Abstract
Insulin plays a central role in the regulation of vertebrate metabolism. The hormone, the post-translational product of a single-chain precursor, is a globular protein containing two chains, A (21 residues) and B (30 residues). Recent advances in human genetics have identified dominant mutations in the insulin gene causing permanent neonatal-onset DM2 (1–4). The mutations are predicted to block folding of the precursor in the ER of pancreatic β-cells. Although expression of the wild-type allele would in other circumstances be sufficient to maintain homeostasis, studies of a corresponding mouse model (5–7) suggest that the misfolded variant perturbs wild-type biosynthesis (8, 9). Impaired β-cell secretion is associated with ER stress, distorted organelle architecture, and cell death (10). These findings have renewed interest in insulin biosynthesis (11–13) and the structural basis of disulfide pairing (14–19). Protein evolution is constrained not only by structure and function but also by susceptibility to toxic misfolding.
Insulin plays a central role in the regulation of vertebrate metabolism. The hormone, the post-translational product of a single-chain precursor, is a globular protein containing two chains, A (21 residues) and B (30 residues). Recent advances in human genetics have identified dominant mutations in the insulin gene causing permanent neonatal-onset DM2 (1–4). The mutations are predicted to block folding of the precursor in the ER of pancreatic β-cells. Although expression of the wild-type allele would in other circumstances be sufficient to maintain homeostasis, studies of a corresponding mouse model (5–7) suggest that the misfolded variant perturbs wild-type biosynthesis (8, 9). Impaired β-cell secretion is associated with ER stress, distorted organelle architecture, and cell death (10). These findings have renewed interest in insulin biosynthesis (11–13) and the structural basis of disulfide pairing (14–19). Protein evolution is constrained not only by structure and function but also by susceptibility to toxic misfolding.
Biosynthesis of Insulin
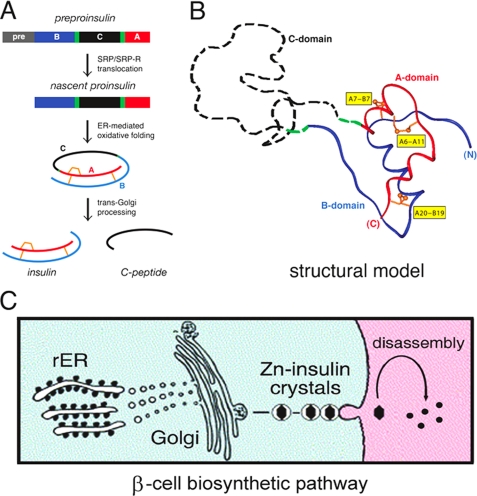
The insulin gene encodes a single-chain precursor, preproinsulin (Fig. 1A, upper). The signal peptide (gray bar) is cleaved upon ER translocation to yield proinsulin. The translocated polypeptide is reduced and unfolded. Preproinsulin and proinsulin contain a connecting domain (black in Fig. 1A) between the B- and A-domains (blue and red, respectively) (20). Folding in the ER is coupled to specific pairing of three disulfide bridges (Fig. 1A, center). These bridges (A6-A11, A7-B7, and A20-B19) (gold in Fig. 1B) are essential for stability and bioactivity (17, 21–29). Proinsulin consists of a folded insulin-like moiety and disordered connecting peptide (the C-domain) (dashed black line in Fig. 1B) (30–33). Conserved within the insulin superfamily, the cystines provide interior struts in the hydrophobic core (A19-B20 and A6-A11) and an external staple (A7-B7). The structure of insulin (Fig. 1A, lower) requires maintenance of each bridge. Disulfide isomers exhibit molten structures of marginal stability and low biological activity (34–36).
FIGURE 1.
Proinsulin and its biosynthetic pathway. A, pathway of insulin biosynthesis beginning with preproinsulin (upper): signal peptide (gray), B-domain (blue), dibasic BC junction (green), C-domain (red), dibasic CA junction (green), and A-domain (red). In the ER, the unfolded prohormone undergoes specific disulfide pairing to yield native proinsulin (center). Cleavage of BC and CA junctions by prohormone convertases (PC1 and PC2) and carboxypeptidase E leads to mature insulin and the C-peptide (lower). SRP/SRP-R, signal recognition particle/signal recognition particle receptor. B, structural model of insulin-like moiety and disordered connecting peptide (dashed black line). The A- and B-domains are shown in red and blue, respectively; the disordered connecting domain is shown by the dashed black line. Cystines are labeled in yellow boxes. C, cellular pathway of insulin biosynthesis. Nascent proinsulin folds as a monomer in the rough ER (rER; left), wherein zinc ion concentration is low; in post-Golgi granules, proinsulin is processed by cleavage of the connecting peptide to yield mature insulin, and zinc-stabilized hexamers begin to assemble. Zinc-insulin crystals are observed in secretory granules. Upon secretion into the portal circulation (right), hexamers dissociate to yield bioactive insulin monomers. Although the structure of an isolated monomer resembles that of a crystallographic protomer, marked changes in conformation may be required for receptor binding. Native hexamer assembly and induced fit of the insulin monomer may provide complementary structural adaptations to the threat of toxic protein misfolding (49, 75, 76).
Proinsulin binds only weakly to the insulin receptor; the bioactive hormone is liberated by proteolytic processing (12, 37). Upon transit through the Golgi apparatus and entry into immature secretory granules (38), the C-peptide is excised by specific prohormone convertases (39). Cleavage occurs at conserved dibasic sites (BC and CA junctions) (green in Fig. 1, A and B). The mature hormone is stored as Zn2+-stabilized hexamers within specialized secretory granules (Fig. 1C) (40). Hexamers dissociate upon secretion into the portal circulation (Fig. 1C, right). Because the monomer is exquisitely susceptible to fibrillation (41), its zinc-mediated assembly within β-cells may represent a defense against toxic misfolding in the secretory granule (13).
Although insulin biosynthesis occurs via a single-chain precursor, in vitro chemical synthesis employs isolated A- and B-peptides (42). The fidelity of chain combination implies that chemical folding information is contained within these sequences (43). Many insulin analogs have been prepared by this protocol, facilitating pharmaceutical applications (44, 45). Despite the general robustness of insulin chain combination, certain substitutions impede yield (16, 46–52). The genetics of neonatal DM highlights such synthetic failures as models of impaired folding, providing structural insight into a disease of toxic protein misfolding.
Mechanism of Disulfide Pairing
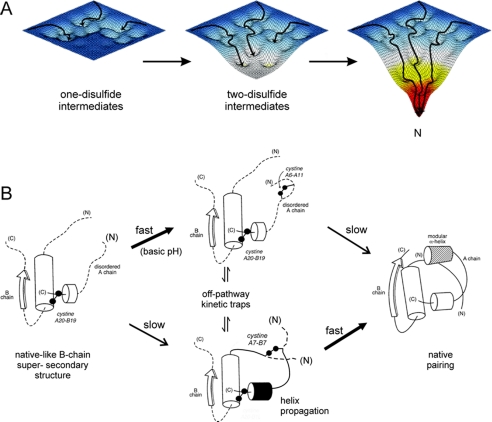
Oxidative folding of proteins may be probed by chemical trapping of populated disulfide intermediates (53). Such studies of proinsulin-related polypeptides are notable for the transient accumulation of one- and two-disulfide intermediates (14, 15, 54, 55). Their partial folding may be represented by a series of trajectories on successive free-energy landscapes (Fig. 2A). Each landscape governs the dynamics of an ensemble of accessible conformations in the presence of a specific subset of disulfide bridges. Because the folding chain acquires structure stepwise upon successive disulfide pairing, the landscapes proceed from shallow to steep. The preferred sequence of disulfide intermediates, as defined by chemical trapping, hence provides a framework for visualizing a progression of multiple folding trajectories on funnel-shaped landscapes. This perspective integrates the classical disulfide-centered paradigm (56) with biophysical models of protein folding (57, 58).
FIGURE 2.
Energy landscape view of proinsulin folding and disulfide pairing. A, formation of successive disulfide bridges may be viewed as enabling a sequence of folding trajectories on a succession of steeper funnel-shaped free-energy landscapes. B, preferred pathway of disulfide pairing begins with cystine A20-B19 (left), whose pairing is directed by a nascent hydrophobic core formed by the central B-domain α-helix (residues B9–B19), part of the C-terminal B-chain β-strand (residues B24–B26), and part of the C-terminal A-domain α-helix (residues A16–A20). Alternative pathways mediate formation of successive disulfide bridges (middle) en route to the native state (right). The mechanism of disulfide pairing is perturbed by clinical mutations associated with misfolding of proinsulin. Sites of non-cysteine-related mutations causing neonatal DM (supplemental Fig. S2) highlight native structural features critical to foldability (supplemental Fig. S3–S5).
A structural pathway of disulfide pairing has been proposed based on equilibrium models (Fig. 2B) (17, 21–23, 25–29, 59). A key role is played by initial formation of cystine A20-B19, which in the native state connects the C-terminal α-helix of the A-domain to the central α-helix of the B-domain. This bridge, which packs within a cluster of conserved aliphatic and aromatic side chains in the hydrophobic core, is proposed to contribute to stabilization of a specific folding nucleus (17, 59, 60). The structural role of cystine A20-B19 in populated one- and two-disulfide intermediates has been investigated through construction of analogs containing pairwise substitution of the other cysteines with Ala or Ser (17, 21–29). Such analogs exhibit partial folds with attenuated but non-negligible α-helix content. Mutations near A20-B19 impair chain combination and biosynthesis of single-chain precursors in Saccharomyces cerevisiae (25, 50, 60–62). After A20-B19 pairing, folding proceeds through multiple alternative channels (see supplemental text). As successive disulfide bridges are introduced in model domains, 1H NMR spectra exhibit progressively increased chemical shift dispersion, suggesting stepwise stabilization of structure in accord with the landscape perspective. On-pathway two-disulfide intermediates interconvert with non-native disulfide isomers as off-pathway kinetic traps (Fig. 2B, center).
Non-native disulfide isomers of proinsulin have been observed in isolated islets and cell culture (10, 16, 63–65). Although such isomers are generally not secreted, amino acid substitutions in human proinsulin can enhance the fraction of mispairing in the ER (64, 65). Because propensity to misfold in this assay does not correlate with effects of substitutions on thermodynamic stability in vitro, its mechanism is not well understood. It is possible that the substituted side chains perturb the relative stabilities or kinetic accessibility of disulfide intermediates disproportionately to their effects on the native state. Alternatively, these residues may contribute to interactions of the nascent polypeptide with ER chaperones and oxidative machinery (66). Engagement of cell type-specific chaperones and foldases in insulin biosynthesis is likely to underlie the failure (due to aggregation) of diverse transfected mammalian cell lines to support efficient folding and secretion of proinsulin (67).
Diabetes-associated Mutations
Neonatal DM develops prior to immune system maturation and so presents as an autoantigen negative form of DM. This presentation may be due to mutations in any of several genes (68). The most common cause is a heterozygous activating mutation in a subunit of the β-cell voltage-gated potassium channel, either KCNJ11 (encoding the Kir6.2 subunit) or ABCC8 (encoding the SUR1 subunit) (69, 70). The resulting diabetic phenotype may be transient or permanent. Recognition of this syndrome is important as such patients may be treated with oral agents that inhibit the channel (sulfonylureas) rather than insulin (68).
Dominant mutations in the insulin gene have recently been recognized as the second most common cause of permanent neonatal DM (1–4). Such mutations occur in each region of preproinsulin: its signal peptide and B-C-A domains (supplemental Fig. S1). The majority of mutations result in addition or removal of a cysteine, leading in either case to an odd number of potential pairing sites (supplemental Fig. S2). This imbalance is thought to lead to misfolding and aggregation. Remarkably, one human mutation (CysA7 → Tyr) is the same as previously observed in the Mody4 mouse model (the Akita mouse) (5–7), in which a dominant mutation in the Ins2 gene leads to progressive postnatal β-cell failure (9, 10). Physicochemical studies of the variant murine proinsulin indicate partial unfolding with increased aggregation (71). Such perturbations are in accord with structural studies of human insulin and proinsulin analogs lacking cystine A7-B7 (28, 60). Heterozygous expression of a variant Ins2 allele encoding substitution CysA6 → Ser (uncovered in the course of an N-ethyl-N-nitrosourea mouse mutagenesis screen) likewise causes β-cell dysfunction and progressive DM (72).
Identification of identical human and murine mutations at position A7 strongly suggests that the pathogenesis of neonatal DM in humans is similar to that extensively characterized in the Akita mouse (5–7, 9, 10). Although uncertainties remain in the precise time course and mechanism of β-cell degeneration, the β-cells of Akita islets exhibit an early defect in the folding and trafficking of both wild-type and variant proinsulins, elevated markers of ER stress, progressive deposition of electron-dense material in the ER and Golgi apparatus associated with morphological abnormalities, mitochondria swelling, and eventual loss of β-cell mass due to apoptosis or other forms of cell death (9, 10).
Perturbation of disulfide pairing in nascent proinsulin can in principle range from severe or mild, depending on the site of mutation and the properties of the substituted side chain (supplemental Figs. S3 and S4). Key sites in the structure of insulin required for the foldability of proinsulin are discussed in the supplemental text. Whereas an odd number of cysteines or compromise of a key non-cysteine site presumably imposes a severe block to folding, mutations causing less marked impairment would be expected to present later in life as autoantibody-negative presumed Type 1 or 2 DM. One such mutation, presenting in the second decade as maturity-onset diabetes of the young, has been described (4). Chronic elevation of ER stress in β-cells presumably leads to a slow but progressive loss of β-cell mass. ER stress may likewise contribute to the pathogenesis of insulin Los Angeles (PheB24 → Ser), a classical insulinopathy with partial retention of activity (73). It is not known whether or to what extent subtle perturbations of insulin biosynthesis (due to either variant insulin genes or mutations in the ER folding machinery) contribute to the pathogenesis of nonsyndromic Type 2 DM.
Concluding Remarks
The evolution of insulin is enjoined by multiple biological constraints, reflecting sequence requirements of biosynthesis, structure, and function (supplemental Fig. S3A). We thus imagine that conserved residues contribute to one or more of the following processes: foldability in the β-cell, protection from intra- or extracellular toxic misfolding, self-assembly within secretory granules, and receptor binding. The overlapping nature of these constraints may account for the limited sequence diversity among vertebrate insulins (74). An intrinsic tension between folding-competent and active conformations, only partially resolved by induced fit, may underlie the role of chronic ER stress (8) in the progression of β-cell dysfunction in Type 2 DM (49, 75, 76).
The discovery of DM-associated mutations in proinsulin highlights general principles of protein folding. The native state of a globular protein may be viewed as a coalescence of discrete subdomains consistent with classical diffusion-collision and framework models of protein folding (77). Funnel-like energy landscapes suggest the importance of parallel events in folding (57) even in the presence of preferred trajectories (78). Disulfide trapping studies of insulin-related polypeptides have defined predominant intermediates, enabling structural interpretation of many of the clinical mutations. Sites of mutation reflect mechanisms of oxidative folding not fully revealed by structural features of the native state, once achieved. Toxic protein misfolding provides an implicit constraint governing the evolution of proinsulin.
Supplementary Material
Acknowledgments
I thank Q.-x. Hua for assistance with figures; J. Williamson for landscape images; N. F. Phillips for advice; and P. Arvan, G. G. Dodson, M. Liu, and D. F. Steiner for discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants DK40949 and DK0697674 (to M. A. W.). This work was also supported by the American Diabetes Association. This minireview is a contribution from the Cleveland Center for Membrane and Structural Biology. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, references, and Figs. S1–S5.
- DM
- diabetes mellitus
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Støy J., Edghill E. L., Flanagan S. E., Ye H., Paz V. P., Pluzhnikov A., Below J. E., Hayes M. G., Cox N. J., Lipkind G. M., Lipton R. B., Greeley S. A., Patch A. M., Ellard S., Steiner D. F., Hattersley A. T., Philipson L. H., Bell G. I. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15040–15044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colombo C., Porzio O., Liu M., Massa O., Vasta M., Salardi S., Beccaria L., Monciotti C., Toni S., Pedersen O., Hansen T., Federici L., Pesavento R., Cadario F., Federici G., Ghirri P., Arvan P., Iafusco D., Barbetti F.Early Onset Diabetes Study Group of the Italian Society of Pediatric Endocrinology and Diabetes (2008) J. Clin. Investig. 118, 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edghill E. L., Flanagan S. E., Patch A. M., Boustred C., Parrish A., Shields B., Shepherd M. H., Hussain K., Kapoor R. R., Malecki M., MacDonald M. J., Støy J., Steiner D. F., Philipson L. H., Bell G. I., the Neonatal Diabetes International Collaborative Group. Hattersley A. T., Ellard S. (2008) Diabetes 57, 1034–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molven A., Ringdal M., Nordbø A. M., Raeder H., Støy J., Lipkind G. M., Steiner D. F., Philipson L. H., Bergmann I., Aarskog D., Undlien D. E., Joner G., Søvik O., the Norwegian Childhood Diabetes Study Group. Bell G. I., Njølstad P. R. (2008) Diabetes 57, 1131–113518192540 [Google Scholar]
- 5.Yoshioka M., Kayo T., Ikeda T., Koizumi A. (1997) Diabetes 46, 887–894 [DOI] [PubMed] [Google Scholar]
- 6.Wang J., Takeuchi T., Tanaka S., Kubo S. K., Kayo T., Lu D., Takata K., Koizumi A., Izumi T. (1999) J. Clin. Investig. 103, 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oyadomari S., Koizumi A., Takeda K., Gotoh T., Akira S., Araki E., Mori M. (2002) J. Clin. Investig. 109, 525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ron D. (2002) J. Clin. Investig. 109, 443–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izumi T., Yokota-Hashimoto H., Zhao S., Wang J., Halban P. A., Takeuchi T. (2003) Diabetes 52, 409–416 [DOI] [PubMed] [Google Scholar]
- 10.Liu M., Hodish I., Rhodes C. J., Arvan P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15841–15846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steiner D. F., Cunningham D., Spigelman L., Aten B. (1967) Science 157, 697–700 [DOI] [PubMed] [Google Scholar]
- 12.Steiner D. F., Clark J. L., Nolan C., Rubenstein A. H., Margoliash E., Aten B., Oyer P. E. (1969) Recent Prog. Horm. Res. 25, 207–282 [DOI] [PubMed] [Google Scholar]
- 13.Dodson G., Steiner D. (1998) Curr. Opin. Struct. Biol. 8, 189–194 [DOI] [PubMed] [Google Scholar]
- 14.Miller J. A., Narhi L. O., Hua Q. X., Rosenfeld R., Arakawa T., Rohde M., Prestrelski S., Lauren S., Stoney K. S., Tsai L., Weiss M. A. (1993) Biochemistry 32, 5203–5213 [DOI] [PubMed] [Google Scholar]
- 15.Qiao Z. S., Guo Z. Y., Feng Y. M. (2001) Biochemistry 40, 2662–2668 [DOI] [PubMed] [Google Scholar]
- 16.Hua Q. X., Liu M., Hu S. Q., Jia W., Arvan P., Weiss M. A. (2006) J. Biol. Chem. 281, 24889–24899 [DOI] [PubMed] [Google Scholar]
- 17.Hua Q. X., Mayer J. P., Jia W., Zhang J., Weiss M. A. (2006) J. Biol. Chem. 281, 28131–28142 [DOI] [PubMed] [Google Scholar]
- 18.Qiao Z. S., Guo Z. Y., Feng Y. M. (2006) Protein Pept. Lett. 13, 423–429 [DOI] [PubMed] [Google Scholar]
- 19.Guo Z. Y., Qiao Z. S., Feng Y. M. (2008) Antioxid. Redox Signal. 10, 127–139 [DOI] [PubMed] [Google Scholar]
- 20.Steiner D. F. (1967) Trans. N.Y. Acad. Sci. 30, 60–68 [DOI] [PubMed] [Google Scholar]
- 21.Narhi L. O., Hua Q. X., Arakawa T., Fox G. M., Tsai L., Rosenfeld R., Holst P., Miller J. A., Weiss M. A. (1993) Biochemistry 32, 5214–5221 [DOI] [PubMed] [Google Scholar]
- 22.Hua Q. X., Hu S. Q., Frank B. H., Jia W., Chu Y. C., Wang S. H., Burke G. T., Katsoyannis P. G., Weiss M. A. (1996) J. Mol. Biol. 264, 390–403 [DOI] [PubMed] [Google Scholar]
- 23.Dai Y., Tang J. G. (1996) Biochim. Biophys. Acta 1296, 63–68 [DOI] [PubMed] [Google Scholar]
- 24.Hober S., Uhlén M., Nilsson B. (1997) Biochemistry 36, 4616–4622 [DOI] [PubMed] [Google Scholar]
- 25.Weiss M. A., Hua Q. X., Jia W., Chu Y. C., Wang R. Y., Katsoyannis P. G. (2000) Biochemistry 39, 15429–15440 [DOI] [PubMed] [Google Scholar]
- 26.Guo Z. Y., Feng Y. M. (2001) Biol. Chem. 382, 443–448 [DOI] [PubMed] [Google Scholar]
- 27.Feng Y., Liu D., Wang J. (2003) J. Mol. Biol. 330, 821–837 [DOI] [PubMed] [Google Scholar]
- 28.Jia X. Y., Guo Z. Y., Wang Y., Xu Y., Duan S. S., Feng Y. M. (2003) Protein Sci. 12, 2412–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan H., Guo Z. Y., Gong X. W., Xi D., Feng Y. M. (2003) Protein Sci. 12, 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pekar A. H., Frank B. H. (1972) Biochemistry 11, 4013–4016 [DOI] [PubMed] [Google Scholar]
- 31.Snell C. R., Smyth D. G. (1975) J. Biol. Chem. 250, 6291–6295 [PubMed] [Google Scholar]
- 32.Brems D. N., Brown P. L., Heckenlaible L. A., Frank B. H. (1990) Biochemistry 29, 9289–9293 [DOI] [PubMed] [Google Scholar]
- 33.Weiss M. A., Frank B. H., Khait I., Pekar A., Heiney R., Shoelson S. E., Neuringer L. J. (1990) Biochemistry 29, 8389–8401 [DOI] [PubMed] [Google Scholar]
- 34.Sieber P. S., Eisler K., Kamber B., Riniker B., Rittel W., Marki F., deGasparo M. (1978) Hoppe-Seyler's Z. Physiol. Chem. 359, 113–123 [DOI] [PubMed] [Google Scholar]
- 35.Hua Q. X., Gozani S. N., Chance R. E., Hoffmann J. A., Frank B. H., Weiss M. A. (1995) Nat. Struct. Biol. 2, 129–138 [DOI] [PubMed] [Google Scholar]
- 36.Hua Q. X., Jia W., Frank B. H., Phillips N. F., Weiss M. A. (2002) Biochemistry 41, 14700–14715 [DOI] [PubMed] [Google Scholar]
- 37.Galloway J. A., Hooper S. A., Spradlin C. T., Howey D. C., Frank B. H., Bowsher R. R., Anderson J. H. (1992) Diabetes Care 15, 666–692 [DOI] [PubMed] [Google Scholar]
- 38.Huang X. F., Arvan P. (1994) J. Biol. Chem. 269, 20838–20844 [PubMed] [Google Scholar]
- 39.Steiner D. F. (1998) Curr. Opin. Chem. Biol. 2, 31–39 [DOI] [PubMed] [Google Scholar]
- 40.Huang X. F., Arvan P. (1995) J. Biol. Chem. 270, 20417–20423 [DOI] [PubMed] [Google Scholar]
- 41.Brange J., Andersen L., Laursen E. D., Meyn G., Rasmussen E. (1997) J. Pharm. Sci. 86, 517–525 [DOI] [PubMed] [Google Scholar]
- 42.Katsoyannis P. G. (1966) Science 154, 1509–1514 [DOI] [PubMed] [Google Scholar]
- 43.Wang C. C., Tsou C. L. (1991) Trends Biochem. Sci. 16, 279–281 [DOI] [PubMed] [Google Scholar]
- 44.Brange J. (1997) Diabetologia 40, S48–S53 [DOI] [PubMed] [Google Scholar]
- 45.Hirsch I. B. (2005) N. Engl. J. Med. 352, 174–183 [DOI] [PubMed] [Google Scholar]
- 46.Hu S. Q., Burke G. T., Schwartz G. P., Ferderigos N., Ross J. B., Katsoyannis P. G. (1993) Biochemistry 32, 2631–2635 [DOI] [PubMed] [Google Scholar]
- 47.Weiss M. A., Nakagawa S. H., Jia W., Xu B., Hua Q. X., Chu Y. C., Wang R. Y., Katsoyannis P. G. (2002) Biochemistry 41, 809–819 [DOI] [PubMed] [Google Scholar]
- 48.Hua Q. X., Chu Y. C., Jia W., Phillips N. F., Wang R. Y., Katsoyannis P. G., Weiss M. A. (2002) J. Biol. Chem. 277, 43443–43453 [DOI] [PubMed] [Google Scholar]
- 49.Nakagawa S. H., Zhao M., Hua Q. X., Hu S. Q., Wan Z. L., Jia W., Weiss M. A. (2005) Biochemistry 44, 4984–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakagawa S. H., Hua Q. X., Hu S. Q., Jia W., Wang S., Katsoyannis P. G., Weiss M. A. (2006) J. Biol. Chem. 281, 22386–22396 [DOI] [PubMed] [Google Scholar]
- 51.Hua Q. X., Nakagawa S., Hu S. Q., Jia W., Wang S., Weiss M. A. (2006) J. Biol. Chem. 281, 24900–24909 [DOI] [PubMed] [Google Scholar]
- 52.Wan Z. L., Huang K., Hu S. Q., Whittaker J., Weiss M. A. (2008) J. Biol. Chem. 283, 21198–21210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baldwin T. O., Ziegler M. M., Chaffotte A. F., Goldberg M. E. (1993) J. Biol. Chem. 268, 10766–10772 [PubMed] [Google Scholar]
- 54.Hober S., Forsberg G., Palm G., Hartmanis M., Nilsson B. (1992) Biochemistry 31, 1749–1756 [DOI] [PubMed] [Google Scholar]
- 55.Huang Y., Liang Z., Feng Y. (2001) Sci. China 44, 593–600 [DOI] [PubMed] [Google Scholar]
- 56.Creighton T. E. (1997) Biol. Chem. 378, 731–744 [DOI] [PubMed] [Google Scholar]
- 57.Dill K. A., Chan H. S. (1997) Nat. Struct. Biol. 4, 10–19 [DOI] [PubMed] [Google Scholar]
- 58.Oliveberg M., Wolynes P. G. (2005) Q. Rev. Biophys. 38, 245–288 [DOI] [PubMed] [Google Scholar]
- 59.Hua Q. X., Narhi L., Jia W., Arakawa T., Rosenfeld R., Hawkins N., Miller J. A., Weiss M. A. (1996) J. Mol. Biol. 259, 297–313 [DOI] [PubMed] [Google Scholar]
- 60.Hua Q. X., Nakagawa S. H., Jia W., Hu S. Q., Chu Y. C., Katsoyannis P. G., Weiss M. A. (2001) Biochemistry 40, 12299–12311 [DOI] [PubMed] [Google Scholar]
- 61.Chu Y. C., Burke G. T., Chanley J. D., Katsoyannis P. G. (1987) Biochemistry 26, 6972–6975 [DOI] [PubMed] [Google Scholar]
- 62.Kristensen C., Kjeldsen T., Wiberg F. C., Schäffer L., Hach M., Havelund S., Bass J., Steiner D. F., Andersen A. S. (1997) J. Biol. Chem. 272, 12978–12983 [DOI] [PubMed] [Google Scholar]
- 63.Liu M., Ramos-Castañeda J., Arvan P. (2003) J. Biol. Chem. 278, 14798–14805 [DOI] [PubMed] [Google Scholar]
- 64.Zhang B. Y., Liu M., Arvan P. (2003) J. Biol. Chem. 278, 3687–3693 [DOI] [PubMed] [Google Scholar]
- 65.Liu M., Li Y., Cavener D., Arvan P. (2005) J. Biol. Chem. 280, 13209–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noiva R. (1999) Semin. Cell Dev. Biol. 10, 481–493 [DOI] [PubMed] [Google Scholar]
- 67.Zhu Y. L., Abdo A., Gesmonde J. F., Zawalich K. C., Zawalich W., Dannies P. S. (2004) Endocrinology 145, 3840–3849 [DOI] [PubMed] [Google Scholar]
- 68.Murphy R., Ellard S., Hattersley A. T. (2008) Nat. Clin. Pract. Endocrinol. Metab. 4, 200–213 [DOI] [PubMed] [Google Scholar]
- 69.Slingerland A. S., Hattersley A. T. (2005) Ann. Med. 37, 186–195 [DOI] [PubMed] [Google Scholar]
- 70.Babenko A. P., Polak M., Cavé H., Busiah K., Czernichow P., Scharfmann R., Bryan J., Aguilar-Bryan L., Vaxillaire M., Froguel P. (2006) N. Engl. J. Med. 355, 456–466 [DOI] [PubMed] [Google Scholar]
- 71.Yoshinaga T., Nakatome K., Nozaki J., Naitoh M., Hoseki J., Kubota H., Nagata K., Koizumi A. (2005) Biol. Chem. 386, 1077–1085 [DOI] [PubMed] [Google Scholar]
- 72.Herbach N., Rahtkolb B., Kemter E., Pichl L., Klaften M., de Angelis M. H., Halban P. A., Wolf E., Aigner B., Wanke R. (2007) Diabetes 56, 1268–1276 [DOI] [PubMed] [Google Scholar]
- 73.Shoelson S. E., Polonsky K. S., Zeidler A., Rubenstein A. H., Tager H. S. (1984) J. Clin. Investig. 73, 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baker E. N., Blundell T. L., Cutfield J. F., Cutfield S. M., Dodson E. J., Dodson G. G., Hodgkin D. M., Hubbard R. E., Isaacs N. W., Reynolds C. D. (1988) Philos. Trans. R. Soc. Lond. B Biol. Sci. 319, 369–456 [DOI] [PubMed] [Google Scholar]
- 75.Hua Q. X., Xu B., Huang K., Hu S. Q., Nakagawa S., Jia W., Wang S., Whittaker J., Katsoyannis P. G., Weiss M. A. (2009) J. Biol. Chem. 284, 14586–14596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu B., Huang K., Chu Y. C., Hu S. Q., Nakagawa S., Wang S., Wang R. Y., Whittaker J., Katsoyannis P. G., Weiss M. A. (2009) J. Biol. Chem. 284, 14597–14608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ptitsyn O. B. (1991) FEBS Lett. 285, 176–181 [DOI] [PubMed] [Google Scholar]
- 78.Lazaridis T., Karplus M. (1997) Science 278, 1928–1931 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.