Abstract
The integration of semiconductor nanoparticle quantum dots (QDs) into a modular, microfluidic biosensor for the multiplexed quantitation of three important cancer markers, carcinoembryonic antigen (CEA), cancer antigen 125 (CA125), and Her-2/Neu (C-erbB-2) was achieved. The functionality of the integrated sample processing, analyte capture and detection modalities was demonstrated using both serum and whole saliva specimens. Here, nano-bio-chips that employed a fluorescence transduction signal with QD-labeled detecting antibody were used in combination with antigen capture by a microporous agarose bead array supported within a microfluidics ensemble so as to complete the sandwich-type immunoassay. The utilization of QD probes in this miniaturized biosensor format resulted in signal amplification 30 times relative to that of standard molecular fluorophores as well as affording a reduction in observed limits of detection by nearly 2 orders of magnitude (0.02 ng/mL CEA; 0.11 pM CEA) relative to enzyme-linked immunosorbent assay (ELISA). Assay validation studies indicate that measurements by the nano-bio-chip system correlate to standard methods at R2 = 0.94 and R2 = 0.95 for saliva and serum, respectively. This integrated nano-bio-chip assay system, in tandem with next-generation fluorophores, promises to be a sensitive, multiplexed tool for important diagnostic and prognostic applications.
Keywords: Salivary Diagnostics, Cancer Biomarkers, Quantum Dots, Nano-Bio-Chip, Lab-on-a-Chip, Point-of-Care
1. INTRODUCTION
Protein biomarkers show promise in diagnosis and treatment planning of cancer early in its development through screening as well as staging, metastasis evaluation, and determining response to pharmacologic intervention (Lee et al., 2008). Occult blood tests, PAP smears, and prostate specific antigen (PSA) are established screening tools for cancer detection in asymptomatic individuals (ACS 2008). Breast cancer companion diagnostic tests based on Her-2/Neu and prognostic assays utilizing alpha-fetoprotein in testicular cancers have also been developed (Soper et al., 2006). Additional progress has been made using protein biomarkers to monitor colon cancer response to treatment via carcinoembryonic antigen (CEA) or ovarian cancer via cancer antigen 125 (CA125) (Das and Bast, 2008). In spite of these advances, widespread use of protein biomarkers remains elusive due to methodological and technological obstacles.
Early stage detection of cancer frequently requires the use of four or more biomarkers to identify at-risk individuals with adequate confidence and currently used methods, based on ELISA, are not easily multiplexed (Harris et al., 2007). Furthermore, in order to diagnose a cancer in its nascency, physicians recommend regular and frequent screening events—an arrangement incompatible with the high cost, lengthy turnaround times, and intrusive sampling procedures of current assay designs. In addition, most of the currently used screening methods have limits of detection (LODs) near the diagnostic decision values that provide optimal separation between clinical categories, creating difficulties in accurately evaluating patients in early stages of neoplastic disease, when biomarker concentrations are only slightly altered (Bhasin et al., 2008).
To satisfy these unmet clinical needs, integrated microfluidic systems have become an increasingly attractive alternative to the centralized laboratory paradigm. Although these so-called “lab-on-a-chip” (LOC) systems have many potential advantages including small sample size, low cost, and short analysis times, their most significant contribution to disease prevention lies in their ability to complete multiplexed analyses at the point-of-care (POC). These POC systems allow clinicians to have early, routine, and frequent access to a wide array of diagnostic test results, information that is conventionally only available at large oncology-focused hospitals or via lengthy and expensive reference laboratories. To supplement further the potential for facile screening events, an increasing number of POC methods have begun using saliva as a diagnostic medium (Wong, 2008).
Indeed, there is growing evidence that saliva serves as a “mirror of the body” and contains biomarkers and components of the serum proteome that offer important information on both oral and systemic disease (Mandel, 1993, Malamud, 2006). Important studies completed by Wong, Oppenheim, Malamud, Streckfus and others have resulted in critical information related to the genomic and proteomic profiles that are present in saliva and other glandular secretions (Hu et al., 2007, Oppenheim et al., 2007). Whole saliva is simple to collect and store and is obtained easily in sufficient quantities for testing. For patients, the non-invasive collection method of oral fluid sampling reduces anxiety and discomfort, thereby ameliorating testing antipathy and promoting more frequent screening events. In addition, as a constantly regenerated fluid, saliva offers a better ‘physiological snapshot’ and work by Wong and Singh and others has explored the use of saliva for POC analyses (Herr et al., 2007, Tan et al., 2008). Despite the obvious advantages offered by saliva, there are also distinct challenges associated with its use. The saliva sample matrix is more variable and heterogeneous than serum, and contains high levels of mucins and proteolytic enzymes (Streckfus et al., 2006). In addition, disease-relevant biomarkers may be expressed at concentrations several orders of magnitude lower than in serum, although they still reflect nicely the expression profiles of biomarkers found in blood that are of diagnostic significance (Streckfus et al., 2001). Thus, because of the low levels of salivary biomarkers, it sometimes becomes difficult to distinguish between background and target-specific signal in these low concentration samples.
The use of advanced detection methodologies, such as semiconductor nanoparticle quantum dot (QD) fluorophores, promises to help overcome some of the challenges inherent to using saliva in POC devices. These nanoparticles are composed of a CdSe core and ZnS coat followed by a polymer passivation layer resulting in particles ranging from 5 to 20 nm (Liu et al., 2008). The long term photostability, strong intensity, and size tunable, narrow emission profile of QDs, in tandem with their broad UV excitation spectra have made them useful qualitative and quantitative probes for a number of applications both on the macro and micro size regime. Despite this work, no universal approach for linking QDs to bioligands for POC cancer diagnostics/prognostics has emerged, a challenge addressed in Soper’s recent review (Soper et al., 2006). Although many excellent examples of QDs as qualitative imaging tools exist in the literature, complete integration of QDs as quantative probes, especially in tandem with similarly small analyzer platforms has yet to be demonstrated (Resch-Genger et al., 2008).
Contributing towards this effort to develop new systems for early detection of neoplastic diseases, recent work in our lab developed miniaturized devices that serve as “nano-bio-chip” (NBC) sensors amenable to analysis at the POC (Ali et al., 2003, Christodoulides et al., 2007, Weigum et al., 2007). The NBC ensemble employs a size-tunable nano-net within agarose microspheres, and a fluorescent transduction signal arising from nanoparticles (nano) to isolate and quantify biological analytes (bio) from complex matrices within a closed, miniaturized system (chip). Importantly, the performance metrics of these miniaturized sensor systems correlate well with and, in many cases, out-perform, established gold standard macroscopic analysis methods, making them ideally suited for the analysis of complex fluids, such as oral samples.
In this study, we describe the incorporation of QDs as detection elements in the NBC system for the measurement of three well-characterized cancer biomarkers: CEA, CA125, and Her-2/Neu (C-erbB-2). These NBC assay systems are explored in their capacity to serve as sensitive and specific cancer diagnostic tools for use with both serum and saliva samples. Here, the advantages of QDs relative to gold standard molecular fluorophores, are explored in a POC-compatible device using a completely integrated system including all optics, mechanical and microfluidic elements in tandem with clinical samples.
2. MATERIALS AND METHODS
2.1. Immunoreagents
Phosphate buffered saline (PBS) from Thermo Scientific (Waltham, MA) at pH 7.4, used as a wash buffer and diluent of the protein standards, was prepared fresh daily and filtered with a 0.2 µm membrane filter. To maintain protein stability and reduce non-specific binding, bovine serum albumin from Sigma Aldrich (St. Louis, MO) (BSA) was added at 1% by weight to PBS. Detection and capture monoclonal antibody pairs, specific to CEA and CA125, as well as an isotype control antibody were purchased from Fitzgerald (Concord, MA); Her-2/Neu-specific antibodies were acquired from R&D Biosystems (Minneapolis, MN). Recombinant CEA, CA125, and Her-2/Neu antigen were obtained from Biodesign (Saco, MA), Fitzgerald, and R&D Biosystems, respectively. These proteins were used without further purification and diluted in PBS/BSA to appropriate concentrations. Nanoparticle QDs with emission maximum at 565 nm (QD 565) and 655 nm (QD 655) were purchased from Invitrogen (Eugene, OR) as were reagents for a succinimidyl 4-(N-maleimidomethyl) cyclohexanecarboxylate (SMCC) facilitated antibody conjugation. Additional QDs were purchased from Evident Technologies (Troy, NY). Alexa Fluor 488 and reagents for immunolabeling were purchased from Invitrogen along with purification supplies. Absorbance and fluorescence spectroscopy were performed on a SpectraMax Plus and SpectraMax GeminiXS microtiter spectrophotometer, respectively, both from Molecular Devices (Sunnyvale, CA).
Passivated CdSe/ZnS QDs were linked to antibody by established techniques (Xing et al., 2007). Briefly, intact IgG molecules were subjected to dithiothreitol (DTT) treatment to cleave disulfide bonds. Incubation of SMCC-activated QD particles with the reduced antibody fragments linked them to the surface. The conjugate was purified by size exclusion chromatography to remove unbound antibody and concentrated with 50 kD cutoff centrifuge tubes to approximately 1 µM. Monoclonal detection antibodies were labeled with Alexa Fluor 488 via a tetrafluorophenyl activated form of the dye. Please see Supplementary Information S.1 and S.2 for additional details regarding bead reactor construction and sample processing.
2.2. Data Interpretation
Photomicrographs were captured both before and after detection bioconjugate introduction to allow for background corrected images. These were saved as 12-bit colorized TIFF files and analyzed via NIH ImageJ software (Bethesda, MD) with bead fluorescence signal intensity correlating to the concentration of analyte in the sample. Automated deconvolution macros converted the image to a grayscale 8-bit image with areas of interest (AOIs) drawn corresponding to individual beads. The highest pixel intensity of successive line profiles across the AOIs yielded a mean signal, and beads with signal more than three standard deviations away from this mean, occasionally caused by either air bubbles or fluorescent bioconjugate precipitate, were excluded from further pixel analysis. For 8-bit images, these pixel bit depth values ranged from 0 to 255. Intensity versus concentration calibration curves were constructed with best-fit regression analysis for determination of unknown sample concentration.
3. RESULTS AND DISCUSSION
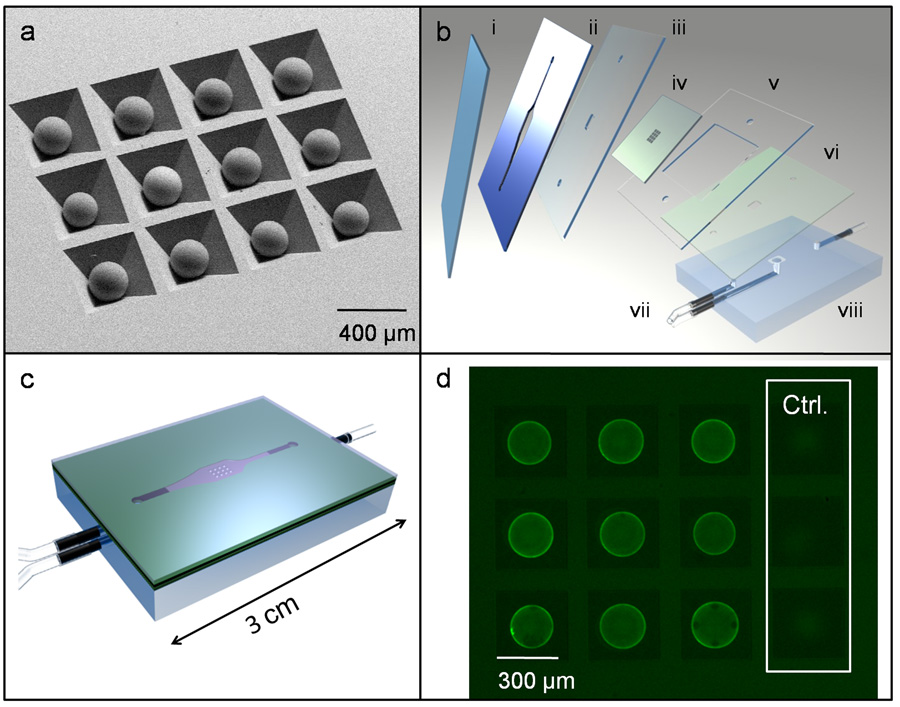
A micro-electro-mechanical systems (MEMS) platform with integrated microfluidics elements and bead containers, prepared from anisotropically etched Si-100 wafers, as described previously (Goodey et al., 2001), supports the integrated NBC assay investigations. The main objective of this new study is to characterize the analytical performance of cancer biomarker assays employing QDs as a detection moiety in the NBC. The QD-based platform includes all fundamental components of sandwich immunoassay, with the inclusion of agarose beads as solid phase support. Different clones of monoclonal antibody serve as capture and detection bodies. Each bead is individually addressable by location of well within the silicon chip and bead specificity determined by type of antibody covalently bound to bead (Fig. 1a). The beads are layered within the NBC between precision-cut layers of laminate adhesive designed for reagent delivery (Fig. 1b). These fluids are delivered through the top inlet of the flow cell apparatus, allowed to pass through and around the sensitized beads with nonreactive components passed on to waste through the lower port (Fig. 1c). The NBC is then used in tandem with benchtop microscope and peristaltic pump. Fluorophore conjugated detection antibody completes the immunocomplex and yields signal output digitally recorded by CCD camera. Day to day differences in microscope optics, camera, and excitation source power are monitored via fluorescent calibration slides and may be accounted for by tuning the light intensity via condenser optics; however, we have found our optical arrangement to be stable until approximately 250 hours of bulb lifetime. Thus, the intensity of fluorescence signal, embedded in a digital photomicrograph, relates directly the amount of antigen originally present in the sample (Fig. 1d). Please see Supplementary S.3 for additional details of NBC construction and use.
Figure 1.
(a) SEM photomicrograph of beads in anisotropically etched silicon chip. (b) Chip (iv) is fitted between double-sided adhesive layer (ii) and cover slip (i) with laminate layers (iii, v, vi) included to direct fluid flow through PMMA base (viii) and inlet and outlet ports (vii). (c) Sealed LOC assembly. (d) Fluorescent image of beads after immunoassay including negative controls as imaged with one second of CCD camera integration (exposure) time.
3.1 QD Integration into NBC
To use QDs for immunoassay, stabilized nanoparticles are linked to a recognition element, i.e. antibody, through a variety of covalent and non-covalent strategies with all techniques emphasizing the importance of retaining QD optical properties and hydrophilicity as well as recognition moiety specificity (Xing et al., 2007). Characterization of QDs in the transduction pathway of the integrated NBC employed such figures of merit as background, signal, and non-specific signal (noise). In typical arrangements, 9 bead replicates, with a concentration of 1.0 mg/mL capture antibody, reported specific signal and 3 beads with an irrelevant, isotype antibody served as negative reference controls (Fig. 1d). With a 4X objective, the deconvolution macro resulted in the averaging of 104 successive line scans across each 280 µm bead or a resolution of 2.69 µm/pixel. The mean of the 9 beads was defined as signal (S), with the non-specific signal emanating from 3 isotype control beads termed noise (N); background (B) was defined as output from biomarker-specific beads in the absence of antigen. After determining the most intense commercially available QDs (see Supplementary S.4), non-specific interactions were examined. First, QDs unbound to antibody, were examined without antigen present and yielded a S/N ratio of 1.05. When 100 ng/mL of CEA antigen was included and experiment repeated with bare QD particles, S/N was 0.70. These ratios, in addition to low signal output (S < 2), for these two experiments indicates that the minimal background and noise later observed should be attributed to non-specific biological interactions, and not to aberrant binding of nanoparticle to immobilized antigen or bead scaffold. Next, this investigation explored the most appropriate method to prepare QD-antibody bioconjugates.
Previously, excellent labeling was observed in the NBC via a secondary antibody strategy in which QD was thiol bound to secondary F(ab’)2 which was then linked to primary antibody on the cell surface (Jokerst et al., 2008). This scheme is incompatible with the bead-based assays, as the secondary antibody binds to the capture as well as detecting molecules. Other groups report using the avidin/biotin pathway for labeling biological analytes with QDs (Zajac et al., 2007). Although biotinylated detecting antibody and avidin-coated QDs successfully reported mid-level antigen values at S > 60, it unfortunately resulted in non-specific noise and yielded S/N values of 0.82. The poor response observed for this application may be due to the inability to remove the relatively large unbound QD-avidin-biotin-antibody immunocomplex from the bead’s nano-net pores. For the CEA assay, this structure consists of a total immunocomplex length of 74 nm and molecular weight 550 kD. This size is on the order of the 100 to 300 nm pore diameters typical of 2% agarose gels making these conditions consistent with the clogging of bead apertures or QD probe trapped non-specifically in bead interior (Narayanan et al., 2006).
To engineer bioconjugate more compatible with the agarose environment, the protein capture elements were tailored to a size roughly half the QD diameter, or 7.5 nm. By reducing whole antibody via DTT and cleaving the disulfide IgG backbone, steric crowding within the bead was reduced while retaining the analyte recognition portion of the antibody. Preparation of the QD component of the bioconjugate proceeded via SMCC activation of PEG-carboxy coated nanoparticles (Pathak et al., 2007). The exposed free sulfhydryls on cleaved IgGs then reduced onto activated QDs yielding an intense, specific transduction bioconjugate that was stable up to one year from preparation.
Through titration studies in the NBC using a constant amount of antigen (50 ng/mL) assayed with a varying amount of detecting antibody (data not shown), 3.7 nM of the QD element of the detection body (0.47 µg/mL of antibody) was determined to be optimal for labeling efficiency while keeping background signal to a minimum. As inherently hydrophobic materials, the tendency of QDs to aggregate at high concentration can be a challenge for use. Detecting antibody bioconjugates at 3.7 nM in PBS/BSA, in tandem with 0.2 µm nylon filtration of antibody-QD conjugate immediately prior to introduction into the NBC eliminated flocculation. To verify specificity of the QD-antibody bioconjugate and determine B, an antigen free sample (buffer only), was analyzed. After washing steps, B and N values below 1 resulted. Upon inclusion of the CEA antigen, a signal on the CEA sensitized beads developed proportional to amount of antigen introduced. For example, at 10 ng/mL antigen, S = 52.85 and S/N = 17.17. Importantly, the S values drop by 97.4% when a Her-2/Neu detecting antibody replaces the QD-CEA bioconjugate, demonstrating specificity of the reporter probe. Values for N were 3.1% of S at mid level antigen concentrations, further authenticating immunoassay specificity; N values remain consistently low across the range of antigen concentrations used in these experiments. Results of background and isotype-controlled experiments for the Her-2/Neu and CA125 antigen were similar to those of CEA, with N typically less than 10% of S, and B values below 2.
3.2 The QD Advantages Relative to Molecular Fluorochromes
The next objective was to compare the performance of QDs to traditional fluorophores. Such comparisons have challenges including the maintenance of similar excitation energies and antibody/fluorophore ratios (Resch-Genger et al., 2008). To perform a stringent comparison, this study utilized Alexa Fluor 488 (AF488) as it is one of the brightest fluorophores available for contrast versus QD probes; see Supplementary S.4 (Panchuk-Voloshina et al., 1999). To characterize the exact amount of both antibody and fluorophore in the AF488 and QD bioconjugate stock, quantitative absorption spectroscopy was employed. First, calibration curves using the monoclonal detecting antibody were constructed via a Bradford assay. These curves, in tandem with values for the fluorophore labeled bioconjugate, gave amount of IgG in the two detecting antibody preparations. Specifically, 5.9 × 10−6 M IgG was present in the AF488 conjugate and 7.6 × 10−7 M antibody was found for the QD case. To calculate amount of fluorophore, absorbance measurements were used with the extinction coefficients provided by fluorophore manufacturers; concentrations of 3.7 × 10−7 M QD and 2.5 × 10−5 M AF were measured. Since both bioconjugates were extensively purified with size exclusion chromatography, the fluorophore and IgG concentrations determined here reflected the amount of labeled detecting antibody. Thus, by taking the ratios of fluorophore and IgG, this specific batch of QD-antibody conjugate had an antibody to fluorophore ratio of 2:1. Due to the difference in size between QD (10 nm) and Alexa Fluor (1nm), more than one AF molecule may be bound to each antibody. Using the ratios of fluorophore to antibody as determined above, approximately four Alexa Fluor elements attach to each whole IgG antibody.
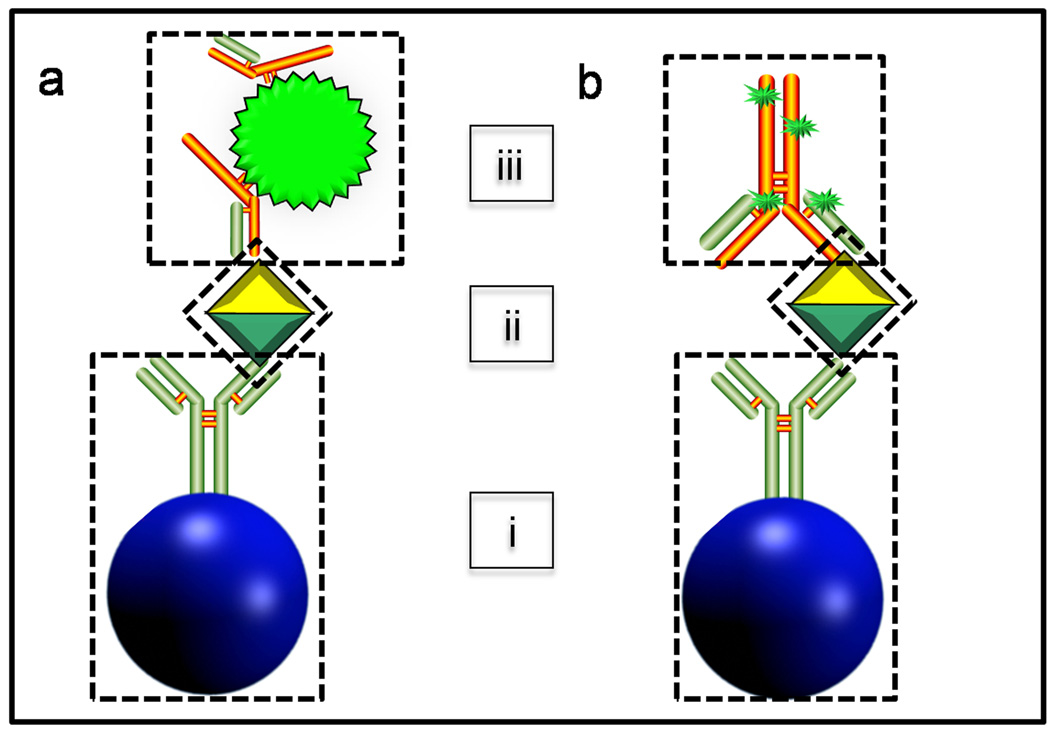
For comparison purposes, isomolar concentrations of recombinant CEA antigen (100 ng/mL) was analyzed under identical experimental conditions except for fluorophore on the detecting antibody. Two aliquots (one QD and one AF) of detecting antibody with isomolar concentrations of fluorophore in the bioconjugate were prepared (3.7 nM AF488 or 3.7 nM QD 565) and used to label antigen, and images for both QD (Fig. 2a) and AF488 (Fig. 2b) recorded. This required a 1:100 dilution of the stock QD bioconjugate and a 0.015:100 dilution for stock AF488. Due to the larger size of the quantum dot (~15 nm with passivation layer) relative to the molecular AF dye, a greater number of antibodies may be bound to its surface (Immunoschematics in Fig. 2a and 2b). This necessitated an additional assay at isomolar concentration (0.47 µg/mL) of antibody in the AF488 bioconjugate.
Figure 2.
Immunoschematics employing QDs (a) or Alexa Fluor (b) retain similar characteristics including agarose bead as underlying support structure for capture antibody immobilization (i), antigen (ii), and detecting antibody/fluorophore conjugate (iii).
Comparison of S values for the three photomicrographs (1 QD-based and 2 AF-based) indicates values 33.5 times higher for QD versus AF488 at isomolar fluorophore concentration and 6.5 for the isomolar antibody experiment, all at 1 second of exposure time. Similar to QD, AF488 yielded N values less than 10% of S. In addition to whole IgG for AF488 labeling, AF488 bioconjugates consisting of Fab and F(ab’)2 antibody fragments were also explored. These values were lower by a factor of 1.5 relative to whole IgG. To determine if increase in QD signal was due to higher excitation energy, interrogation of the excitation optics with power meter at 400 and 480 nm, corresponding to the QD and Alexa Fluor optics, respectively, indicated a QD illumination strength advantage of 1.3. Thus, even when the relative concentrations of the labels are tracked carefully alongside the excitation powers, there remains a significant enhancement (25 times more signal isomolar fluorophore) of the QDs relative to AF488, one of the most robust commercially available fluorochromes.
3.3 Analytical Performance of NBC Assays
The utility of QDs in the NBC was characterized by determining the assay analytical performance, especially versus ELISA systems, the gold standard protein detection method since 1971 (Crowther, 1995). The first descriptor explored was assay time, which minimized four separate assay events: sample introduction, sample wash, detection bioconjugate incubation, and bioconjugate wash. On the NBC system, sample and bioconjugate incubation times were empirically determined by systemically reducing them from 30 minutes until the 5.0 ng/mL calibrator dropped below S/N of 10. Times of 5 minutes for sample flow (0.2 mL/min) and 15 minutes for bioconjugate (0.1 mL/min) were optimal and further reductions resulted in unacceptable reductions in LOD. For washing, 1 minute of 1.1 mL/min PBS/BSA rinse eliminates the vast majority of unbound immunocomplex. For very low backgrounds, however, a two-stage final washing scheme was critical, which used the stringent 1.1 mL/min flush, followed by a 5 minute, 0.2 mL/min, gentle cleanse cycle. This allowed both diffusion and convection to remove unbound bioconjugate deep within bead core. The assay detailed above results in a total analysis time of 27 minutes, versus the 4 – 24 hours needed for conventional ELISA and calibration slides and standards were again used here to account for any variance in excitation source intensity or microscope optics.
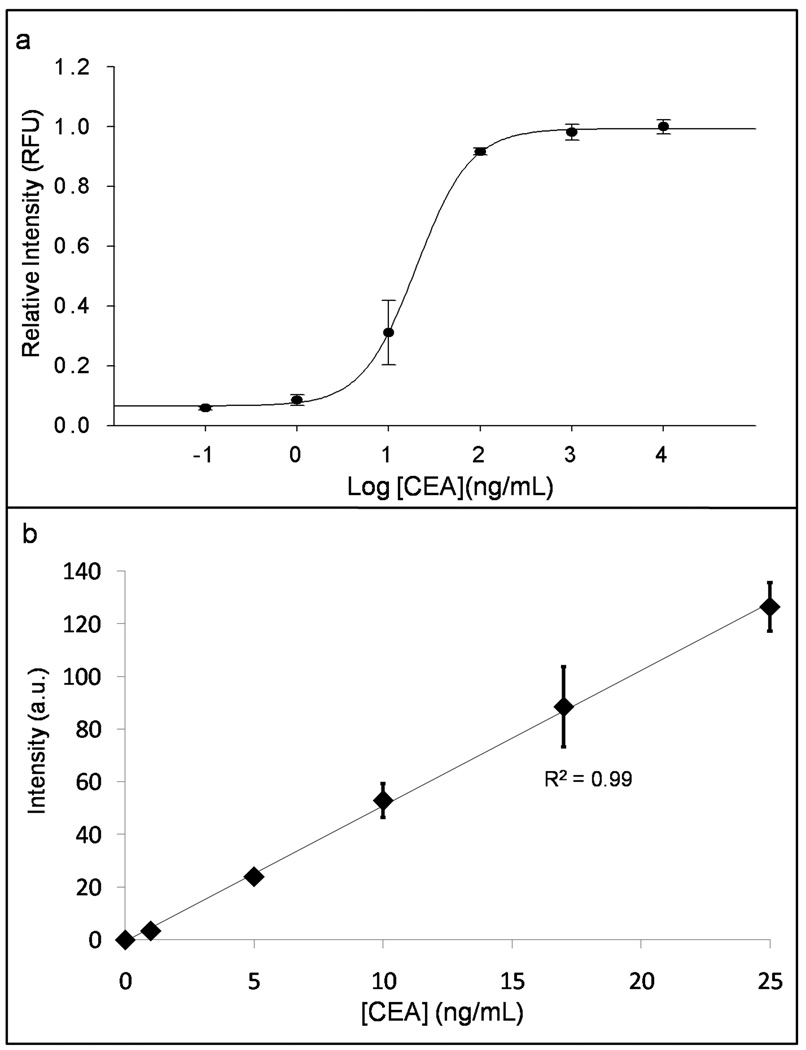
Next, calibration curves for all three analytes were constructed across a range of concentrations using recombinant antigen in buffer as well as a negative control, zero calibrator point. To determine the linear dynamic range, increasing concentrations of analyte across six orders of magnitude from the limit of detection to the saturation point of the assay gave differing intensities as detailed in Figure 3a for CEA. Here, it was critical that the linear dynamic range of the NBC assay overlap with the physiologically relevant range of analyte in biological samples. Commonly accepted disease diagnostic decision thresholds for the three analytes in serum are 2.5 ng/mL for CEA (5 ng/mL for smokers), 15 ng/mL Her-2/Neu, and 35 U/mL CA125 (Baron et al., 2005, Pallud et al., 2005). The three analytes on the NBC system gave linear dose responses across the range of 0 – 100 ng/mL CEA, 0 – 60 ng/mL Her-2/Neu, and 0 – 400 U/mL CA125. Although all had acceptable performance, CEA was the focus for the remainder of the quantitative studies and later optimized to give the very sensitive dose curve seen in Figure 3b. Calibration data above 25 ng/mL was unnecessary for routine analysis, especially upon sample dilution. Importantly, the sensitivity of the NBC for any of the analytes could be attenuated in order to have a wider linear dynamic range by decreasing either capture or detection antibody concentrations. Best-fit line regression analysis for the two methods gives S = 5.144 [CEA] + 11.86 (R2 = 0.99) with the NBC approach, and S = 0.0562 [CEA] + 0.2071 (R2 = 0.98) for ELISA at 0 – 50 ng/mL, where S, signal, is QD fluorescent intensity as described above in the NBC or absorbance at 450 nm (substrate λmax) in ELISA. When S is standardized relative to the highest calibrator, and NBC and ELISA data plotted on the same axis, sensitivity, as determined by the slope of a best-fit line, is 1.9 times larger with the NBC versus ELISA (plot not shown). Importantly, across the above range of concentrations, noise, or signal from isotype control beads is <10% of signal from the analyte-specific beads indicating specific recognition of antigen and antibody.
Figure 3.
CEA dose response curves were constructed by adding recombinant antigen to buffer followed by analysis with the NBC. a) Logarithmic figure spanning six orders of magnitude concentration of CEA with sharpest response overlapping physiologically relevant range from 0.1 to 100 ng/mL. Signal is relative to the maximum intensity produced by highest concentration calibration point and expressed as relative fluorescence units (RFU). b) Linear and sensitive dose response over region of typical sample concentration.
Low LODs are critical when analyzing saliva for biomarkers due to lower analyte concentration versus serum. The use of QDs allows for a high signal output at relatively low concentrations of detection element to maintain the low background signals critical for excellent LOD measurements. The LOD for CEA, defined at 3σ above background, was 0.02 ng/mL in the NBC, 2.61 ng/mL when the QD-based system was replaced with an AF488 signaling method, and 1.20 ng/mL in ELISA. For CEA, a glycosolated protein with a molecular weight of 180 kD, the LOD corresponds to 0.11 pM (Mitchell, 1980). Limit of quantitation (LOQ), defined as 10σ above background, was 0.10 ng/mL CEA. Similarly, low LODs were measured for Her-2/Neu with results of 0.27, 3.70, and 1.50 ng/mL for NBC-QD, NBC-AF488, and ELISA, respectively. Importantly, this order of magnitude improvement in LODs via QD detection compensates for the order of magnitude decrease in concentration of some biomarkers in saliva and also demonstrates the efficacy of the NBC for detecting trace amounts of biomarkers in diluted samples (Aps and Martens, 2005).
Reproducibility of the system was evaluated both from an intra-assay and inter-assay approach. As each analysis contains nine analyte beads, every assay results in a mean concentration with standard deviation (Fig. 3b). The RSD values are <10%, with lower values found at decreasing concentrations of analyte. Bead to bead variations are partly due to pressure-driven flow. Computational fluid dynamic experiments, to homogenize the flow conditions across all bead local environments, are ongoing. To evaluate precision between assays, one serum sample (mean value 8.60 ng/mL) was examined 6 times. The %CV was 6.5; variances for ELISA approach 40% depending upon analyte concentration (Choi et al., 2007). An additional study of linearity and precision utilized a single high volume, high concentration serum sample analyzed unadulterated and at five dilutions down to 1:32. Background corrected intensities were converted to concentration by calibration curve, and multiplied by dilution factors resulting in the corrected concentration. Fluorescence signal as a function of dilution factor was linear at R2 = 0.99.
3.4. Analytical Performance of NBC for Clinical Samples
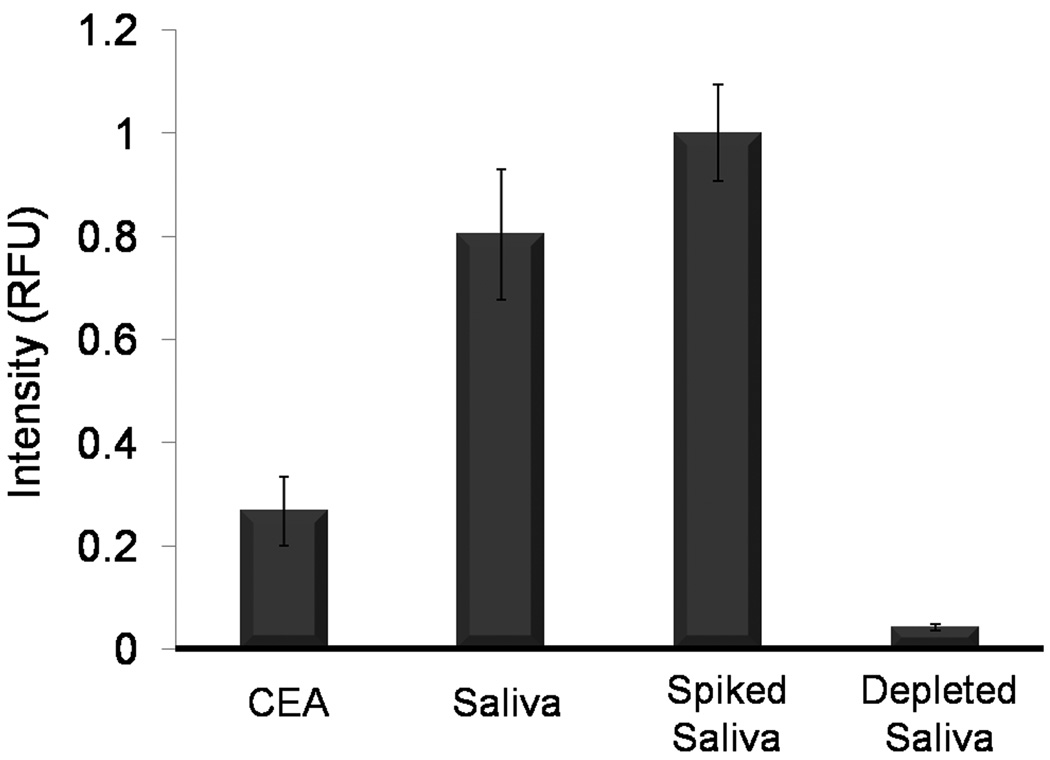
The NBC analytical performance was evaluated in the context of CEA determinations for both saliva and serum samples. For the initial stages of these clinical proof-of-principle studies, signal generated by saliva was compared to recombinant CEA antigen. Beads were exposed to recombinant CEA antigen (50 ng/mL), 1:4 dilutions of saliva in PBS/BSA, saliva (1:4) spiked with recombinant antigen (to 50 ng/mL), and saliva (1:4) depleted of CEA by immunoaffinity chromatography. The presence of signal in saliva, signal increase after spiking with antigen, but most importantly, the 94.84% reduction in signal after CEA depletion, offers powerful evidence for the validity of this saliva-based assay on the NBC and the authenticity of CEA-specific signal in saliva (Fig. 4).
Figure 4.
Validation of authentic CEA signal in saliva. Assay results of recombinant antigen at one half calibration curve maximum (50 ng/mL), saliva at 1:4 dilution, saliva sample spiked with recombinant antigen, and CEA-depleted saliva indicates that signal in saliva is authentic to CEA.
For a correlation study, whole saliva (n = 6) was collected from apparently healthy subjects and these specimens were analyzed with the NBC and ELISA. The methods correlate strongly with R2 value of 0.94. (Please see Supplementary S.5 for correlation data.) The relatively high concentration (> 50 ng/mL) of analyte present in saliva is consistent with previous reports and is reasonable, as CEA is a known epithelial protein (Nagler et al., 2006). In serum (S.5), the NBC platform also correlates nicely with an automated clinical analyzer (Siemens Centaur) and yields a R2 value of 0.95 for 24 samples. Although these results correlate nicely, the absolute values showed differences. These systematic differences are likely due to variations in the values for standards and/or changes in the specimen concentrations that resulted from the differences in sample handling at the two sites. Three serum samples of the 24 were above the 2.5 ng/mL CEA discriminator with the NBC, but below this value via the reference method. Additional studies using larger samples pools and more detailed diagnostic information are required to determine the clinical relevance of the NBC. Nevertheless, these proof-of-principle pilot studies yielded strong correlation results for both serum and saliva analysis using the new NBC relative to established laboratory-based methods and the low LODs observed, illustrate the potential to affect positive patient outcomes through saliva-based measurements.
3.5. Multiplexing Capabilities
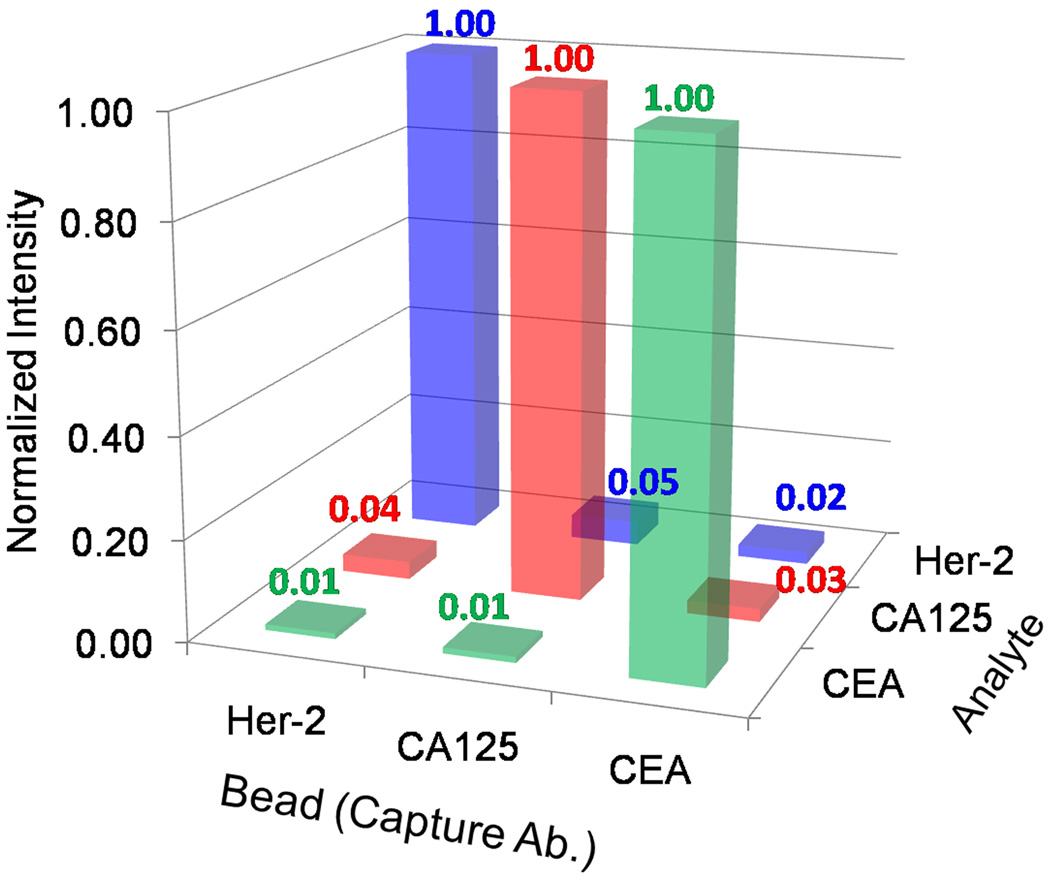
The use of numerous cancer biomarkers increases both the sensitivity and specificity of cancer assays and the NBC demonstrates the capacity for multiplexed tests (Harris et al., 2007). First, beads in redundancies of three and sensitized to either CEA, Her-2/Neu, and CA125, were arranged in a 4 × 5 chip along with isotype control beads. All beads produced signal upon introduction of all three antigens, in buffer, at concentrations near the dose curve maximum, as well as a 9.2 nM cocktail of all detecting antibodies. When only Her-2/Neu antigen and the detecting antibody cocktail was introduced, signal from the specific interaction between immobilized Her-2/Neu antigen and Her-2/Neu detection bioconjugate was S = 20.2 with a small amount of signal cross talk emanating from the CEA (S = 0.03) and CA125 non-specific beads (S = 0.97). This experiment was repeated for CEA and CA125 analytes and the strong, specific signal normalized to a value of unity (Fig. 5). Signal from non-specific beads was less than 5% of the most intense, specific signal for all three experiments. An analogous series of assays in which all three antigens at values near the dose curve maximum, i.e. 25 ng/mL for CEA were introduced concurrently, followed by sequential interrogation by a single detection antibody (3.7 nM) resulted in non-specific interactions that were less than 5% of the specific signal. This data demonstrates the utility of the NBC sensor as a highly specific, multiplexed tool for diagnostic profiles of complex fluids.
Figure 5.
The NBC multiplexes via the spatial arrangement of beads on the chip. In these experiments, cross reactivity was evaluated. Beads specific to Her-2/Neu, CEA, and CA125 express both specific and non-specific signal. Non-authentic output emanating from untargeted beads, plotted as a percentage of specific signal intensity, shows strong specificity across the assay systems. Non-specific signal is less than 5% of signal on specific interaction beads.
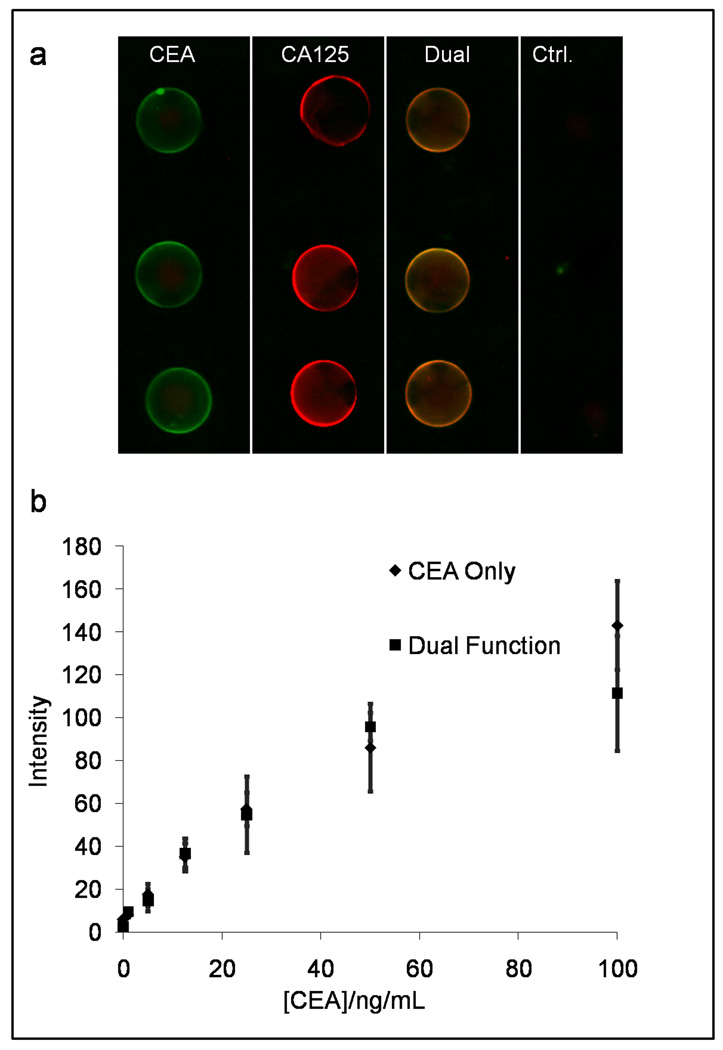
To demonstrate the capacity of the NBC system to perform a multiplexed assay within a single bead, beads functionalized with 1.0 mg/mL CEA or 1.0 mg/mL CA125 specific capture antibodies (or both antibodies at 1.0 mg/mL) were arrayed on a single chip along with beads conjugated to isotype control antibody. Both antigens and a cocktail of relevant fluorescently labeled detection antibodies at (1:100 dilution or 3.7 nM) were introduced into the NBC. Here, detecting antibody specific to CEA fluoresces green (QD 565), and CA125 antibody is red (QD 655). Here, as expected, the CA125 sensitized beads fluoresced red, while the CEA sensitized beads fluoresced green as collected by the appropriate filter cubes specific to each color (see Supplementary S.3 for optical consideration). As seen in Figure 6, the dual-function beads, which fluoresced orange after this assay, demonstrate efficient detection of both captured analytes. For assays at concentrations near the dose curve maximum (25 ng/mL CEA and 300 U/mL CA125), the S/N values for the dual function and singly selective CA 125 beads are not significantly different at 1.59 ± 0.25 and 1.79 ± 0.30, respectively. Similarly, S/N values for single and doubly selective CEA beads are 130 ± 12 and 104 ± 22. Differences between the two assays systems are due to non-specific binding of detecting antibody.
Figure 6.
Multiplexing capabilities of the NBC also includes single-bead duplexing in which dually functionalized beads measure two analytes concurrently (a). Beads sensitized to CEA, CA125, and both biomarkers are used simultaneously on a single chip. (b) Across the physiologically relevant range of concentrations no statistical difference is seen in fluorescent signal output between sole CEA and dual CEA/CA125 beads.
The CEA-specific signal response achieved by both CEA-sensitized and dual-function beads is statistically identical across the physiologically interesting region and is independent of CA125 antigen (0–400 U/mL) presence (Fig. 6b). This is the first demonstration of multianalyte detection on a single bead with this NBC system type. The additional element of multiplexed capacity demonstrated here via multiple colors on individual beads, allow an increased number of independent assay systems to achieve robust measurements in the same miniaturized space. In addition, as UV light easily excites different color QDs (corresponding to unique analytes), this novel multiplexing scheme, in tandem with a dual bandpass emission filter and color CCD detector, would not increase analyzer optical requirements, a design critical for POC systems.
4. CONCLUSIONS
The construction and validation of a microfluidic biosensor incorporating the intensity and fluorescence attributes of QDs makes important progress toward the completion of a harmonized nano-bio analysis system. Although some reports have recently demonstrated quantitative immunoassay of cancer biomarkers such as CEA with QDs via both avidin and SMCC facilitated bond formation, very few studies have utilized QDs to measure human specimens versus gold standard methods (Zajac et al., 2007). These prior clinical studies rely on QDs in tandem with a chaperon structure such as a latex bead (Klostranec et al., 2007). To the best of our knowledge, this is the first paper in which QDs provide quantitative cancer biomarker information using blood or salivary clinical samples. The multiplexed and programmable NBC system has clinically demonstrated advantages including intense signal, low LODs necessary for saliva analysis, and short assay times. The potential for “nanotyping”, or multiplexed profiling of disease fingerprints via QDs, is an exciting application of this class of fluorophores that is closer to realization after this study (Xing et al., 2007). The integrated, disposable NBC described herein is highly compatible with the prototype analyzer described previously that combines optics, battery power, optics, and image analysis into a compact tool eliminating lab-based infrastructure (Jokerst et al., 2008). Such an arrangement is among the most integrated use of applied nanomaterials reported to date and expands further the utility of our compact NBC analyzer for true POC analyses.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge funding provided by the National Institutes of Health through the National Institute of Dental and Craniofacial Research (U01 DE017793). The content is solely the responsibility of the authors and does not necessarily represent or reflect views of the NIH or the Federal Government. Support was also provided by a private gift to UTHSCSA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- ACS. Cancer Facts and Figures 2008. Atlanta, GA: American Cancer Society; 2008. [Google Scholar]
- Ali MF, Kirby R, Goodey AP, Rodriguez MD, Ellington AD, Neikirk DP, Mcdevitt JT. Analytical Chemistry. 2003;75:4732–4739. doi: 10.1021/ac034106z. [DOI] [PubMed] [Google Scholar]
- Aps JK, Martens LC. Forensic Sci. Int. 2005;150:119–131. doi: 10.1016/j.forsciint.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Baron T, Boardman Cecelia H, Lafky Jacqueline M, Rademaker A, Liu D, Fishman David A, Podratz Karl C, Maihle Nita J. Cancer Epidemiol. Biomarkers Prev. 2005;14:306–318. doi: 10.1158/1055-9965.EPI-04-0423. [DOI] [PubMed] [Google Scholar]
- Bhasin S, Zhang A, Coviello A, Jasuja R, Ulloor J, Singh R, Vesper H, Vasan RS. Steroids. 2008;73:1311–1317. doi: 10.1016/j.steroids.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Choi DH, Katakura Y, Matsuda R, Hayashi Y, Ninomiya K, Shioya S. J. Biosci. Bioeng. 2007;103:427–431. doi: 10.1263/jbb.103.427. [DOI] [PubMed] [Google Scholar]
- Christodoulides N, Floriano PN, Miller CS, Ebersole JL, Mohanty S, Dharshan P, Griffin M, Lennart A, Ballard KLM, King CP, Jr, Langub MC, Kryscio RJ, Thomas MV, Mcdevitt JT. Ann. N. Y. Acad. Sci. 2007;1098:411–428. doi: 10.1196/annals.1384.035. [DOI] [PubMed] [Google Scholar]
- Crowther JR, editor. Method Mol.Biol. Totowa, NJ: 1995. ELISA: Theorgy and Practice. [DOI] [PubMed] [Google Scholar]
- Das PM, Bast RC., Jr Biomarkers in Medicine. 2008;2:291–303. doi: 10.2217/17520363.2.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodey A, Lavigne JJ, Savoy SM, Rodriguez MD, Curey T, Tsao A, Simmons G, Wright J, Yoo SJ, Sohn Y, Anslyn EV, Shear JB, Neikirk DP, Mcdevitt JT. J. Am. Chem. Soc. 2001;123:2559–2570. doi: 10.1021/ja003341l. [DOI] [PubMed] [Google Scholar]
- Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC., Jr J. Clin. Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, Singh AK. PNAS. 2007;104:5268–5273. doi: 10.1073/pnas.0607254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Loo JA, Wong DT. Annals of the New York Academy of Sciences. 2007;1098:323–329. doi: 10.1196/annals.1384.015. [DOI] [PubMed] [Google Scholar]
- Jokerst JV, Floriano PN, Christodoulides N, Simmons GW, Mcdevitt JT. Lab Chip. 2008;8:2079–2090. doi: 10.1039/b817116e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klostranec JM, Xiang Q, Farcas GA, Lee JA, Rhee A, Lafferty EI, Perrault SD, Kain KC, Chan WCW. Nano Letters. 2007;7:2812–2818. doi: 10.1021/nl071415m. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Wark AW, Corn RM. Analyst. 2008;133:975–983. doi: 10.1039/b717527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Howarth M, Greytak AB, Zheng Y, Nocera DG, Ting AY, Bawendi MG. J. Am. Chem. Soc. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamud D. J. Am. Dent. Assoc. 2006;137:284–286. doi: 10.14219/jada.archive.2006.0158. [DOI] [PubMed] [Google Scholar]
- Mandel ID. J. Am. Dent. Assoc. 1993;124:85–87. doi: 10.14219/jada.archive.1993.0007. [DOI] [PubMed] [Google Scholar]
- Mitchell KF. Cancer Immunology Immunotherapy. 1980;10:1–5. [Google Scholar]
- Nagler R, Bahar G, Shpitzer T, Feinmesser R. Clin. Cancer Res. 2006;12:3979–3984. doi: 10.1158/1078-0432.CCR-05-2412. [DOI] [PubMed] [Google Scholar]
- Narayanan J, Xiong J-Y, Liu X-Y. Journal of Physics: Conference Series. 2006;28:83–86. [Google Scholar]
- Oppenheim FG, Salih E, Siqueira Walter L, Zhang W, Helmerhorst Eva J. Ann. N.Y. Acad. Sci. 2007;1098:22–50. doi: 10.1196/annals.1384.030. [DOI] [PubMed] [Google Scholar]
- Pallud C, Guinebretiere JM, Guepratte S, Hacene K, Neumann R, Carney W, Pichon MF. Anticancer Res. 2005;25:1433–1440. [PubMed] [Google Scholar]
- Panchuk-Voloshina N, Haugland RP, Bishop-Stewart J, Bhalgat MK, Millard PJ, Mao F, Leung W-Y, Haugland RP. Journal of Histochemistry and Cytochemistry. 1999;47:1179–1188. doi: 10.1177/002215549904700910. [DOI] [PubMed] [Google Scholar]
- Pathak S, Davidson MC, Silva GA. Nano Letters. 2007;7:1839–1845. doi: 10.1021/nl062706i. [DOI] [PubMed] [Google Scholar]
- Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Nat. Meth. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- Soper SA, Brown K, Ellington A, Frazier B, Garcia-Manero G, Gau V, Gutman SI, Hayes DF, Korte B, Landers JL, Larson D, Ligler F, Majumdar A, Mascini M, Nolte D, Rosenzweig Z, Wang J, Wilson D. Biosens. Bioelectron. 2006;21:1932–1942. doi: 10.1016/j.bios.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Streckfus C, Bigler L, Dellinger T, Dai X, Cox WJ, Mcarthur A, Kingman A, Thigpen JT. Oral Surg Oral Med Oral Pathol Oral Radiol Endol. 2001;91:174–179. doi: 10.1067/moe.2001.111758. [DOI] [PubMed] [Google Scholar]
- Streckfus CF, Bigler LR, Zwick M. J. Oral. Path. Med. 2006;35:292–300. doi: 10.1111/j.1600-0714.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- Tan W, Sabet L, Li Y, Yu T, Klokkevold PR, Wong DT, Ho CM. Biosens. Bioelectron. 2008;24:266–271. doi: 10.1016/j.bios.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigum SE, Floriano PN, Christodoulides N, Mcdevitt JT. Lab Chip. 2007;7:995–1003. doi: 10.1039/b703918b. [DOI] [PubMed] [Google Scholar]
- Wong DT, editor. Salivary Diagnostics. Hoboken, NJ: Wiley-Blackwell; 2008. [Google Scholar]
- Xing Y, Chaudry Q, Shen C, Kong KY, Zhau HE, Chung LW, Petros JA, O'regan RM, Yezhelyev MV, Simons JW, Wang MD, Nie S. Nat. Protoc. 2007;2:1152–1165. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- Zajac A, Song D, Qian W, Zhukov T. Colloids Surf. B. 2007;58:309–314. doi: 10.1016/j.colsurfb.2007.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.