Abstract
STIM1 and STIM2 are dynamic transmembrane endoplasmic reticulum Ca2+ sensors, coupling directly to activate plasma membrane Orai Ca2+ entry channels. Despite extensive sequence homology, the STIM proteins are functionally distinct. We reveal that the short variable N-terminal random coil sequences of STIM1 and STIM2 confer profoundly different activation properties. Using Orai1-expressing HEK293 cells, chimeric replacement of the 43-amino-acid STIM1 N terminus with that of STIM2 attenuates Orai1-mediated Ca2+ entry and drastically slows store-induced Orai1 channel activation. Conversely, the 55-amino-acid STIM2 terminus substituted within STIM1 strikingly enhances both Orai1-mediated Ca2+ entry and constitutive coupling to activate Orai1 channels. Hence, STIM N termini are powerful coupling modifiers, functioning in STIM2 to “brake” the otherwise constitutive activation of Orai1 channels afforded by its high sensitivity to luminal Ca2+.
The transmembrane ER4 proteins STIM1 and STIM2 function as sensors of Ca2+ within ER stores (1, 2). Depletion of luminal Ca2+ within the ER triggers aggregation and translocation of STIMs into junctions closely associated with the plasma membrane, where they activate the highly Ca2+-selective Orai family of store-operated channels (SOCs) via conformational coupling (3–8). Recent investigations of the cytoplasmic portion of STIM1 revealed that it alone is sufficient to activate Orai (9–12) via a short (∼100 amino acids) region centered around the second coiled-coil domain (see Fig. 1) (13–15). However, although activation of Orai1 is mediated entirely within the C-terminal portion of STIM, physiological control of STIM1 and STIM2 is exerted via their N-terminal ER-luminal Ca2+-sensing domains. The extent to which structural differences between these domains in STIM1 and STIM2 contribute to their distinct properties (16–19) remains poorly understood. Although STIM2 has the capacity to sense ER Ca2+ and activate SOCs (16, 17, 19), overexpressed STIM2 inhibits endogenous SOCs (18). Moreover, the kinetics of SOC activation by STIM2 are much slower than STIM1 (17). STIM2 was recently revealed to have a decreased Ca2+-sensing affinity when compared with STIM1 by virtue of three amino acid substitutions in the Ca2+-binding EF-hand domain (16). Although the lower affinity of the STIM2 EF-hand accounts for differences in the activation thresholds of STIM1 and STIM2 (16, 20, 21), it does not explain the slow kinetics of STIM2 nor its dominance over endogenous SOC activation. However, recent investigations reveal similar abilities of the cytosolic portions of STIM1 and STIM2 to activate Orai1 (12). Hence, although activation of Orai1 is mediated entirely within the C-terminal portion of STIM, physiological control of STIM1 and STIM2 is exerted via their N-terminal ER-luminal Ca2+-sensing domains.
FIGURE 1.
Schematic diagram depicting the domain structure of STIM1, STIM2, and STIM chimeras. The currently defined domains of STIM1 and STIM2 are depicted as canonical (cEF) and hidden (hEF) EF-hands, SAM domains, transmembrane domains (TM), coiled-coil structures, a proline-rich domain (P), and a polybasic tail (K). The sequences of the STIM1 and STIM2 N-terminal domains were aligned using the lalign program and depicted with red indicating identical amino acids and blue indicating similarity.
The initial triggering events for STIM1 and STIM2 proteins involve the unfolding and aggregation of the N-terminal domains resulting from dissociation of Ca2+ from the luminal EF-hand Ca2+ binding domains (20–23). Recent evidence reveals that this unfolding is much slower for the N terminus of STIM2 than for STIM1 (21). Although most of the N termini of STIM1 and STIM2 are highly homologous, significant variability exists in the first 60 N-terminal amino acids upstream from the EF-hands, comprising a flexible random coil domain (21). Intriguingly, these upstream sequences appear to markedly modify the stability of the N-terminal domains of STIM1 and STIM2 (21). We reveal here that these sequences confer profound distinctions between STIM1 and STIM2 in their coupling to activate SOCs. In STIM2, this domain acts as a powerful “brake” to restrict constitutive activation of SOCs, occurring as a result of its high sensitivity to luminal Ca2+.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK293 cells overexpressing Orai1 were developed as described previously (24) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (37 °C; 5% CO2) and G418 (250 μg/ml).
DNA Constructs and Transfections
Single fusion chimeras of STIM1 and STIM2 were generated by Mutagenex (Piscataway, NJ). Briefly, the signal peptide and N-terminal amino acids of STIM1 (Met-1–Phe-65) were fused with STIM2 (Glu-70–Lys-746) to generate STIM2 (S1NT). Similarly, the signal peptide and N-terminal amino acids of STIM2 (Leu-1–Leu-69) were fused with STIM1 (Glu-66–Lys-685) to generate STIM1 (S2NT). These constructs were inserted into pIRES Neo (Clontech) and introduced into HEK293 cells stably expressing Orai1 by electroporation using the Gene Pulser II electroporation system (Bio-Rad) at 350 V, 960 microfarads, and infinite resistance followed by 48 h in culture.
Cell Lysis and Western Blot
Cells were lysed in Nonidet P-40 lysis buffer (1% w/v Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl, pH 8.0 with protease inhibitors), cleared by centrifugation, and normalized for protein content. Proteins were resolved on 6–8% SDS-PAGE gels transferred to nitrocellulose paper and analyzed with the indicated antibodies as described previously (25). Each experiment was repeated at least three times.
Cytosolic Ca2+ Measurements
Ratiometric imaging of intracellular Ca2+ using fura-2 was as described previously (25). Cells grown on coverslips were placed in cation-safe solution (107 mm NaCl, 7.2 mm KCl, 1.2 mm MgCl2, 11.5 mm glucose, 20 mm Hepes-NaOH, pH 7.2) and loaded with fura-2/AM (2 μm) for 30 min at 20 °C. Cells were washed, and dye was allowed to de-esterify for a minimum of 30 min at 20 °C. Approximately 95% of the dye was confined to the cytoplasm as determined by the signal remaining after saponin permeabilization (26). Ca2+ measurements were made using a Leica DMI 6000B fluorescence microscope controlled by the Slidebook software (Intelligent Imaging Innovations; Denver, CO). Fluorescence emission at 505 nm was monitored while alternating between 340 and 380 nm excitation wavelengths at a frequency of 0.67 Hz; intracellular Ca2+ measurements are shown as 340/380 nm ratios obtained from groups (35–45) of single cells. Measurements shown are representative of a minimum of three and, in most cases, a larger number of independent experiments.
Electrophysiology
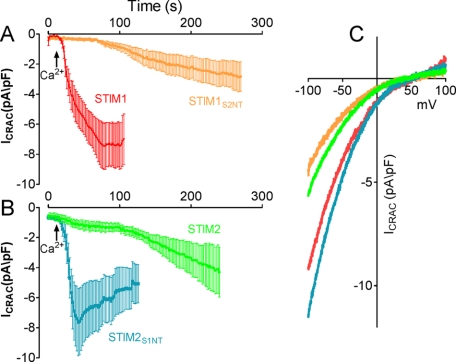
Electrophysiological recordings were generated using conventional whole cell recordings as described previously (19). Cells grown on coverslips were placed into the recording chamber. Immediately after establishing the whole cell configuration, linear voltage ramps of 50-ms duration spanning the voltage range of −100 to +100 mV were delivered from a holding potential of 0 mV at a rate of 0.5 Hz. We used automatic capacitive and series resistance compensation of the EPC-10 amplifier (HEKA Electronics). PatchMaster and Origin software were used for acquisition and analysis. The temporal development of inward (at −100 mV) current was measured from the individual ramps. The intracellular solution contained 145 mm CsGlu, 10 mm HEPES, 10 mm BAPTA, 8 mm NaCl, 10 mm Mg2+, 2 mm Mg2+-ATP, pH 7.2; osmolarity was adjusted to ∼300 mosm. The excess Mg2+ and ATP were used to ensure the inhibition of the endogenous TRPM7 channels. At the time of break-in, the extracellular solution contained 145 mm NaCl, 10 mm CsCl, 2.8 mm KCl, 10 mm HEPES, 10 mm glucose, pH 7.4; osmolarity was adjusted to ∼300 mosm. Ca2+ (10 mm) was added ∼50 s after break-in to avoid quenching of BAPTA when indicated in Fig. 3A. All I/V curves recorded are shown at the time of maximum activation. The maximal Ca2+ release-activated Ca2+ (CRAC) currents at −100 mV after leak subtraction were used for statistical analysis.
FIGURE 3.
Orai1 activation kinetics are controlled by the N-terminal flexible random coil domains of STIM1 and STIM2. HEK293 stably expressing Orai1 were transiently transfected with STIM1, STIM2, STIM1S2NT, or STIM2S1NT. A and B, currents were measured at −100 mV, normalized by their respective cell size, averaged, and plotted versus time ± S.E. Data are presented as normalized average time courses of BAPTA-induced ICRAC in HEK293 cells expressing STIM1 or STIM1S2NT (A) or STIM2 or STIM2S1NT (B). pF, picofarads. C, representative I/V curve measured at peak levels of CRAC current in panels A and B. Data depicted in this figure are based on between 9 and 18 independent measurements.
RESULTS AND DISCUSSION
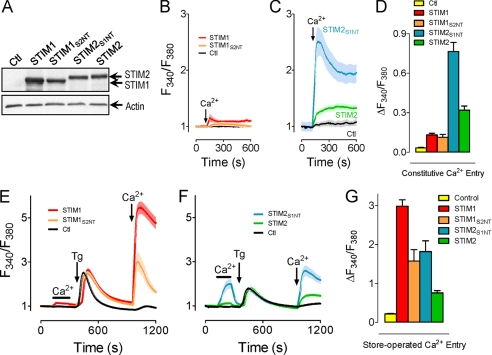
The STIM1 and STIM2 proteins are highly homologous through most of their structure (Fig. 1). However, this homology decreases considerably within a short 50–60-amino-acid N-terminal sequence upstream of the EF-hand and within the long C-terminal tails. To identify subtype-specific differences between STIM1 and STIM2, we examined the roles of the two variable regions by chimeric exchange. Initially, we exchanged the longer variable C-terminal regions of STIM1 (485–685) and STIM2 (488–746), examining their ability to modify endogenous activation of SOCs. Although expressed STIM1 resulted in slight enhancement, expression of STIM2 caused a profound loss of SOC-mediated Ca2+ entry (supplemental Fig. S1) in agreement with prior studies (18). Surprisingly, the inhibitory effect of expressed STIM2 on endogenous SOC activation was still observed using a STIM2 protein within which the STIM1-C terminus had been chimerically introduced (STIM2S1CT) (supplemental Fig. S1B). Consistent with this, the reciprocal substitution of the STIM2-CT within STIM1 (STIM1S2CT) resulted in no modification of endogenous SOC function (supplemental Fig. S1C).
Because the inhibitory action of STIM2 was independent of its C terminus, we turned our attention to the short variable N-terminal region. Chimeric exchange of the short N-terminal flexible random coil domains had profound effects on Orai1 activation characteristics. STIM1/STIM2 chimeras with the N-terminal 69 amino acids of STIM2 replacing the N-terminal 65 amino acids of STIM1 and vice versa are referred to as STIM1S2NT and STIM2S1NT, as described in Fig. 1. The two segments also contain signal peptides (22 amino acids for STIM1; 14 amino acids for STIM2), cleavage of which results in a 43-amino-acid N-terminal STIM1 segment (residues 23–66) and a 55-amino-acid STIM2 segment (residues 15–69). The chimeras were each tagged with yellow fluorescent protein for detection in transfected cells, as described previously (24). Expression levels of each of the STIM chimeric constructs after transfection into HEK293 cells stably expressing Orai1-CFP were unaltered from expression of wild-type STIM proteins as determined by Western analysis using anti-green fluorescent protein antibodies (Fig. 2A). The larger size of STIM2WT and STIM2S1NT primarily reflects the greater length of the STIM2 C terminus (63 additional amino acids) (27). We compared Orai1 activation by each of these two STIM chimeras with activation by STIM1WT or STIM2WT, examining their roles in mediating both constitutive and store-operated Ca2+ entry. As shown in Fig. 2B, little constitutive Ca2+ entry was observed in cells expressing either the STIM1WT or the STIM1S2NT construct, yet considerable constitutive entry was observed after transfection with either STIM2WT or STIM2S1NT (Fig. 2C). The effectiveness of the STIM2 constructs was consistent with a recent proposal that the STIM2 EF-hand has lower affinity for Ca2+ and hence a propensity to be more constitutively active (16). However, not explained by possible differences in Ca2+ affinity was the dramatic increase in the magnitude of Ca2+ entry in cells transfected with STIM2S1NT as opposed to STIM2WT (Fig. 2, C and D).
FIGURE 2.
The N-terminal flexible random coil domains of STIM1 and STIM2 modify Orai1-mediated Ca2+ entry. A, expression of yellow fluorescent protein-tagged STIM constructs as demonstrated by Western blot. Ctl, control. B–F, cells plated on coverslips were loaded with 2.5 μm fura-2/AM. B and C, Ca2+ (1 mm) was added to store-replete cells expressing STIM1 or STIM1S2NT (B) or STIM2 or STIM2S1NT (C) that had briefly (∼5 min) been maintained in nominally Ca2+-free medium. D, differences in constitutive Ca2+ entry at the plateau in cells treated in panels B and C were quantified and compared (n ≥ 13). E and F, cells expressing STIM1 or STIM1S2NT (E) or STIM2 or STIM2S1NT (F) were treated with Ca2+ (1 mm) before (store-replete) or after (store-depleted) treatment with thapsigargin (2 μm) in the absence of extracellular Ca2+ (10 min). G, differences in SOC activity in cells treated in panels E and F were quantified and compared (n ≥ 13).
To further investigate the role of the short N-terminal domains of STIM1 and STIM2, we examined how replacement of the N terminus of STIM1 with that of STIM2 would affect store-dependent Ca2+ entry. As shown in Fig. 2B, without store depletion, neither STIM1WT nor STIM1S2NT was activated, which would be consistent with a lower sensitivity (higher affinity) STIM1 EF-hand requiring store depletion. To examine SOC activation after store depletion, stable CFP-Orai1-expressing HEK293 cells transfected with each of the STIM constructs (Fig. 2, E–G) were treated with thapsigargin (2 μm) under nominally Ca2+-free conditions. In Orai1-overexpressing cells without STIM protein expression, SOC activity was greatly attenuated (Fig. 2, E and F), consistent with the dominant negative action of overexpressed Orai1 described previously (19). No significant differences in ER Ca2+ release were observed in cells expressing the different STIM constructs (Fig. 2, E and F), indicating no effect of STIM proteins on ER Ca2+ leak or pumping. Further, no differences in STIM clustering or association with CFP-Orai1 were observed by fluorescence microscopy (data not shown). Intriguingly, the two chimeras had opposite effects; expression of STIM2S1NT increased SOC activity over STIM2WT (Fig. 2F), whereas STIM1S2NT decreased SOC activity when compared with STIM1WT (Fig. 2D). In other words, the STIM1 N-terminal domain confers increased Ca2+ entry, whereas the STIM2 N-terminal domain confers decreased Ca2+ entry. We also noted that constructs containing the STIM1 C termini stimulated greater SOC activity (STIM1S2NT versus STIM2WT; STIM1WT versus STIM2S1NT; Fig. 2, E and F). A similar difference was noted between STIM2WT and STIM2S1CT (supplemental Fig. S1B), although not between STIM1WT and STIM1S2CT (supplemental Fig. S1C). Hence, there may be differences in the STIM/Orai coupling efficiency in the C termini of STIM1 and STIM2. Irrespectively, the current observations are the first to reveal distinct functional properties for the N-terminal flexible random coil domains of STIM1 and STIM2.
To further characterize the effect of the N-terminal domains of STIM1 and STIM2 on the activation of Orai1, we examined the characteristics of CRAC current activation in stable Orai1-expresssing HEK293 cells transfected with each of the holo-proteins and chimeric constructs. Analysis of CRAC channel activation revealed that the N-terminal domains imposed remarkable differences in the kinetics of CRAC channel activation (Fig. 3). CRAC channel analysis was performed following intracellular perfusion of 10 mm BAPTA to initiate store depletion. To differentiate constitutively active CRAC current and leak currents, experiments were initiated with 50 μm external Ca2+ to prevent CRAC current. As shown in Fig. 3A, CRAC current in STIM1-expressing cells developed between 50 and 100 s after break-in, a time period typical for BAPTA-induced store depletion in HEK293 cells (28). In contrast, cells expressing the STIM1S2NT construct exhibited a profound delay in the activation of ICRAC (Fig. 3A). We considered whether this difference could have reflected altered rates of store depletion. However, there were no differences between thapsigargin-induced Ca2+ release rates for cells expressing STIM1WT or STIM1S2NT (Fig. 2D). Thus, these observations reveal that simple substitution of the short STIM1 N terminus with the corresponding STIM2 sequence confers a profound delay in the development of CRAC.
Significantly, the rate of CRAC channel activation in the STIM1S2NT-expressing cells was almost identical to that observed in cells transfected with STIM2WT (Fig. 3B). In the converse experiment, exchange of the STIM2 N terminus with the N-terminal domain from STIM1 profoundly increased the rate of onset of ICRAC (Fig. 3B), resulting in peak CRAC current only a few seconds after the addition of Ca2+ to the extracellular solution. The absence of any delay in reaching full activation of CRAC indicates that CRAC is constitutively active under these conditions, exactly consistent with the constitutive Ca2+ entry result for STIM2S1NT shown in Fig. 2C. Lastly, we observed no differences in the I/V relationship in cells expressing these different STIM constructs (Fig. 3C), indicating that expression of STIM chimeras led to currents with typical CRAC channel characteristics.
In recent studies, Ikura and colleagues (21, 22) examined the unfolding and aggregation of a segment of the N terminus of STIM1 containing only the two EF-hands and sterile alpha motif (SAM) domains, designated the “EF-SAM” domain, changes believed to reflect the early activation steps for STIM1 following ER-luminal Ca2+. Recently, these authors revealed that the same segment of STIM2 underwent a 70-fold slower rate of aggregation upon Ca2+ withdrawal (21), consistent with the far slower kinetics of CRAC activation seen with STIM2 when compared with STIM1 (Fig. 3, A and B). Importantly, it was revealed that the upstream short N-terminal sequences of each STIM protein (residues 23–57 for STIM1 and 15–61 for STIM2) conferred a large increase in stability when attached to the EF-SAM domain, greatly altering the Ca2+ dissociation-induced destabilization that results in STIM protein activation. Our results reveal that these exact same N-terminal regions exert profound, STIM subtype-specific alterations on the kinetics of activation of SOCs. These regions were described as having considerable random coil character (21), and we may conclude that their major influence on the stability of the EF-SAM domains results from strong interactions with and significant alteration of the structure of the entire N-terminal domains of STIM1 and STIM2. We propose that differences in the rate of development of CRAC induced by STIM1 and STIM2 reflect not only their relative rates of aggregation after loss of Ca2+ but also the profound retarding effect of the N-terminal random coil domain of STIM2.
In a prior study, we interpreted the delay in the development of CRAC in cells expressing STIM2 and Orai1 as reflecting clearance of a cytoplasmic inhibitor (17). Because calmodulin added to the pipette solution blocked current development, we proposed calmodulin as a mediator of this inhibition. We now reveal that the slow kinetics are recapitulated precisely by swapping the STIM1 and STIM2 N termini. Indeed, the ER-luminal location of the STIM N terminus precludes any possible modification by cytosolic factors such as calmodulin. Instead, rather than an extrinsic control, we conclude that the distinct kinetics of CRAC development between STIM1 and STIM2 reflect intrinsic differences in the molecules. The recent findings on the increased stability of the luminal portion of STIM2 in the Ca2+-free state relative to STIM1 (21) provide compelling support for the concept that the delay in STIM2-induced CRAC current development reflects a delay in Ca2+ disassociation-induced aggregation kinetics.
Overall, the results redefine the role of STIM2 as a sensitive but highly constrained activator of SOCs. Although its EF-hand is primed to become activated with minimal store depletion (16), its luminal configuration limits its activation, the N-terminal flexible random coil domain serving as a powerful brake in the activation of Orai1. This prevents the otherwise constitutive activation of Orai1 channels due to their high sensitivity to luminal Ca2+. The results also explain an apparent controversy: the inhibition of endogenous SOC activity by overexpressed STIM2 (18) despite its well described ability to couple with Orai1. Clearly, the intrinsically restrained action of STIM2 dominates. Although cells generally express more STIM1 than STIM2 (29), expression of STIM2 prevails over endogenous STIM1. This dominant action of STIM2 indicates that it can have a decisive physiological role in the control of Ca2+ signal generation.
Supplementary Material
This work was supported by grants from the Pennsylvania Department of Health (to J. S.), American Heart Association Grant 0730184N (to J. S.), and National Institutes of Health Grant AI058173 (to D. L. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ER
- endoplasmic reticulum
- BAPTA
- 1,2-bis(2-amino-phenoxy)ethane-N,N,N,N-tetraacetic acid
- CRAC
- Ca2+-release activated Ca2+ (current)
- HEK293
- human embryonic kidney 293
- SOC
- store-operated channel
- WT
- wild type
- CT
- C terminus
- NT
- N terminus
- CFP
- cyan fluorescent protein
- SAM
- sterile alpha motif.
REFERENCES
- 1.Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. (2005) J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S. H., Tanasa B., Hogan P. G., Lewis R. S., Daly M., Rao A. (2006) Nature 441, 179–185 [DOI] [PubMed] [Google Scholar]
- 4.Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. (2006) Science 312, 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S. L., Yeromin A. V., Zhang X. H., Yu Y., Safrina O., Penna A., Roos J., Stauderman K. A., Cahalan M. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9357–9362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hewavitharana T., Deng X., Soboloff J., Gill D. L. (2007) Cell Calcium 42, 173–182 [DOI] [PubMed] [Google Scholar]
- 7.Hogan P. G., Rao A. (2007) Trends Biochem. Sci. 32, 235–245 [DOI] [PubMed] [Google Scholar]
- 8.Luik R. M., Lewis R. S. (2007) Trends Mol. Med. 13, 103–107 [DOI] [PubMed] [Google Scholar]
- 9.Zhang S. L., Kozak J. A., Jiang W., Yeromin A. V., Chen J., Yu Y., Penna A., Shen W., Chi V., Cahalan M. D. (2008) J. Biol. Chem. 283, 17662–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang G. N., Zeng W., Kim J. Y., Yuan J. P., Han L., Muallem S., Worley P. F. (2006) Nat. Cell Biol. 8, 1003–1010 [DOI] [PubMed] [Google Scholar]
- 11.Li Z., Lu J., Xu P., Xie X., Chen L., Xu T. (2007) J. Biol. Chem. 282, 29448–29456 [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Deng X., Zhou Y., Hendron E., Mancarella S., Ritchie M. F., Tang X. D., Baba Y., Kurosaki T., Mori Y., Soboloff J., Gill D. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7391–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E., Lewis R. S. (2009) Cell 136, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muik M., Fahrner M., Derler I., Schindl R., Bergsmann J., Frischauf I., Groschner K., Romanin C. (2009) J. Biol. Chem. 284, 8421–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan J. P., Zeng W., Dorwart M. R., Choi Y. J., Worley P. F., Muallem S. (2009) Nat. Cell Biol. 11, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandman O., Liou J., Park W. S., Meyer T. (2007) Cell 131, 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parvez S., Beck A., Peinelt C., Soboloff J., Lis A., Monteilh-Zoller M., Gill D. L., Fleig A., Penner R. (2008) FASEB J. 22, 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soboloff J., Spassova M. A., Hewavitharana T., He L. P., Xu W., Johnstone L. S., Dziadek M. A., Gill D. L. (2006) Curr. Biol. 16, 1465–1470 [DOI] [PubMed] [Google Scholar]
- 19.Soboloff J., Spassova M. A., Tang X. D., Hewavitharana T., Xu W., Gill D. L. (2006) J. Biol. Chem. 281, 20661–20665 [DOI] [PubMed] [Google Scholar]
- 20.Zheng L., Stathopulos P. B., Li G. Y., Ikura M. (2008) Biochem. Biophys. Res. Commun. 369, 240–246 [DOI] [PubMed] [Google Scholar]
- 21.Stathopulos P. B., Zheng L., Ikura M. (2009) J. Biol. Chem. 284, 728–732 [DOI] [PubMed] [Google Scholar]
- 22.Stathopulos P. B., Li G. Y., Plevin M. J., Ames J. B., Ikura M. (2006) J. Biol. Chem. 281, 35855–35862 [DOI] [PubMed] [Google Scholar]
- 23.Stathopulos P. B., Zheng L., Li G. Y., Plevin M. J., Ikura M. (2008) Cell 135, 110–122 [DOI] [PubMed] [Google Scholar]
- 24.Hewavitharana T., Deng X., Wang Y., Ritchie M. F., Girish G. V., Soboloff J., Gill D. L. (2008) J. Biol. Chem. 283, 26252–26262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soboloff J., Spassova M., Xu W., He L. P., Cuesta N., Gill D. L. (2005) J. Biol. Chem. 280, 39786–39794 [DOI] [PubMed] [Google Scholar]
- 26.Ma H. T., Patterson R. L., van Rossum D. B., Birnbaumer L., Mikoshiba K., Gill D. L. (2000) Science 287, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 27.Williams R. T., Manji S. S., Parker N. J., Hancock M. S., Van Stekelenburg L., Eid J. P., Senior P. V., Kazenwadel J. S., Shandala T., Saint R., Smith P. J., Dziadek M. A. (2001) Biochem. J. 357, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peinelt C., Vig M., Koomoa D. L., Beck A., Nadler M. J., Koblan-Huberson M., Lis A., Fleig A., Penner R., Kinet J. P. (2006) Nat. Cell Biol. 8, 771–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh-Hora M., Yamashita M., Hogan P. G., Sharma S., Lamperti E., Chung W., Prakriya M., Feske S., Rao A. (2008) Nat. Immun. 9, 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.