Abstract
Almost all of the chlorine-containing gas emitted from natural sources is methyl chloride (CH3Cl), which contributes to the destruction of the stratospheric ozone layer. Tropical and subtropical plants emit substantial amounts of CH3Cl. A gene involved in CH3Cl emission from Arabidopsis was previously identified and designated HARMLESS TO OZONE LAYER (hereafter AtHOL1) based on the mutant phenotype. Our previous studies demonstrated that AtHOL1 and its homologs, AtHOL2 and AtHOL3, have S-adenosyl-l-methionine-dependent methyltransferase activities. However, the physiological functions of AtHOLs have yet to be elucidated. In the present study, our comparative kinetic analyses with possible physiological substrates indicated that all of the AtHOLs have low activities toward chloride. AtHOL1 was highly reactive to thiocyanate (NCS−), a pseudohalide, synthesizing methylthiocyanate (CH3SCN) with a very high kcat/Km value. We demonstrated in vivo that substantial amounts of NCS− were synthesized upon tissue damage in Arabidopsis and that NCS− was largely derived from myrosinase-mediated hydrolysis of glucosinolates. Analyses with the T-DNA insertion Arabidopsis mutants (hol1, hol2, and hol3) revealed that only hol1 showed increased sensitivity to NCS− in medium and a concomitant lack of CH3SCN synthesis upon tissue damage. Bacterial growth assays indicated that the conversion of NCS− into CH3SCN dramatically increased antibacterial activities against Arabidopsis pathogens that normally invade the wound site. Furthermore, hol1 seedlings showed an increased susceptibility toward an Arabidopsis pathogen, Pseudomonas syringae pv. maculicola. Here we propose that AtHOL1 is involved in glucosinolate metabolism and defense against phytopathogens. Moreover, CH3Cl synthesized by AtHOL1 could be considered a byproduct of NCS− metabolism.
Methyl chloride (CH3Cl) is the most abundant halohydrocarbon emitted into the atmosphere and constitutes about 17% of the chlorine currently in the stratosphere (1). CH3Cl is derived mainly from natural sources and contributes to the destruction of the stratospheric ozone layer. As the total abundance of ozone-depleting gases such as chlorofluorocarbons in the atmosphere has begun to decrease in recent years as a result of The Montreal Protocol on Substances That Deplete the Ozone Layer, the impact of CH3Cl emission from natural sources will become greater on the atmospheric chemistry. CH3Cl emission into the atmosphere has been estimated at 1,700–13,600 Gg/year (1), which underscores the great uncertainty of the estimation. Oceans (2), biomass burning (3), wood-rotting fungi, and coastal salt marshes (4) are the major sources of CH3Cl production. Recently, it was reported that large amounts of CH3Cl are emitted from tropical and subtropical plants, which are hence considered as the major sources of CH3Cl (5–7). It was estimated that the CH3Cl emission from tropical plants could account for 30–50% of the global CH3Cl emission (8). To accomplish an accurate estimation of CH3Cl production in the atmosphere through “bottom-up” approaches, elucidating the mechanisms and physiological functions of CH3Cl emission from plants will be important.
The biological synthesis of methyl halides has been demonstrated mainly by biochemical analyses. The enzymatic activities that transfer a methyl group from S-adenosyl-l-methionine (SAM)2 to halide ions (Cl−, Br−, I−), which synthesize methyl halides, were first discovered in cell-free extracts of Phellinus pomaceus (a white rot fungus), Endocladia muricata (a marine red alga), and Mesembryanthemum crystallinum (ice plant, a halophytic plant) (9). Enzyme purification and cDNA cloning of the methyl chloride transferase (MCT) was first reported with Batis maritima, a halophytic plant that grows abundantly in salt marshes. As high concentrations of ions such as Cl− are often detrimental to plants, halophytic plants are considered to possess various salt tolerance mechanisms. MCT was hypothesized to control and regulate the internal concentration of Cl−, rich in the habitat in which halophytic plant grows (10, 11).
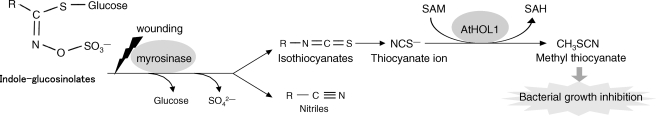
In the meantime, purification of thiol methyltransferase (TMT), which methylates bisulfide (HS−) and halide (Cl−, Br−, I−) ions was reported with cabbage, Brassica oleracea (12). The purified and recombinant TMTs were later shown to also methylate the thiocyanate ion (NCS−), which is called pseudohalide because of its chemical properties similar to halide ions (13, 14). NCS− is a hydrolysis product found in some glucosinolates, which are secondary metabolites found mainly in the order Brassicales including the model plant Arabidopsis thaliana (15). Upon tissue damage such as by insect or herbivore attack, glucosinolates are hydrolyzed by myrosinase (β-thioglucosidase) into biologically active compounds including isothiocyanates. Isothiocyanates derived from indole glucosinolates and 4-hydroxybenzyl glucosinolates are reported to be highly unstable and yield NCS− upon reacting with various nucleophiles (15–17). Based on the enzymatic activity, the physiological role of TMT was speculated to metabolize glucosinolate breakdown products (14). However, there are no reported studies that examine these MCT and TMT hypotheses through in vivo experiments.
An Arabidopsis homolog of MCT was also identified, and its T-DNA insertion Arabidopsis mutants were analyzed (18). Because the gene disruption eliminated almost all of the methyl halide emissions from the mutants, the gene was revealed to be involved in methyl halide synthesis and was designated HOL (HARMLESS TO OZONE LAYER; denoted as AtHOL1 in our studies) based on the mutant phenotype (18). Recently, we identified AtHOL1 homologs AtHOL2 and AtHOL3 in Arabidopsis, and we demonstrated biochemically that the three recombinant AtHOLs have SAM-dependent methyltransferase activities (19). In this study, reverse genetic and biochemical analyses of all AtHOL isoforms revealed that AtHOL1 in vivo is involved in the methylation of NCS− produced by glucosinolate hydrolysis. Although there are several studies that have examined the biological activities of glucosinolate hydrolysis products, the mechanisms of NCS− synthesis and its methylation to methyl thiocyanate (CH3SCN) have yet to be reported in detail. The biological activity and physiological function of CH3SCN synthesized by AtHOL1 was also examined.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Wild-type Arabidopsis (A. thaliana ecotype Col-0), three T-DNA insertion Arabidopsis mutants (hol1, hol2, hol3), and the three AtHOL1-overexpressing Arabidopsis lines were used in this study. Seeds were sterilized and grown on half-strength Murashige-Skoog (MS) agar medium or soil under controlled conditions (12 h light/12 h dark cycle at 22 ± 1 °C).
Isolation of T-DNA Insertion Arabidopsis Mutants
T-DNA insertion Arabidopsis mutants (stock names AtHOL1, SALK_005204C; AtHOL2, SALK_021226C; AtHOL3, SALK_ 014648C) were obtained from the Nottingham Arabidopsis Stock Centre. Homozygous mutant plants were screened by two sets of PCR using genomic DNA as templates. In the first PCR, T-DNA insertion was confirmed using a T-DNA left border primer (LBb1) and a gene-specific primer (AtHOL1-2, AtHOL2-3, or AtHOL3-2). In the second PCR, homozygosity of the T-DNA insertions was determined using gene-specific primers (AtHOL1-2 and AtHOL1-3; AtHOL2-2 and AtHOL2-3; AtHOL3-2 and AtHOL3-3) (supplemental Table 1) that flank the inserted T-DNAs. mRNA accumulation in the screened homozygous mutant plants was examined by RT-PCR using gene-specific primer sets (AtHOL1-2 and AtHOL1/2-1; AtHOL2-8 and AtHOL2-9; AtHOL3-2 and AtHOL3-3). AtPP2AA3 (At1g13320) was amplified with primers AtPP2AA3-1 and AtPP2AA3-2 as an internal control (20). Total RNA was purified from 4-week-old mutant plants, and cDNA was synthesized by Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) using a poly(A) primer, attB2T19VN. RT-PCR conditions were as follows: denaturation at 98 °C for 2 min and then 35 cycles of 94 °C for 15 s, 55 °C for 30 s, and 72 °C for 50 s.
Phylogenetic Analysis
The amino acid sequences of HOL homologs deduced from the completed genomic sequences, cDNA sequences, and assembled expressed sequence tag sequences from various photosynthetic organisms were extracted through BLAST search (blast.ncbi.nlm.nih.gov/Blast.cgi) of the available nucleic acid sequence data bases. The unrooted phylogenetic tree was constructed based on the obtained amino acid sequences using the ClustalW (21) and Njplot programs (22).
Overexpression of AtHOL1 in Arabidopsis
The AtHOL1 cDNA fragment containing the open reading frame was prepared by PCR (19) and cloned between the XbaI and XhoI sites in the binary vector pBI121 under the control of the cauliflower mosaic virus 35S promoter. The constructed binary vector (pBI121-AtHOL1) was introduced into Agrobacterium tumefaciens (LBA4404) by electroporation, which was then used to transform wild-type Arabidopsis by floral dipping. Transgenic T1 Arabidopsis were selected on half-strength MS agar medium containing 25 mg/liter kanamycin sulfate.
High Performance Liquid Chromatography (HPLC) Analyses
SAM-dependent methyltransferase activities were analyzed by ion pair reverse-phase HPLC using a Waters system (600 pump, 600E system controller, 486 tunable absorbance detector; Waters, Milford, MA). Samples were separated by the octadecylsilyl (ODS) column (4.6 × 150-mm; ODS-80Ts, Tosoh, Tokyo) protected by a guard column (ODS-80Ts guardgel, Tosoh) with 70% solvent A (8 mm octanesulfonic acid sodium salt, 50 mm NaH2PO4, pH 3.0 adjusted by H3PO4) and 30% solvent B (100% methanol) at an isocratic flow rate of 1.0 ml/min. S-adenosyl-l-homocysteine (SAH) was detected by UV absorption at 254 nm. The SAH was identified by comparison with the retention time of pure SAH.
GC-ECD and GC-MS Analyses
The quantitative determination of CH3SCN was carried out by gas chromatography equipped with an electron capture detector (GC-ECD; GC-9A, Shimadzu, Kyoto, Japan). The samples in 2.0-ml glass vials were incubated at 70 °C for 30 min, and each headspace was sampled using a gas-tight syringe followed by an injection into a 200 × 0.3-cm inner diameter stainless column packed with Porapak Q (Waters) in the GC-ECD. The column and injection port temperatures were 180 and 250 °C, respectively, and the flow rate of the carrier gas (N2) was 40 ml/min. The product was identified by comparison with the retention time of pure CH3SCN (Fluorochem, Glossop, UK) and quantified by peak area. The structural identification of CH3SCN produced by Arabidopsis was performed by gas chromatography/mass spectrometry (GC-MS; 5973 MSD, Agilent Technologies, Wilmington DE) using the HP-5ms semi-volatile column (30 m × 0.25 mm inner diameter, 0.25-μm film thickness, Agilent Technologies). The samples in 2.0-ml glass vials were incubated at 70 °C for 30 min, and each sampled 500-μl head space was injected into the column. The carrier gas (helium) flow was 0.9 ml/min. The column temperature was 30 °C, and the mass spectrometer was operated in electron impact mode at 70 eV. The CH3SCN was identified by comparison with pure CH3SCN in selected ion-monitoring mode at mass to charge ratios (m/z) of 73, 72, and 46 (supplemental Fig. S1).
Kinetic Analyses of Recombinant AtHOL Proteins
For kinetic analyses of AtHOL1, AtHOL2, and AtHOL3, glutathione S-transferase-tagged recombinant AtHOLs were expressed in Escherichia coli strain BL21, and tag-deleted AtHOLs were purified from soluble fractions as described (19). Each of the recombinant AtHOLs had 15 extra amino acids (GSTSLYKKAGSEFAL) at the N terminus. The assay mixture volume was 75 μl containing 0.1 m Tris acetate (pH 7.5), up to 1.58 μg of each of the AtHOLs, and varying concentrations of each substrate. S-(5′-Adenosyl)-l-methionine chloride (Wako, Osaka, Japan), KSCN (Nacalai Tesque, Kyoto, Japan), KCl (Nacalai Tesque), and (NH4)2S (Nacalai Tesque) were used as substrates. Because the reaction rates were linear for at least 1 h (data not shown), all of the assay mixtures were incubated for 1 h at 25 °C. To stop the reaction, 1 m HClO4 (75 μl) was added to the assay mixture. After centrifugation (16,100 × g for 10 min), 50 μl of the supernatant was injected into the HPLC system. The methyltransferase activities of AtHOLs were measured by quantifying the SAH produced by the enzymatic reactions.
Quantitative Analyses of NCS− Synthesis in Arabidopsis
In the assay method that was established to sensitively and specifically quantify NCS− in Arabidopsis, NCS− was methylated by the recombinant AtHOL1 protein and converted to CH3SCN, which was then quantified using GC-ECD.3 The quantification was not influenced by ingredients in Arabidopsis extracts. To quantify NCS− in homogenized Arabidopsis, ∼4-week-old wild-type and T-DNA mutant lines were homogenized and centrifuged at 16,100 × g for 10 min. Each supernatant (20 μl) was analyzed. To investigate the effects of lower degrees of wounding on NCS− synthesis, ∼20 mg (fresh weight) of 4-week-old wild-type Arabidopsis seedlings were nicked by scissors along the mid-vein of all the leaves. Then the seedlings were homogenized on ice with 100 μl of 2.5% 5-sulfosalicylic acid to denature proteins contained in the samples, and the mixtures were neutralized by adding 0.2 n NaOH (100 μl). The homogenates were centrifuged at 16,100 × g for 10 min, and the supernatant (80 μl) was analyzed. Following the same procedure, basal level NCS− concentration was determined with the intact Arabidopsis seedlings that were homogenized under denaturing conditions. To examine the influence of myrosinase on NCS− production, 0.025 unit of myrosinase (thioglucosidase, EC 3.2.1.147; Sigma-Aldrich) was added to the extracts of the intact Arabidopsis seedlings.
Quantitative Analyses of CH3SCN Synthesis in Arabidopsis
About 400 mg (fresh weight) of 4-week-old wild-type and T-DNA insertion mutant Arabidopsis were homogenized on ice. The samples were centrifuged at 16,100 × g for 10 min, and each supernatant (200 μl) was transferred to a 2.0-ml glass vial sealed with a screw cap fitted with a Teflon-lined septum and incubated overnight at 25 °C followed by the head-space analyses of CH3SCN by GC-ECD. To examine the rate-limiting substrate in CH3SCN production, the supernatants (180 μl) were mixed with 10 mm KSCN (20 μl) or 10 mm SAM (20 μl), incubated overnight, and analyzed by GC-ECD as indicated above. To examine the involvement of wounding in CH3SCN production, 3-week-old wild-type Arabidopsis grown on soil were used. Wild-type Arabidopsis (∼200 mg) was wounded as performed in the analyses of NCS− synthesis indicated above. The samples were incubated overnight in 2.0-ml glass vials at 25 °C, and the 500-μl head space was analyzed for CH3SCN by GC-ECD.
In Vitro Assay of Antibacterial Activity
Two bacterial pathogens, Pseudomonas syringae pv. maculicola (MAFF 302539) and Erwinia carotovora subsp. carotovora (MAFF 301879), which were obtained from the Genebank of the National Institute of Agrobiological Sciences, Japan, and a nonpathogenic bacterium, E. coli strain DH5α, were used for antibacterial activity assay. All of the bacteria were cultured in LB medium at 28 °C for P. syringae and E. carotovora and at 37 °C for E. coli. The bacteria were cultured overnight for E. carotovora and E. coli and for 2 days for P. syringae. The concentrations of KSCN and CH3SCN causing 50% bacterial growth inhibition (IC50) were determined by monitoring the cell populations at A600 (23, 24). The assay cultures (200 μl) were prepared from the bacterial suspension cultures at an A600 of 0.2, and KSCN and CH3SCN were dissolved in water and ethanol, respectively. To avoid volatilization of CH3SCN, each assay culture was incubated in a 2.0-ml glass vial sealed with a screw cap fitted with a Teflon-lined septum. The cultures were incubated at temperatures appropriate for each bacterium and cultured by shaking at 150 rpm for 15 h followed by measuring the A600. The corresponding amounts of solvents used to dissolve KSCN and CH3SCN did not affect bacterial growth (data not shown).
Pathogen Inoculation
P. syringae pv. maculicola was cultured in LB medium as described above and harvested by centrifugation at 4000 × g for 10 min. The bacterial pellet was rinsed once by 10 mm MgCl2 and resuspended in 10 mm MgCl2 at an A600 of 0.5 (25). The wild-type and hol1 Arabidopsis seeds (∼400 seeds each) grown on half-strength MS agar medium were inoculated with droplets of the bacterial suspension (3 μl). For mock treatment, 10 mm MgCl2 solution was used instead of the bacterial suspension. The survival rate of the seedlings grown for 19 days was evaluated based on the emergence of true leaves.
RESULTS
Phylogenetic Analysis of HOL Genes
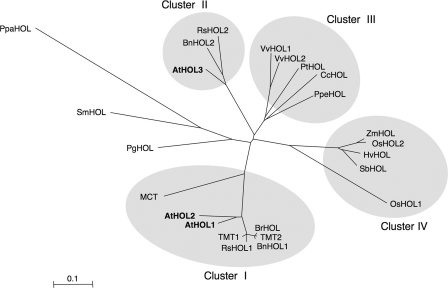
BLAST searches of nucleotide sequence data bases were performed using the amino acid sequence of AtHOL1 as a query. Twenty-five HOL homologs from 19 different plant species were distributed from unicellular green algae to gymnosperms and angiosperms (supplemental Fig. S2). All of the plant species such as Arabidopsis, rice, poplar, wine grape, milo, moss (Physcomitrella patens), and red alga (Cyanidioschyzon merolae), for which the genomes have been completely sequenced, contained at least one HOL homolog, suggesting that HOL is a gene conserved in photosynthetic organisms. A phylogenetic tree was constructed based on the amino acid sequences of HOL homologs from multicellular species only (Fig. 1). The tree reflected the genealogy of the derived species. One of the most remarkable aspects was that HOL homologs from Brassicales plants including Arabidopsis were grouped into two clusters, cluster I and cluster II. However, HOL homologs from dicotyledonous plants other than Brassicales plants were all grouped in a single cluster, cluster III, and those from monocotyledonous plants were also all grouped in a single cluster, cluster IV. Because AtHOL1 and AtHOL2 formed a cluster together with TMT and MCT, we hypothesized that AtHOL proteins would have enzymatic activities similar to those of TMT and MCT. However, a detailed enzymatic characterization of the AtHOL proteins had yet to be reported.
FIGURE 1.
Phylogenetic tree of HOL homologous proteins. The amino acid sequences were aligned using the ClustalW program, and the phylogenetic tree was built using the Njplot program (unrooted). Bar = 0.1 amino acid substitutions/site. Sequences from A. thaliana (AtHOL1, AtHOL2, and AtHOL3: NM_129953, NM_129954, and NM_180072, respectively), B. oleracea (TMT1 and TMT2: AAK69760 and AAK69761, respectively), Raphanus sativus (EW721469 and ED955532 for RsHOL1; EW738453 for RsHOL2), Brassica rapa (BrHOL: ABL86248), Brassica napus (BnHOL1 and BnHOL2: TA24304_3708 and EV084135.1, respectively), B. maritima (MCT: AAD26120), Vitis vinifera (VvHOL1 and VvHOL2: GSVIVT00034750001 and GSVIVT00034749001, respectively), Populus trichocarpa (PtHOL: gw1.XVII.90.1), Coffea canephora (CcHOL: TA6982_49390), Prunus persica (PpeHOL: TA5314_3760), Oryza sativa (OsHOL1 and OsHOL2: NP_001051867 and NP_001056843, respectively), Zea mays (ZmHOL: TA188770_4577), Hordeum vulgare (HvHOL: TA43367_4513), Sorghum bicolor (SbHOL: Sb10g003780.1), Picea glauca (PgHOL: DR569353.1), Selaginella moellendorfii (SmHOL1: e_gw1.23.943.1), and Physcomitrella patens (PpaHOL: BY959091.1 and BY959091.1) were from the GenBankTM data base, the TIGR data base (www.tigr.org), and the U. S. Department of Energy Joint Genome Institute (www.jgi.doe.gov/).
Kinetic Characterization of Recombinant AtHOL Proteins
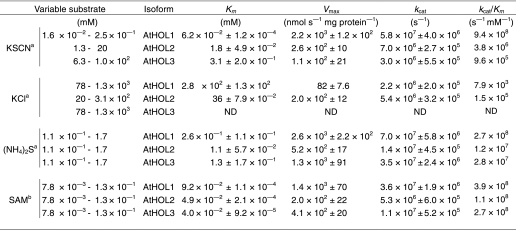
We first examined the substrate specificities and kinetic characteristics of the three AtHOL proteins. Among the compounds that worked as in vitro substrates for MCT or TMT (11, 14), three substrates (Cl−, NCS−, and HS−) that were known to exist in Arabidopsis were analyzed. Glutathione S-transferase-tagged recombinant AtHOL proteins were expressed in E. coli, and tag-deleted AtHOL proteins were purified from soluble fractions (supplemental Fig. S3), which permitted more proper folding of the proteins than those prepared from insoluble fractions as done in previous studies (11, 13). Enzymatic activities for the three substrates were largely distinct among AtHOL isoforms (Table 1). The catalytic efficiencies (kcat/Km) of all AtHOL proteins for Cl− (AtHOL1, 7.9 × 103; AtHOL2, 1.5 × 105; AtHOL3, not detected) were the lowest among the three analyzed substrates. On the other hand, AtHOL1 was highly reactive to NCS− with a markedly high kcat/Km value (9.4 × 108). Based on the kcat/Km values, the most preferred substrate for both AtHOL2 and AtHOL3 was HS−.
TABLE 1.
Kinetic parameters for methyltransferase activities of recombinant AtHOLs
ND, not detected. Data are the average of two technical repeats, and listed values represent S.E.
a Constant substrate was 0.5 mM SAM.
b Constant substrate was 1.7 mM (NH4)2S.
Isolation of AtHOL T-DNA Insertion Mutants
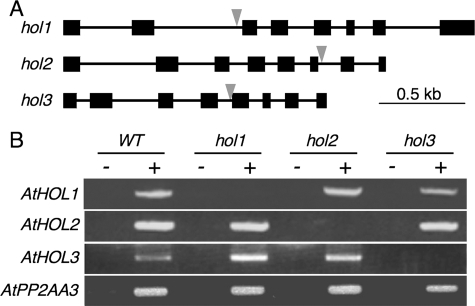
To investigate the physiological functions of the three AtHOL genes through in vivo experiments, we obtained T-DNA insertion Arabidopsis mutants for AtHOL1, AtHOL2, and AtHOL3. As shown in Fig. 2A, the mutants for AtHOL1 (SALK_005204C), AtHOL2 (SALK_021226C) and AtHOL3 (SALK_014648C) possessed T-DNA inserts within introns 2, 6, and 4, respectively. Semiquantitative RT-PCR analyses with the obtained homozygous mutant plants, designated hol1, hol2, and hol3, indicated no transcript accumulation of the T-DNA-inserted AtHOL genes in each mutant (Fig. 2B). The obtained hol1 plant was the same line used to show that AtHOL1 is responsible for methyl halide production (18). There were no morphological defects in these mutant plants under normal growth conditions (data not shown).
FIGURE 2.
T-DNA insertion Arabidopsis mutants of AtHOL1, AtHOL2, and AtHOL3. A, schematic representation of the three AtHOLs with the T-DNA insertions in the mutants used in this study. Black boxes and lines represent exons and introns, respectively. Gray arrows represent locations of the T-DNA insertion. B, RT-PCR analysis of each AtHOL transcript in the wild type (WT) and the three mutant Arabidopsis. PCR templates were prepared with (+) or without (−) reverse transcriptase in cDNA synthesis reactions. PP2A was used as an internal control.
Responses of Wild-type, AtHOL-disrupted, and AtHOL1-overexpressing Arabidopsis to NCS−
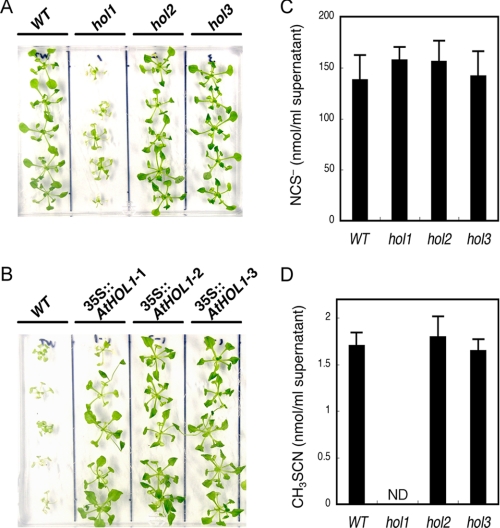
Based on our prediction that the substrate preferences of AtHOLs shown in our kinetic analyses reflect the in vivo responses to the substrates, we examined the response of the wild type and the three hol mutant Arabidopsis toward NCS−, HS−, and Cl−. All of the analyzed Arabidopsis seedlings showed the same growth under various concentrations of (NH4)2S (as HS−), NaCl (as Cl−), and KCl (as Cl−) (data not shown). On the other hand, among the three mutants, only hol1 showed an increased sensitivity to 1 mm KSCN (as NCS−) (Fig. 3A). AtHOL1-overexpressing Arabidopsis were grown on the medium containing a higher concentration of KSCN (2.5 mm), at which concentration the growth of wild-type Arabidopsis was inhibited (26, 27) (Fig. 3B). Compared with wild-type Arabidopsis, ∼4-week-old AtHOL1-overexpressing plants showed clearer resistance to KSCN on the medium (Fig. 3B) than the seedlings just after germination (28). These results provided the first evidence that, among the three isoforms, only AtHOL1 metabolized NCS− in vivo.
FIGURE 3.
Phenotypic analyses of hol mutants and AtHOL1-overexpressing Arabidopsis. A, susceptibility of the wild type (WT) and the three hol mutant Arabidopsis to 1 mm KSCN. ∼2 week-old seedlings grown on half-strength MS agar medium were transferred to medium containing 1 mm KSCN and grown for an additional 12 days. B, susceptibility of wild-type and AtHOL1-overexpressing Arabidopsis (35S::AtHOL1-1, 35S::AtHOL1-2, and 35S::AtHOL1-3) to 2.5 mm KSCN. ∼4-week-old AtHOL1-overexpressing T2 seedlings were selected on half-strength MS agar medium containing 25 μg/ml kanamycin sulfate. Wild-type and screened AtHOL1-overexpressing seedlings were grown on half-strength MS agar medium containing 2.5 mm KSCN for 12 days. C and D, NCS− (C) and CH3SCN (D) synthesized in supernatants of the homogenized wild type and each hol mutant Arabidopsis. Four-week-old seedlings were analyzed. Data are the average of three technical repeats, and bars indicate S.E. ND, not detected. The NCS− concentration was not significantly different (p > 0.1 by t test) among wild-type and the three mutant plants. The content of synthesized CH3SCN was not significantly different (p > 0.1 by t test) among wild-type, hol2 and hol3 plants.
NCS− and CH3SCN Synthesis in Wild-type and AtHOL-disrupted Arabidopsis
Thereafter, we focused on the functional analyses of AtHOL1, which was highly reactive to NCS−. To date, NCS− has been detected in crushed glucosinolate-containing plants such as Brassica vegetables (16). Other reports showed that in vitro hydrolysis of indole glucosinolates by myrosinase produces NCS− (16). However, to our knowledge, there have been no detailed in vivo studies demonstrating that NCS− is derived solely from myrosinase-catalyzed hydrolysis of glucosinolates. Myrosinases and glucosinolates are stored separately in intact plant tissues. Upon tissue wounding, such as caused by chewing insects, these enzymes come into contact followed by glucosinolate hydrolysis (29–31). To investigate NCS− synthesis, we first established an assay to sensitively and specifically quantify NCS− in the supernatant of homogenized, “thoroughly wounded” Arabidopsis seedlings. In the assay, NCS− in the reaction mixture was converted to CH3SCN by recombinant AtHOL1, which specifically methylates NCS−, and then CH3SCN was quantified by GC-ECD (see “Experimental Procedures”). By utilizing the established method, we demonstrated that NCS− accumulated to ∼140 μm in homogenized wild-type Arabidopsis seedlings, and the accumulation was unaffected by the disruption of each AtHOL gene (Fig. 3C). Because the concentration of accumulated NCS− (ca. 140 μm) was much higher than the Km value (62 μm) for NCS− of the recombinant AtHOL1, we hypothesized that NCS− was methylated to CH3SCN by AtHOL1 in homogenized Arabidopsis seedlings. Indeed, GC-MS analyses confirmed the synthesis of CH3SCN in the supernatant of homogenized wild-type Arabidopsis seedlings (supplemental Fig. S1). To investigate the involvement of the three AtHOL genes in CH3SCN production, we quantified CH3SCN synthesized in the supernatant of each homogenized AtHOL-disrupted mutant, hol1, hol2, and hol3. As a result, CH3SCN synthesis in hol1 was undetectable, whereas both hol2 and hol3 showed the same levels of CH3SCN synthesis as wild-type Arabidopsis (Fig. 3D).
Involvement of Myrosinase in NCS− and CH3SCN Synthesis in Arabidopsis
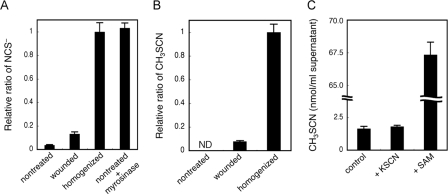
We then verified the involvement of myrosinase in NCS− and CH3SCN production in Arabidopsis. The amount of NCS− and CH3SCN in the seedlings of leaves uniformly wounded by scissors was 13 and 7.7%, respectively, that of the homogenized ones (Fig. 4, A and B). In the unwounded seedlings, NCS− accumulation was only 3.7% of the homogenized ones, and CH3SCN synthesis was undetectable. These results suggested that both NCS− and CH3SCN synthesis varied based on the degree of wounding. When purified myrosinase was added to the assay mixture of unwounded seedlings, the NCS− concentration increased to the same level as that in the homogenized seedlings. This result revealed that myrosinase was involved in the in vivo NCS− production, and almost all of the accumulated NCS− in homogenized Arabidopsis seedlings was actually derived from glucosinolates.
FIGURE 4.
Analysis of NCS− and CH3SCN synthesis in Arabidopsis. A, role of wounding and myrosinase in NCS− synthesis in wild-type Arabidopsis. The relative ratios of NCS− accumulated in intact (nontreated), partially wounded (wounded), and thoroughly wounded (homogenized) Arabidopsis seedlings were examined. Purified myrosinase was added to the nontreated sample (nontreated + myrosinase). Data are the average of three to four technical repeats, and bars indicate S.E. The NCS− concentration was significantly increased (p < 0.001 by t test) in wounded and homogenized samples compared with nontreated and wounded samples, respectively. The NCS− concentration was not significantly different (p > 0.1 by t test) between homogenized and nontreated + myrosinase samples. B, role of wounding in CH3SCN synthesis in wild-type Arabidopsis. The relative ratio of CH3SCN synthesized in intact (nontreated), partially wounded (wounded), and thoroughly wounded (homogenized) Arabidopsis seedlings were examined. Data are the average of two technical repeats, and bars indicate S.E. ND means not detected. The content of synthesized CH3SCN was significantly increased (p < 0.003 by t test) in homogenized samples compared with wounded samples. C, the rate-limiting substrate on CH3SCN synthesis in Arabidopsis. CH3SCN synthesis in the supernatants of the homogenized wild-type Arabidopsis (control) and the supernatants with 1 mm KSCN or with 1 mm SAM were determined. Data are the average of three technical repeats, and bars indicate S.E. The content of synthesized CH3SCN was not significantly different (p > 0.1 by t test) between the control and +KSCN samples, and it was significantly increased (p < 0.001 by t test) in +SAM samples compared with the control.
Rate-limiting Substrate for CH3SCN Synthesis in Arabidopsis
We then determined the rate-limiting substrate for CH3SCN synthesis. The addition of KSCN to the supernatant of homogenized Arabidopsis seedlings did not increase CH3SCN synthesis, indicating that all the SAM was exhausted in the supernatant. Meanwhile the addition of SAM increased the synthesis by ∼40-fold (Fig. 4C). These results indicated that the supply of SAM may be rate-limiting for CH3SCN synthesis, although CH3SCN synthesis was triggered by wounding that first induced NCS− synthesis.
In Vitro Antibacterial Activities of NCS− and CH3SCN
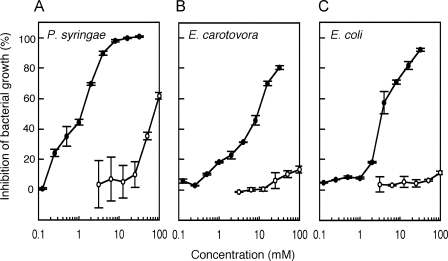
Thus far, there have been several studies reporting on the biological activities of glucosinolate-hydrolyzed products, typically isothiocyanates and nitriles (29). As for NCS−, none of the reported studies could detect biological activities on the analyzed bacteria and fungi (15, 32). On the other hand, a limited number of studies have demonstrated that bacteria belonging to Pseudomonas genus showed a negative chemotaxis to CH3SCN (33, 34). Based on this finding, we hypothesized that CH3SCN synthesized by Arabidopsis also had biological activities toward bacteria. To answer this question, we examined the influence of CH3SCN on bacterial growth. P. syringae pv. maculicola and E. carotovora subsp. carotovora, which are pathogenic to Arabidopsis and normally invade the wound site, and non-pathogenic E. coli strain DH5α were selected, and their growth was investigated under various concentrations of KSCN and CH3SCN. The KSCN concentration that caused 50% inhibition of bacterial growth (IC50) was over 50 mm for all of the bacteria analyzed (Fig. 5). On the other hand, the IC50 of CH3SCN for P. syringae pv. maculicola, E. carotovora subsp. carotovora, and E. coli was dramatically lowered to 1.2, 9.4, and 3.6 mm, respectively. These results suggested, for the first time, that the conversion of NCS− into CH3SCN catalyzed by AtHOL1 increased the toxicity of NCS− to the bacteria.
FIGURE 5.
In vitro antibacterial activity of KSCN and CH3SCN. Growth inhibition of the plant bacterial pathogens P. syringae pv. maculicola (A) and E. carotovora subsp. carotovora (B) and a non-pathogenic bacterium, E. coli strain DH5α (C), were measured at varying concentrations of KSCN (open circles) and CH3SCN (filled circles). The ratio of growth inhibition was recorded after 15 h. Data are the average of two technical repeats, and bars indicate S.E.
Enhanced Susceptibility of AtHOL1-disrupted Arabidopsis to P. syringae pv. maculicola
To investigate whether the pathogens were affected by CH3SCN synthesis in planta, we tested the wild-type and hol1 Arabidopsis on their susceptibility to P. syringae pv. maculicola. Because younger wild-type seedlings accumulate larger amounts of AtHOL1 mRNA than 4-week-old seedlings (18), we hypothesized that the differences in susceptibility between wild-type and hol1 Arabidopsis might be observed with younger seedlings. Seeds of wild-type and hol1 Arabidopsis were infected with P. syringae pv. maculicola and grown for 19 days on half-strength MS agar medium (supplemental Fig. S4). Indeed, the percentage of surviving seedlings was significantly lower for hol1 (7.5%) compared with wild-type Arabidopsis (21%) (Fig. 6), suggesting that hol1 Arabidopsis increased the susceptibility toward P. syringae pv. maculicola.
FIGURE 6.
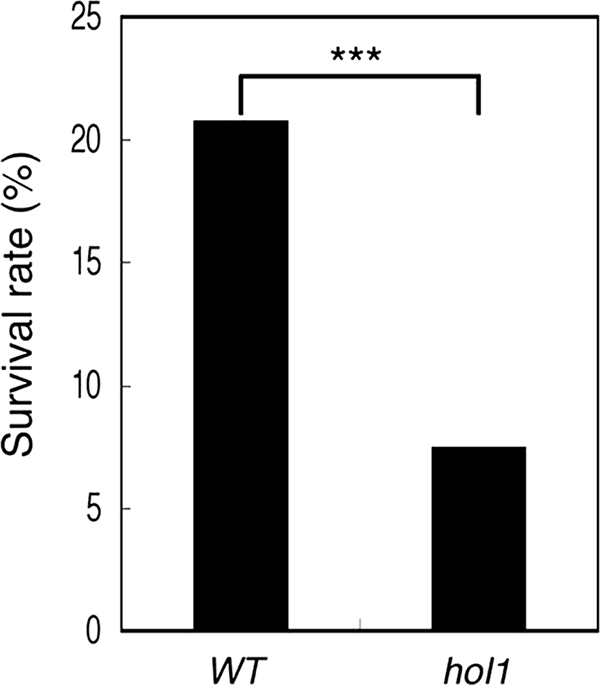
Susceptibility of wild-type and AtHOL1-disrupted Arabidopsis toward P. syringae pv. maculicola. The survival rate of the seedlings was evaluated based on the emergence of true leaves and was normalized by the seed germination rate of wild-type (WT) and hol1 Arabidopsis. The germination rate between wild-type and hol1 seeds was not significantly different (data not shown). The survival rate was significantly different (***, p < 0.001, χ2 = 31.79 by chi-square test) between wild-type (n = 452) and hol1 (n = 428) Arabidopsis. The data are derived from three independent experiments for both seeds.
DISCUSSION
AtHOL1 Was Highly Reactive to NCS− rather than Cl− in Arabidopsis
We demonstrated, for the first time, the comparative kinetic analyses of all the HOL isoforms in a single plant species and found that AtHOL1 was highly reactive to NCS−. AtHOL1 was originally reported to be involved in the production of methyl halides in Arabidopsis (18). However, the kinetic analyses of recombinant AtHOL1 showed that the kcat/Km value for Cl− was lowest among the analyzed substrates, and the Km value (280 mm) was much higher than the normal intracellular Cl− concentration (Table 1). The activity of AtHOL3 for Cl− was undetectable, whereas that of AtHOL2 was the highest among the three isoforms. However, the contribution of AtHOL2 to CH3Cl production was hypothesized to be subtle because methyl halide emissions depend mostly on AtHOL1 (18), and AtHOL2 mRNA accumulation is much lower than AtHOL1 (19). Ni and Hager (11) proposed that the physiological function of MCT, an AtHOL1 homolog in a Brassicales plant, is to regulate the intracellular Cl− concentration. However, the amount of CH3Cl emitted from Arabidopsis was so low that intracellular Cl− concentrations could not be reduced in Arabidopsis tissues. This hypothesis is supported by the results of our kinetic analyses showing the lower activities of AtHOLs toward Cl− and by the results showing that none of the hol mutants was more sensitive than wild-type Arabidopsis toward 0–100 mm KCl or NaCl in the half-strength MS agar medium (data not shown). These results together suggest that Cl− is not the physiological substrate for AtHOL proteins.
HS− was the most preferred substrate for AtHOL2 and AtHOL3 and the second most preferred substrate for AtHOL1 among the analyzed substrates. Therefore, the activity of AtHOL proteins for HS− may also contribute to the synthesis of CH3SH (35, 36), which is reported also to be synthesized from methionine hydrolysis by methionine γ-lyase in Arabidopsis (37).
On the other hand, NCS− was highly reactive for AtHOL1 with a kcat/Km that was ∼250- and 980-fold higher than that for AtHOL2 and AtHOL3, respectively. The NCS− concentration (∼140 μm) in the homogenized Arabidopsis tissue was higher than the Km value (62 μm) of AtHOL1 for NCS− (Table 1). Furthermore, CH3SCN synthesis was dependent on AtHOL1 (Fig. 3). These results suggest that NCS− is the physiological substrate for AtHOL1. Through reverse genetic and biochemical analyses, we have demonstrated that AtHOL1 is responsible for CH3SCN production in vivo (Fig. 3).
NCS− Was Derived from Glucosinolate Hydrolysis in Arabidopsis
We demonstrated that NCS− and CH3SCN synthesis varied based on the degree of wounding; the synthesis of almost all NCS− was derived from glucosinolates through hydrolysis by myrosinase (Fig. 4). Our in vivo studies demonstrated a novel metabolic pathway of a glucosinolate breakdown product catalyzed by AtHOL1. Indole glucosinolate breakdown products have also been detected on intact leaf surfaces (38). Glucosinolates in intact tissues of Arabidopsis are constantly renewed in the cell (39, 40). Sulfur deficiency also induces a conditional turnover of glucosinolates in Arabidopsis (41). Hence the basal level of NCS− in Arabidopsis tissues observed in Fig. 4A was possibly derived from the turnover of glucosinolates.
Role of CH3SCN Production in Plant Defense
Only about 1% of NCS− could be converted to CH3SCN, possibly because of the exhaustion of SAM in the supernatant of homogenized Arabidopsis seedlings (Fig. 3, C and D). Thus, the primary physiological role of AtHOL1 may not be the detoxification of NCS− by volatilization (14). Rather, CH3SCN synthesis (by methylation of NCS−) markedly increased its toxicity toward the bacteria (Fig. 5). To date, the biological activities of glucosinolate breakdown products such as nitriles and isothiocyanates have been studied in detail (29). For example, isothiocyanates contain antibacterial activities (15), and indole-3-acetonitrile has antifungal activity (42). Indole-3-carbinol and indole-3-acetonitrile act as a stimulant and a deterrent, respectively, for an ovipositing butterfly (38). CH3SCN reported herein may be considered a novel member of such biologically active compounds derived from glucosinolates. Although further studies are needed to understand how CH3SCN inhibits bacterial growth, CH3SCN indeed showed an inhibitory effect on the analyzed bacteria ranging from plant pathogens to a bacterium unrelated to plant pathogens. The observed effect on the different classes of bacteria implies that CH3SCN synthesis is involved in a broad spectrum antibacterial defense in Arabidopsis. Indeed, our in vivo studies have demonstrated that the disruption of AtHOL1 increases the susceptibility of the mutant seedlings toward P. syringae pv. maculicola (Fig. 6). These results imply that CH3SCN synthesis catalyzed by AtHOL1 is at least in part responsible for the resistance to P. syringae pv. maculicola.
To date, indole glucosinolates are the only glucosinolates reported to generate NCS− through a myrosinase-catalyzed hydrolysis that also exists in Arabidopsis. Indole glucosinolate synthesis is induced by methyl jasmonate (43), elicitors derived from the bacterial pathogen E. carotovora (32), and the fungal pathogen Alternaria brassicae (44) and by an interaction with a mycorrhizal fungus (45). PEN2 in Arabidopsis is responsible for efficient entry by the non-host fungus Blumeria graminis f. sp. hordei (46). PEN2 may preferentially hydrolyze indole glucosinolates producing defensive compounds at incipient fungal entry sites without tissue damage (40, 46). These studies (40, 42–46) imply that CH3SCN production by AtHOL1 might also be induced by pathogens, in which case NCS− synthesis and a continuous supply of SAM within an intact cell would cause a higher level of CH3SCN synthesis at the pathogen entry site. These observations may explain the difference between the CH3SCN concentration in homogenized wild-type Arabidopsis (Fig. 3D) and the CH3SCN concentration that inhibited bacterial growth (Fig. 5).
Methyl Halide Production by AtHOL1
In conclusion, our study suggests that AtHOL1 metabolizes glucosinolate-derived NCS−, which induces the plant defense mechanism (CH3SCN synthesis) (Fig. 7). Hence, methyl halide production by AtHOL1 could be considered a byproduct of NCS− metabolism in Arabidopsis. Glucosinolate-containing plants generally show a high level of methyl halide synthesis (47), which might be due to HOL genes belonging to cluster I (Fig. 1). On the other hand, we have shown that HOL homologs are widespread among photosynthetic organisms (Fig. 1). Hence, HOL homologs not found in cluster I may function differently than AtHOL1 and function commonly in photosynthetic organisms. Further analyses are needed to elucidate whether there is a common function for HOL homologs and their involvement in methyl halide emission, which is an important issue in global atmospheric chemistry.
FIGURE 7.
Proposed model of AtHOL1 function based on the results demonstrated in this study.
Supplementary Material
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre for the SALK T-DNA lines and the Genebank of the National Institute of Agrobiological Sciences, Japan, for the plant pathogens.
This work was supported by Grant-in-aid for Scientific Research 19780249 from the Ministry of Education, Science, Sports, and Culture of Japan (to T. N.), a Sasagawa Scientific Research Grant from the Japan Science Society (to Y. N.), and a research grant from the ESPEC Foundation for Global Environmental Research and Technology (to T. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S4.
Y. Nagatoshi and T. Nakamura, unpublished data.
- SAM
- S-adenosyl-l-methionine
- GC-ECD
- gas chromatography equipped with an electron capture detector
- GC-MS
- gas chromatography/mass spectrometry
- HOL
- harmless to ozone layer
- HPLC
- high performance liquid chromatography
- MCT
- methyl chloride transferase
- MS
- Murashige-Skoog
- RT
- reverse transcription
- SAH
- S-adenosyl-l-homocysteine
- TMT
- thiol methyltransferase.
REFERENCES
- 1.World Meteorological Organization (2007) Scientific Assessment of Ozone Depletion: 2006, Global Ozone Research and Monitoring Project-Report 50, World Meteorological Organization, Geneva [Google Scholar]
- 2.Khalil M. A., Moore R. M., Harper D. B., Lobert J. M., Erickson D. J., Koropalov V., Sturges W. T., Keene W. C. (1999) J. Geophys. Res. 104, 8333–8346 [Google Scholar]
- 3.Lobert J. M., Keene W. C., Logan J. A., Yevich R. (1999) J. Geophys. Res. 104, 8373–8390 [Google Scholar]
- 4.Rhew R. C., Miller B. R., Bill M., Goldstein A. H., Weiss R. F. (2002) Biogeochemistry 60, 141–161 [Google Scholar]
- 5.Saito T., Yokouchi Y. (2006) Atmos. Environ. 40, 2806–2811 [Google Scholar]
- 6.Harper D. B., Hamilton J. T., Ducrocq V., Kennedy J. T., Downey A., Kalin R. M. (2003) Chemosphere 52, 433–436 [DOI] [PubMed] [Google Scholar]
- 7.Yokouchi Y., Ikeda M., Inuzuka Y., Yukawa T. (2002) Nature 416, 163–165 [DOI] [PubMed] [Google Scholar]
- 8.Saito T., Yokouchi Y. (2008) Geophys. Res. Lett. 35, in press [Google Scholar]
- 9.Wuosmaa A. M., Hager L. P. (1990) Science 249, 160–162 [DOI] [PubMed] [Google Scholar]
- 10.Ni X., Hager L. P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni X., Hager L. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3611–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Attieh J. M., Hanson A. D., Saini H. S. (1995) J. Biol. Chem. 270, 9250–9257 [DOI] [PubMed] [Google Scholar]
- 13.Attieh J., Djiana R., Koonjul P., Etienne C., Sparace S. A., Saini H. S. (2002) Plant Mol. Biol. 50, 511–521 [DOI] [PubMed] [Google Scholar]
- 14.Attieh J., Kleppinger-Sparace K. F., Nunes C., Sparace S. A., Saini H. S. (2000) Plant Cell Environ. 23, 165–174 [Google Scholar]
- 15.Brown J., Morra M. J. (2005) Glucosinolate-containing Seed Meal as a Soil Amendment to Control Plant Pests, 2000–2002, University of Idaho, Moscow, ID [Google Scholar]
- 16.Agerbirk N., Olsen C. E., Sorensen H. (1998) J. Agric. Food Chem. 46, 1563–1571 [Google Scholar]
- 17.Agerbirk N., De Vos M., Kim J., Jander G. (2008) Phytochem. Rev. 8, 101–120 [Google Scholar]
- 18.Rhew R. C., Østergaard L., Saltzman E. S., Yanofsky M. F. (2003) Curr. Biol. 13, 1809–1813 [DOI] [PubMed] [Google Scholar]
- 19.Nagatoshi Y., Nakamura T. (2007) Plant Biotechnol. 24, 503–506 [Google Scholar]
- 20.Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005) Plant Physiol. 139, 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perrière G., Gouy M. (1996) Biochimie 78, 364–369 [DOI] [PubMed] [Google Scholar]
- 23.Feder R., Dagan A., Mor A. (2000) J. Biol. Chem. 275, 4230–4238 [DOI] [PubMed] [Google Scholar]
- 24.Cammue B. P., De Bolle M. F., Terras F. R., Proost P., Van Damme J., Rees S. B., Vanderleyden J., Broekaert W. F. (1992) J. Biol. Chem. 267, 2228–2233 [PubMed] [Google Scholar]
- 25.Kabisch U., Landgraf A., Krause J., Bonas U., Boch J. (2005) Microbiology 151, 269–280 [DOI] [PubMed] [Google Scholar]
- 26.Stiehl B., Bible B. B. (1989) HortScience 24, 99–101 [Google Scholar]
- 27.Ju H. Y., Bible B. B., Chong C. (1983) J. Chem. Ecol. 9, 1255–1262 [DOI] [PubMed] [Google Scholar]
- 28.Midorikawa K., Nagatoshi Y., Nakamura T. (2009) Plant Biotechnol. 26, in press [Google Scholar]
- 29.Wittstock U., Halkier B. A. (2002) Trends Plant Sci. 7, 263–270 [DOI] [PubMed] [Google Scholar]
- 30.Bones A. M., Rossiter J. T. (2006) Phytochemistry 67, 1053–1067 [DOI] [PubMed] [Google Scholar]
- 31.Grubb C. D., Abel S. (2006) Trends Plant Sci. 11, 89–100 [DOI] [PubMed] [Google Scholar]
- 32.Brader G., Tas E., Palva E. T. (2001) Plant Physiol. 126, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohga T., Masduki A., Kato J., Ohtake H. (1993) FEMS Microbiol. Lett. 113, 63–66 [DOI] [PubMed] [Google Scholar]
- 34.Masduki A., Nakamura J., Ohga T., Umezaki R., Kato J., Ohtake H. (1995) J. Bacteriol. 177, 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bates T. S., Lamb B. K., Guenther A., Dignon J., Stoiber R. E. (1992) J. Atmos. Chem. 14, 315–337 [Google Scholar]
- 36.Bentley R., Chasteen T. G. (2004) Chemosphere 55, 291–317 [DOI] [PubMed] [Google Scholar]
- 37.Goyer A., Collakova E., Shachar-Hill Y., Hanson A. D. (2007) Plant Cell Physiol. 48, 232–242 [DOI] [PubMed] [Google Scholar]
- 38.De Vos M., Kriksunov K. L., Jander G. (2008) Plant Physiol. 146, 916–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen B. L., Chen S., Hansen C. H., Olsen C. E., Halkier B. A. (2002) Planta 214, 562–571 [DOI] [PubMed] [Google Scholar]
- 40.Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., Humphry M., Consonni C., Panstruga R., Sanchez-Vallet A., Molina A., Schulze-Lefert P. (2009) Science 323, 101–106 [DOI] [PubMed] [Google Scholar]
- 41.Hirai M. Y., Yano M., Goodenowe D. B., Kanaya S., Kimura T., Awazuhara M., Arita M., Fujiwara T., Saito K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedras M. S., Nycholat C. M., Montaut S., Xu Y., Khan A. Q. (2002) Phytochemistry 59, 611–625 [DOI] [PubMed] [Google Scholar]
- 43.Sasaki-Sekimoto Y., Taki N., Obayashi T., Aono M., Matsumoto F., Sakurai N., Suzuki H., Hirai M. Y., Noji M., Saito K., Masuda T., Takamiya K., Shibata D., Ohta H. (2005) Plant J. 44, 653–668 [DOI] [PubMed] [Google Scholar]
- 44.Doughty K. J., Porter A. J. R., Morton A. M., Kiddle G., Bock C. H., Wallsgrove R. (1991) Ann. Appl. Biol. 118, 469–477 [Google Scholar]
- 45.Rostás M., Bennett R., Hilke M. (2002) J. Chem. Ecol. 28, 2449–2463 [DOI] [PubMed] [Google Scholar]
- 46.Lipka V., Dittgen J., Bednarek P., Bhat R., Wiermer M., Stein M., Landtag J., Brandt W., Rosahl S., Scheel D., Llorente F., Molina A., Parker J., Somerville S., Schulze-Lefert P. (2005) Science 310, 1180–1183 [DOI] [PubMed] [Google Scholar]
- 47.Saini H. S., Attieh J. M., Hanson A. D. (1995) Plant Cell Environ. 18, 1027–1033 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.