with the Gα and Gγ-like proteins.
Figure 3. The WD domain of Vps15 is sufficient but not necessary to bind Atg14.
Detergent-solubilized extracts (Total) from cells expressing the indicated Flag fusion proteins and Atg14 fused to a triple Myc epitope were incubated with Flag resin, eluted with 3X Flag peptide (Flag IP), resolved by 10% SDS-PAGE, and analyzed by immunoblotting with antibodies against Flag, Myc, and G6PDH (Load control). IP, immunoprecipitation. WD, WD domain. KD, kinase domain. ID, intermediate domain. *, indicates protein of interest.
Figure 4. Atg14 is not necessary to mediate the interaction of Vps15 and Gpa1.
Detergent-solubilized extracts from wildtype (WT) and atg14Δ mutant cells expressing the indicated Flag fusion proteins and Gpa1 fused to a Myc epitope were incubated with Flag resin, eluted with 3X Flag peptide, resolved by 10% SDS-PAGE, and analyzed by immunoblotting with antibodies against Flag, Myc, and G6PDH (Load control). IP, immunoprecipitation. WD, WD domain. KD, kinase domain. ID, intermediate domain. *, indicates protein of interest.
Although the crystal structure of the WD-like domain of Vps15 shows that it is comprised of a seven-bladed propeller structure, similar to that of canonical Gβ proteins, this analysis also revealed a lack of conservation in the predicted Gα-binding interface. The lack of a canonical Gα-binding site within Vps15 indicates that interaction must occur in a distinct manner. Accordingly we undertook a molecular evolution analysis to identify potential binding residues within Vps15. We first generated a multiple-sequence alignment of Vps15 orthologs from several fungi, arthropods, mammals and amphibians (Supplemental Data). Invariant and highly conserved residues within this alignment were placed in a spatial context by mapping them onto the crystal structure of Vps15 (far right panel of Figure 1A). This analysis revealed two residues, Arg-1261 and Phe-1262, that are invariant across all orthologs, implying a strong evolutionary pressure to preserve function. In addition, the side chains protrude into the solvent, making them available for protein-protein interactions. Two additional invariant residues were found on the opposite side of the Vps15 propeller, Glu-60 and Ser-104. However these two residues form multiple hydrogen bonds to each other and may therefore be important for structural constraints. We observed no other invariant residues on the surface of Vps15.
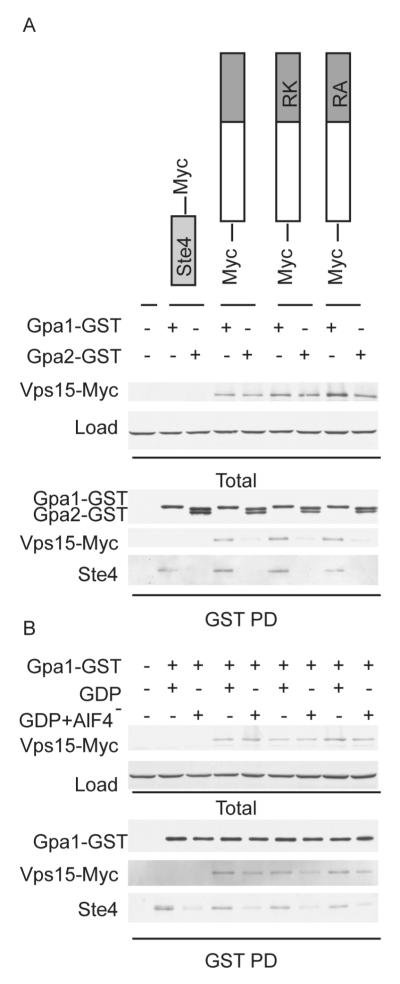
To determine the importance of the conserved Arg-1261, we examined the functional consequences of replacing this residue within Vps15. For these experiments we purified Gpa1 fused to glutathione S-transferase (GST) and monitored co-purification of Vps15 or various Vps15 truncations. Gpa1-GST was used in this case to corroborate the results of the Vps15-immunopreciptation experiments shown above. Additionally, the GST purification method is in our experience better suited for analysis of nucleotide-dependent interactions of G protein α and βγ subunits. Accordingly, the WD domain of Vps15 was co-expressed with Gpa1-GST or with a second G protein α subunit Gpa2-GST, used here as a negative control (Figure 5). Cell lysates were mixed with glutathione-Sepharose resin, washed extensively, eluted with reduced glutathione, and analyzed by immunoblotting using antibodies against GST (to detect purified Gpa1 or Gpa2) and Myc (to detect any co-purifying Vps15). Although Ste4 was tagged with Myc, it did not bind well with an anti-Myc antibody, and was instead detected using an anti-Ste4 antibody (data not shown, see Figure 7). As shown in Figure 5, the Vps15 WD domain copurified with Gpa1 but did not copurify with Gpa2.
Figure 5. Arg-1261 is necessary for the WD domain of Vps15 to bind efficiently to Gpa1.
Detergent-solubilized extracts (Total) from cells expressing the indicated Myc fusion proteins and Gpa1 or Gpa2 fused to GST were incubated with glutathione-Sepharose resin, eluted with glutathione (GST PD, pulldown), resolved by 10% SDS-PAGE, and analyzed by immunoblotting with antibodies against GST, Myc and G6PDH (Load control). PD, pulldown. GST, glutathione S-transferase. WD, WD domain. RA, Arg-1261 to Ala. RK, Arg-1261 to Lys.
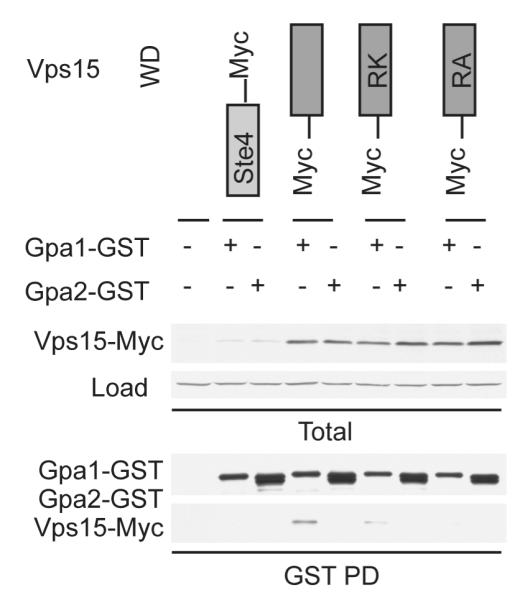
Figure 7. Arg-1261 is not necessary for larger truncations of Vps15 to bind to Gpa1.
(A) Detergent-solubilized extracts (Total) from cells expressing the indicated Myc fusion proteins and Gpa1 or Gpa2 fused to GST were incubated with glutathione-Sepharose resin, eluted with glutathione (GST PD), resolved by 10% SDS-PAGE, and analyzed by immunoblotting with antibodies against Myc, GST, Ste4 and G6PDH (Load control). (B) Detergent-solubilized extracts from cells expressing the indicated Myc fusion proteins and Gpa1 fused to GST were lysed in the presence of either GDP or GDP-AlF4-, incubated with glutathione-Sepharose resin, eluted with glutathione, and analyzed by immunoblotting with antibodies against Myc, GST, Ste 4, and G6PDH. PD, pulldown. GST, glutathione S-transferase. RA, Arg-1261 to Ala. RK, Arg-1261 to Lys.
We then investigated the functional role of the conserved Arg (Figure 1). This residue was of particular interest because of a similarly-conserved arginine “finger” present in most G proteins (46-52). In G protein α subunits the arginine finger acts in cis to stabilize the transition state, promote GTP hydrolysis, and turn off the transmitted signal (46, 53). Thus we substituted Arg-1261 in both a conservative (to Lys) and non-conservative (to Ala) manner, and determined the ability of each mutant to bind to Gpa1 as described above. As shown in Figure 5, substitution of Arg-1261 to either Lys or Ala resulted in diminished co-purification with Gpa1-GST. This result indicates that Arg-1261 is necessary for Vps15 to bind efficiently to Gpa1. These mutants are likely to be properly folded given that other functions are preserved (see below).
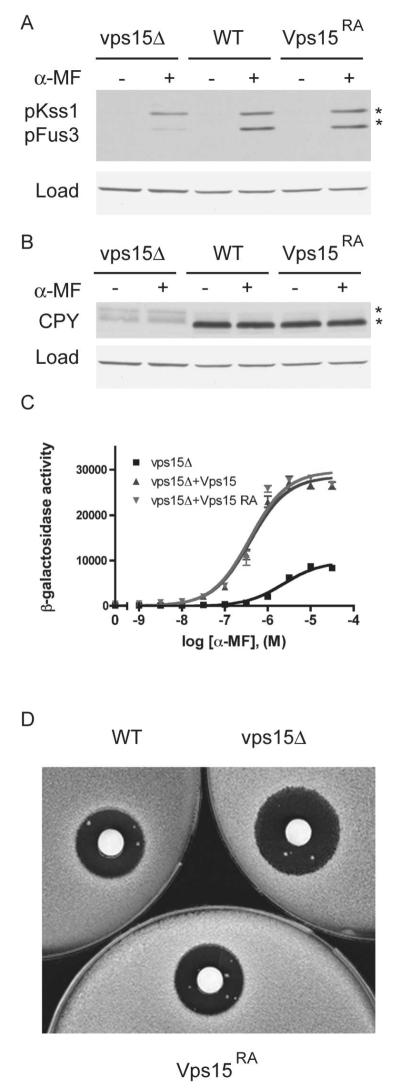
We have shown previously that Vps15 is needed for proper pheromone responses, including pheromone-dependent MAP kinase activation, gene transcription, and cell-division arrest. Thus we investigated the role of Arg-1261 in pheromone signaling. To monitor MAP kinase activity, cell extracts were analyzed by immunoblotting using antibodies against the dually phosphorylated, fully active forms of Fus3 and Kss1 (54). As shown previously, cells lacking VPS15 exhibit diminished Fus3 phosphorylation after pheromone stimulation; in contrast, cells expressing Vps15R1261A exhibit wildtype levels of MAP kinase phosphorylation (Figure 6A). To monitor transcriptional activation, we employed a reporter comprised of a pheromone-inducible promoter and β-galactosidase (FUS1-lacZ). As shown previously, cells lacking VPS15 exhibit dampened induction and a >3-fold rightward shift in the EC50 for pheromone stimulation, whereas cells expressing Vps15R1261A exhibited wildtype transcription activity (Figure 6C). Finally using the growth arrest plate assay (halo assay), we monitored the ability of the cells to undergo cell division arrest in response to pheromone. Once again, Vps15R1261A-expressing cells produced wildtype zones of growth inhibition (Figure 6D). Therefore, while it contributes to Gpa1 binding, Arg-1261 does not contribute to Vps15-mediated signaling.
Figure 6. The conserved Arg-1261 is not necessary for G protein signaling at the endosome.
(A) Wildtype (WT), vps15Δ, and VPS15R1261A (Vps15RA) strains were treated with 3μM α-mating factor (MF) for 30 minutes and analyzed by immunoblotting using antibodies against p44/p42 and G6PDH (Load control). (B) The same samples as in panel (A) analyzed using antibodies against carboxypeptidase Y (CPY) and G6PDH. (C) The same strains were transformed with a plasmid containing a pheromone-inducible FUS1-lacZ reporter; transcriptional activation was measured by monitoring β-galactosidase activity in response to pheromone. (D) The same strains were plated and treated with 45 micrograms of α factor to induce cell division arrest. α-MF, alpha mating factor. RA, Arg-1261 to Ala.
In addition to its role in pheromone signaling, Vps15 has well-established functions in promoting autophagy and at least two types of protein trafficking (6-8, 10, 12, 42, 45, 55, 56). First, Vps15 is needed for newly synthesized vacuolar proteins, like carboxypeptidase Y (CPY), to move from the endoplasmic reticulum, through the Golgi, to the endosome, and finally to the vacuole compartment. During transport the newly-synthesized CPY becomes modified in the ER to the pro-CPY1 form (67 kDa), then processed in the Golgi to the pro-CPY2 form (69 kDa), and finally it is modified in the vacuole to yield the mature protein product (61 kDa) (10, 12, 42, 55). To determine if Arg-1261 has any role in CPY sorting, we prepared extracts from vps15R1261A cells, treated with or without pheromone, and monitored the maturation of CPY by immunoblotting. As shown in Figure 6B, vacuolar sorting of carboxypeptidase Y is unaffected by substitution of Arg-1261 in Vps15.
We have shown above that the WD domain of Vps15 is sufficient to bind Gpa1 in vitro, and does so in a manner that depends on Arg-1261. Nevertheless, Vps15 is capable of promoting the G protein signal in vivo even when Arg-1261 is mutated. Based on these findings we postulated that the kinase and/or intermediate domains of Vps15 might contribute more substantially to the signaling or trafficking functions of the protein. For instance we showed above that the Vps15 intermediate domain as well as the WD domain contributes to Gpa1 binding. Thus we investigated the functionality of Arg-mutant forms of Vps15 containing the intermediate domain as well as the WD domain. The wildtype and arginine-substituted forms of Vps15 were co-expressed with either Gpa1-GST or Gpa2-GST, purified, and analyzed by immunoblotting using antibodies against GST and Myc (to detect any co-purifying Vps15). In this case, Gpa1 bound equally well to wildtype and mutant forms of Vps15 (Figure 7a). These findings indicate that the intermediate domain can compensate for the absence of the conserved arginine present in the WD domain, at least with respect to Gpa1 binding. Our hypothesis was that Arg-1261 functions in the manner of an arginine finger, to promote G protein binding and GTP hydrolysis. However we have shown that this residue is a relatively minor contributor to binding. It remains to be determined if the adjacent conserved Phe-1262 likewise contributes to binding.
Previously, we showed that Vps15 binds preferentially to the unactivated, GDP-bound, form of Gpa1 (5). Thus we sought to determine if Arg-1261 contributes to the nucleotide-dependent interaction of Vps15 with Gpa1. Wildtype and mutant Vps15 were co-expressed with either Gpa1-GST or Gpa2-GST, then lysed in the presence of either GDP or GDP plus AlF4-, a transition state mimic that induces the activated conformation of Gα. As observed previously for full-length Vps15, the WD domain with the intermediate domain of Vps15 bound preferentially to GDP-bound (unactivated) Gpa1. The arginine mutants likewise bound preferentially to unactivated Gpa1 (Figure 7B). These results indicate that neither the kinase domain nor the conserved Arg-1261 is necessary to maintain the nucleotide-specificity of Vps15 binding to Gpa1.
The data presented above reveal that the intermediate and WD domains of Vps15 both contribute to Gpa1 binding, and are sufficient to confer nucleotide-specificity of interaction. We have shown previously that Vps15 is required for G protein signaling at the endosome (5). However we have already ruled out a role for the conserved arginine in sustaining the G protein signaling function of Vps15. These findings suggest that domains necessary for binding and for signaling functions may not fully overlap. Thus we investigated which domains of Vps15 contribute to pheromone signaling, using the same signaling assays described above in Figure 6. First we determined which domains of Vps15 are necessary to confer MAP kinase activation. As shown in Figure 8A, the kinase domain with the intermediate domain is sufficient to sustain MAP kinase activation in response to pheromone. Likewise, by the transcription-reporter assay the kinase domain with the intermediate domain is sufficient to confer wildtype transcriptional activation (Figure 8C) and growth-inhibition responses (Figure 8D). Finally, the kinase domain with the intermediate domain of Vps15 is sufficient to sustain at least some degree of carboxypeptidase Y maturation (Figure 8B). Neither the WD domain alone or the WD domain with the intermediate domain is sufficient to sustain the signaling functions of Vps15 (Figure 8A-D).
Figure 8. The kinase domain with the intermediate domain of Vps15 is necessary to promote G protein signaling at the endosome.
(A) WT and vps15Δ cells expressing the indicated Flag fusion proteins or empty vector were treated with 3 μM α-mating factor (MF) for 30 minutes and analyzed by immunoblotting with antibodies against p44/p42 and G6PDH (Load control). (B) The same samples analyzed using antibodies against carboxypeptidase Y (CPY) and G6PDH. (C) The same strains were transformed with a plasmid containing a pheromone-inducible FUS1-lacZ reporter; transcriptional activation was measured by monitoring β-galactosidase activity in response to pheromone. (D) The same strains were plated and treated with 45 micrograms of α factor to induce cell division arrest. α-MF, alpha mating factor.
Taken together the findings presented above indicate that the WD domain of Vps15 has a Gβ-like structure and is sufficient to bind Gpa1, but cannot by itself sustain signaling. Whereas the conserved arginine contributes to Gpa1 binding, it is not required for Gpa1 signaling. By comparison, the intermediate domain also contributes to Gpa1 binding, and together with the kinase domain is both necessary and sufficient for pheromone signaling and vacuolar sorting activities.
DISCUSSION
The yeast pheromone response pathway represents one of the best-characterized models for the study of G protein signaling and regulation. Upon pheromone activation, Gα as well as Gβγ subunits activate downstream effectors. Release of Gβγ at the plasma membrane leads to activation of the MAP kinases Fus3 and Kss1 (2). At the endosomal membrane, activated Gα transmits a signal that requires the PtdIns 3-kinase and results in activation of Fus3 (5, 6). The endosomal PtdIns 3-kinase Vps34 is a direct binding partner for the GTP-activated form of Gpa1, and this interaction leads to elevated production of PtdIns 3-P. Thus G protein signaling can occur at the endosomal membrane as well as at the plasma membrane.
Given that cell surface receptors and the Gβγ subunits Ste4 and Ste18 are concentrated at the plasma membrane, we considered whether there is an alternative Gβγ pair at the endosomal membrane. Here we have examined the extent to which Vps15 resembles more typical Gβ proteins such as Ste4. We have shown that Vps15 indeed possesses many structural and functional characteristics of canonical Gβ proteins. Both Vps15 and Ste4 are direct binding partners of Gpa1, and both bind preferentially to the unactivated (GDP-bound) form of the protein. Here we have demonstrated that Vps15 contains a seven-bladed propeller structure resembling that of known Gβ proteins. We showed further that the Gβ-like domain is sufficient to bind to Gpa1, as well as to the putative Gγ protein Atg14. As documented previously for typical Gγ subunits, Atg14 is not necessary to bridge the interaction of the Gα (Gpa1) and Gβ-like (Vps15) proteins.
While these findings highlight similarities between Vps15 and canonical Gβ proteins, there are also many important differences. Ste4 is predominantly expressed at the plasma membrane. Vps15 was previously shown by cell fractionation and fluorescence microscopy methods to be located at the endosome, Golgi and vacuolar membranes (11, 57). Typical Gβγ pairs are anchored to the plasma membrane through prenylation (and in some cases palmitoylation) of Gγ (58), while the Vps15/Atg14 complex is anchored to the endosomal membrane through myristoylation of Vps15 (12).
We have documented additional important differences between Ste4 and Vps15. The Gβ protein Ste4 is necessary to transmit the signal to downstream effectors at the plasma membrane (2). In contrast, the Gβ-like WD domain of Vps15 is neither necessary nor sufficient to transmit the G protein signal at the endosome. Our efforts to dock Gpa1 onto the β propeller domain of Vps15 (using a similar orientation to that of known Gα-Gβ pairs) revealed no conserved interactions, indicating that the Gpa1-Vps15 interface must differ from that of more typical G protein subunits. Indeed we have shown here that binding also entails the intermediate domain. Moreover, the intermediate domain and kinase domain are necessary and sufficient to restore full pheromone responsiveness to a vps15 mutant strain. While binding and signaling activities of Vps15 require the intermediate domain, the sequence of this region provides no clues as to its function. Thus we limited our analysis to the intermediate domain in conjunction with the well-defined kinase and/or WD domains only. These observations lead us to conclude that the Gβ-like WD domain of Vps15 serves primarily as a scaffold that promotes assembly of key signaling pathway components, including Gpa1 and Atg14, whereas the kinase domain and the intermediate domain transmit the signal. The same domains that mediate Vps34 interaction (14) also mediate G protein signaling and vacuolar sorting.
Taken together our observations highlight the close interplay of Vps15 in G protein signaling and vacuolar sorting, and are consistent with our earlier suggestion that PtdIns 3-P production is required for both processes (5). Thus just as Gpa1 promotes PtdIns 3-P production, this second messenger could in turn be required for proper trafficking of Gpa1 or some other critical signaling protein, whether or not that protein actually binds to PtdIns 3-P. The signaling phenotypes exhibited by vps15 and vps34 are not the result of some global defect in membrane trafficking, however; deletion of VPS30 or VPS38 also result in a strong vacuolar sorting defect similar to vps15 and vps34 (59), but neither component is required for Gpa1 signaling (5). Indeed of the dozens of vacuolar trafficking mutants that have been described, only vps15 and vps34 attenuate Gpa1 signaling (59).
In summary, we have defined the signaling and binding functions of specific domains within Vps15. Our findings highlight important similarities and differences between canonical Gβ-proteins at the plasma membrane and Gβ-like proteins located within the cell.
Supplementary Material
Table 3.
Primer sequences.
| Primer Name | Sequence (5′-3′) |
|---|---|
| Vps15fn | GAG CTC ATG GGG GCA CAA TTA TCA C |
| Vps15ksrn | GAG CTC TTA AAT GCC ACG ATA TTT ATT CAG |
| Vps15klrn | GAG CTC TTA ATA GCT GTT GCT GAC GAC TAC C |
| Vps15wdlfn | GAG CTC TAT CGT GGC ATT TTC TTC CC |
| Vps15rn | GAG CTC TTA TTG GAA GAT TCC AAT AAG |
| Vps15wdsfn | GAG CTC GAA GGC GAC GTT GAA AGC |
| Vps15fc | CCC GGG ATG GGG GCA CAA TTA TCA C |
| Vps15rc | CCC GGG TTG GAA GAT TCC AAT AAG |
| Vps15wdlfc | CCC GGG ATG TAT CGT GGC ATT TTC TTC C |
| Vp15wdsfc | CCC GGG ATG GAA GGC GAC GTT GAA AG |
| Atg14fn | GAG CTC CAT TGC CCA ATT TGC CAC CAT AG |
| Atg14rn | GAG CTC CTA GCC TAC CAC GTA CCA TCG |
| 2myc | TC GAC ATG GAA CAA AAA TTA ATT TCT GAA GAA GAT TTA GCT GAA CAA AAG TTA ATT TCT GAA GAA GAC TTA G |
| Vps15R1261A | GAT ATA TGG GAT ATC GCT TTC AAC GTG CTG ATA AGG |
| Vps15R1261K | GAT ATA TGG GAT ATC AAA TTC AAC GTG CTG ATA AGG |
ACKNOWLEDGEMENTS
We thank Sveta Gershburg for preparation of the baculovirus and expression of the protein and Zhongmin Jin for assistance with data collection at the Advanced Photon Source. We thank Jonathan Backer for valuable advice and feedback.
ABBREVIATIONS
- α-MF
alpha mating factor
- CPY
carboxypeptidase Y
- G6PDH
glucose-6-phosphate dehydrogenase
- GDP
Guanosine diphosphate
- GST
glutathione S-transferase
- GTP
Guanosine-5′-triphosphate
- ID
intermediate domain
- IP
immunoprecipitation
- KD
kinase domain
- MAP
mitogen-activated protein
- PD
pulldown
- PtdIns
phosphatidylinositol
- SDS-PAGE
sodium dodecyl sulfate — polyacrylamide gel electrophoresis
- VPS
vacuolar protein sorting
- WD
tryptophan-aspartate
Footnotes
Sources of Funding: This work was supported by National Institutes of Health Grants GM080739 (H.G.D.) and GM081881 (J.E.S.) and a T32 Cancer Cell Biology Training Grant (E.J.H.)
SUPPORTING INFORMATION AVAILABLE. Supporting Table 1 shows the alignment of Vps15 used to select conserved residues in the WD domain of Vps15. Representative electron density is shown in Supporting Figure 1. This material is available free of charge via the Internet at http://pubs.acs.org.
The coordinates and structure factors for the Vps15 WD repeat domain crystal structure have been deposited in the Protein Data Bank (accession code 3GRE) and will be released upon publication of this manuscript.
REFERENCES
- 1.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 2.Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: Paradigms and principles. Annu. Rev. Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 3.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 4.Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 5.Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 6.Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. Embo J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stack JH, Emr SD. Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J Biol Chem. 1994;269:31552–31562. [PubMed] [Google Scholar]
- 8.Brown WJ, DeWald DB, Emr SD, Plutner H, Balch WE. Role for phosphatidylinositol 3-kinase in the sorting and transport of newly synthesized lysosomal enzymes in mammalian cells. J Cell Biol. 1995;130:781–796. doi: 10.1083/jcb.130.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan Y, Backer JM. Regulation of class III (Vps34) PI3Ks. Biochem Soc Trans. 2007;35:239–241. doi: 10.1042/BST0350239. [DOI] [PubMed] [Google Scholar]
- 10.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 11.Herman PK, Stack JH, DeModena JA, Emr SD. A novel protein kinase homolog essential for protein sorting to the yeast lysosome-like vacuole. Cell. 1991;64:425–437. doi: 10.1016/0092-8674(91)90650-n. [DOI] [PubMed] [Google Scholar]
- 12.Herman PK, Stack JH, Emr SD. A genetic and structural analysis of the yeast Vps15 protein kinase: evidence for a direct role of Vps15p in vacuolar protein delivery. Embo J. 1991;10:4049–4060. doi: 10.1002/j.1460-2075.1991.tb04981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray JT, Panaretou C, Stenmark H, Miaczynska M, Backer JM. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic. 2002;3:416–427. doi: 10.1034/j.1600-0854.2002.30605.x. [DOI] [PubMed] [Google Scholar]
- 14.Budovskaya YV, Hama H, DeWald DB, Herman PK. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J Biol Chem. 2002;277:287–294. doi: 10.1074/jbc.M109263200. [DOI] [PubMed] [Google Scholar]
- 15.Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 16.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of x-ray diffaction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 19.Cowtan K. DM: An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 20.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Zeller CE, Parnell SC, Dohlman HG. The RACK1 ortholog Asc1 functions as a G-protein beta subunit coupled to glucose responsiveness in yeast. J Biol Chem. 2007;282:25168–25176. doi: 10.1074/jbc.M702569200. [DOI] [PubMed] [Google Scholar]
- 24.Song J, Hirschman J, Gunn K, Dohlman HG. Regulation of membrane and subunit interactions by N-myristoylation of a G protein α subunit in yeast. J Biol Chem. 1996;271:20273–20283. doi: 10.1074/jbc.271.34.20273. [DOI] [PubMed] [Google Scholar]
- 25.Chasse SA, Flanary P, Parnell SC, Hao N, Cha JY, Siderovski DP, Dohlman HG. Genome-scale analysis reveals Sst2 as the principal regulator of mating pheromone signaling in the yeast Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:330–346. doi: 10.1128/EC.5.2.330-346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman GA, Garrison TR, Dohlman HG. Endoproteolytic Processing of Sst2, a Multidomain Regulator of G Protein Signaling in Yeast. J Biol Chem. 2000;275:37533–37541. doi: 10.1074/jbc.M005751200. [DOI] [PubMed] [Google Scholar]
- 27.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouali M, King RD. Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 2000;9:1162–1176. doi: 10.1110/ps.9.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 32.Meiler J. PROSHIFT: protein chemical shift prediction using artificial neural networks. J Biomol NMR. 2003;26:25–37. doi: 10.1023/a:1023060720156. [DOI] [PubMed] [Google Scholar]
- 33.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto- oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman G, Garrison TR, Dohlman HG. Analysis of RGS proteins in Saccharomyces cerevisiae. Methods Enzymol. 2002;344:617–631. doi: 10.1016/s0076-6879(02)44744-1. [DOI] [PubMed] [Google Scholar]
- 36.Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 37.Collaborative Computational Project The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 38.Lu G. A WWW service system for automatic comparison of protein structures. Protein Data Bank Quarterly Newsletter. 1996;78:10–11. [Google Scholar]
- 39.Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [see comments] [DOI] [PubMed] [Google Scholar]
- 40.Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, Russell RB. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- 41.De Camilli P, Emr SD, McPherson PS, Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- 42.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. GTPase mechanism of G proteins from the 1.7-A crystal structure of transducin α-GDP-AIF-4. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 47.Bourne HR. G proteins. The arginine finger strikes again. Nature. 1997;389:673–674. doi: 10.1038/39470. [DOI] [PubMed] [Google Scholar]
- 48.Kotting C, Kallenbach A, Suveyzdis Y, Wittinghofer A, Gerwert K. The GAP arginine finger movement into the catalytic site of Ras increases the activation entropy. Proc Natl Acad Sci U S A. 2008;105:6260–6265. doi: 10.1073/pnas.0712095105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmadian MR, Hoffmann U, Goody RS, Wittinghofer A. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry. 1997;36:4535–4541. doi: 10.1021/bi962556y. [DOI] [PubMed] [Google Scholar]
- 50.Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol. 1997;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- 51.Mittal R, Ahmadian MR, Goody RS, Wittinghofer A. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science. 1996;273:115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- 52.Rensland H, Lautwein A, Wittinghofer A, Goody RS. Is there a rate-limiting step before GTP cleavage by H-ras p21? Biochemistry. 1991;30:11181–11185. doi: 10.1021/bi00110a023. [DOI] [PubMed] [Google Scholar]
- 53.Bourne HR. How receptors talk to trimeric G proteins. Curr Opin Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 54.Sabbagh W, Jr., Flatauer LJ, Bardwell AJ, Bardwell L. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol Cell. 2001;8:683–691. doi: 10.1016/s1097-2765(01)00322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung G, Ueno H, Hayashi R. Carboxypeptidase Y: structural basis for protein sorting and catalytic triad. J Biochem. 1999;126:1–6. doi: 10.1093/oxfordjournals.jbchem.a022408. [DOI] [PubMed] [Google Scholar]
- 56.Obara K, Noda T, Niimi K, Ohsumi Y. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells. 2008;13:537–547. doi: 10.1111/j.1365-2443.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- 57.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 58.Hirschman JE, Jenness DD. Dual lipid modification of the yeast ggamma subunit Ste18p determines membrane localization of Gbetagamma. Mol Cell Biol. 1999;19:7705–7711. doi: 10.1128/mcb.19.11.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.