SYNOPSIS
Rho family small GTPases are critical regulators of multiple cellular functions. Dbl homology domain-containing proteins are the classical guanine nucleotide exchange factors (GEFs) responsible for activation of Rho proteins. Zizimin1 is a Cdc42-specific GEF that belongs to a second family of mammalian Rho-GEFs, CZH proteins, which possess a novel type of GEF Domain. CZH proteins can be divided into a subfamily related to DOCK180 and a subfamily related to zizimin1. The two groups share two conserved regions named the CZH1 (or DHR1) domain and the CZH2 (DHR2 or DOCKER) domains, the latter exhibiting GEF activity. We now show that limited proteolysis of zizimin1 suggests the existence of structural domains that do not correspond to those identified on the basis of homologies. We demonstrate that the N-terminal half binds to the GEF domain through three distinct areas, including the CZH1, to inhibit the interaction with Cdc42. The N-terminal Pleckstrin homology (PH) domain binds phosphoinositides and mediates zizimin1 membrane targeting. These data define two novel functions for the N-terminal region of zizimin1.
Keywords: Dock9, zizimin, structure, limited proteolysis
INTRODUCTION
The Rho family of low molecular weight GTPases includes twenty-two human genes, the best known members being RhoA, Rac1 and Cdc42 [1, 2]. Rho GTPases control fundamental processes common to all eukaryotes, including morphogenesis, polarity, movement, and cell division [3, 4]. Each GTPase acts downstream of multiple cell surface receptors and upstream of multiple effector proteins. Cdc42 mediates processes including, cell polarity, regulation of the actin and microtubules cytoskeletons, intracellular trafficking, gene expression, cell cycle progression and cell-cell contacts [5–7].
Rho proteins are active when bound to GTP and inactive when bound to GDP. Conversion to the active state is catalyzed by guanine nucleotide exchange factors (GEFs). For nucleotide exchange, the GEF first binds with low affinity to the GDP-bound protein and induces dissociation of GDP from this complex, which leads to formation of a higher affinity intermediate. This intermediate dissociates upon binding of GTP [8]. GEFs can therefore be distinguished from other GTPase interacting proteins by their ability to bind preferentially to the nucleotide-depleted (ND) state [9, 10].
The classical GEFs for Rho GTPases share a common motif, designated the Dbl-homology (DH) domain that mediates nucleotide exchange [11]. However, a second family of mammalian Rho-GEFs, CZH proteins, possess a novel type of GEF domain named by various groups as the CZH2, DOCKER or DHR2 domain [10, 12, 13]. Sixty nine DH-domain-containing proteins and eleven CZH2-domain-containing proteins have been identified [10, 12, 14–16]. Mammalian Rho GEFs therefore highly outnumber their substrates. The high number of Rho-GEFs enables a wide variety of cell-surface receptors to control the activity of Rho proteins. It appears that GEFs can also modulate the pathways downstream of Rho GTPases by scaffolding effectors [17]. The CZH family includes CDM (Ced-5, DOCK180, Myoblast city) proteins that activate Rac, zizimin proteins that activate Cdc42, and additional members [10, 12, 13, 16, 18–20]. CZH proteins are implicated in cell migration, phagocytosis of apoptotic cells, neurite outgrowth and lymphocytes homing, activation and development [16]. Based on sequence homology, domain structure and phylogenetic analysis, CZH family can be divided into a subfamily related to DOCK180 and a subfamily related to zizimin1 [16]. The two groups share two conserved regions that we named CZH1 and CZH2, for CDM-zizimin homology 1 and 2, respectively [10]. All of the mammalian CZH proteins and the vast majority of homologs identified in lower eukaryotes contain both the CZH1 and CZH2 regions. The CZH1 always precedes the CZH2 and proteins containing CZH1 but lacking CZH2 have not been identified [16]. While mechanisms of activation of Dbl family proteins and DOCK180 have been extensively investigated, how zizimin1 and related proteins are regulated is largely unknown.
The zizimin-related proteins can be further divided into two groups: zizimin (or DockD) and zir (or DockC) [12, 16]. These two groups share ∼30% identity over most of their sequences but only the zizimin proteins possess pleckstrin homology (PH) domains near their N-terminus. PH domains are best known for their ability to target proteins to membranes by binding to phosphoinositides [21, 22]. Some PH domains interact at high specificity and affinity with second messengers produced by PI 3-kinases (Ptd(3,4,5)P3 or Ptd(3,4)P2)) to facilitate transient membrane translocation. The majority of PH domains, however, bind in vitro with lower affinity and specificity to a variety of phosphoinositides, and most of these are apparently incapable of independent membrane targeting [21, 22]. Some of the promiscuous PH domains require protein oligomerization, which bring together several PH domains, or cooperate with additional elements within the same polypeptide to mediate membrane targeting. Alternatively, some promiscuous PH domains bind to membranes via interactions with both phosphoinositides and additional (non-phosphoinositide) factors including proteins [23].
These considerations prompted us to investigate the function of the zizimin1 N-terminus. We found that not only the CZH1 domain but also two other sequences in the N-teminus bind the CZH2 domain to inhibit Cdc42 binding. We also found that the PH domain exhibits promiscuous phosphoinositide binding and mediates membrane targeting. Additionally, proteolytic studies to identify stable zizimin1 fragments suggest that the structural domains are distinct from those identified by comparing sequences across the CZH family.
EXPERIMENTAL
Cell culture and transfections
Cos7 cells and NIH 3T3 cells were grown in DMEM supplemented with 10% fetal bovine serum, penicillin and streptomycin (all from Invitrogen, Carlsbad, CA). 24h before transfection, cells were subcultured so as to reach 50–70% confluency the next day for transfection. Cells were transfected in 100 mm tissue-culture dishes with 1.3 µg of total DNA using Effectene (Qiagen; Valencia, CA) according to the manufacturer’s instructions. In the experiments presented in figure 1C, figure 1E, figure 2, figure 4 and figure 5C, cells were transfected by electroporation. 6×106 cells were electroporated in 400 µl Ca2+/Mg2+-free PBS with 20 µg total DNA in 0.4 ml cuvets at 250 volts and 950 µF. Cells were collected 48 hours post transfection.
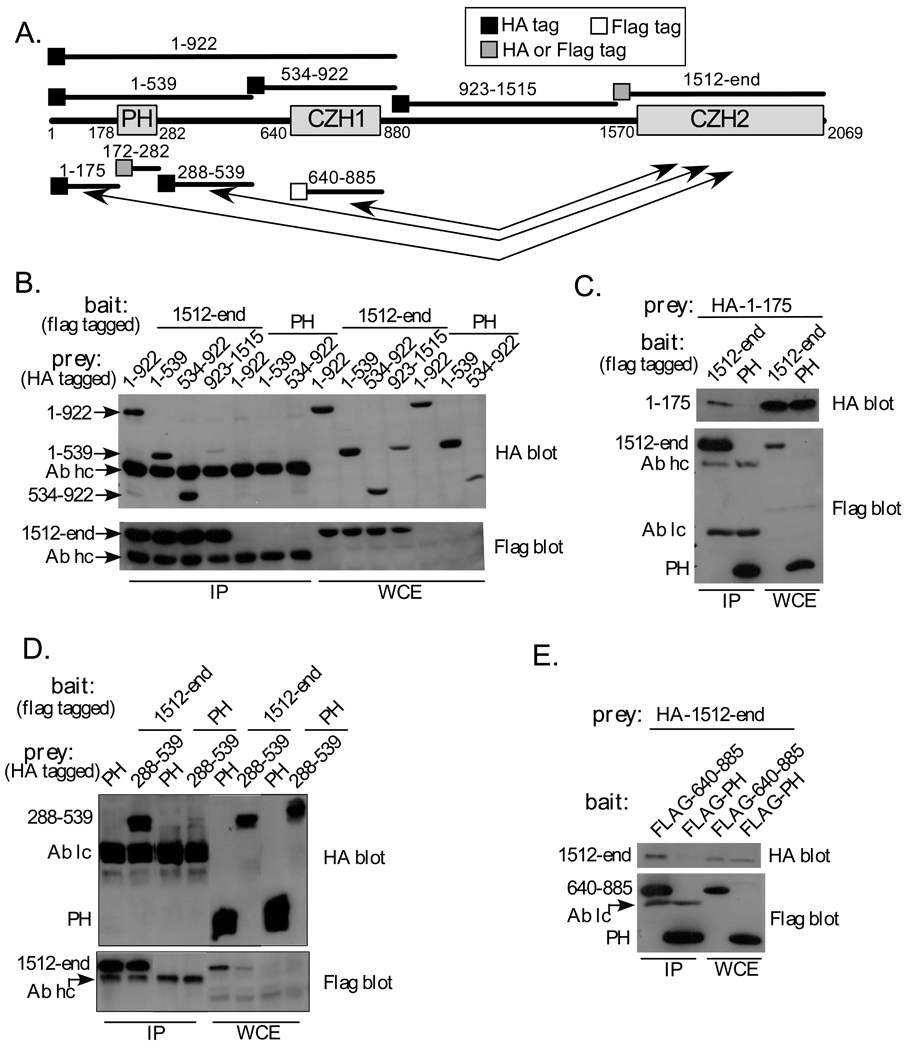
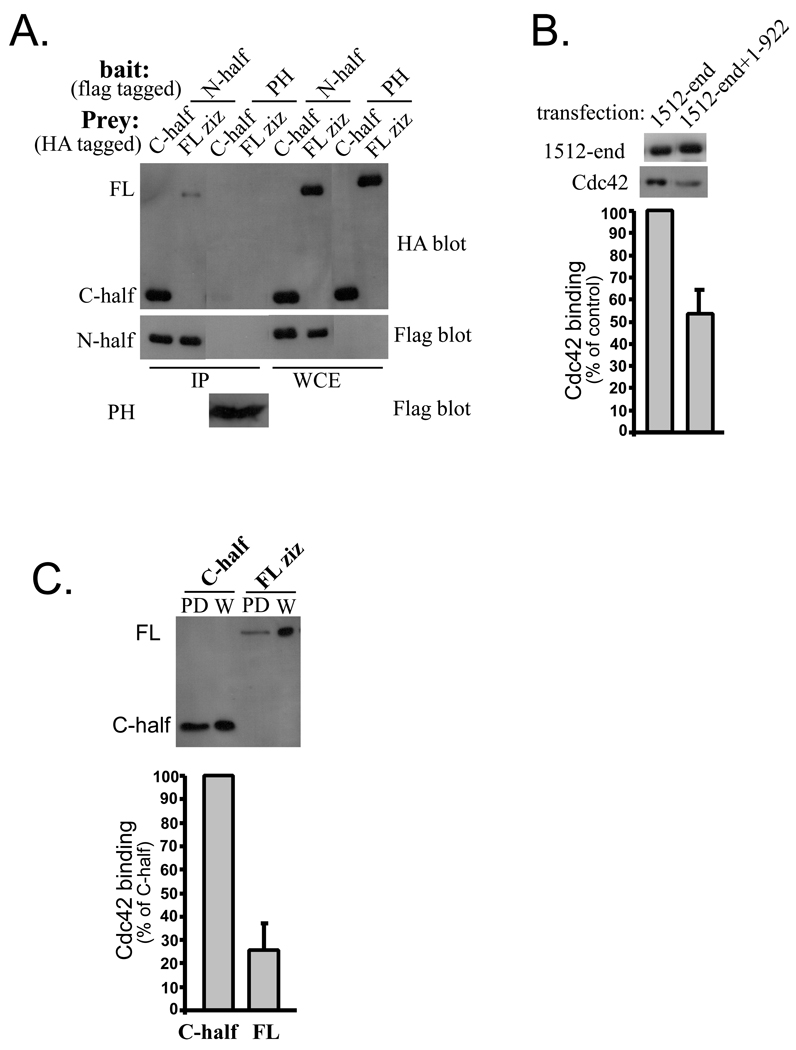
Fig. 1. Elements from zizimin1 N-terminal half bind to the CZH2 domain.
A Schematic of zizimin1 structure and fragments used in Figure 1. The Pleckstrin homology (PH), CDM-zizimin homology 1 (CZH1) and CDM-ziziminin homology 2 (CZH2) areas are displayed and amino acid boundaries are indicated. Arrows highlight the three N-terminal areas that interact with the CZH2. Location of HA or Flag tags is indicated by black or white boxes, respectively, gray boxes indicate fragments that are tagged by HA or Flag in different experiments.
B HA-tagged zizimin1 fragments 1–922, 1–539, 534–922 or 923–1515 were expressed in Cos7 cells together with Flag zizimin11512-end or Flag zizimin1 PH domain (specificity control) as indicated. 48 hours post transfection, cells were lysed and immunoprecipitated (IPed) with anti-Flag antibody. The IPs and the corresponding whole cell extracts (WCE; representing 9% of the total) were analyzed by Western blotting with anti-HA Ab (top panel) or anti-Flag Ab (bottom panels). Arrows indicate the position of each fragment on the gels, Ab hc is the heavy chain of the Flag Ab used for IP. The PH domain is not visible due to its low molecular weight but expresses at similar levels to the other fragments (see 1C).
C, D. The experiments were done as in B. Ab lc is the light chain of the Flag Ab used for IP. The CZH2 domain associates with the N-terminal subfragments 1–175 and 288–539 but not the PH domain (172–282).
E HA zizimin11512-end was expressed in Cos7 cells together with the Flag-tagged minimal CZH1 domain (zizimin1640–885) or with Flag-PH domain (specificity control) as indicated. Samples were immunoprecipitated with anti-Flag and HA-tagged fragments detected as in B.
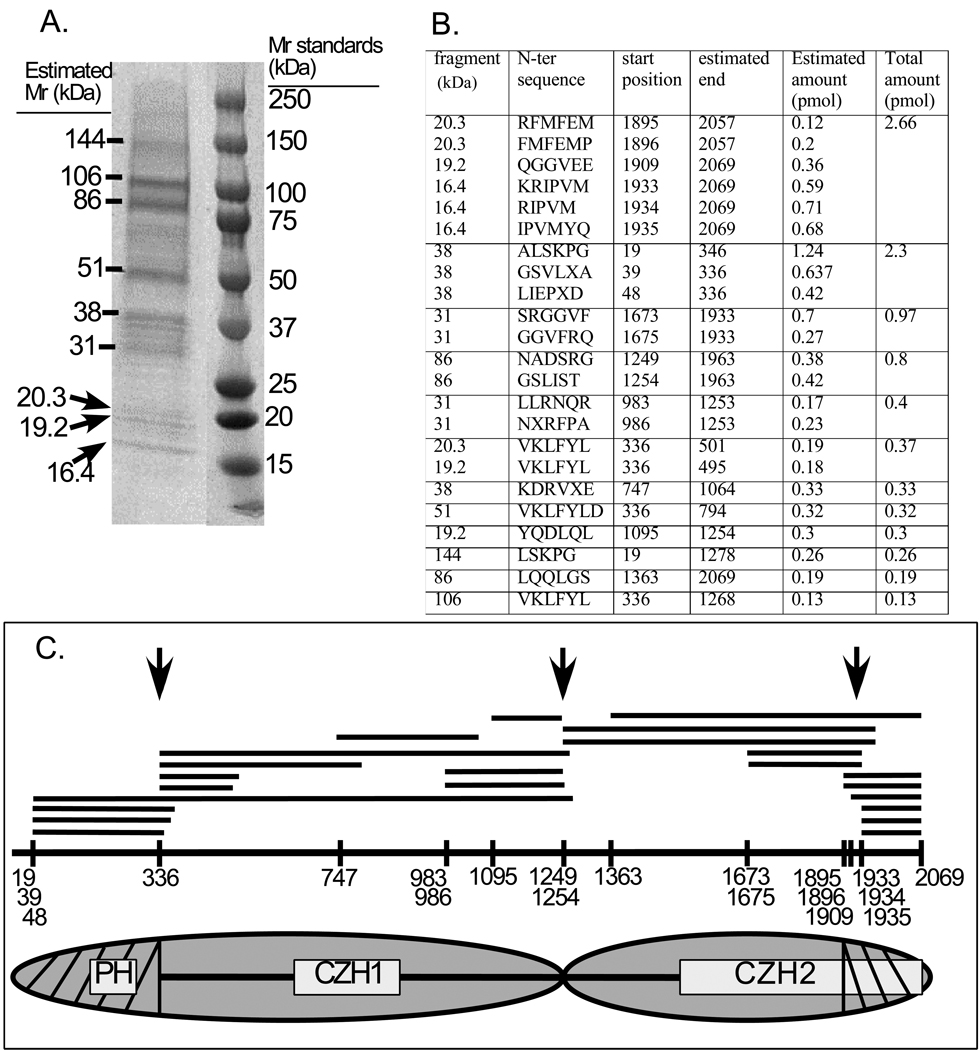
Fig. 2. Limited proteolysis of zizimin1.
Zizimin1 was digested with trypsin as described in Materials and Methods and the digests resolved on SDS-PAGE, transferred to PVDF membrane and sequenced by Edman degradation. A) Coomassie brilliant blue R stain of the PVDF membrane. The position of the fragments and their estimated molecular mass (Mr) is indicated on the left; Mr standards on the right. B) Summary of sequenced fragments. N-terminal sequences and start positions within zizimin1 are indicated. Fragment ends were estimated based on predicted size and availability of tryptic sites. Amount of the sequenced peptides were estimated as described in Materials and Methods; total amount represent the sum amounts of fragments of similar size and position. C) Top: Schematic representation of fragments and their start positions within the zizimin1 sequence. Arrows mark potential cleavage sites in zizimin1. Bottom: postulated domain structure of zizimin1 based on limited proteolysis and fragments expression data. The CDM-zizimin sequence homology areas (CZH1 & CZH2) and pleckstrin homology (PH) domain are boxed. Zizimin1 structure as composed of two large structural domains (N-half & C-half; marked by ovals) and its subdomains (hatched areas) is displayed.
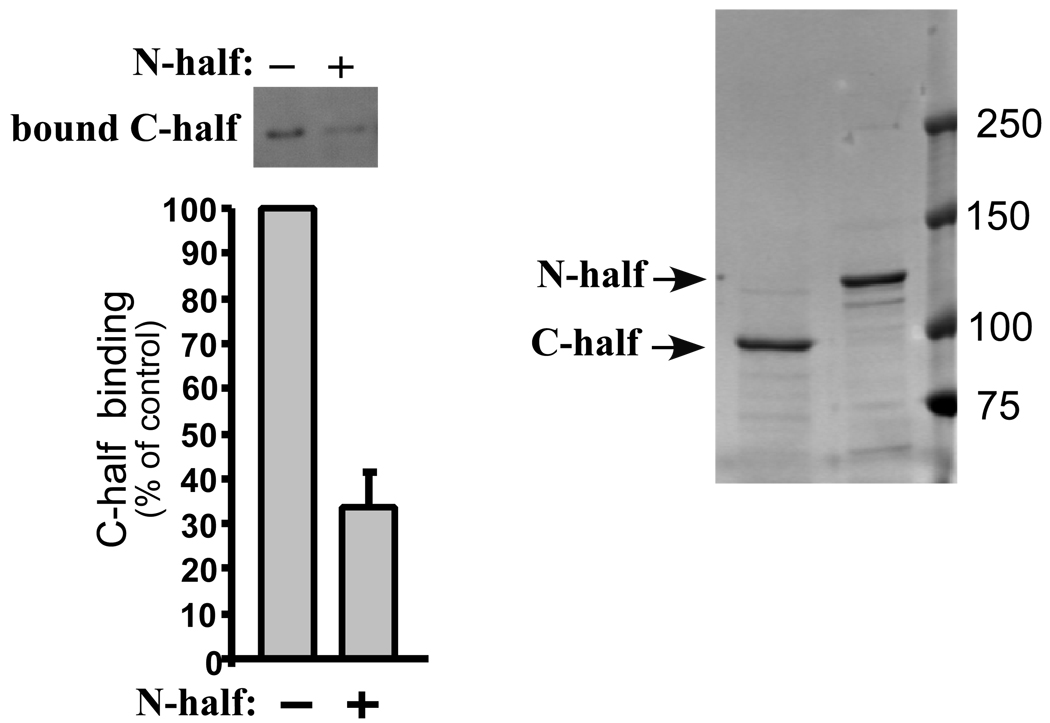
Fig. 4. Zizimin1 N-terminal half inhibits binding of the C-terminal half to Cdc42.
Flag-tagged zizimin1 N- or C-terminal halves were purified from Cos7 cells by Flag IP and peptide elution (coomassie-stained gel is displayed on the right). Binding of the C-terminal half (at 20 nM) to Cdc42 (at 200 nM) was assayed in the absence or presence of 100 nM N-terminal half. Bound C-terminal half was detected by Western blotting with anti-Flag and quantified by densitometry (left panel). Values are mean ± SD (n=3) relative to control (without N-half).
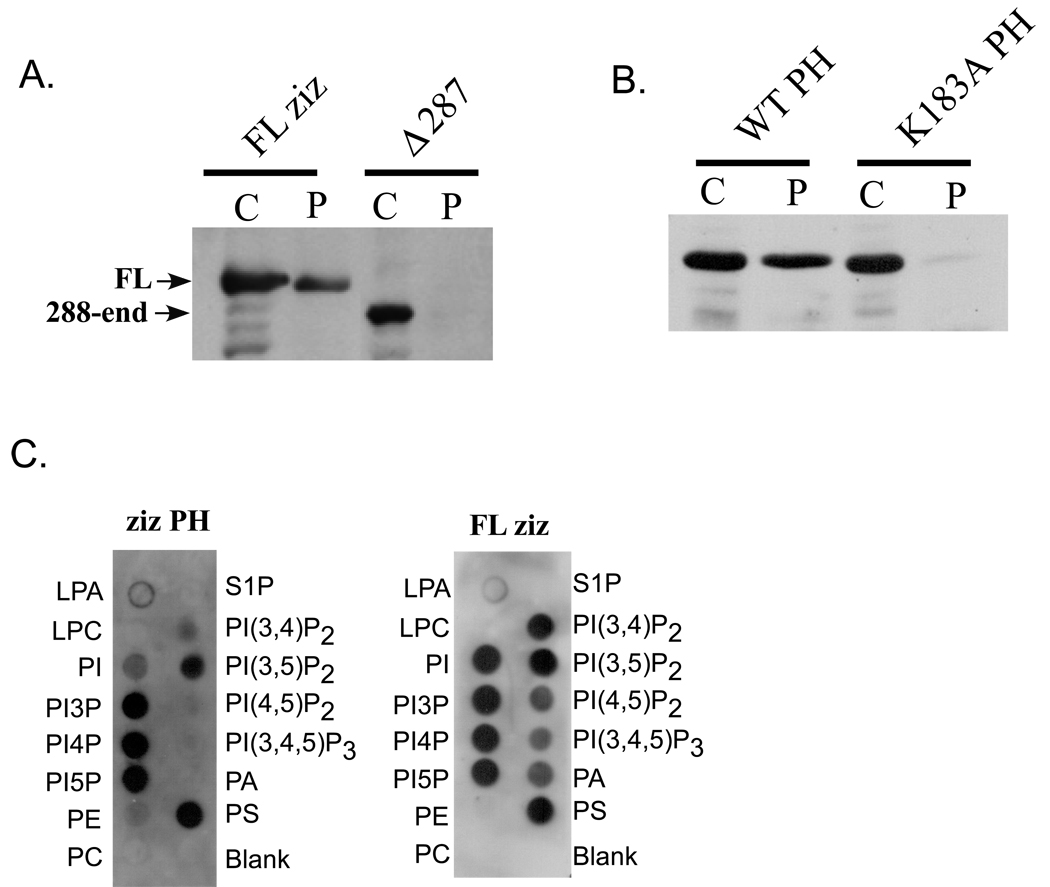
Fig. 5. Zizimin1 PH domain mediates membrane targeting.
A NIH 3T3 cells were transfected with HA-tagged full-length (FL) zizimin1 or a mutant lacking the PH domain (zizimin1Δ287). Cells were lysed in hypotonic buffer and separated into cytoplasmic (c) or particulate (p) fractions, which were analyzed by Western blotting with anti-HA. The particulate fractions represent 5x more cell equivalents than the cytoplasmic fractions.
B GFP-tagged zizimin1 PH domain (zizimin1172–282) or K183A mutated PH domain were expressed in NIH 3T3 cells and their distribution between the cytoplasmic and particulate fractions analyzed as in A. The blot was probed with anti-GFP Ab. The particulate fractions analyzed represent 10x more cell equivalents than the cytoplasmic fractions.
C Lipid dot blots (100 pmol/spot) were probed with 0.5 µg/ml Flag-tagged zizimin1 PH domain (35 nM) or the full-length protein (2.1 nM). Proteins were detected using anti Flag Ab. Control blots probed with unrelated flag-tagged fragments displayed no signal (not shown). LPA – lysophosphatidic acid, LPC – lysophosphatidylcholine, PI – phosphatidylinositol, PI3P – phosphatidylinositol-3-phosphate, PI4P - phosphatidylinositol-4-phosphate, PI5P - phosphatidylinositol-5-phosphate, PE – phosphatidylethanolamine, PC – phosphatidylcholine, S1P – sphingosine-1-phosphate, PI(3,4)P2 - phosphatidylinositol-3,4-biphosphate, PI(3,5)P2 - phosphatidylinositol-3,4-biphosphate, PI(4,5)P2 - phosphatidylinositol-4,5-biphosphate, PI(3,4,5)P3 - phosphatidylinositol-3,4,5-triphosphate. PA - phosphatidic acid, PC – phosphatidylcholine.
Immunoprecipitation experiments
Transfected cells were collected by trypsinization, washed twice in cold PBS and lysed for 10 min on ice in 50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, and 1% protein inhibitors mixture (final concentration 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), 15 µM pepstatinA, 14 µM E-64, 40 µM bestatin, 20 µM leupeptin, and 0.8 µM aprotinin; Sigma # P8340; St. Louis, MO). Lysates were cleared by 10 min centrifugation at 16,000 × g. A portion of these whole cell extracts (WCE) was reserved and the rest incubated with M2 anti-FLAG affinity beads (Sigma # F2426) for 3 hours, on a rocker at 4°C. Beads were washed 3X in lysis buffer (LB) at 4°C, and eluted in 2x SDS-PAGE loading buffer (0.125 M Tris, pH 6.8, 4% SDS, 20% glycerol 10% β͂ mercaptoethanol). Eluates were resolved by SDS-PAGE and fragments detected by western blotting. Experiments testing lipids effects on zizmin1 C-half association with the N-half were done as in Fig. 3A but in some cases cell lysates were supplemented with brain PI4P (Avanti Polar Lipds, Alabaster, AL, #840045X) or with egg phosphatidic acid (Avanti Polar Lipds, #840101) to a final concentration of 200_µM. Immunoprecipitation, washing and western blotting were done as described above. Additional controls included immunoprecipitation of HA-C-half with unrelated FLAG tagged protein (E47) in the presence of the above lipids, which detected no nonspecific binding.
Fig. 3. Inhibition of Cdc42 binding by the N-terminus of zizimin1.
A Association between zizimin1 N- and C-terminal halves. HA full-length (FL) zizimin1 or HA C-terminal half were expressed in Cos7 cells together with Flag zizimin1 N-terminal half or with Flag zizimin1 PH domain (specificity control) as indicated. HA-tagged proteins in the Flag IPs detected as in Fig. 1B. WCE represents 26% of the lysate used for IP, thus, approximately one quarter of the C-half associated with the N-half. Schematic presentation of zizimin1 N- and C-halves is brought in Fig. 2C.
B Reduced Cdc42 binding by CZH2: zizimin11–922 complex. Cos7 cells were transfected with Flag CZH2 domain (1512-end) plus HA-zizimin11–922 and immunoprecipitated with anti-HA resin to isolate the CZH2:zizimin1–922 complexes. Cells were also transfected with zizimin11512-end alone and immunoprecipitated with anti-Flag resin. The IPs were incubated for 1 hour with nucleotide-depleted (ND) GST-Cdc42 and washed. Resins were eluted with GTPγS containing buffer to release Cdc42, then with SDS-PAGE loading buffer to release the CZH2 domain. Eluates were analyzed by Western blotting with anti-GST or anti-Flag to detect Cdc42 or the CZH2 domain respectively. Bands were quantified by densitometry. Bound Cdc42 was normalized to the amount of CZH2 in the IPs. Values are mean ± range (n=2) expressed as percent relative to control.
C Higher Cdc42 binding by the isolated C-terminal half. HA-tagged FL Zizimin1 or its C-terminal half were expressed in Cos7 cells and lysates prepared. ND-Cdc42 was used to pull down (PD) the proteins. Binding was assessed by Western blotting with anti-HA and quantified by densitometry. Values (mean ± SD, n=5) for bound protein were normalized for expression level and presented as percent binding relative to the C-half. The whole cell extracts represent 10% of the amount used for PD.
Limited proteolysis
Flag-zizimin1 was expressed in Cos7 cells and immunoprecipitated by M2 anti-FLAG affinity beads as above. Beads were washed 3X in LB, 30 min each time and once for 30 min in 50 mM Tris, pH 7.5, 500 mM NaCl, 1% triton X-100. The protein was eluted 30 min at 4°C in 50 mM Tris, pH 7.5, 500 mM NaCl, 1% Triton X-100 and 0.2 mg/ml FLAG peptide (Sigma # F3290). The eluate was dialyzed to a buffer containing 20 mM Tris, pH 7.5, 80 mM NaCl, 0.1% Triton X-100, 1 mM DTT and kept at 4°C before digestion. To setup the conditions for zizimin1 digestion the protein was treated at 37°C with increasing doses of trypsin (protemics grade; Sigma # T6567) for various incubation periods. At 1/25 enzyme/substrate ratio, relatively stable fragments were obtained between 20’ and 120’ (as detected by SDS-PAGE and silver staining; not shown). For fragments identification, 25 µg zizimin1 at 1.7 µM (400 ng/µl), was incubated at 37°C for 60 min with 68 nM trypsin, in dialysis buffer. The reaction was stopped by addition of 15 µl 5x SDS-PAGE loading buffer followed by 5’ incubation at 95°C. Digests were resolved on 4–12% polyacrylamid gels and transferred to PVDF membranes. Membranes were stained by coomassie brilliant blue R and individual bands excised. N-terminal sequencing was performed with an Applied Biosystems Procise 494 sequencer using the manufacturer’s PL-PVDF protein cycle and data analyzed with the associated model 610 data system. Estimation of peptides amounts was done by comparing peak area of sequenced amino acids with amino acids standards (4 pmole). The estimation procedure does not correct for differential transfer efficiency of different fragments, larger fragments tend to transfer less efficiently.
Cdc42 binding assays
Zizimin1 fragments were expressed in Cos7 cells. Lysates were prepared as above and supplemented with either 5 mM EDTA (which removes the nucleotides bound to GTPases by chelating Mg2+ ions that are essential for nucleotide binding) or 5 mM MgCl2 (GTP-or GDP-loaded condition). Bacterially expressed GST−Cdc42 attached to glutathione agarose beads (Sigma, St Louis, MO) was incubated for 5 min at 30 °C in 50 mM Tris pH 7.5, 1 mM DTT, 5 mM EDTA with 100 µM GTPγS, 100 µM GDP or without nucleotides (nucleotide-depleted condition). Cdc42 beads were then added to the cell lysates and incubated 1 h at 4 °C with shaking. Beads were washed 3X and eluted for 5 min at room temperature in 100 µM GTPγS in 50 mM Tris pH 7.5, 5 mM MgCl2, 1 mM DTT. For Fig. 6, beads were eluted in 2x SDS-PAGE loading buffer. The eluates were resolved by SDS−polyacrylamide gel electrophoresis (PAGE) and analysed by Western blotting. For Fig. 3B, lysates were incubated for 2 hours with anti HA (Covance #MMS-101P; Princeton, NJ) or M2 anti-Flag (Sigma # F3165) bound to protein G resin (GE healthcare # 17–0885-01). Recombinant GST-Cdc42 was added (final concentration 200 nM) and incubated for an additional 1h. Resins were washed 2X in LB with 5 mM EDTA and once with LB. Cdc42 was eluted in LB containing 100 µM GTPγS and 5 mM MgCl2 for 5’ at RT. Beads were then eluted with SDS-PAGE loading buffer to release remaining zizimin1 fragments. For Fig. 3C, lysates were incubated for 1 hour at 4°C with 25 µg GST-Cdc42 bound to glutathione beads (Sigma # E2404), beads were washed and eluted with GTPγS as above. For Fig. 4, zizimin1 fragments were purified as in the limited proteolysis experiment. Flag eluates were diluted and the buffer adjusted to 50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 2 mg/ml BSA, 5 mM EDTA and 20 nM zizimin1 C-terminal half ± 100 nM zizimin1 N-terminal half. The reaction mixture was incubated 1 hour at 4°C, GST-Cdc42 was added to a 200 nM final concentration and incubated for an additional hour. GST-Cdc42 was precipitated by glutathione beads during the final (3rd) hour of incubation, beads were washed and zizimin1 eluted with GTPγS.
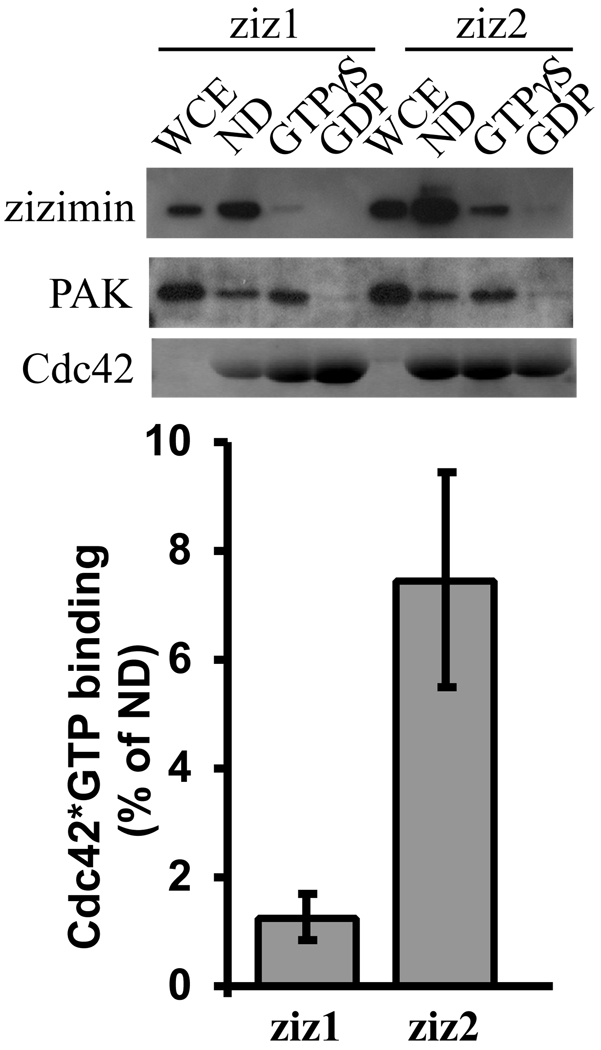
Fig. 6. Analysis of zizimin1 & 2 binding to Cdc42.
Lysates from cells transfected with HA-tagged full-length zizimin1 or zizimin2 were incubated with nd-, GTPγS-loaded or GDP-loaded GST-Cdc42 beads. Beads were washed then eluted in SDS-PAGE loading buffer. Samples were analyzed by Western blotting with anti-HA. Whole cell extracts (WCE) represent 5% of the total. Bands were quantified by densitometry. Zizimin bound to GTPγS-Cdc42 is presented as percent of zizimin bound to nd-Cdc42. Values are means ± SD (n=3). PAK binding to the Cdc42 beads was assessed with anti S141-PAK Ab (Biosource). Gels were stained with Coomassie blue to assess GST-Cdc42 protein.
Cell fractionation
NIH 3T3 cells on tissue culture plates were washed twice in cold PBS, rinsed in 20 mM HEPES, pH 7.5, 5 mM KCl and scraped on ice in hypotonic lysis buffer containing 5 mM HEPES, pH 7.5, 1% protein inhibitors mixture (see above), 1mM NaVO4, 1mM sodium pyrosphosphate and 1mM NaF. Cell suspensions were lysed using Dounce homogenizer (30 strokes) and spun 10’ at 400 × g to pellet the nuclei. The supernatant was collected and spun 30 min at 20,000 × g. Supernatants (cytoplasmic fractions) were collected and brought to 20 mM HEPES, pH 7.5, 1% Triton X-100, 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS final concentrations. Pellets (particulate fractions) were suspended for 10 min on ice in 1/5 or 1/10 volume of the cytoplasmic fractions, in buffer identical to the final cytosolic buffer. The particulate fractions were clarified centrifugation at 20,000 × g for 10’. Final fractions were supplemented with 25% volume of 5x SDS-PAGE loading buffer and analyzed by Western blotting.
Lipids binding assay
Lipids dot blots (Echelon Biosciences; Salt Lake City, UT) were done according to the manufacturers instructions. Briefly, membranes were blocked in TBS, 0.1% tween-20, 3% fatty acid free BSA (Sigma # A7030). Zizimin1 PH domain or the full-length protein were added for 2h at 0.5 µg/ml in blocking buffer. Blots were washed and incubated with anti-Flag mAb followed by goat anti mouse HRP, all in blocking buffer. All procedures were carried out at RT, signals were developed by ECL chemiluminescence (Amersham Biosciences).
DNA constructs
Construction of pEF4-HA-Kpn and pEF4-Flag-Kpn vectors were previously described [10, 24]. pEF-HA-zizimin1 WT, 288-end (Δ287), 1–539, 1–922, 1512-end and pEF-Flag zizimin1 WT, PH (172–282), 1512-end were also previously described [10, 24]. Zizimin1 fragments 534–922, 923–1515, PH (172–282), 1–175, 288–539, 640–885, 1–1184, 1–1215, 1–1253, 1183-end and 1254-end were amplified by PCR using human zizimin1 templates and cloned in pEF-FLAG-Kpn or pEF-HA-Kpn using KpnI and NotI sites. pEGFP Zizimin1172–282 & pEGFP Zizimin1172–282 K183A: human zizimin1 in pBluescript [10] was mutated (K183A) using site directed mutagenesis kit QickChange XL (Stratagene; La Jolla, CA) and the primers 5’- GCA TGG CTG GCT GTA CGC CGG CAA CAT GAA CAG TGC C −3’ AND 5’- GGC ACT GTT CAT GTT GCC GGC GTA CAG CCA GCC ATG C-3’. The mutation creates an NgoM1 site. The area coding for zizimin1 PH domain (amino acids 172–282) was amplified by PCR using the templates pBluescript zizimin1 WT or pBluescript zizimin1 K183A mutant and cloned in pEGFP-C2 (Clontech Laboratories, Mountain View, CA) using the EcoRI and KpnI sites. pKH3-zizimin2 (full-length; [25]) is a gift from Dr. Rick Cerione (Department of Molecular Medicine, Cornell University, Ithaca, NY). All constructs were confirmed by sequencing.
RESULTS
Multiple N-terminal elements interact with the CZH2 region
To test potential interactions between zizimin1 CZH1 and CZH2 domains, fragments containing these areas were prepared and their association tested by co-immunoprecipitation. HA-tagged zizimin1534–922 (CZH1) was co-transfected into Cos7 cells with Flag-tagged zizimin11512-end (CZH2) or with Flag zizimin1 PH domain (fragments described in Fig 1A). When cell lysates were immunoprecipitated with antibody to Flag, HA-zizimin1 CZH1 was efficiently precipitated from cells co-expressing Flag-CZH2 but not from cells expressing Flag-PH (Fig. 1B). As additional specificity controls, association of the CZH2 with other zizimin1 fragments was tested in parallel; these included zizimin1923–1515, which contains the area between the CZH1 and CZH2, and zizimin11–539, which contains the N-terminal portion (see Fig. 1A). Very little if any zizimin1923–1515 co-precipitated with Flag-CZH2, demonstrating specificity (Fig. 1B). Unexpectedly, the N-terminal fragment (zizimin11–539), also specifically precipitated with the CZH2 fragment (Fig. 1B).
To further analyze this interaction, the 1–539 region was split into three smaller fragments (zizimin11–175, zizimin1172–282 (PH domain) and zizimin1288–539) and their association with the CZH2 region tested. The CZH2 fragment precipitated zizimin11–175 (Fig. 1C) and zizimin1288–539 (Fig. 1D), whereas the PH domain did not bind (Fig. 1D). Analysis of whole cell extracts demonstrated in all cases similar expression of these constructs in the CZH2 and control (PH) transfected cells (Fig. 1B,C,D). The results therefore suggest that the CZH2 region interacts specifically with at least three different sequences within the first 922 amino acids of zizimin1 (see schematic description of the interactions in Fig. 1A).
The CZH1 sequence homology domain is contained within zizimin1 residues 640–880 but zizimin1 fragments starting at or around amino acid 640 expressed poorly (not shown). Hence, a larger fragment (zizimin1534–922), which is better expressed, was used in the experiment presented in Fig.1B. To test if the minimal CZH1 homology area is sufficient for binding the CZH2, we used electroporation to obtain higher expression. As shown in Fig. 1E, HA-zizimin1-CZH2 co-immunoprecipitated specifically with Flag- zizimin1640–885. The CZH2-containing fragment utilized above (zizimin11512-end; Fig. 1) is also larger than the region identified by sequence homology (homology between the zizimin and DOCK180 proteins start at or around zizimin1 residue 1612 and is higher from 1717-end [16]). We showed previously that while the zizimin1 CZH2 fragment (zizimin11693-end) is sufficient for Cdc42 binding, zizimin11512-end bound Cdc42 with much higher affinity [10], suggesting that the added sequence is part of the functional domain. However, zizimin11693-end bound specifically to the zizimin1 N-terminal elements tested above (not shown). The minimal CZH1 and CZH2 homology regions are therefore sufficient for mutual binding.
Analysis of zizimin1 structure by limited proteolysis
The poor expression of the domains based on sequence homology suggested that the structural unit/s within zizimin1 may differ from the minimal CZH2 or CZH1 areas. Limited proteolysis has often been used to identify stable domains within a protein, based on the observation that proteolysis occurs preferentially at exposed and flexible loops, whereas tightly folded domains are resistant [26]. Limited digestion of zizimin1 by trypsin resulted in relatively stable fragments (not shown, but see Materials and Methods), which were resolved by SDS-PAGE and electroblotted to PVDF membranes (Fig. 2A). Their N-terminal sequences were determined by Edman degradation and approximate C-termini were calculated based on migration in SDS PAGE. The relative abundance among the fragments was estimated by comparing amino acid amounts in the different peptides sequenced.
Tryptic digestion of zizimin1, a 236 kDa protein, yielded multiple bands, which upon sequencing were often found to contain a few different fragments (Fig.2A,B). One fragment of high apparent abundance contained the N-terminus, PH domain and ∼50 additional residues past the PH domain, while a second highly abundant fragment contained the C-terminal ∼170 amino acids. The results also identified a major cleavage site in the vicinity of residue 1250, which split the molecule in two slightly unequal halves (Fig. 2C). The C-terminal 170 aa fragment is included in the CZH2 homology domain (Fig. 2C). A similar fragment (zizimin11876-end) is required, but not sufficient, for Cdc42 binding ([10] and Meller,et al. unpublished). We therefore hypothesize that the 170 aa C-terminal fragment represents a subdomain within the GEF domain, while the full domain may begin at the central cleavage site (at the vicinity of residue 1250). We also hypothesize that the CZH1 homology area is part of a larger domain that includes either the entire N-terminal half of the protein, (1–1250) or from the cleavage site at residue 335 to around 1250.
Based on these predictions, several zizimin1 constructs were made and relative expression in Cos7 cells tested (not shown). High expression was observed for fragments containing either the C-terminal half of the protein (zizimin11254-end and even higher for zizimin11183-end) or the N-terminal half (zizimin11–1184, zizimin11–1215 or zizimin11–1253). The N-terminal unit (zizimin11–335) also expressed well. However, deletion of residues 1–334 resulted in fragments (zizimin1335–1253 or zizimin1335–986) that expressed poorly. This small N-terminal region may therefore constitute a subdomain within a larger N-terminal domain.
High affinity binding between the N- and C-terminal fragments
The above data identify two large structural domains, an N-terminal half that includes the PH and CZH1 domains and a C-terminal half that includes the GEF domain. The N-terminal region contains at least three binding sites for the CZH2 domain. Association between the two halves was further tested by co-immunoprecipitation. HA-tagged zizimin11183-end (C-terminal half) was co-transfected into Cos7 cells with Flag-tagged zizimin11–1253 (N-terminal half) or with Flag-PH domain. Immunoprecipitation with antibody to Flag revealed specific association of the two halves of the molecule (Fig. 3A). Comparison of whole cell extracts and immunoprecipitates suggests that the two halves bind very efficiently (see Fig. 3A legend). As an additional control, binding of full-length zizimin1 to the N-terminal half was also tested. A specific interaction was observed, however, full-length zizimin1 bound much less than the N-terminal half (Fig. 3A). These data suggest that in the full-length protein, the C-terminal domain is preferentially occupied by the adjacent N-terminal-half through intramolecular interactions.
Interaction between zizimin1 N- and C-terminal halves inhibit Cdc42 binding
Zizimin1 binds and activates Cdc42 through the CZH2 domain [10]. To test if the interaction between the N-terminal elements and the CZH2 region affects Cdc42 binding, we first examined Cdc42 binding by the CZH2 construct with and without bound zizimin11–922. Cos7 cells were co-transfected with Flag-CZH2 and HA-zizimin11–922 and cell lysates were immunoprecipitated with anti-HA antibodies to isolate only CZH2 fragments that are associated with zizimin11–922. In parallel, an equal amount of CZH2 was immunoprecipitated using anti-Flag antibodies from cells expressing only Flag-CZH2. These immunoprecipitates were then analyzed for binding nucleotide-depleted (ND)-Cdc42. The zizimin11–922:CZH2 complexes bound about 50% as much Cdc42 as the free CZH2 (Fig. 3B). However, the CZH2 exists as a dimer [24] so that up to 50% of the CZH2 fragment in the zizimin11–922 immunoprecipitates may still be available for Cdc42 binding. The inhibitory effect may therefore be underestimated.
To further explore this idea, we examined how the interaction between zizimin1 N- and C-terminal halves affected Cdc42 binding. Cos7 cells were transfected with full-length zizimin1 or with the isolated C-terminal half. Binding of these constructs to ND-Cdc42 showed that the C-terminal half bound ∼4X better than full-length zizimin1 (Fig. 3C). Deletion of the N-terminal half therefore enhances Cdc42 binding, again suggesting that the N-terminus inhibits Cdc42 binding by the GEF domain.
As an additional approach, zizimin1 N- and C-terminal halves were purified and binding of the C-terminus to Cdc42 tested in the absence or presence of the N-terminus. The zizimin1 N-terminus inhibited Cdc42 binding to the C-terminus by ∼70% (Fig. 4), despite the modest (5 fold) molar excess of the N-terminal half over the C-terminal half. These results support an autoinhibitory mechanism whereby elements from the N-terminal half inhibit interaction of the C-terminal GEF domain with Cdc42.
Zizimin1 PH domain mediates membrane targeting
We next tested whether the predicted PH domain [10] contributes to zizimin1 membrane targeting. Full-length zizimin1 or a deletion mutant lacking the PH domain (zizimin1Δ287) were expressed in NIH 3T3 cells. Cells were separated into cytoplasmic and particulate fractions and zizimin1 distribution tested by Western blotting. As shown in Fig.5A, the proportion of ziziminΔ287 in the particulate fraction was markedly reduced when compared to wild type (WT) protein. To determine whether the PH domain is sufficient for membrane targeting, WT PH domain or PH domain with mutation of a conserved lysine (K183A) were expressed in NIH 3T3 cells and cells fractionated. As shown in Fig. 5B, the WT PH domain localized significantly to the particulate fraction whereas the mutated domain was hardly detected. The results therefore suggest that the PH domain is important for zizimin1 membrane targeting.
We next investigated the binding of zizimin1 PH domain to phosphoinositides and phospholipids. Full-length zizimin1 or the isolated PH domain were purified from Cos7 cells. Lipids dot blots were overlaid with purified proteins and binding detected by immuno-blotting. The PH domain bound to a variety of lipids, with highest binding to mono-phosphorylated inositides and lower but detectable binding to di-phosphoinositides and phosphatidylserine (Fig. 5C). The full-length protein displayed similar promiscuous binding (Fig. 5C). However, its affinity appeared to be higher, as 17 fold less protein yielded similar or higher signals (Fig. 5C). We hypothesize that zizimin1 dimerization [24] may account for the higher binding.
Analysis of zizimin interaction with active Cdc42
The zizimin subfamily include three genes that we named zizimin1, zizimin2 and zizimin3 [10, 16, 20]. Lin et al. recently described an interaction of zizimin2 (also named DOCK11 or ACG) with active Cdc42 and proposed that it may provide a positive feedback for zizimin2 activation [25]. Full-length zizimin2 bound preferentially to active Cdc42 mutant (Q61L) compared to the Cdc42 mutant with reduced affinity for nucleotides (T17N), whereas the zizimin2 CZH2 domain bound preferentially to T17N Cdc42 [25]. We evaluated zizimin1 interactions with GTP-loaded (active) Cdc42. As a positive control, zizimin2 was analyzed in parallel. Lysates of cells overexpressing full-length zizimin1 or zizimin2 were incubated with GDP-, GTP- or nd-Cdc42 beads and binding assayed. GTP loading of Cdc42 was confirmed by probing for the Cdc42 effector PAK. Zizimin1 bound strongly to nd-Cdc42 while binding to GTP-Cdc42 was hardly detected and binding to GDP-Cdc42 is not detected at all (Fig. 6). Zizimin2 also bound preferentially to nd-Cdc42, however its interaction, with GTP-Cdc42 was substantially higher than for zizimin1 (Fig. 6).
DISCUSSION
Among CZH family unconventional GEFs, significant progress has been made towards understanding regulation of DOCK180 (reviewed in [16]). DOCK180 is activated by binding ELMO proteins, which can be regulated by RhoG [27–30]. DOCK180 can also be recruited to the membrane through Crk adapter proteins [31–33]. Much less, however, is known about how zizimin and zir subfamily members are regulated.
Our data show that the N-terminal half of zizimin1 interacts with the CZH2 GEF domain through three distinct regions. This interaction inhibits Cdc42 binding to the GEF domain, as indicated by both direct inhibition and by enhanced Cdc42 binding to the isolated C-terminus. Altogether, our results suggest that zizimin1 interaction with Cdc42 is autoinhibited. The mechanism for zizimin1 activation is not known. As the PH domain bind lipids we tested effects of lipid binding on zizimin1 autoinhibition by adding lipids to co-IPs of the N- and C-terminal domains. The results, however, demonstrated no effect of lipid binding on association of zizimin1 C-half with the N-half (not shown). Detailed elucidation of activation mechanisms for zizimin1 must await identification of upstream receptors and pathways that control its activity. However, identification of the intermolecular interactions described here may facilitate the search for upstream receptors as well as the elucidation of detailed biochemical mechanisms.
The CZH1 and CZH2 domains were identified as areas of sequence homology between the DOCK180- and zizimin-related proteins [10, 12]. These sequence homologies suggested that these regions may carry out common functions in the two subfamilies. In support of this idea, the CZH2 area was shown to overlap with a GEF domain in both DOCK180 and zizimin proteins [16]. We note, however, that in both cases fragments ∼100 residues larger than the CZH2 homology area were substantially more active [10, 12, 13]. Though we cannot exclude secondary effects on conformational stability, this result could indicate that the functional GEF domains are larger than the homology areas. If so, they would include sequences not conserved between DOCK180 and zizimin proteins. Evidence for differential mechanism of action also exists as the DOCK180 GEF domain cooperates with ELMO to facilitate nucleotide exchange [29] while by all indications zizimin1 does not.
These results point toward some interesting similarities and differences between DOCK180 and zizimin. An autoinhibitory mechanism in DOCK180 was described recently whereby the N-terminal SH3 domain interacts with its C-terminal GEF domain to inhibit Rac binding [30]. We also note a study reporting binding of phosphatidylinositol (3,4,5)P3 to the DOCK180 CZH1/DHR1 region and its requirement for DOCK180 targeting to membranes [34]. However, this result might be taken to suggest that the PH domain from ELMO could replace the zizimin PH domain, the ELMO PH domain is required for the interaction of the complex with Rac, rather than membranes [29].
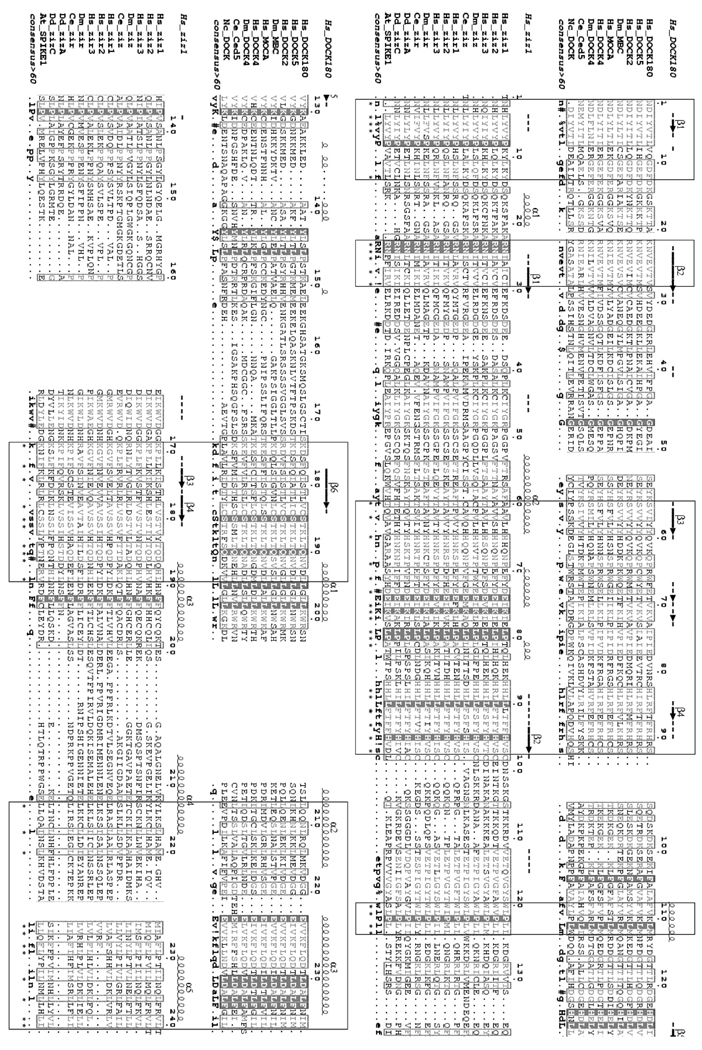
Though limited proteolysis data must be interpreted with caution, these results are consistent with the CZH1 and 2 domains being larger than the domains identified by sequence homology. The fragments exhibiting highest expression and function were made using domain boundaries that were not evident from the sequence homologies, suggesting the existence of substantially larger structural, if not functional, domains. Furthermore, comparison of multiple alignments of CZH1 sequences from DOCK180- and zizimin-related proteins demonstrate great variability between the two subfamilies (Fig. 7). Thus, the minimal CZH1 domain defined by sequence homology may not be an independent domain. Further assessment of zizimin domain structure awaits crystallization and high resolution analysis.
Fig. 7. Sequence alignment of CZH1 domains.
CZH1 sequences of zizimin-related and DOCK180-related proteins were aligned separately using the Multalin program and consensus sequences were generated [35]. Residues sharing >60% similarity are typed in gray while residues sharing 100% identity are presented on dark background. Secondary structure predictions for zizimin1 and DOCK180 CZH1 domains were made by the PHD program [36] and are shown on top of the alignments. The two resulting alignments were aligned together. Stretches showing resemblance to the consensus sequences are boxed and similar columns are marked by asterisks. ! represents I or V; # represents any one of N, D, Q or E; and $ represents Y or F. The numbering is according to zizimin1, or DOCK180, with 1 being residue 640 or 426, respectively. The approximate boundaries of the domain were determined as follows: The zizimin1 sequence was truncated into overlapping 200–500 amino acid segments and each segment was searched against the NCBI NR database using PSI-BLAST at an E-value threshold of 1e−2. BLAST hits were retrieved and separate multiple alignments were constructed for each segment using CLUSTALW [37]. Areas of homology between zizimin and DOCK180 proteins were identified and confined on the basis of alignment quality and secondary structure prediction. Hs, Homo sapiens; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Nc, Neurospora crassa; Dd, Dictyostelium discoideum; At, Arabidopsis thaliana.
Lin et al. recently described preferential binding of zizimin2 to constitutively active Cdc42 mutant (Q61L) when compared to a Cdc42 mutant with reduced affinity for nucleotides (T17N) [25]. However, our results demonstrate highest binding of zizimin1 and zizimin2 to nd-Cdc42 (Fig. 6). We note also that we cloned zizimin1 using a biochemical screen for proteins that bound nd-Cdc42 and were released upon addition of GTP, parameters that characterize GEFs [10]. In the same screen we detected also zizimin2 [10]. Currently we have no explanation for the apparent discrepancy between our results those of Lin et al. though differential affinity of zizimin proteins to nd-Cdc42 vs. N17 Cdc42 most likely account for it.
REFRENCES
- 1.Sorokina EM, Chernoff J. Rho-GTPases: new members, new pathways. J. Cell. Biochem. 2005;94:225–231. doi: 10.1002/jcb.20327. [DOI] [PubMed] [Google Scholar]
- 2.Wennerberg K, Der CJ. Rho-family GTPases: it’s not only Rac and Rho (and I like it) J. Cell Sci. 2004;117:1301–1312. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
- 3.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 4.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 5.Cerione RA. Cdc42: new roads to travel. Trends Cell Biol. 2004;14:127–132. doi: 10.1016/j.tcb.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- 7.Etienne-Manneville S. Cdc42 - the centre of polarity. J. Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 8.Cherfils J, Chardin P. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- 9.Hart MJ, Sharma S, elMasry N, Qiu RG, McCabe P, Polakis P, Bollag G. Identification of a novel guanine nucleotide exchange factor for the Rho GTPase. J. Biol. Chem. 1996;271:25452–25458. doi: 10.1074/jbc.271.41.25452. [DOI] [PubMed] [Google Scholar]
- 10.Meller N, Irani-Tehrani M, Kiosses WB, Del Pozo MA, Schwartz MA. Zizimin1, a novel Cdc42 activator, reveals a new GEF domain for Rho proteins. Nat. Cell Biol. 2002;4:639–647. doi: 10.1038/ncb835. [DOI] [PubMed] [Google Scholar]
- 11.Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr. Opin. Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 12.Cote JF, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 2002;115:4901–4913. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- 13.Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, Ravichandran KS. Unconventional Rac-GEF activity is mediated through the Dock180 ELMO complex. Nat. Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 14.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 16.Meller N, Merlot S, Guda C. CZH proteins: a new family of Rho-GEFs. J. Cell Sci. 2005;118:4937–4946. doi: 10.1242/jcs.02671. [DOI] [PubMed] [Google Scholar]
- 17.Marinissen MJ, Gutkind JS. Scaffold proteins dictate Rho GTPase-signaling specificity. Trends Biochem Sci. 2005;30:423–426. doi: 10.1016/j.tibs.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Miyamoto Y, Yamauchi J, Sanbe A, Tanoue A. Dock6, a Dock-C subfamily guanine nucleotide exchanger, has the dual specificity for Rac1 and Cdc42 and regulates neurite outgrowth. Exp. Cell Res. 2007;313:791–804. doi: 10.1016/j.yexcr.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 19.Watabe-Uchida M, John KA, Janas JA, Newey SE, Van Aelst L. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron. 2006;51:727–739. doi: 10.1016/j.neuron.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 20.Nishikimi A, Meller N, Uekawa N, Isobe K, Schwartz MA, Maruyama M. Zizimin2: a novel, DOCK180-related Cdc42 guanine nucleotide exchange factor expressed predominantly in lymphocytes. FEBS Lett. 2005;579:1039–1046. doi: 10.1016/j.febslet.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 22.Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 23.Lemmon MA. Pleckstrin homology domains: not just for phosphoinositides. Biochem. Soc. Trans. 2004;32:707–711. doi: 10.1042/BST0320707. [DOI] [PubMed] [Google Scholar]
- 24.Meller N, Irani-Tehrani M, Ratnikov BI, Paschal BM, Schwartz MA. The novel Cdc42 guanine nucleotide exchange factor, zizimin1, dimerizes via the Cdc42-binding CZH2 domain. J. Biol. Chem. 2004;6 doi: 10.1074/jbc.M404535200. in press. [DOI] [PubMed] [Google Scholar]
- 25.Lin Q, Yang W, Baird D, Feng Q, Cerione RA. Identification of a DOCK180-related guanine nucleotide exchange factor that is capable of mediating a positive feedback activation of Cdc42. J. Biol. Chem. 2006;281:35253–35262. doi: 10.1074/jbc.M606248200. [DOI] [PubMed] [Google Scholar]
- 26.Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim. Pol. 2004;51:299–321. [PubMed] [Google Scholar]
- 27.DeBakker CD, Haney LB, Kinchen JM, Grimsley C, Lu M, Klingele D, Hsu PK, Chou BK, Cheng LC, Blangy A, Sondek J, Hengartner MO, Wu YC, Ravichandran KS. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr. Biol. 2004;14:2208–2216. doi: 10.1016/j.cub.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 28.Katoh H, Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 2003;424:461–464. doi: 10.1038/nature01817. [DOI] [PubMed] [Google Scholar]
- 29.Lu M, Kinchen JM, Rossman KL, Grimsley C, deBakker C, Brugnera E, Tosello-Trampont AC, Haney LB, Klingele D, Sondek J, Hengartner MO, Ravichandran KS. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat. Struct. Mol. Biol. 2004;11:756–762. doi: 10.1038/nsmb800. [DOI] [PubMed] [Google Scholar]
- 30.Lu M, Kinchen JM, Rossman KL, Grimsley C, Hall M, Sondek J, Hengartner MO, Yajnik V, Ravichandran KS. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr. Biol. 2005;15:371–377. doi: 10.1016/j.cub.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 31.Li L, Guris DL, Okura M, Imamoto A. Translocation of CrkL to focal adhesions mediates integrin-induced migration downstream of Src family kinases. Mol. Cell Biol. 2003;23:2883–2892. doi: 10.1128/MCB.23.8.2883-2892.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiyokawa E, Hashimoto Y, Kurata T, Sugimura H, Matsuda M. Evidence that DOCK180 up-regulates signals from the CrkII-p130(Cas) complex. J. Biol. Chem. 1998;273:24479–24484. doi: 10.1074/jbc.273.38.24479. [DOI] [PubMed] [Google Scholar]
- 33.Kiyokawa E, Mochizuki N, Kurata T, Matsuda M. Role of Crk oncogene product in physiologic signaling. Crit Rev Oncog. 1997;8:329–342. doi: 10.1615/critrevoncog.v8.i4.30. [DOI] [PubMed] [Google Scholar]
- 34.Cote JF, Motoyama AB, Bush JA, Vuori K. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 2005;7:797–807. doi: 10.1038/ncb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. Improved sensitivity of profile searches through the use of sequence weights and gap excision. Comput. Appl. Biosci. 1994;10:19–29. doi: 10.1093/bioinformatics/10.1.19. [DOI] [PubMed] [Google Scholar]