Abstract
Nitric-oxide synthases (NOSs) are calmodulin-dependent flavoheme enzymes that oxidize l-Arg to nitric oxide (NO) and l-citrulline. Their catalytic behaviors are complex and are determined by their rates of heme reduction (kr), ferric heme-NO dissociation (kd), and ferrous heme-NO oxidation (kox). We found that point mutation (E762N) of a conserved residue on the enzyme's FMN subdomain caused the NO synthesis activity to double compared with wild type nNOS. However, in the absence of l-Arg, NADPH oxidation rates suggested that electron flux through the heme was slower in E762N nNOS, and this correlated with the mutant having a 60% slower kr. During NO synthesis, little heme-NO complex accumulated in the mutant, compared with ∼50–70% of the wild-type nNOS accumulating as this complex. This suggested that the E762N nNOS is hyperactive because it minimizes buildup of an inactive ferrous heme-NO complex during NO synthesis. Indeed, we found that kox was 2 times faster in the E762N mutant than in wild-type nNOS. The mutational effect on kox was independent of calmodulin. Computer simulation and experimental measures both indicated that the slower kr and faster kox of E762N nNOS combine to lower its apparent Km,O2 for NO synthesis by at least 5-fold, which in turn increases its V/Km value and enables it to be hyperactive in steady-state NO synthesis. Our work underscores how sensitive nNOS activity is to changes in the kox and reveals a novel means for the FMN module or protein-protein interactions to alter nNOS activity.
Nitric oxide (NO)2 is a biological mediator that is produced in animals by three NO synthase isozymes (NOS, EC 1.14.13.39): inducible NOS (iNOS), neuronal NOS (nNOS), and endothelial NOS (eNOS) (1, 2). The NOS are modular enzymes composed of an N-terminal oxygenase domain and a C-terminal flavoprotein domain, with a calmodulin (CaM)-binding site connecting the two domains (3). During NO synthesis, the flavoprotein domain transfers NADPH-derived electrons through its FAD and FMN cofactors to a heme located in the oxygenase domain. The FMN-to-heme electron transfer enables heme-dependent oxygen activation and a stepwise conversion of l-Arg to NO and citrulline (4, 5). Heme reduction also requires that CaM be bound to NOS and is rate-limiting for NO biosynthesis (6–9).
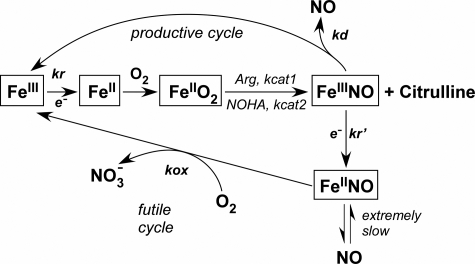
NOS enzymes operate under the constraint of having their newly made NO bind to the ferric heme before it can exit the enzyme (10). How this intrinsic heme-NO binding event impacts NOS catalytic cycling is shown in Fig. 1 and has previously been discussed in detail (10–13). The l-Arg to NO biosynthetic reaction (FeIII to FeIIINO in Fig. 1) is limited by the rate of ferric heme reduction (kr), because all biosynthetic steps downstream are faster than kr. However, once the ferric heme-NO complex forms at the end of each catalytic cycle, it can either dissociate to release NO into the medium (at a rate kd as shown in Fig. 1) or become reduced by the flavoprotein domain (at a rate k′r in Fig. 1; equal to kr) to form the enzyme ferrous heme-NO species (FeIINO), which releases NO very slowly (11, 12). Consequently, two cycles compete during steady-state NO synthesis (Fig. 1); NO dissociation from the ferric heme (kd) is part of a “productive cycle” that releases NO and is essential for NOS bioactivity, whereas reduction of the ferric heme-NO complex (kr′) channels the enzyme into a “futile cycle” that actually represents a NO dioxygenase activity. The rate of futile cycling is also determined by the rate of O2 reaction with the ferrous heme-NO complex (at a rate kox in Fig. 1), which regenerates the ferric enzyme. Surprisingly, NOS enzymes have evolved to have a broad range of kr (varies 40×), kox (varies 15×), and kd (varies 30×) values (Table S1) (12). This causes each NOS to distribute quite differently during steady-state NO synthesis and gives each NOS a unique catalytic profile (12).
FIGURE 1.
Global kinetic model for NOS catalysis. Ferric enzyme reduction (kr) is rate-limiting for the biosynthetic reactions (central linear portion). kcat1 and kcat2 are the conversion rates of the enzyme FeIIO2 species to products in the l-Arg and Nω-hydroxy-l-arginine (NOHA) reactions, respectively. The ferric heme-NO product complex (FeIIINO) can either release NO (kd) or become reduced (k′r) to a ferrous heme-NO complex (FeIINO), which reacts with O2 (kox) to regenerate ferric enzyme. Enzyme partitioning and NO release are determined by the relative rates of kr, kox, and kd. This figure is adapted from Ref. 12.
The enzyme physical and electronic factors that may set and regulate each of the three kinetic parameters (kr, kox, and kd) in NOS enzymes remain to be fully described. At present, the composition of the NOS flavoprotein domain and CaM appear to be primarily responsible for determining the kr (14–17), whereas the composition of the NOS oxygenase domain is presumed to determine the kd and kox (18, 19). Indeed, our recent point mutagenesis study identified a patch of electronegative residues on the FMN subdomain that are required to maintain a normal kr and NO synthesis activity in nNOS, suggesting that subdomain electrostatic interactions are important in the process (20). We found particularly large effects when the negative charge at Glu762 was neutralized via mutation to Asn. Remarkably, the NO synthesis activity of E762N nNOS was double that of wild-type nNOS, despite the mutant displaying a slow kr that was half of wild type. In the current report, we show that the E762N mutation has an additional, unsuspected effect on the kox kinetic parameter of nNOS. How this effect alters distribution of the nNOS enzyme during steady-state catalysis, impacts the apparent Km,O2, and leads to hyperactive NO synthesis is described. Our finding that the nNOS flavoprotein domain can tune a key kinetic parameter that defines the rate of a heme-based reaction in the nNOS oxygenase domain is unusual and suggests a means by which protein-protein interactions could regulate the catalytic behavior of nNOS.
EXPERIMENTAL PROCEDURES
General Methods and Materials
All reagents and materials were obtained from Sigma, Amersham Biosciences, or other sources as previously reported (17, 20, 21). Absorption spectra and steady-state kinetic data were obtained using a Shimadzu UV-2401PC spectrophotometer. Stopped-flow experiments were performed using a Hi-Tech Scientific KinetAsyst SF-61DX2 stopped-flow system equipped with anaerobic set-up and rapid scanning photodiode array detector. Data from multiple identical stopped-flow experiments were averaged to improve the signal/noise ratio. The spectral traces were fit according to single or multiple exponential equations using software provided by the instrument manufacturer. The best fit was determined when adding further exponentials did not improve the fit as judged from the residuals. All plots and some additional curve fitting were done using Origin® 7.5 (OriginLab, Northampton, MA). All experiments were repeated two or more times with at least two independently prepared batches of proteins to ensure consistent reproducibility of the results. Data were analyzed and are expressed as mean ± S.D. Anaerobic samples were prepared in an air-tight cuvette using repeated cycles of vacuum followed by a positive pressure of catalyst-deoxygenated nitrogen gas.
Molecular Biology
Wild-type and mutant nNOS proteins containing a His6 tag attached to their N termini were overexpressed in Escherichia coli strain BL21(DE3) using a modified pCWori vector as described (20, 22). Restriction digestions, cloning, and bacterial growth were performed using standard procedures. Transformations were done using a TransformAid bacterial transformation kit (Fermentas). Oligonucleotides used to construct site-directed mutants in nNOS were obtained from Integrated DNA Technologies (Coralville, IA). Mutations (in boldface type) and their corresponding oligonucleotides were as follows: E762N sense, 5′-TACGCCACAAACACAGGCAAATCACAGGCCTATGCC-3′; E762N antisense, 5′-GGCATAGGCCTGTGATTTGCCTGTGTTTGTGGCGTA-3′. Site-directed mutagenesis was done using a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Incorporated mutations were confirmed by DNA sequencing at the Cleveland Clinic Genomics Core.
Expression and Purification of Wild-type and Mutant Proteins
All proteins were purified in the presence of H4B and l-Arg as described previously (14, 20). The ferrous heme-CO adduct absorbing at 444 nm was used to measure hemeprotein content with an extinction coefficient of ϵ444 = 74 mm−1 cm−1 (A444–A500) (20). Purity of each protein was assessed by SDS-PAGE and spectral analysis.
NO Synthesis, NADPH Oxidation, and Flux Measurement
Steady-state activities of wild-type and E762N nNOS proteins were determined at 10 or 25 °C using the spectrophotometric oxyhemoglobin assay (20). Cuvettes contained 0.1–0.2 μm nNOS, 40 mm EPPS (pH 7.6), 150 mm NaCl, 0.3 mm dithiothreitol, 4 μm FAD, 4 μm FMN, 10 μm H4B, 5 mm l-Arg, 1 mg/ml bovine serum albumin, 1 mm Ca2+, 0.5 mm EDTA, 1 μm CaM, 100 units/ml catalase, 25 units/ml superoxide dismutase, and 5 μm oxyhemoglobin. The reaction was initiated with 300 μm NADPH in a total reaction volume of 500 μl and was run for 2 min. The NO-mediated conversion of oxyhemoglobin to methemoglobin was monitored at 401 nm and converted to a rate of NO synthesis using a difference extinction coefficient of ϵ401 = 38 mm−1 cm−1. NADPH oxidation rates were similarly measured at 340 nm in the presence of oxyhemoglobin under identical conditions, and the rate of NADPH oxidation was calculated using an extinction coefficient of ϵ340 = 6.2 mm−1 cm−1. For the electron flux measurement through the NOS heme during steady-state catalysis, we measured the rate of NADPH oxidation by each CaM-bound enzyme in the absence of l-Arg (17).
Measurement of Apparent Km,O2
Apparent Km and Vmax values were calculated by fitting the initial rates of NO synthesis measured at 10 or 25 °C and at various oxygen concentrations to a hyperbolic equation using the Origin® version 7.5 software (OriginLab) (17). Reactions were run in septum-sealed cuvettes that contained 0.2 μm NOS (nNOS wild type or E762N nNOS) and the additives described above for NO synthesis assay diluted in various ratios of N2-, air-, or O2-saturated buffer solutions. The reaction was initiated with 300 μm NADPH in a total reaction volume of 800 μl and was run for 2 min. The initial O2 concentration in each reaction was calculated based on the solution mixing ratio and the O2 concentration of air- or O2-saturated buffer at 10 or 25 °C (∼0.35, 1.7, 0.26, and 1.26 mm, respectively).
Spectroelectrochemical Measurement of Flavin Midpoint Reduction Potentials
Sample preparation and redox titrations were carried out in a glove box (Bell Technology) under nitrogen atmosphere with oxygen levels below 5 ppm as described previously (23). The E762N nNOSr protein concentration was 30–40 μm containing either EDTA (1 mm) or CaCl2 (2 mm) plus CaM (60–80 μm) in 40 mm EPPS buffer, pH 7.6, 10% glycerol, and 150 mm NaCl. Absorption spectra were recorded in a Cary 50 spectrophotometer using a dip probe detector, and the potentials were monitored using Accumet AB15 coupled to a silver/silver chloride electrode saturated with 4 m KCl. Measurements were done in a custom-made glass beaker kept in a water bath at 15 ± 1 °C. A reductive titration was performed by the stepwise addition of sodium dithionite, whereas an oxidative titration was done by adding potassium ferricyanide. The redox mediators with midpoint potentials in the range of flavin potentials used were 0.5–1 μm phenazine methasulfate (+80 mV), indigo carmine (−125 mV), 2-hydroxy-1,4-naphthoquinone (−152 mV), anthraquinone-2,6-disulfide (−184 mV), anthraquinone-2-sulfonate (−225 mV), phenosafranin (−252 mV), safranin O (−280 mV), benzyl viologen (−348 mV), and methyl viologen (−443 mV). The electrode was calibrated against phenosafranin (−252 mV) and the potential of a 5 mm solution of ferricyanide/ferrocyanide in 0.1 m potassium phosphate (pH 7.0), at 25 °C (+425 mV). A correction factor of (+199 mV) was obtained, which is in good agreement with the reported potential of the electrode. Based on the temperature dependence of the electrode potential, a correction factor of (+209 mV) at 15 °C was used. The absorption changes at 457 and 600 nm were plotted with electrochemical potentials (mV). The midpoint potentials were calculated using the four-electron Nernst equation (Equation S1), where A represents the absorbance; a–e are the relative absorbance values contributed by the diflavin in each of five nondegenerate oxidation states; E is the observed system potential, and E′1–E′4 are the four midpoint potentials, two for each flavin.
Anaerobic Heme Reduction Measurements
The kinetics and extent of heme reduction were analyzed at 10 °C, as described previously (14, 20, 24, 25), using a stopped-flow apparatus and diode array detector (Hi-Tech Scientific KinetAsyst SF-61DX2) equipped for anaerobic analysis. Ferric heme reduction was followed by formation of the ferrous heme-CO complex at 444 nm. Reactions were initiated by rapidly mixing an anaerobic, buffered, CO-saturated solution containing either 100 μm NADPH or 5 mm CaCl2 with an anaerobic, buffered, CO-saturated solution containing wild-type or mutant NOS (8 μm), 100 mm EPPS (pH 7.6), 100 mm NaCl, 20 μm H4B, 5 mm l-Arg, 0.3 mm dithiothreitol, 40 μm CaM, and either 1 mm Ca2+ when triggered with NADPH or 50 μm NADPH when triggered with Ca2+. Signal/noise ratios were improved by averaging 8–10 individual mixing experiments. The rate of heme reduction was determined by fitting the time course of the absorbance increase at 444 nm to a single exponential equation using a nonlinear least squares method provided by Hi-Tech Ltd. The extent of heme reduction was determined from the concurrent absorbance decrease at 650 nm.
Kinetics of Enzyme Ferrous Heme-NO Complex Formation
Enzyme spectral transitions during the initial phase of NO synthesis were studied at 10 °C in the Hi-Tech Scientific KinetAsyst SF-61DX2 stopped-flow spectrophotometer, as previously described (26). To initiate NO synthesis, an air-saturated solution containing 40 mm EPPS, pH 7.6, 10% glycerol, 150 mm NaCl, 4–7 μm nNOS or mutant, 20 μm H4B, 0.4 mm dithiothreitol, 3 mm l-Arg, 0.5 mm EDTA, 2 mm Ca2+, and 40 μm CaM was rapidly mixed with an air-saturated buffered solution containing 100 μm NADPH. Absorbance at 436 nm was monitored to follow ferrous heme-NO formation, and absorbance at 340 nm was monitored to follow NADPH oxidation (10, 18). Signal/noise ratios were improved by averaging at least 10 individual mixing experiments. Each experiment was performed with two enzyme preparations.
Reaction of Ferrous Heme-NO Complexes with Oxygen
Protein solutions containing 8 μm full-length ferric nNOS (or, in some cases, nNOSoxy) were prepared in 40 mm EPPS buffer, pH 7.6, 10% glycerol, 150 mm NaCl, 20 μm H4B, 3 mm l-Arg, 1 mm EDTA, and 0.5 mm dithiothreitol. In some cases, the NaCl concentration was changed to 50 or 250 mm. Samples were made anaerobic in sealed cuvettes by several cycles of nitrogen/vacuum. Then Fe(III)-heme was titrated with sodium dithionite to produce the ferrous enzyme. Reduction of the enzyme was monitored in either a Cary 100 or a Shimadzu UV-2401 PC spectrophotometer. Titration was stopped as soon as the shift of the Soret band and the decrease of the 650 nm band were complete to avoid the addition of excess dithionite. Ferrous enzyme was then titrated with an NO-saturated buffer solution. NO titration was stopped when the red shift of the Soret band was complete. The resulting protein Fe(II)-NO complexes were rapidly mixed with air-saturated buffer (oxygen concentration 280 μm) with the same composition as the protein buffer at 10 °C. In some cases, the syringe containing air-saturated buffer also contained the E762N nNOS reductase domain protein at the concentrations indicated in the text. Reactions were carried out in a Hi-Tech Scientific KinetAsyst SF-61DX2 stopped-flow system equipped with anaerobic set-up and rapid scanning photodiode array detector. The rates of absorbance decrease at 436 nm and increase at 393 nm were used to determine ferrous-NO complex disappearance and ferric enzyme formation, respectively (18, 27).
Simulations of nNOS Distribution During Steady-state NO Synthesis
The distribution of the different NOS species in steady-state conditions was calculated by the global model in MathCAD version 7.0 (11, 28) or Fortran source code (13). Simulations assume constant values for [O2] = 280 μm and [NADPH] = 40 μm. The values for heme reduction (kr), NO dissociation (koff or kd), and ferrous heme-NO oxidation (kox) were kox = 0.18 s−1 (2 times the half air-saturated value), kr = 4.6 s−1, and kd = 5 s−1 for wild-type nNOS and kox = 0.32 s−1 (2 times the half air-saturated value), kr = 1.7 s−1, and kd = 5 s−1 for E762N nNOS. The other values were the same for both simulations: k2 = k5 = 9 × 105 m−1 s−1, k3 = k6 = 26 s−1, and k9 = 1 × 10−4 s−1. Experimentally observed oxidation rates (kox) for wild-type and mutant ferrous heme-NO complexes, which we derived at half air-saturated condition, were converted to bimolecular rate constants proportional to O2 concentration for use in the simulations for deriving apparent Km,O2. For simulation of the NO synthesis reactions, the concentration of O2 was typically set at 280 μm but was also varied in the simulations to derive the apparent Km,O2 value. The NADPH and chosen O2 concentrations were kept constant during each simulation to avoid secondary effects due to their exhaustion. Simulated NO synthesis rates were fit using a hyperbolic equation in Origin® version 7.5 software (OriginLab) to estimate the apparent Km,O2.
RESULTS
Steady-state NO Synthesis and NADPH Oxidation
Table 1 compares the NO synthesis activities and NADPH oxidation rates that we obtained for wild-type nNOS and the E762N mutant. The NO synthesis activity of E762N nNOS was nearly twice that of wild type, confirming our previous report (20). The rate of NADPH oxidation by the mutant also exceeded that of wild type during NO synthesis, such that it oxidized about 2.9 NADPH per NO formed compared with 2.0 NADPH per NO formed by wild-type nNOS. However, a good proportion of the excess NADPH consumption by E762N nNOS could be attributed to an increased rate of flavin auto-oxidation (29, 30), as was indicated by the mutant having a higher rate of NADPH oxidation in the absence of CaM (Table 1). If one subtracts this rate from the rate of NADPH consumption observed during NO synthesis, it shows that the E762N mutant and wild-type nNOS are similar regarding their ratio of NADPH consumed per NO synthesized.
TABLE 1.
Steady-state NO synthesis and NADPH oxidation rates of wild-type and mutant nNOS
Rates were measured at 25 and 10 °C, as described under “Experimental Procedures.” The NO synthesis rate was measured using l-Arg as substrate. Values (min−1) represent the mean ± S.D. of three independent measurements with two preparations of each enzyme. WT, wild type.
| Enzyme | NO synthesis | NADPH oxidation rates |
||
|---|---|---|---|---|
| +CaM +Arg | −CaM +Arg | +CaM −Arg | ||
| min−1 | min−1 | |||
| Activities at 25 °C | ||||
| nNOS WT | 56 ± 2.5 | 115 ± 7 | 8 ± 0.2 | 346 ± 6 |
| E762N | 105 ± 8.7 | 308 ± 16 | 94 ± 7 | 142 ± 3 |
| Activities at 10 °C | ||||
| nNOS WT | 21 ± 1.5 | 40 ± 4 | 5 ± 0.5 | 124 ± 10 |
| E762N | 34 ± 3 | 107 ± 11 | 27 ± 3 | 47 ± 4 |
We also wished to examine how the E762N mutation impacts steady-state electron flux through the nNOS heme in the absence of NO synthesis. This can be determined from the rate of NADPH oxidation by each CaM-bound enzyme in the absence of l-Arg (17, 31). Under these conditions, NADPH-derived electrons pass from the flavins to the NOS heme, which then quickly reduces O2 and releases superoxide or H2O2 instead of generating NO (17). Thus, the steady-state NADPH oxidation rate will primarily reflect the electron flux through the heme and will be proportional to the rate of heme reduction, which remains the slow step during catalysis. Table 1 shows that the NADPH oxidation rate of CaM-bound nNOS increased about 2-fold in the l-Arg-free condition, consistent with previous reports (32, 33). The rate of NADPH oxidation by E762N nNOS was more than 2-fold slower in the l-Arg-free condition. These data show that electron flux through the heme is actually slower in the E762N mutant, except when NO synthesis from l-Arg is taking place.
Flavin Midpoint Reduction Potentials
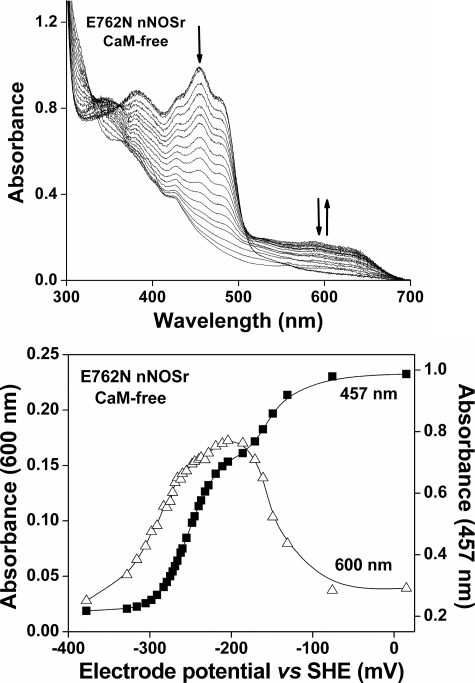
In the crystal structure of nNOSr (34), Glu762 is positioned on the surface and relatively close (∼8 Å) to the isoalloxazine ring of bound FMN. We performed spectro-electrochemical titrations with E762N nNOSr to determine if the mutation has an effect on the flavin midpoint potentials. The fully oxidized E762N nNOSr protein was sequentially titrated under anaerobic conditions with dithionite and ferricyanide, and spectra were recorded after each addition once the system reached thermodynamic equilibrium (35, 36). The collected spectra and the absorbance changes at 457 and 600 nm versus potential for the CaM-free and CaM-bound mutant enzyme are shown in Fig. 2 and Fig. S1, respectively. FAD and FMN midpoint potentials were calculated by fitting the absorbance changes, as done before (23, 37). Table 2 contains the calculated midpoint reduction potentials for the bound FAD and FMN. The E762N nNOS shows an approximately 90-mV increase in its FMN sq/hq couple and a corresponding 110-mV decrease in its FMN ox/sq couple, relative to wild-type nNOS. This result was the same for the CaM-free and CaM-bound enzymes (Table 2). The mutant FAD midpoint potentials remained similar to wild-type, consistent with the mutation being located in the FMN subdomain. Thus, the E762N mutation alters the FMN midpoint potentials of nNOS so as to thermodynamically stabilize the FMN hq and destabilize the FMN sq, relative to wild-type.
FIGURE 2.
Potentiometric titration of E762N nNOSr flavins in the absence of CaM at 15 °C. Top, representative set of visible spectra obtained during potentiometric titration of CaM-free E762N nNOSr with sodium dithionite. Bottom, plots of absorbance at 457 nm (solid squares) and at 600 nm (open triangles) versus the electrochemical potential (mV) for CaM-free E762N nNOSr along with the lines of best fit as calculated using the four-electron Nernst equation described under “Experimental Procedures.” Data are representative of at least two experiments.
TABLE 2.
Equilibrium midpoint potentials (Em) versus standard hydrogen electrode of nNOSr proteins
Midpoint potentials of each flavin (ox/sq and sq/hq) were determined for the nNOSr proteins by potentiometric titration at 15 ± 1 °C, as described under “Experimental Procedures.” WT, wild type.
| Sample | Condition | FMN |
FAD |
||
|---|---|---|---|---|---|
| ox/sq | sq/hq | ox/sq | sq/hq | ||
| WT nNOSr | −CaM | −49 ± 5 | −274 ± 5 | −232 ± 7 | −280 ± 6 |
| +CaM | −30 ± 4 | −267 ± 5 | −234 ± 6 | −284 ± 9 | |
| E762N nNOSr | −CaM | −156 ± 9 | −187 ± 8 | −253 ± 4 | −275 ± 9 |
| +CaM | −153 ± 8 | −174 ± 10 | −250 ± 7 | −274 ± 8 | |
Rate and Extent of Heme Reduction
We next measured the rate and extent of heme reduction in anaerobic stopped-flow reactions that monitored the formation of the enzyme ferrous heme-CO complex at 444 nm. The reactions were initiated at 10 °C either by mixing CaM-bound nNOS enzymes with excess NADPH or by mixing NADPH-reduced NOS enzymes with Ca2+ to trigger CaM binding and heme reduction (14). The spectral and kinetic traces are shown in Fig. S2. The E762N nNOS had a ∼60% slower heme reduction rate (Table 3) and also had a decreased extent of heme reduction at equilibrium (29% versus 71% for wild type). A slower and less extensive heme reduction in E762N nNOS is consistent with its having a more positive FMN sq/hq midpoint potential. It may also help to explain why the mutant has slower electron flux through the heme during NADPH oxidation in the CaM-bound, l-Arg-free state.
TABLE 3.
Heme reduction and ferrous heme-NO buildup of wild-type and mutant nNOS
All reactions were carried out at 10 °C in a stopped-flow spectrophotometer as described under “Experimental Procedures.” The values for heme reduction in wild type and E762N are the means ± S.D. of 7–10 individual reactions and are representative of experiments done with two enzyme preparations. Rate constants for heme-NO complex buildup were determined during the initial phase of NO synthesis. The numbers in parentheses indicate the percentage of absorbance change for each phase. ND, not detected; WT, wild type.
| Enzyme | Heme reduction |
Heme-NO complex buildup | |
|---|---|---|---|
| Ca2+-initiated | NADPH-initiated | ||
| s−1 | s−1 | ||
| nNOS WT | 4.2 ± 0.3 | 4.6 ± 0.4 | k1 = 9.4 (60%) |
| k2 = 1.5 (40%) | |||
| E762N | 1.7 ± 0.16 | 1.2 ± 0.1 | k1 = 0.34 (100%) |
| k2 = ND | |||
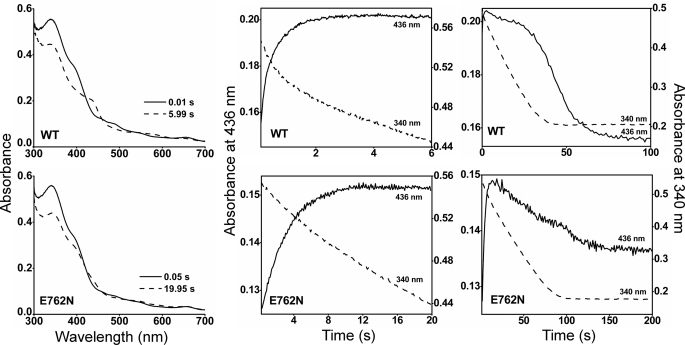
Ferrous Heme-NO Complex Formation during NO Synthesis
When a NOS initiates NO synthesis, the newly made NO binds to the ferric enzyme heme before it exits the enzyme (Fig. 1). In nNOS, this process leads the enzyme to exist primarily as its ferrous heme-NO species during steady-state NO synthesis and greatly attenuates the steady-state activity of nNOS (10). A diminished heme-NO complex buildup could conceivably enable the greater NO synthesis activity of the E762N mutant. We utilized rapid scanning stopped flow spectroscopy to compare heme-NO complex formation in the E762N and wild-type nNOS after they initiate their NO synthesis from l-Arg. Air-saturated solutions containing enzyme, l-Arg, cofactors, Ca2+, and CaM were rapidly mixed at 10 °C with an air-saturated solution containing NADPH to trigger NO synthesis. As shown in Fig. 3, a majority of wild-type nNOS quickly converted to the ferrous heme-NO complex, as judged by the buildup of heme Soret absorbance at 436 nm, a single broad visible band near 570 nm, and loss of absorbance at 395 nm, consistent with previous reports (10, 38). In the E762N nNOS reaction, much less enzyme partitioned into a heme-NO complex, and the majority of the enzyme remained in its ferric form, as judged by the continued strong Soret absorbance at 395 nm (Fig. 3). The absorbance values at 436 and 340 nm were plotted versus time to follow the kinetics of heme-NO complex formation and concurrent NADPH oxidation in either reaction (18, 27), and the kinetic traces are shown at two different time scales in Fig. 3. In both the wild-type and E762N nNOS, the level of heme-NO complex reached a plateau and then began to decrease as more of the NADPH became oxidized. Heme-NO complex buildup in wild-type nNOS was rapid and best described as biphasic, as reported previously (18), whereas heme-NO complex buildup in E762N was more gradual and was best described as monophasic (Table 3). From the slope of the absorbance changes at 340 nm, we estimate that the initial rate of NADPH oxidation prior to heme-NO complex buildup was about 2-fold faster in the wild-type nNOS (1.38 min−1) than in the E762N mutant (0.66 min−1), consistent with the mutant having a slower rate of heme reduction and electron flux through the heme. Heme-NO complex buildup in the wild-type nNOS reaction caused its NADPH oxidation rate to decrease by about 10-fold (from 1.38 to 0.12 min−1), as seen previously (18, 39). In contrast, the rate of NADPH consumption in the E762N nNOS reaction decreased only 1.3-fold (from 0.66 to 0.50 min−1) upon heme-NO complex buildup. This left the E762N mutant catalyzing a ∼4-fold faster rate of NADPH oxidation than wild-type nNOS during the steady-state phase of the reactions, consistent with the mutant being hyperactive for NO synthesis in the steady state. Our data indicate that the greater NO synthesis activity of E762 N nNOS is associated with a reduced buildup of the ferrous heme-NO species during the steady state.
FIGURE 3.
Kinetics of heme-NO complex buildup, decay, and simultaneous NADPH oxidation. All reactions were run at 10 °C in a stopped-flow spectrophotometer, as described under “Experimental Procedures.” The left panels depict absorbance traces collected at the indicated times after mixing. The middle and right panels depict the kinetics of absorbance change at 436 nm for formation of the heme-NO complex (solid lines) and kinetics of absorbance change at 340 nm for following NADPH oxidation (dashed lines). The traces for wild-type (WT) or mutant nNOS are the means ± S.D. of 7–10 individual reactions and are representative of experiments done with two enzyme preparations.
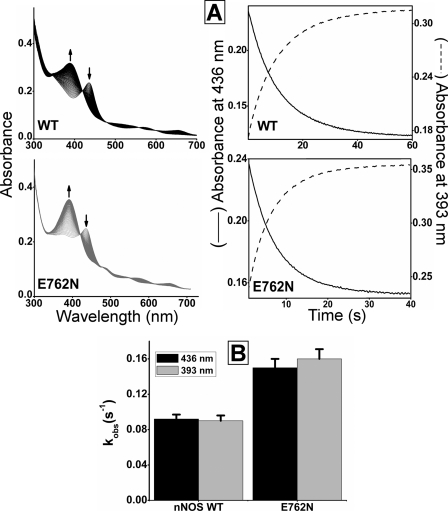
Reaction of O2 with the Enzyme Ferrous Heme-NO Complex
The rate of this reaction, termed kox, is an intrinsic kinetic parameter (Fig. 1) that can greatly impact NOS enzyme distribution during catalysis and consequently the steady-state NO synthesis activity (12, 18, 27). A faster kox is one way for a NOS enzyme to have less ferrous heme-NO complex buildup in the steady state. We determined kox values of E762N and wild-type nNOS by mixing anaerobic solutions of the ferrous heme-NO enzymes with an air-saturated solution in the stopped-flow spectrophotometer at 10 °C and following the reactions using the diode array detector. Overlay of the consecutive scans recorded during reactions of either wild-type or E762N nNOS (Fig. 4, left panels) shows an absorbance decrease occurred between 415 and 445 nm along with an absorbance increase between 360 and 410 nm as the reactions progressed. These absorbance changes indicate the loss of the ferrous heme-NO complex and generation of ferric enzyme, respectively. Kinetic traces of absorbance change at 436 and 393 nm (Fig. 4, right panels) were best fit to monophasic transitions in all cases, and the observed kox rates are compared in Fig. 4B. The kox of E762N nNOS was about 2 times faster than that of wild-type nNOS. This difference was maintained at three different salt concentrations in the reaction buffer (50, 150, and 250 mm added NaCl; Table S2). In addition, we performed control experiments in which the kox of the ferrous heme-NO complex of the nNOSoxy domain was determined in the presence or absence of a molar excess of added E762N nNOS reductase domain protein. We found that the added E762N reductase domain had no impact on the magnitude of kox under this circumstance (data not shown). Together, our results indicate that the E762N mutation affects the kox of nNOS but only when its reductase and oxygenase domains are covalently linked.
FIGURE 4.
Reaction of ferrous heme-NO complexes with O2. Anaerobic ferrous-NO enzymes were reacted with air-saturated solution in the stopped flow instrument at 10 °C. A, the left panels contain rapid scanning spectra recorded during the reaction of ferrous-NO enzymes with air-saturated buffer. The right panels show the kinetics of spectral change at 436 and 393 nm. B, observed rate constants for reaction of ferrous-NO complexes with O2. The values are the mean ± S.D. of 7–10 reactions and representative of two separate experiments. WT, wild-type nNOS.
Modeling Enzyme Distribution during Steady-state NO Synthesis
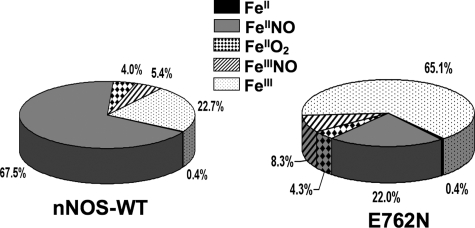
We next incorporated the kr and kox values we measured for E762N nNOS in a computer simulation of the global kinetic model (Fig. 1) (28) in order to understand the catalytic behavior of the mutant relative to wild-type nNOS. The model focuses on the five main enzyme species that are present during steady-state NO synthesis, namely the ferric, ferrous, ferrous-O2 (or ferric-superoxy), ferric-NO, and ferrous-NO forms (Fig. 1). The kinetic values and rates that we used in the simulations are described under “Experimental Procedures.” For wild-type nNOS, its fast kr relative to its slower kd and kox values causes it to exist predominantly as the ferrous-NO species during the steady state (12, 28). Indeed, the enzyme distribution we modeled (Fig. 5) indicated that ∼23% of nNOS is present as ferric enzyme and about 68% as the ferrous heme-NO species in the steady state (Fig. 5), with the remaining 9% of nNOS mainly distributed between the FeIIO2 and FeIIINO species. This falls within estimates reported in literature (10, 12, 28, 31, 38), which range between 67 and 85% ferrous heme-NO complex, and is also consistent with the spectrum that we obtained for wild-type nNOS during steady-state NO synthesis (Fig. 3).
FIGURE 5.
NOS enzyme distribution patterns during steady-state NO synthesis. NOS distributions were calculated by simulation of the global kinetic model, assuming constant concentrations of O2 (280 μm) and NADPH (40 μm). WT, wild type.
The decreased kr and faster kox values of E762N nNOS cause a shift in the calculated enzyme distribution during steady-state NO synthesis (Fig. 5). The ferrous heme-NO species now represents only 22% of the total enzyme, whereas the ferric species becomes predominant and represents 65% of the total enzyme. The remaining 13% of E762N nNOS distributes mainly between the FeIIO2 and FeIIINO species (Fig. 5). This calculated distribution is consistent with the spectrum that we obtained for the E762N mutant during steady-state NO synthesis (Fig. 3). The computer simulations confirm that the slower kr and faster kox in E762N nNOS can explain its having less ferrous heme-NO complex buildup during NO synthesis.
NO Synthesis Activity versus O2 Concentration
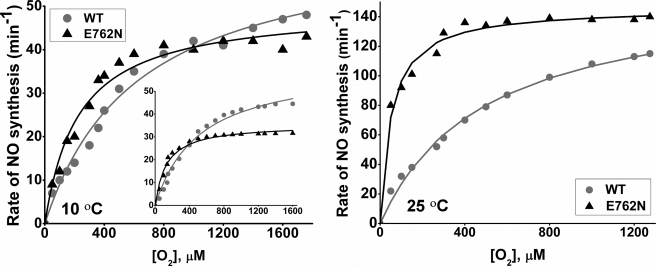
The wild-type nNOS has a very high apparent Km,O2 for NO synthesis (300–500 μm) (12, 38). This is due to its kr, kox, and kd parameters being set to favor buildup of the ferrous heme-NO complex during NO synthesis, which in turn makes the O2 concentration response of the kox reaction the major determinant of the apparent Km,O2 (17, 38).3 Changes in kr or kox can significantly change the apparent Km,O2 of a NOS (12, 17, 25). We therefore measured the apparent Km,O2 values for E762N and wild-type nNOS by determining their NO synthesis rates at a range of different O2 concentrations (Fig. 6). These experiments were run at 10 °C as well as 25 °C so we could directly compare the results of our kinetic and steady-state measures that were taken at the same temperature (10 °C).
FIGURE 6.
Oxygen activity response curves for NO synthesis. The initial rates of NO synthesis were determined at each designated O2 concentration. Left, experimental data measured at 10 °C; right, experimental values measured at 25 °C. Simulated values at 10 °C are shown in the left panel (inset). The conditions are described under “Experimental Procedures.” WT, wild-type nNOS.
The E762N mutant had greater NO synthesis activity than wild-type nNOS across a wide range of O2 concentrations. The difference between mutant and wild-type was more pronounced at 25 °C than at 10 °C. The apparent Km,O2 and Vmax values that we calculated from this data for E762N nNOS and wild-type nNOS are listed in Table 4. The E762N mutant had a lower apparent Km,O2 than wild-type nNOS at both 25 and 10 °C. In addition, the apparent Vmax for the E762N mutant was lower than that of wild-type, consistent with only the kr values limiting the NO synthesis activity at very high [O2]. Overall, the V/K values for E762N mutant were higher than for wild-type nNOS, particularly so at 25 °C. This indicates that the mutational effect on Km,O2 offsets the effect on Vmax and enables the E762N mutant to be a more efficient NO synthase in the steady state.
TABLE 4.
Apparent Km,O2 values for NO synthesis of wild-type and mutant nNOS
Rates were determined at 25 and 10 °C, as described under “Experimental Procedures.” The NO synthesis rate was measured using l-Arg as substrate. Values (min−1) represent the mean ± S.D. of three independent measurements with two preparations of each enzyme. Simulated values were calculated by the global model in MathCAD 7.0 at 10 °C, as described under “Experimental Procedures.” Values at each O2 concentration were plotted and fit according to Michaelis-Menten kinetics. V/K is the ratio of Vmax and corresponding Km values. WT, wild type.
| Enzyme | Measured values, 25 °C |
Measured values, 10 °C |
Simulated values, 10 °C |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Km,O2 | Vmax | V/K | Km,O2 | Vmax | V/K | Km,O2 | Vmax | V/K | |
| μm | min−1 | μm | min−1 | μm | min−1 | ||||
| nNOS WT | 493 ± 45 | 160 ± 15 | 0.32 | 628 ± 64 | 66 ± 3 | 0.10 | 532 | 64 | 0.12 |
| E762N | 52 ± 6 | 147 ± 14 | 2.83 | 227 ± 25 | 49 ± 2 | 0.22 | 124 | 36 | 0.29 |
We next ran computer simulations of the global kinetic model at a range of O2 concentrations, using the kinetic parameters that we measured at 10 °C, and plotted the calculated NO synthesis activities versus O2 concentration (Fig. 6, inset) to obtain the predicted apparent Km,O2 values for the E762N mutant and wild-type nNOS. In the simulations, we held the kr and kd values constant, whereas the kox value was assumed to vary in direct proportion to the O2 concentration, based on related data we have obtained with wild-type nNOS (25, 38).3 The simulated data gave an apparent Km,O2 of 532 μm for wild-type and a value of 124 μm for E762N nNOS (Table 4). The simulated results closely mimic the experimental data that we obtained at 10 °C (Fig. 6, left, inset) and the apparent Km,O2 and Vmax values. The simulations confirm that the altered kr and kox values of E762N nNOS will lower its apparent Km,O2 and make it hyperactive for NO synthesis in air-saturated buffer, where the O2 concentration is ∼200 μm.
DISCUSSION
Basis for Hyperactive NO Synthesis
Our results suggest that E762N nNOS has less ferrous heme-NO complex buildup during NO synthesis due to the mutation affecting both the kr and kox kinetic parameters. A slower kr decreases the probability of the mutant enzyme forming the ferrous heme-NO complex during NO synthesis, whereas the faster kox causes any ferrous heme-NO complex that does form to react twice as quickly with O2. These two effects combine to cause a significant decrease in the apparent Km,O2 for NO synthesis, which in turn increases the enzyme V/K value and enables the mutant to have supernormal NO release rates over a wide range of O2 concentrations.
There are three points to note regarding our proposed model. First, the different levels of ferrous heme-NO complex buildup in the E762N mutant and wild-type nNOS reactions are independent of any difference in the concentration of solution NO during the measurement. Previous studies had shown that ferrous heme-NO complex buildup in wild-type nNOS is practically independent of solution NO concentrations that are achieved during the measurement (10). In our current study, we added superoxide dismutase to our reactions to nullify any differences due to concurrent superoxide formation by the enzymes (particularly in the E762N nNOS reaction, which has greater uncoupled NADPH oxidation than wild-type). Moreover, our computer simulation of the reactions did not factor in an external NO concentration (i.e. solution [NO] was fixed at 0), yet it was still able to correctly model the unique enzyme distribution and catalytic behavior of E762N nNOS exclusively based on its altered kr and kox values. As a second point, the slower kr of E762N nNOS cannot, on its own, cause the mutant to be hyperactive for NO synthesis and instead would cause it to have a lower NO synthesis activity than wild-type at any given O2 concentration. How the NO synthesis activity of nNOS changes as a function of kr alone is well documented in studies that used CaM structural analogs or point mutations that alter only kr and in accompanying computer simulations (25, 40, 41). Only an increase in kox (either alone or together with a change in the kr) could cause nNOS to increase its V/K value and have hyperactive NO release, as we observed for the E762N mutant. Indeed, the “positive” effect that a faster kox has on the enzyme distribution (via decreased ferrous heme-NO complex buildup) overcomes the “negative” effect of a slower kr on the NO biosynthesis rate and enables the E762N nNOS to have a larger V/K value and supernormal NO release rates, although it is actually synthesizing NO more slowly than wild type. Essentially, the hyperactive NO synthesis activity results from the apparent Km,O2 being shifted to the left. As a third point, nNOS is unique among the three mammalian NOS regarding its sensitivity to relatively small changes in the kox parameter. This is because only nNOS has its kr, kox, and kd kinetic parameters set to enable significant buildup of the ferrous heme-NO species during NO synthesis. In comparison, iNOS has a kox that is faster than its kr. Thus, increasing the kox rate 2-fold would not greatly alter iNOS distribution and so would not alter its catalytic behavior (see Fig. 1). Similarly, the kr of eNOS is so slow relative to its kd that an extremely small proportion of enzyme partitions into the ferrous heme-NO species during NO synthesis. This greatly diminishes the impact of any change in kox for eNOS. These differences among the NOS isoforms have been previously reviewed (12, 13).
Basis for the E762N Mutational Effects on kr and kox
During nNOS catalysis, the FMN subdomain must oscillate between an interaction with the FNR module to receive electrons and an interaction with the NOSoxy domain to deliver electrons. This involves two separate conformational equilibrium of the FMN subdomain and has been described as a tethered or shuttle mechanism for electron transfer as discussed in detail elsewhere (34, 35, 37, 42–45). On the basis of previous studies (20, 34, 46), an electronegative patch on the FMN subdomain, which includes Glu762, is hypothesized to interact with a complementary electropositive patch on the surface of nNOSoxy in order to align the domains and facilitate the FMN-to-heme electron transfer (20). This is the structural context in which the E762N mutation has its effects.
The catalytic behavior of E762N nNOS is reminiscent of W409F nNOS, which also has a faster kox, a slower kr, and hyperactive NO synthesis activity compared with wild-type nNOS (18, 27), but in the case of W409F nNOS, the mutation lies within the nNOSoxy domain on the proximal side of the heme. The W409F mutation eliminates a hydrogen bond between Trp409 and the cysteine sulfur atom that binds to the heme iron to form a heme-thiolate bond (18, 31). The W409F mutation causes a 70-mV drop in the midpoint potential of the ferric/ferrous heme couple relative to wild type (31, 47). This effect is consistent with the Trp409 mutation diminishing the hydrogen bonding properties of the heme thiolate (18, 48) and in turn slowing the kr and speeding kox in W409F nNOS.
In comparison, the E762N mutation is located on the surface of the FMN subdomain and causes an approximately 90-mV increase in the FMN sq/hq couple. This makes the FMN hq a poorer reducing agent toward the nNOS heme. The relative change in FMN and heme thermodynamics can almost certainly explain the less extensive and slower heme reduction that we observed in E762N nNOS. Specifically, the midpoint potential of the heme ferrous-CO/ferric couple of nNOS, which is reported to be −230 mV (49), is about 50 mV more negative than the measured FMN sq/hq couple in our E762N nNOS. This makes heme reduction as measured by CO binding thermodynamically uphill to an extent that can explain the estimated 7:3 ratio of ferric/ferrous-CO enzyme species that we observed for E762N nNOS after equilibrium was achieved in our stopped-flow heme reduction reactions (Fig. S2). Conceivably, the slower kr of E762N nNOS could be due to an additional effect; namely the mutation at Glu762 might diminish productive complex formation between the FMN subdomain and the NOSoxy domain as required for electron transfer, as we previously suggested (20). However, given the mutant's clear effect on the thermodynamics of the FMN-to-heme electron transfer, there is no need to invoke this alternative mechanism at this time.
Regarding the faster kox of E762N nNOS, our data imply that it manifests through a physical interaction between the FMN subdomain and nNOSoxy. An alternative possibility is that the mutant's higher rate of flavin auto-oxidation could be involved. In this circumstance, the superoxide generated by the increased flavin auto-oxidation might react with or otherwise cause the ferrous heme-NO complex to oxidize faster to the ferric enzyme, leading to an increase in the observed kox. Two results argue against this possibility. First, the mutational effect on kox is insensitive to added superoxide dismutase and catalase. Second, other nNOS mutants that have comparably increased flavin auto-oxidation rates do not have hyperactive NO synthesis (50).
Our current results strongly argue that some type of domain-domain interaction must occur between the FMN subdomain and NOSoxy in E762N nNOS in order to alter kox, even in the absence of CaM. But this concept is puzzling initially, because most models for nNOS function propose that CaM must bind before the FMN subdomain can physically interact with NOSoxy within the enzyme dimer. However, this aspect of the “model” has never been tested by any direct measure. Although NO synthesis clearly requires an interaction between the FMN and NOSoxy domains in nNOS, an absence of NO synthesis (or heme reduction) does not necessarily mean that no domain-domain interaction is allowed. Instead, it may just mean that a specific subset of interactions that are essential for heme reduction are not allowed. Recent published data support this concept. For example, we know that the FMN subdomain of nNOS exists in an approximately 1:1 conformational equilibrium between its FNR-associated (FMN-shielded) and dissociated states (FMN-deshielded) when the enzyme is in the fully reduced, CaM-free form (35, 37). Because our kox measurements were also performed with fully reduced nNOS proteins, these conformational equilibrium measures have direct relevance to our current study. Moreover, we know that the CaM-free E762N nNOS has a greater cytochrome c reductase activity than wild-type nNOS (20). This implies that an even greater proportion of its FMN subdomain may exist in the dissociated conformational state.4 Thus, 50% (or more) of the FMN subdomains in a given CaM-free, reduced E762N nNOS population exist free from their partner FNR subdomains and are presumably available to interact with the NOSoxy domain. A second point is that the complementary groups of charged residues that may enable the FMN-NOSoxy interaction are still present (with the exception of Glu762) on the two domains whether CaM is bound or not. Given these circumstances, it is easy to imagine that interactions could still occur between the FMN subdomain and NOSoxy in the CaM-free nNOS. In any case, our data provide the first evidence that the FMN subdomain can influence kox in nNOS, despite kox being a kinetic parameter that describes a heme-based reaction that takes place fully within the NOSoxy domain.5
Beyond its involving a domain-domain interaction, it is unclear how the E762N mutation might alter kox. We believe that the change it causes in FMN midpoint potentials, on its own, is unlikely to be responsible, because (i) there is no redox reaction occurring between the heme and the FMN during the kox reaction, and (ii) a distinct mutation at Gly810 in nNOS (15) that caused a very similar change in FMN thermodynamics did not result in hyperactive NO synthesis. Rather, we suspect that the important effect of the E762N mutation is charge neutralization and how this may effect nNOSoxy electronic parameters that impact the heme. This would have to be structurally specific, because neutralization of adjacent surface acidic residues on the FMN subdomain did not result in nNOS hyperactivity (or in a change in kox; see below) (20). Further work on the mechanism of action is in progress.
Relationship to Other Work and Potential Biological Implications
Subdomain protein-protein interactions are known to affect flavin or heme properties within some multidomain enzymes. These include changes in flavin midpoint potential and in the kinetics of flavin reduction or heme-NO dissociation (51–53). In our case, the intraprotein effect we observed on kox in E762N nNOS has a clear and significant impact on enzyme catalytic behavior, namely that it decreases the extent of heme-NO complex buildup, which in turn diminishes the apparent Km,O2, increases the enzyme V/K, and enables a supernormal NO synthesis activity across a broad O2 concentration range. One implication of our results is they suggest a way that exogenous proteins could interact with nNOS to alter its activity, through causing similar modest changes in the kox parameter. Indeed, nNOS may be ideally poised to precisely tune its NO synthesis activity via protein-protein interactions that change its apparent Km,O2 through this mechanism. This could also be a way to control the ability of nNOS to serve as a pulse NO generator, a function recently suggested by Salerno (13). Although proteins that alter the kox of nNOS remain to be identified, a good precedent exists given that nNOS engages in many protein-protein interactions, some known to change its NO synthesis activity. These include heat shock protein 90, which enhances NO synthesis from nNOS (54–56), or Caveolin-1 and the bradykinin receptor, which down-regulate the activity of nNOS (57, 58). Our current work may encourage studies along this line.
Supplementary Material
Acknowledgments
We thank Dr. Jesus Tejero for helpful advice and discussions and Deborah Durra for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants GM51491, CA53914, and HL58883 (to D. J. S.).

The on-line version of this article (available at http://www.jbc.org) contains Equation S1, Tables S1 and S2, and Figs. S1 and S2.
Tejero, J., Santolini, J., and Stuehr, D. J. (2009) FEBS J., in press.
Work with other nNOS mutants that undergo a similar change in their conformational equilibrium (50) shows that this change alone is not enough to alter kox in nNOS (i.e. in the absence of the E762N mutation). Nevertheless, the shift in the conformational equilibrium would conceivably increase (up to a factor of 2) the possibility of an interaction between the FMN and NOSoxy domains in the CaM-free E762N mutant.
Our results are consistent with a paper that appeared while this manuscript was in revision, showing that a similar mutation at Glu546 in the iNOS FMN subdomain alters the MCD spectrum of the iNOS heme (59).
- NO
- nitric oxide
- NOS
- nitric-oxide synthase(s)
- eNOS
- endothelial nitric-oxide synthase
- iNOS
- inducible nitric-oxide synthase
- nNOS
- neuronal nitric-oxide synthase
- CaM
- calmodulin
- EPPS
- 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- H4B
- (6R)-5,6,7,8-tetrahydro-l-biopterin
- sq
- semiquinone
- hq
- hydroquinone
- ox
- oxidized
- nNOSr
- reductase domain of nNOS.
REFERENCES
- 1.Griffith O. W., Stuehr D. J. (1995) Annu. Rev. Physiol. 57, 707–736 [DOI] [PubMed] [Google Scholar]
- 2.Knowles R. G., Moncada S. (1994) Biochem. J. 298, 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masters B. S., McMillan K., Sheta E. A., Nishimura J. S., Roman L. J., Martasek P. (1996) FASEB J. 10, 552–558 [DOI] [PubMed] [Google Scholar]
- 4.Andrew P. J., Mayer B. (1999) Cardiovasc. Res. 43, 521–531 [DOI] [PubMed] [Google Scholar]
- 5.Wei C. C., Crane B. R., Stuehr D. J. (2003) Chem. Rev. 103, 2365–2383 [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi K., Tagawa S., Daff S., Sagami I., Shimizu T. (2001) J. Biol. Chem. 276, 39864–39871 [DOI] [PubMed] [Google Scholar]
- 7.Panda K., Ghosh S., Stuehr D. J. (2001) J. Biol. Chem. 276, 23349–23356 [DOI] [PubMed] [Google Scholar]
- 8.Sagami I., Daff S., Shimizu T. (2001) J. Biol. Chem. 276, 30036–30042 [DOI] [PubMed] [Google Scholar]
- 9.Siddhanta U., Presta A., Fan B., Wolan D., Rousseau D. L., Stuehr D. J. (1998) J. Biol. Chem. 273, 18950–18958 [DOI] [PubMed] [Google Scholar]
- 10.Abu-Soud H. M., Wang J., Rousseau D. L., Fukuto J. M., Ignarro L. J., Stuehr D. J. (1995) J. Biol. Chem. 270, 22997–23006 [DOI] [PubMed] [Google Scholar]
- 11.Santolini J., Meade A. L., Stuehr D. J. (2001) J. Biol. Chem. 276, 48887–48898 [DOI] [PubMed] [Google Scholar]
- 12.Stuehr D. J., Santolini J., Wang Z. Q., Wei C. C., Adak S. (2004) J. Biol. Chem. 279, 36167–36170 [DOI] [PubMed] [Google Scholar]
- 13.Salerno J. C. (2008) FEBS Lett. 582, 1395–1399 [DOI] [PubMed] [Google Scholar]
- 14.Adak S., Aulak K. S., Stuehr D. J. (2001) J. Biol. Chem. 276, 23246–23252 [DOI] [PubMed] [Google Scholar]
- 15.Li H., Das A., Sibhatu H., Jamal J., Sligar S. G., Poulos T. L. (2008) J. Biol. Chem. 283, 34762–34772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishida C. R., Ortiz de Montellano P. R. (1998) J. Biol. Chem. 273, 5566–5571 [DOI] [PubMed] [Google Scholar]
- 17.Haque M. M., Panda K., Tejero J., Aulak K. S., Fadlalla M. A., Mustovich A. T., Stuehr D. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9254–9259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adak S., Wang Q., Stuehr D. J. (2000) J. Biol. Chem. 275, 17434–17439 [DOI] [PubMed] [Google Scholar]
- 19.Wang Z. Q., Wei C. C., Sharma M., Pant K., Crane B. R., Stuehr D. J. (2004) J. Biol. Chem. 279, 19018–19025 [DOI] [PubMed] [Google Scholar]
- 20.Panda K., Haque M. M., Garcin-Hosfield E. D., Durra D., Getzoff E. D., Stuehr D. J. (2006) J. Biol. Chem. 281, 36819–36827 [DOI] [PubMed] [Google Scholar]
- 21.Panda K., Adak S., Konas D., Sharma M., Stuehr D. J. (2004) J. Biol. Chem. 279, 18323–18333 [DOI] [PubMed] [Google Scholar]
- 22.Ghosh D. K., Wu C., Pitters E., Moloney M., Werner E. R., Mayer B., Stuehr D. J. (1997) Biochemistry 36, 10609–10619 [DOI] [PubMed] [Google Scholar]
- 23.Konas D. W., Takaya N., Sharma M., Stuehr D. J. (2006) Biochemistry 45, 12596–12609 [DOI] [PubMed] [Google Scholar]
- 24.Adak S., Ghosh S., Abu-Soud H. M., Stuehr D. J. (1999) J. Biol. Chem. 274, 22313–22320 [DOI] [PubMed] [Google Scholar]
- 25.Adak S., Santolini J., Tikunova S., Wang Q., Johnson J. D., Stuehr D. J. (2001) J. Biol. Chem. 276, 1244–1252 [DOI] [PubMed] [Google Scholar]
- 26.Panda K., Adak S., Aulak K. S., Santolini J., McDonald J. F., Stuehr D. J. (2003) J. Biol. Chem. 278, 37122–37131 [DOI] [PubMed] [Google Scholar]
- 27.Adak S., Stuehr D. J. (2001) J. Inorg. Biochem. 83, 301–308 [DOI] [PubMed] [Google Scholar]
- 28.Santolini J., Adak S., Curran C. M., Stuehr D. J. (2001) J. Biol. Chem. 276, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 29.Roman L. J., Martásek P., Miller R. T., Harris D. E., de La Garza M. A., Shea T. M., Kim J. J., Masters B. S. (2000) J. Biol. Chem. 275, 29225–29232 [DOI] [PubMed] [Google Scholar]
- 30.Stuehr D., Pou S., Rosen G. M. (2001) J. Biol. Chem. 276, 14533–14536 [DOI] [PubMed] [Google Scholar]
- 31.Adak S., Crooks C., Wang Q., Crane B. R., Tainer J. A., Getzoff E. D., Stuehr D. J. (1999) J. Biol. Chem. 274, 26907–26911 [DOI] [PubMed] [Google Scholar]
- 32.Abu-Soud H. M., Stuehr D. J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10769–10772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Presta A., Weber-Main A. M., Stankovich M. T., Stuehr D. (1998) J. Am. Chem. Soc. 120, 9460–9465 [Google Scholar]
- 34.Garcin E. D., Bruns C. M., Lloyd S. J., Hosfield D. J., Tiso M., Gachhui R., Stuehr D. J., Tainer J. A., Getzoff E. D. (2004) J. Biol. Chem. 279, 37918–37927 [DOI] [PubMed] [Google Scholar]
- 35.Ilagan R. P., Tiso M., Konas D. W., Hemann C., Durra D., Hille R., Stuehr D. J. (2008) J. Biol. Chem. 283, 19603–19615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noble M. A., Munro A. W., Rivers S. L., Robledo L., Daff S. N., Yellowlees L. J., Shimizu T., Sagami I., Guillemette J. G., Chapman S. K. (1999) Biochemistry 38, 16413–16418 [DOI] [PubMed] [Google Scholar]
- 37.Ilagan R. P., Tejero J., Aulak K. S., Ray S. S., Hemann C., Wang Z. Q., Gangoda M., Zweier J. L., Stuehr D. J. (2009) Biochemistry 48, 3864–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abu-Soud H. M., Rousseau D. L., Stuehr D. J. (1996) J. Biol. Chem. 271, 32515–32518 [DOI] [PubMed] [Google Scholar]
- 39.Wang Z. Q., Wei C. C., Santolini J., Panda K., Wang Q., Stuehr D. J. (2005) Biochemistry 44, 4676–4690 [DOI] [PubMed] [Google Scholar]
- 40.Gachhui R., Abu-Soud H. M., Ghosha D. K., Presta A., Blazing M. A., Mayer B., George S. E., Stuehr D. J. (1998) J. Biol. Chem. 273, 5451–5454 [DOI] [PubMed] [Google Scholar]
- 41.Tiso M., Konas D. W., Panda K., Garcin E. D., Sharma M., Getzoff E. D., Stuehr D. J. (2005) J. Biol. Chem. 280, 39208–39219 [DOI] [PubMed] [Google Scholar]
- 42.Feng C., Tollin G., Holliday M. A., Thomas C., Salerno J. C., Enemark J. H., Ghosh D. K. (2006) Biochemistry 45, 6354–6362 [DOI] [PubMed] [Google Scholar]
- 43.Feng C., Tollin G., Hazzard J. T., Nahm N. J., Guillemette J. G., Salerno J. C., Ghosh D. K. (2007) J. Am. Chem. Soc. 129, 5621–5629 [DOI] [PubMed] [Google Scholar]
- 44.Ghosh D. K., Holliday M. A., Thomas C., Weinberg J. B., Smith S. M., Salerno J. C. (2006) J. Biol. Chem. 281, 14173–14183 [DOI] [PubMed] [Google Scholar]
- 45.Konas D. W., Zhu K., Sharma M., Aulak K. S., Brudvig G. W., Stuehr D. J. (2004) J. Biol. Chem. 279, 35412–35425 [DOI] [PubMed] [Google Scholar]
- 46.Shimanuki T., Sato H., Daff S., Sagami I., Shimizu T. (1999) J. Biol. Chem. 274, 26956–26961 [DOI] [PubMed] [Google Scholar]
- 47.Crane B. R., Arvai A. S., Gachhui R., Wu C., Ghosh D. K., Getzoff E. D., Stuehr D. J., Tainer J. A. (1997) Science 278, 425–431 [DOI] [PubMed] [Google Scholar]
- 48.Couture M., Adak S., Stuehr D. J., Rousseau D. L. (2001) J. Biol. Chem. 276, 38280–38288 [DOI] [PubMed] [Google Scholar]
- 49.Ost T. W., Daff S. (2005) J. Biol. Chem. 280, 965–973 [DOI] [PubMed] [Google Scholar]
- 50.Tiso M., Tejero J., Panda K., Aulak K. S., Stuehr D. J. (2007) Biochemistry 46, 14418–14428 [DOI] [PubMed] [Google Scholar]
- 51.Dunford A. J., Rigby S. E., Hay S., Munro A. W., Scrutton N. S. (2007) Biochemistry 46, 5018–5029 [DOI] [PubMed] [Google Scholar]
- 52.Nisimoto Y., Motalebi S., Han C. H., Lambeth J. D. (1999) J. Biol. Chem. 274, 22999–23005 [DOI] [PubMed] [Google Scholar]
- 53.Sam K. A., Fairhurst S. A., Thorneley R. N., Allen J. W., Ferguson S. J. (2008) J. Biol. Chem. 283, 12555–12563 [DOI] [PubMed] [Google Scholar]
- 54.Bender A. T., Silverstein A. M., Demady D. R., Kanelakis K. C., Noguchi S., Pratt W. B., Osawa Y. (1999) J. Biol. Chem. 274, 1472–1478 [DOI] [PubMed] [Google Scholar]
- 55.Kone B. C. (2000) Acta Physiol. Scand. 168, 27–31 [DOI] [PubMed] [Google Scholar]
- 56.Song Y., Zweier J. L., Xia Y. (2001) Am. J. Physiol. Cell Physiol. 281, C1819–C1824 [DOI] [PubMed] [Google Scholar]
- 57.Golser R., Gorren A. C., Leber A., Andrew P., Habisch H. J., Werner E. R., Schmidt K., Venema R. C., Mayer B. (2000) J. Biol. Chem. 275, 5291–5296 [DOI] [PubMed] [Google Scholar]
- 58.Sato Y., Sagami I., Shimizu T. (2004) J. Biol. Chem. 279, 8827–8836 [DOI] [PubMed] [Google Scholar]
- 59.Sempombe J., Elmore B. O., Sun X., Dupont A., Ghosh D. K., Guillemette J. G., Kirk M. L., Feng C. (2009) J. Am. Chem. Soc. 131, 6940–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.