Abstract
The C1 complex of complement is assembled from a recognition protein C1q and C1s-C1r-C1r-C1s, a Ca2+-dependent tetramer of two modular proteases C1r and C1s. Resolution of the x-ray structure of the N-terminal CUB1-epidermal growth factor (EGF) C1s segment has led to a model of the C1q/C1s-C1r-C1r-C1s interaction where the C1q collagen stem binds at the C1r/C1s interface through ionic bonds involving acidic residues contributed by the C1r EGF module (Gregory, L. A., Thielens, N. M., Arlaud, G. J., Fontecilla-Camps, J. C., and Gaboriaud, C. (2003) J. Biol. Chem. 278, 32157–32164). To identify the C1q-binding sites of C1s-C1r-C1r-C1s, a series of C1r and C1s mutants was expressed, and the C1q binding ability of the resulting tetramer variants was assessed by surface plasmon resonance. Mutations targeting the Glu137-Glu-Asp139 stretch in the C1r EGF module had no effect on C1 assembly, ruling out our previous interaction model. Additional mutations targeting residues expected to participate in the Ca2+-binding sites of the C1r and C1s CUB modules provided evidence for high affinity C1q-binding sites contributed by the C1r CUB1 and CUB2 modules and lower affinity sites contributed by C1s CUB1. All of the sites implicate acidic residues also contributing Ca2+ ligands. C1s-C1r-C1r-C1s thus contributes six C1q-binding sites, one per C1q stem. Based on the location of these sites and available structural information, we propose a refined model of C1 assembly where the CUB1-EGF-CUB2 interaction domains of C1r and C1s are entirely clustered inside C1q and interact through six binding sites with reactive lysines of the C1q stems. This mechanism is similar to that demonstrated for mannan-binding lectin (MBL)-MBL-associated serine protease and ficolin-MBL-associated serine protease complexes.
The classical pathway of complement, a major component of innate immune defense against pathogens and altered self, is triggered by C1, a 790-kDa Ca2+-dependent complex assembled from a recognition protein C1q and C1s-C1r-C1r-C1s, a tetramer of two modular proteases, C1r and C1s, that respectively mediate activation and proteolytic activity of the complex (1–3). C1q has the overall shape of a bunch of tulips and comprises six heterotrimeric collagen-like triple helices that assemble through their N-terminal moieties to form a “stalk” and then diverge to form individual “stems,” each prolonged by a C-terminal globular recognition domain (4). C1r and C1s are homologous modular proteases each comprising, starting from the N-terminal end, a C1r/C1s, sea urchin EGF2 (uEGF), bone morphogenetic protein (CUB) module (5), an EGF-like module (6), a second CUB module, two complement control protein modules (7), and a serine protease domain. This modular structure is shared by the mannan-binding lectin-associated serine proteases (MASPs), a group of enzymes that associate with mannan-binding lectin (MBL) and the ficolins and thereby trigger activation of the lectin pathway of complement (8).
Assembly of the C1s-C1r-C1r-C1s tetramer involves Ca2+-dependent heterodimeric C1r-C1s interactions between the CUB1-EGF segments of each protease (9–12). Similarly, MASP-1, MASP-2, MASP-3, and mannan-binding lectin-associated protein 19 (MAp19), an alternative splicing product of the MASP-2 gene comprising the N-terminal CUB1-EGF segment of MASP-2, all associate as homodimers through their N-terminal CUB1-EGF moieties (13–15). The structures of human C1s CUB1-EGF, human MAp19, human MASP-1/3 CUB1-EGF-CUB2, and rat MASP-2 CUB1-EGF-CUB2 have been solved by x-ray crystallography (16–19), revealing that these domains all associate as head-to-tail homodimers through a highly conserved interface involving interactions between the CUB1 module of one monomer and the EGF module of its counterpart. In addition, all CUB modules contained in these structures were found to contain a hitherto unrecognized Ca2+-binding site involving three conserved acidic residues (Glu45, Asp53, and Asp98 in C1s), defining a novel CUB module subset diverging from the type originally described in the spermadhesins (20).
Mutagenesis studies have recently established that assembly of the MBL- and ficolin-MASP complexes involves a major electrostatic interaction between two acidic Ca2+ ligands from the MASP CUB modules and a conserved lysine located in the collagen fibers of MBL and ficolins (16, 18, 21, 22). In the case of C1, a hypothetical model of the C1q/C1r/C1s interface, involving interaction between acidic residues mainly contributed by the C1r EGF module and unmodified lysine residues also located in the collagen-like stems of C1q, was derived from the x-ray structure of the C1s CUB1-EGF interaction domain (16, 23). The aim of this work was to use site-directed mutagenesis to delineate the sites of C1r and C1s involved in the interaction between C1s-C1r-C1r-C1s and C1q. Our data rule out our previous interaction model and provide evidence that C1 assembly involves the same basic Ca2+-dependent mechanism as demonstrated in the case of MBL-MASP and ficolin-MASP complexes.
EXPERIMENTAL PROCEDURES
Reagents and Proteins
Diisopropyl phosphorofluoridate was purchased from Sigma-Aldrich. C1q was purified from human plasma as described previously (24). The VentR DNA polymerase and restriction enzymes were from New England Biolabs (Beverly, MA). The pHC1r3 and pBS-C1s plasmids containing the full-length human C1r and C1s cDNAs (25, 26) were kindly provided by Dr. Agnès Journet (CEA, Grenoble, France) and. Dr. Mario Tosi (University of Rouen, Rouen, France), respectively. Oligonucleotides were purchased from MWG-BIOTECH (Courtaboeuf, France). Antibiotics and molecular biology reagents were obtained from Fermentas (Burlington, Canada). Cellfectin was from Invitrogen. The concentrations of purified proteins were determined using the following absorption coefficients (A1%, 1 cm at 280 nm) and molecular weights: C1q, 6.8 and 459,300; recombinant C1r (S637A mutant), 12.4 and 83,373; recombinant C1s (wild type), 14.5 and 77,465; and C1s-C1r-C1r-C1s tetramer assembled from recombinant C1r and C1s, 13.45 and 321,676. The molecular weights of recombinant C1r and C1s were determined by mass spectrometry analysis using the matrix-assisted laser desorption ionization technique as described previously (27).
Expression of C1r and C1s Variants
Recombinant C1r and C1s variants were expressed using a baculovirus/insect cells system. The protocol used for C1s has been previously described in detail (28). In the case of C1r, a DNA fragment encoding the signal peptide plus the full-length mature protein (amino acid residues 1–705) was amplified by PCR using the VentR polymerase. The sequences of the sense (5′-CCGGAATTCATGTGGCTCTTGTAC-3′) and antisense (5′-CCCAAGCTTTCAGTCCTCCTCCTCCA-3′) primers introduced an EcoRI restriction site (underlined) at the 5′ end of the PCR product and a HindIII site (underlined) at the 3′ end. The amplified DNA was purified using the gel extraction kit QIAquick (Qiagen) and cloned into the pCR-Script Amp SK(+) intermediate vector (Stratagene) according to the manufacturer's instructions. The fragment was excised by digestion with EcoRI and HindIII and cloned into the corresponding sites of the pFastBac1 baculovirus transfer vector (Invitrogen). The resulting construct was characterized by restriction mapping and checked by double-stranded DNA sequencing (Cogenics, Meylan, France).
The expression plasmids coding for all C1r and C1s mutants were generated using the QuikChangeTM XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The mutagenic oligonucleotides were designed according to the manufacturer's recommendations, and a silent restriction site was introduced in each case for screening of positive clones. The pFastBac1/C1s expression plasmid coding for wild type C1s was used as a template for all C1s variants. The pFastBac1/C1r S637A plasmid coding for the S637A C1r mutant was used as a template for all C1r variants. The sequences of all variants were confirmed by double-stranded DNA sequencing.
The recombinant baculoviruses were generated using the Bac-to-BacTM system (Invitrogen) as described previously (13). The bacmid DNA was purified using the Qiagen midiprep purification system and used to transfect Sf21 insect cells with cellfectin in Sf900 II SFM medium (Invitrogen) as recommended by the manufacturer. Recombinant virus particles were collected 4 days later and amplified as described by King and Possee (29). High Five cells (1.75 × 107 cells/175-cm2 tissue culture flask) were infected with the recombinant viruses at a multiplicity of infection of 2 in SF900 II SFM medium at 28 °C for 48 h (C1s variants) or 55 h (C1r variants). The culture supernatants containing recombinant C1r or C1s were collected by centrifugation, supplemented with 1 mm diisopropyl phosphorofluoridate, and stored frozen at −20 °C until use.
Reconstitution and Purification of the C1s-C1r-C1r-C1s Tetramer Variants
The relative C1r or C1s content of each culture supernatant was estimated by SDS-PAGE analysis (30) followed by Coomassie Blue staining and gel scanning. Based on this estimate, C1r- and C1s-containing supernatants were mixed in proportions appropriate to achieve a C1r:C1s ratio of ∼1:1. The resulting mixture was dialyzed against 25 mm NaCl, 2 mm CaCl2, 50 mm triethanolamine HCl, pH 7.4, and loaded onto a Q-Sepharose Fast Flow column (Pharmacia) (50 ml) equilibrated in the same buffer. Elution was carried out by applying a linear gradient from 25 to 250 mm NaCl in the same buffer. Fractions containing the C1s-C1r-C1r-C1s tetramer were identified by SDS-PAGE analysis, pooled, and concentrated by ultrafiltration to 0.2–0.4 mg/ml. Further purification was achieved by high pressure gel filtration on either a TSK G3000 SW column (7.5 × 60 cm) (Tosoh Bioscience, Tokyo, Japan) or a Superose 6 10/300 GL column (Amersham Biosciences), each equilibrated in 145 mm NaCl, 2 mm CaCl2, 50 mm triethanolamine HCl, pH 7.4. The purified tetramer was concentrated by ultrafiltration to 0.2 mg/ml and stored at 4 °C until use.
Surface Plasmon Resonance Spectroscopy
Analyses were performed using a BIAcore X instrument (GE Healthcare). C1q was immobilized on the surface of a CM5 sensor chip (GE Healthcare) using the amine coupling chemistry, as described previously (31). Binding of the purified C1s-C1r-C1r-C1s tetramer variants was measured over 15,000 resonance units of immobilized C1q, at a flow rate of 20 μl/min in 145 mm NaCl, 2 mm CaCl2, 50 mm triethanolamine HCl, pH 7.4, containing 0.005% surfactant P20 (GE Healthcare). Each sample was injected in parallel over a surface with immobilized bovine serum albumin for subtraction of the bulk refractive index background. Regeneration of the surfaces was achieved by injection of 10 μl of 145 mm NaCl, 5 mm EDTA, 50 mm triethanolamine HCl, pH 7.4. The data were analyzed by global fitting to a 1:1 Langmuir binding model of the association and dissociation phases simultaneously, using the BIAevaluation 3.1 software (GE Healthcare). The apparent equilibrium dissociation constants (KD) were calculated from the ratio of the dissociation and association rate constants (koff/kon). Each C1s-C1r-C1r-C1s variant was analyzed at six different concentrations, ranging from 1 to 10 nm.
Modeling of the C1r/C1s CUB1-EGF-CUB2 Heterodimer
Experimentally determined structures were used in the case of C1s CUB1-EGF (16) and C1r EGF (32), whereas homology models were derived for the modules of unknown structure C1r CUB1 and CUB2 and C1s CUB2. C1r CUB1 was built using the scaffold common to its counterparts in C1s, MAp19, and MASP-1/3 (16–18). A few segments (residues 23–25, 62–67, 74–92, and 107–115) correspond to more variable areas, and their conformation is therefore somewhat arbitrary. The N-terminal residues 1–8 were not included because this stretch shows low structural and sequence homology in the available homologous structures. A Ca2+-binding site, as seen in the C1s, MAp19, and MASP-1/3 structures, was introduced in the model.
Assembly of the head-to-tail C1r/C1s CUB1-EGF heterodimer was carried out using the remarkably conserved intermonomer interface seen in the human C1s, human MAp19, rat MASP-2, and human MASP-1/3 homodimeric structures (16–19). Indeed, there are only a few conservative replacements in C1r compared with C1s, with Phe9, Thr13, Phe17, Gln44, Leu118, Gln122, and Val152 substituting for Tyr5, Leu9, Tyr13, Thr40, Ala109, Val113, and Ile136, respectively (16). The relative positioning of the monomers observed in the C1s structure was used as a template, and assembly of the C1r/C1s heterodimer was achieved by superimposing the C1r CUB1 model and the NMR-derived C1r EGF structure onto one of the C1s monomers. The root mean square deviations calculated for these superimpositions were 0.9 Å (CUB1, 98 Cα pairs) and 1.04 Å (EGF, 33 Cα pairs). Homology models for C1r and C1s CUB2 were derived from the scaffold common to their counterparts in rat MASP-2 and human MASP-1/3 (18, 19). The most arbitrary conformation corresponds to the two-residue insertion at residues 239 and 240 in C1s. Subtle variations may also occur in the C1r segments 242–248 and 254–257. Initial positioning of the C1r and C1s CUB2 modules in the overall C1r-C1s CUB1-EGF-CUB2 heterodimeric structure was achieved using the plane configurations of human MASP-1/3 and rat MASP-2 as templates. The graphics program O (33) was used throughout the modeling procedure.
RESULTS
The objective of this study was to perform point mutations in various areas of the interaction domains of C1r and C1s and to measure the impact of these mutations on the assembly of the C1 complex with a view to delineate the sites of C1r and C1s responsible for the interaction between the C1s-C1r-C1r-C1s tetramer and C1q. For this purpose, C1r and C1s were expressed in a baculovirus/insect cells system and secreted in the culture supernatants, at concentrations of 1–3 and 5–10 mg/liter, respectively. The wild type C1s DNA sequence was used as a template for all of the mutations. In the case of C1r, in contrast, the template sequence used for subsequent mutations contained a Ser to Ala mutation at the active site residue Ser637. The purpose of this modification was to stabilize all C1r variants in the proenzyme form and to prevent spontaneous activation during expression and upon subsequent interaction of the reconstituted C1s-C1r-C1r-C1s tetramer with C1q.
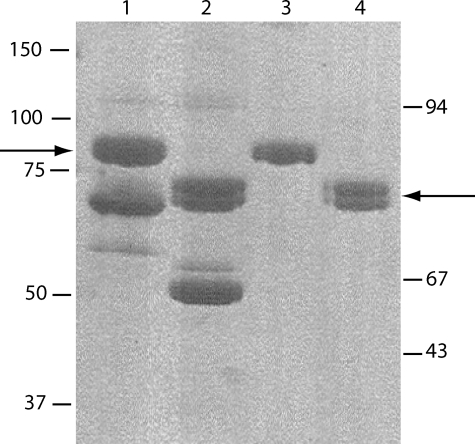
We initially attempted to express and purify recombinant C1r and C1s separately, with a view to subsequently reconstitute the C1s-C1r-C1r-C1s tetramer variants. A major drawback of this protocol arose from the relatively poor expression yield of recombinant C1r, combined with a considerable loss of protein during ion exchange purification. This did not allow us to produce purified C1r variants in sufficient amounts to perform subsequent analyses by surface plasmon resonance spectroscopy. To circumvent this problem, an alternative strategy was used. The culture supernatants containing the C1r and C1s variants were first combined in the presence of Ca2+ ions under appropriate proportions to achieve a C1r:C1s molar ratio of ∼1:1 and thereby reconstitute the C1s-C1r-C1r-C1s tetramer. The resulting mixture was then submitted to ion exchange chromatography on a Q-Sepharose Fast Flow column as described under “Experimental Procedures.” SDS-PAGE analysis of the elution profile provided evidence that C1r and C1s eluted as a single peak containing equimolar amounts of each protein (data not shown), confirming formation of the C1s-C1r-C1r-C1s tetramer. Excess C1s was observed occasionally and eluted in a peak immediately following the tetramer. All of the C1r/C1s combinations tested yielded similar elution profiles, providing a first indication that none of the mutations performed in C1r or C1s had a significant impact on the assembly of the tetramer. The tetramer pool from the ion exchange chromatography column was always contaminated by 40–50% bovine serum albumin, originating from the fetal calf serum contained in the culture medium and partially overlapping with the tetramer (Fig. 1, lanes 1 and 2). Further purification was achieved by high pressure gel permeation, allowing complete elimination of bovine serum albumin and providing further evidence that, in the presence of Ca2+ ions, all of the C1r/C1s combinations tested yielded a peak co-eluting with the “native” tetramer reconstituted from wild type C1s and the S637A C1r mutant (data not shown). As expected, gel filtration analysis in the presence of EDTA resulted in disruption of the tetramer, yielding two peaks corresponding to monomeric C1s and the C1r-C1r dimer. SDS-PAGE analysis of the purified tetramer preparation indicated that C1r and C1s each behaved as single-chain proteins under nonreducing and reducing conditions, indicative of their proenzyme state. Both C1r and C1s had similar apparent molecular masses of 85–90 kDa under reducing conditions (Fig. 1, lanes 3 and 4).
FIGURE 1.
Purification of the recombinant C1s-C1r-C1r-C1s tetramer. The tetramer was reconstituted from wild type C1s and the S637A C1r mutant. SDS-PAGE analysis was performed after ion exchange chromatography (lanes 1 and 2) and after high pressure gel permeation (lanes 3 and 4), under reducing (lanes 1 and 3), and nonreducing conditions (lanes 2 and 4). The arrows indicate the position of recombinant C1r and C1s under both conditions. The positions of reduced and unreduced standard proteins are indicated on the left and right sides of the figure, respectively.
Mutations in the C1r EGF Module Have No Effect on C1 Assembly
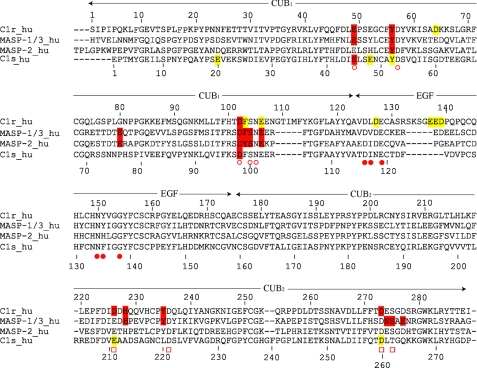
A series of C1s-C1r-C1r-C1s variants were produced, and their ability to associate with C1q was analyzed by surface plasmon resonance spectroscopy, using the tetramer variants as soluble ligands and immobilized C1q. To test the validity of the interaction model derived from the x-ray structure of the C1s interaction domain (16), we initially targeted acidic residues located in the C1r EGF module. Residues Glu137, Glu138, and Asp139, which form an acidic cluster in a large insertion loop of the C1r EGF module (Fig. 2), were each individually mutated to alanine, and the whole Glu137–Asp139 sequence stretch was deleted. As listed in Table 1, none of these modifications significantly decreased the ability of the resulting C1s-C1r-C1r-C1s tetramer variants to associate with C1q, with KD ratios relative to the native tetramer ranging from 0.9 to 1.4. Two other acidic residues of C1r also considered as possible C1q ligands (16), Asp127 (in the EGF module) and Asp64 (in the CUB1 module), were also individually mutated to asparagine. Again, these mutations did not alter the C1q binding ability of C1s-C1r-C1r-C1s. These results were therefore clearly not compatible with a major contribution of the C1r EGF module in the tetramer/C1q interaction, ruling out our previous model of C1 assembly essentially based on this assumption (16).
FIGURE 2.
Sequence alignment of selected CUB1-EGF-CUB2 segments. The amino acid numberings of both C1r (top) and C1s (bottom) are shown, and the approximate domain boundaries are indicated. Residues known to contribute Ca2+ ligands in C1s, MAp19, and MASP-1/3 are marked with open circles (CUB1 site), closed circles (EGF site), and open squares (CUB2 site). C1r and C1s residues interacting with C1q, and MASP residues interacting with MBL and ficolins are colored red. Mutations in C1r and C1s having little or no effect on C1 assembly are colored yellow. The implication of C1s Tyr52 in C1q binding cannot be excluded.
TABLE 1.
Kinetic and dissociation constants for the interaction between C1s-C1r-C1r-C1s variants and immobilized C1q
Shown are the effects of mutations in the C1r EGF module.
| Location of mutation | Tetramer variant | kon | koff | KD (nM) | KD/KD,wt | χ2 |
|---|---|---|---|---|---|---|
| m−1·s−1 | s−1 | nm | ||||
| Wild typea | 0.82 × 106 | 1.21 × 10−3 | 1.47 ± 0.6b | 1.0 | 2.8 | |
| EGF | D127N | 0.89 × 106 | 1.29 × 10−3 | 1.45 | 1.4 | 1.9 |
| E137A | 0.73 × 106 | 1.47 × 10−3 | 2.03 | 1.3 | 5.1 | |
| E138A | 1.09 × 106 | 2.14 × 10−3 | 1.97 | 1.2 | 4.8 | |
| D139A | 0.68 × 106 | 1.17 × 10−3 | 1.72 | 0.9 | 1.3 | |
| Δ137–139 | 1.05 × 106 | 1.42 × 10−3 | 1.35 | 4.4 | ||
| CUB1 | D64N | 0.83 × 106 | 1.27 × 10−3 | 1.53 | 1.0 | 1.7 |
a Tetramer assembled from wild type C1s and the S637A C1r mutant.
b Mean value determined from three separate experiments.
Mutations in the Ca2+-binding Sites of the C1r and C1s CUB Modules Reveal Their Implication in C1q Binding
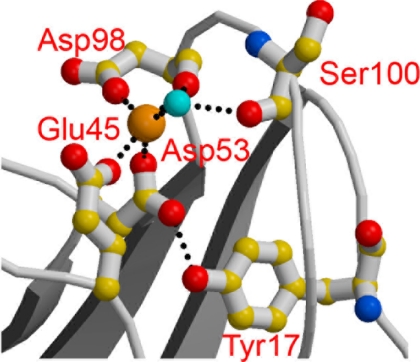
Recent studies on human MAp19 (17) and MASP-3 (18) have provided experimental evidence that both proteins associate with MBL and the ficolins through residues involved in the Ca2+-binding sites contained in their CUB modules. Given the strong structural homology between C1r, C1s, and the MASPs, this prompted us to target residues expected to participate in the Ca2+-binding sites of the C1r and C1s CUB1 and CUB2 modules. We particularly focused our attention on the residues homologous to two of the three conserved acidic residues known to coordinate Ca2+ in C1s CUB1, Glu45 and Asp98 (Fig. 2). The reason for this choice is that, unlike the third Ca2+ ligand (homologous to Asp53 in C1s CUB1), these residues are always positioned on the outer part of the Ca2+-binding site, as shown by the x-ray crystallography analyses performed on C1s (Fig. 3), MAp19 and MASP-1/3 (16–18), and are therefore available for contributing ionic interactions with a protein ligand.
FIGURE 3.
Structure of the Ca2+-binding site of the C1s CUB1 module. Oxygen atoms are shown in red, and nitrogen atoms are in blue. The light blue sphere represents a water molecule. Ionic bonds and hydrogen bonds are represented by dotted lines (adapted from Gregory et al. (16)).
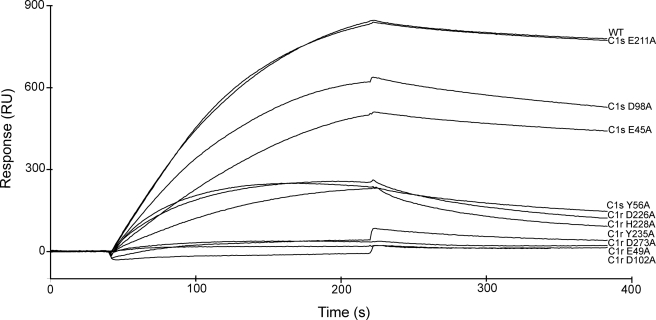
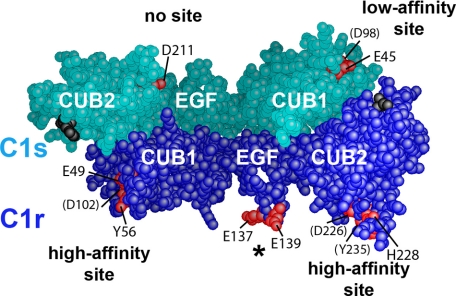
Mutations to alanine of the corresponding residues Glu49 and Asp102 in the C1r CUB1 module each virtually abolished binding of the C1s-C1r-C1r-C1s tetramer to C1q (Fig. 4 and Table 2), providing a clear indication of their implication in the interaction. Mutation of the neighboring residue Tyr56 also had a marked inhibitory effect, in keeping with the observation that replacement of the homologous residues Tyr59 and Tyr56 of human MAp19 and MASP-1/3, respectively, inhibit their interaction with MBL and the ficolins (17, 18). In contrast, mutation of residues Phe103 and Glu106 only had a slight or no significant inhibitory effect, at variance with previous data obtained upon replacement of the corresponding residues of MAp19 and MASP-1/3. We next targeted two of the acidic residues thought to coordinate Ca2+ in the C1r CUB2 module, namely Asp226 and Asp273. Again, mutation of each residue to alanine strongly inhibited the ability of the resulting C1s-C1r-C1r-C1s variants to bind C1q (Fig. 4 and Table 2). Comparable inhibitory effects were observed upon mutation of the neighboring residues His228 and Tyr235, as observed for the homologous residues His218 and Tyr225 of MASP-1/3 (18).
FIGURE 4.
Analysis by surface plasmon resonance spectroscopy of the interaction between selected C1s-C1r-C1r-C1s variants and immobilized C1q. C1q was immobilized on the sensor chip as described under “Experimental Procedures.” The native C1s-C1r-C1r-C1s tetramer reconstituted from wild type C1s and the S637A C1r mutant, and selected variants with mutations in either C1r or C1s were injected at a concentration of 10 nm.
TABLE 2.
Kinetic and dissociation constants for the interaction between C1s-C1r-C1r-C1s variants and immobilized C1q
Shown are the effects of mutations in the Ca2+-binding sites of the C1r and C1s CUB modules.
| Location of mutation(s) | Tetramer variant | kon | koff | KD | KD/KD,wt | χ2 |
|---|---|---|---|---|---|---|
| m−1·s−1 | s−1 | nm | ||||
| Wild typea | 0.98 × 106 | 0.64 × 10−3 | 0.68 ± 0.4b | 3.2 | ||
| C1r CUB1 | E49A | 0.12 × 106 | 9.55 × 10−3 | 82.4 | 121 | 3.9 |
| Y56A | 0.61 × 106 | 3.44 × 10−3 | 5.64 | 8.3 | 1.1 | |
| D102A | NDc | ND | ND | ND | ||
| F103A | 0.75 × 106 | 0.95 × 10−3 | 1.26 | 1.8 | 0.9 | |
| E106A | 1.14 × 106 | 1.18 × 10−3 | 1.03 | 1.5 | 3.9 | |
| C1r CUB2 | D226A | 0.56 × 106 | 4.16 × 10−3 | 7.45 | 10.9 | 1.2 |
| H228A | 0.97 × 106 | 4.86 × 10−3 | 5.02 | 7.4 | 1.8 | |
| Y235A | 0.85 × 106 | 24.3 × 10−3 | 28.6 | 42 | 2.9 | |
| D273A | 0.15 × 106 | 6.12 × 10−3 | 40.8 | 60 | 4.3 | |
| C1s CUB1 | E20A | 0.61 × 106 | 0.71 × 10−3 | 1.15 | 1.7 | 3.1 |
| E45A | 0.35 × 106 | 1.34 × 10−3 | 3.85 | 5.7 | 3.4 | |
| E48A | 0.60 × 106 | 0.80 × 10−3 | 1.33 | 1.9 | 3.7 | |
| Y52A | 0.90 × 106 | 1.40 × 10−3 | 1.55 | 2.3 | 4.2 | |
| D98A | 1.00 × 106 | 1.70 × 10−3 | 1.67 | 2.5 | 4.2 | |
| C1s CUB2 | E211A | 0.77 × 106 | 0.47 × 10−3 | 0.61 | 0.9 | 2.2 |
| D260A | 1.06 × 106 | 0.56 × 10−3 | 0.53 | 0.8 | 3.9 | |
| C1r CUB1/C1s CUB2 | D273A/D98A | ND | ND | ND | ND | |
| D273A/E45A | ND | ND | ND | ND |
a Tetramer assembled from wild type C1s and the S637A C1r mutant.
b Mean value determined from three separate experiments.
c ND, value not measurable due to the weakness of the binding.
A similar strategy was applied to C1s. In the CUB1 module, mutations of the Ca2+-binding residues Glu45 and Asp98 each reproducibly yielded slight inhibitory effects (Fig. 4 and Table 2), suggesting a minor contribution to the tetramer/C1q interaction. Further support for this hypothesis came from the observation that double C1s-C1r-C1r-C1s mutants with an D273A mutation in C1r CUB1 and either a E45A or an D98Ala mutation in C1s CUB1 completely lost their C1q binding ability. In contrast, the single D273A C1r variant retained low, yet measurable binding activity (Table 2). Mutations at Glu20 and Glu48, two acidic residues exposed in the C1s CUB1 structure (16), had no significant inhibitory effect. Mutation at Tyr52 yielded similar results (Table 2), although a contribution of this residue cannot be entirely excluded given the overall low impact of the mutations performed in the C1s CUB1 module. In contrast, the mutations to alanine of the Ca2+-binding residues Glu211 and Asp260 in C1s CUB2 clearly had no effect on the binding of C1s-C1r-C1r-C1s to C1q (Table 2), ruling out an implication of this module in the interaction.
As illustrated on the three-dimensional model of the C1r/C1s CUB1-EGF-CUB2 heterodimer shown in Fig. 5, the above results provided strong experimental evidence for the occurrence of high affinity C1q-binding sites contributed by the CUB1 and CUB2 modules of C1r, and lower affinity sites contributed by the C1s CUB1 module, each of these sites implicating Ca2+-binding residues and residues located in the vicinity of the Ca2+-binding site. In contrast, the mutagenesis data clearly demonstrated that the corresponding site in the C1s CUB2 module, as well as the C1r EGF module, have no significant role in the interaction with C1q.
FIGURE 5.
Three-dimensional space-filling representation of the C1r/C1s CUB1-EGF-CUB2 heterodimer. The head-to-tail C1r/C1s CUB1-EGF-CUB2 heterodimeric structure was built as described under “Experimental Procedures.” C1r and C1s domains are colored dark blue and turquoise, respectively. The Ca2+-binding sites of the C1r and C1s CUB modules are marked by residues Glu49, Tyr56, and Asp102 (C1r CUB1); Asp226, His228, and Tyr235 (C1r CUB2); Glu45 and Asp98 (C1s CUB1); and Glu211 (C1s CUB2), shown in red. Residues barely visible are indicated in parentheses. The acidic cluster Glu137-Glu-Asp139 in the C1r EGF module (indicated by an asterisk) is also shown in red. The C-terminal ends of the C1r and C1s CUB2 modules are shown in black.
DISCUSSION
The aim of this study was to delineate in C1r and C1s the sites mediating interaction between the C1s-C1r-C1r-C1s tetramer and C1q. For this purpose, C1r and C1s were each expressed in a baculovirus/insect cells system, and a series of mutations were carried out in different areas of their interaction domains with a view to measure the ability of the resulting tetramer variants to associate with C1q. In agreement with earlier observations (34), expression of C1r in insect cells resulted in poor yields compared with C1s (28), precluding purification of sufficient amounts of protein for subsequent reconstitution of the tetramer and analysis of its interaction properties. This problem could be overcome in a simple way, by first combining the C1r- and C1s-containing culture supernatants and then purifying the reconstituted C1s-C1r-C1r-C1s tetramer variants by ion exchange and gel permeation chromatography.
Twenty-two mutations were performed in either C1r or C1s, targeting residues located in their CUB1-EGF-CUB2 interaction domains while avoiding those expected to mediate heterodimeric C1r-C1s interaction in the C1s-C1r-C1r-C1s tetramer, as predicted from the homologous homodimeric x-ray structures of human C1s CUB1-EGF (16), rat MASP-2 CUB1-EGF-CUB2 (19), human MAp19 (17), and human MASP-1/3 CUB1-EGF-CUB2 (18). In agreement with these structures, none of the mutations performed in this study had a significant impact on the assembly of the C1s-C1r-C1r-C1s tetramer, as shown by both ion exchange chromatography and gel filtration analyses. Thus, the inhibitions of C1 assembly caused by mutations in C1r or C1s, as observed in this study, solely reflect direct effects on the interactions between C1s-C1r-C1r-C1s and C1q.
A first lesson from this work is that none of the mutations targeting the sequence stretch Glu137-Glu-Asp139 in the C1r EGF module had any detectable effect on C1 assembly. This demonstrates that, contrary to our earlier hypothesis, this acidic cluster has no significant contribution to the interaction with C1q. These data are therefore not compatible with our previous model of C1 assembly (16, 23), where the collagen triple helix of C1q was proposed to bind at the interface of the C1r/C1s CUB1-EGF heterodimer, through ionic interactions with acidic residues mainly contributed by the C1r EGF module. Consistent with these findings, the second round of mutations targeting residues expected to participate in the Ca2+-binding sites of the C1r and C1s CUB modules provided clear experimental evidence for the occurrence of C1q-binding sites contributed by C1r CUB1 and CUB2 and C1s CUB1; each of these sites involving residues also engaged in Ca2+ coordination or located in close vicinity of the Ca2+-binding site. These results are strikingly similar to those obtained previously in the case of MAp19 and MASP-3 (17, 18) and clearly indicate that C1s-C1r-C1r-C1s and the MASPs make use of the same basic mechanism to associate with their partner proteins, C1q, MBL, and the ficolins, respectively. These observations provide indirect evidence that, as already demonstrated by x-ray crystallography in the case of C1s CUB1 (16), both C1r CUB modules contain a Ca2+-binding site. If this hypothesis is correct, then this would mean that substitution of Asp for Glu at position 226 in C1r CUB2 (Fig. 2) is tolerated by the Ca2+-binding site. C1s CUB2 possesses the canonical Glu-Asp-Asp consensus sequence (Fig. 2), and therefore the fact that it is not involved in the interaction with C1q is likely connected to structural and/or functional constraints at the level of the whole C1 complex rather than to a lack of Ca2+-binding site. As proposed for the MASPs (18), a plausible hypothesis is that the two outer acidic Ca2+ ligands in C1r CUB1 and CUB2 and in C1s CUB1 coordinate Ca2+ on one side and mediate interaction with C1q on the other side. However, there are other possibilities, including displacement of the Ca2+ ion upon interaction with C1q. The precise mechanism involved in this interaction therefore remains to be fully elucidated.
In addition, as observed in the MASPs, other interactions are contributed by residues located in the vicinity of the Ca2+-binding sites, such as Tyr56 in C1r CUB1 and His228 and Tyr235 in C1r CUB2 (Table 2). There are, however, subtle differences between the two types of complexes, as exemplified by the fact that, unlike their counterparts in the MASPs (17, 18), Phe103 and Glu106 in C1r CUB1 do not seem to play a major role in the interaction with C1q (Table 2). Notwithstanding these differences, the striking homology, in terms of assembly, between C1 and the MBL/ficolin-MASP complexes lends further credit to the hypothesis that, as demonstrated by site-directed mutagenesis for MBL and L- and H-ficolins (21, 22), interaction in C1 also involves a conserved Lys residue from C1q. As discussed previously (16, 23), the unmodified Lys residues at positions A59, B61, and C58, located about half-way along the C1q collagen stems, are likely candidates for this function.
From a general standpoint, our finding that C1s-C1r-C1r-C1s associates with C1q through sites contributed by both C1r and C1s is fully consistent with a series of earlier data (reviewed in Ref. 35). In the same way, the fact that, unlike C1s CUB1, C1s CUB2 does not contribute a C1q-binding site, is in complete agreement with previous reports showing that the N-terminal CUB1-EGF segment of C1s is sufficient to promote interaction of C1r with C1q (11, 36). Actually, the lack of interaction between C1s CUB2 and C1q is expected to yield more flexibility at the CUB2-CCP1 interface and hence more freedom to the CCP1-CCP2-SP C1s catalytic domain, consistent with its role in the proteolytic activity of C1 (23).
Our mutagenesis data provide evidence that each C1r/C1s CUB1-EGF-CUB2 heterodimer contains two high affinity C1q-binding sites centered on residues Glu49/Asp102 (C1r CUB1) and Asp226/Asp273 (C1r CUB2), and one lower affinity site involving residues Glu45/Asp98 of C1s CUB1. This heterodimer occurs twice in C1s-C1r-C1r-C1s, and therefore the whole tetramer contributes six C1q-binding sites, which is one per each C1q collagen-like stem. Based on the location of these sites within the C1r/C1s CUB1-EGF-CUB2 assembly, we have explored different ways of positioning the two heterodimers with respect to C1q to identify the configuration(s) fulfilling the following constraints: (i) Both heterodimers should interact with C1q in the same way, implying that they be positioned symmetrically about the C1q axis. (ii) Each site contributed by C1r and C1s should make a contact with the corresponding site on a C1q stem, but interaction should be stronger where mutations have the strongest inhibitory effects. Conversely, areas where mutations are ineffective should not be close to a reactive lysine on a C1q stem. Additional constraints arise from the fact that the CUB2 modules of C1r and C1s have to connect with their respective catalytic domains. This question is particularly crucial in the case of C1r, because its catalytic domains are expected to lie inside the C1q cone, underneath the C1r/C1s interaction domains (23). In this regard, if one considers the “plane” configuration described for the homologous MASP-1/3 CUB1-EGF-CUB2 homodimer (18), the most favorable positioning is clearly the one where the C-terminal ends of the CUB2 modules are oriented toward the C1q globular domains.
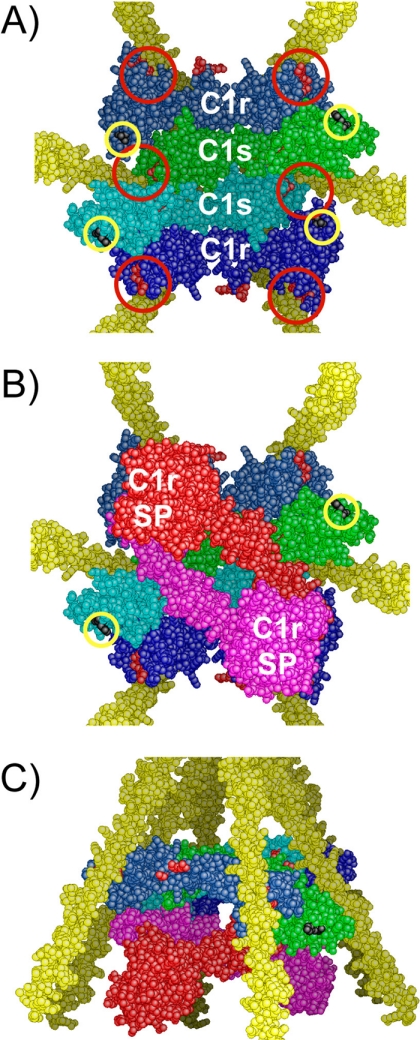
We tested different configurations and found that none of those involving location of the C1r/C1s CUB1-EGF-CUB2 heterodimers on the outside part of the C1q stems, as proposed in our previous model (16, 23), fulfilled the interaction constraints defined above. We next tested the hypothesis that both heterodimers are located inside the cone defined by the C1q stems. In this case, there are only two possible ways of assembling the heterodimers in a head-to-tail manner with respect to the inner molecules, with either CUB1-CUB2 contacts (not shown) or CUB1-EGF contacts (Fig. 6A). In the former configuration, the C1q-binding sites cannot be distributed in a radial fashion because the assembly is too elongated, and the distance between the C-terminal ends of the outer molecules is too large to fit inside the C1q cone. In the other alternative, the configuration is more favorable because the assembly is more compact, and provided that C1s is the inner molecule, then the six binding sites are distributed radially at positions roughly appropriate to make individual contacts with the target lysine residues on the C1q collagen-like stems (Fig. 6A). Consistent with this configuration, C1r is unlikely to be the inner molecule because this would conceal the C1q-binding site contributed by its CUB2 module.
FIGURE 6.
A refined three-dimensional model of C1 assembly. A, bottom view of the assembly between the C1r/C1s CUB1-EGF-CUB2 interaction domains and the C1q collagen stems. C1r and C1s domains are colored dark blue and light blue, respectively. The approximate positioning of the six C1q-binding sites contributed by C1r and C1s is indicated by red circles. The C-terminal ends of the C1r and C1s CUB2 modules are marked by yellow circles. B, bottom view of the assembly featuring the CCP1-CCP2-SP catalytic domains of C1r, shown in red and pink. The C1s catalytic domains, emerging from the C-terminal end of both C1s CUB2 modules (yellow circles), are not shown for clarity. SP, serine protease domain. C, side view of the assembly shown in B.
In this configuration, however, if the C1r-C1s CUB1-EGF-CUB2 heterodimers have the planar orientation seen in the corresponding MASP-1/3 x-ray structure (18), then some of the ineffective mutations are not far from the putative binding sites on the C1q stems. In addition, there would be steric clashes between the C1s CUB2 modules and the C1q stems, and the C termini of the outer molecules (C1r) are still too far away from each other. Nevertheless, the above problems can be overcome by orienting the CUB2 modules toward the C1q globular domains. In other words, the model becomes plausible if the C1r-C1s CUB1-EGF-CUB2 heterodimers deviate from the planar configuration observed in the MASP-1/3 structure (18). Such changes are indeed feasible because (i) only three of the four potential CUB binding sites are functional, and therefore all four sites do not need to lie in the same plane as observed in MASP-1/3, and (ii) the interaction strength varies from one binding site to the other. Thus, a more compact model can be readily obtained by twisting the EGF-CUB2 interfaces and tilting the two CUB1-EGF-CUB2 heterodimers about the C1q axis to bring them closer to each other in their central part. These modifications also modulate the relative positioning of the areas where mutations are ineffective.
In the resulting nonplanar model (Fig. 6A), the interaction domains of C1r and C1s are entirely located inside the C1q cone, in sharp contrast with our previous hypothesis (23). The C1s CUB1-EGF-CUB2 moieties occupy the inner part of the assembly, the interface between them appearing to be sufficiently flat to allow the sliding movements expected to take place upon activation (23). Compared with our previous C1 model (23), the remainder of the complex is essentially unchanged; the C1r catalytic domains occupy the lower part of the C1q cone (Fig. 6, B and C), beneath the interaction domains, the C1s serine protease domains being likely partly located inside the C1q cone, at least transiently during the activation process. Thus, in this refined version of the C1 complex, most of the C1r and C1s domains are located inside C1q, in full agreement with earlier analyses of the C1 complex by electron microscopy (37) and neutron scattering (38). From a functional standpoint, a prominent feature of this model is that each of the six C1q stems is engaged in the interaction with the tetramer. Thus, distortion of any of the C1q stems upon binding of C1 to a target surface is expected to generate part of the mechanical stress thought to trigger C1r activation (39).
From a general standpoint, it should be emphasized that the C1s-C1r-C1r-C1s moiety of C1 undergoes large changes upon C1 assembly and activation. The model arising from this study must therefore be viewed as a ground state representation of a sophisticated machinery undergoing multiple conformational changes.
Acknowledgments
We are grateful to Guy Schoehn and Wai-Li Ling for performing electron microscopy analyses and for helpful discussions.
This work was supported by the Commissariat à l'Energie Atomique, the Centre National de la Recherche Scientifique, and the Université Joseph Fourier, Grenoble.
- EGF
- epidermal growth factor
- MAp19
- mannan-binding lectin-associated protein 19
- MASP
- mannan-binding lectin-associated serine protease
- MBL
- mannan-binding lectin.
REFERENCES
- 1.Cooper N. R. (1985) Adv. Immunol. 37, 151–216 [DOI] [PubMed] [Google Scholar]
- 2.Arlaud G. J., Colomb M. G., Gagnon J. (1987) Immunol. Today 8, 106–111 [DOI] [PubMed] [Google Scholar]
- 3.Arlaud G. J., Gaboriaud C., Thielens N. M., Rossi V., Bersch B., Hernandez J. F., Fontecilla-Camps J. C. (2001) Immunol. Rev. 180, 136–145 [DOI] [PubMed] [Google Scholar]
- 4.Kishore U., Reid K. B. M. (2000) Immunopharmacology 49, 159–170 [DOI] [PubMed] [Google Scholar]
- 5.Bork P., Beckmann G. (1993) J. Mol. Biol. 231, 539–545 [DOI] [PubMed] [Google Scholar]
- 6.Campbell I. D., Bork P. (1993) Curr. Opin. Struct. Biol. 3, 385–392 [Google Scholar]
- 7.Reid K. B. M., Bentley D. R., Campbell R. D., Chung L. P., Sim R. B., Kristensen T., Tack B. F. (1986) Immunol. Today 7, 230–234 [DOI] [PubMed] [Google Scholar]
- 8.Fujita T. (2002) Nat. Rev. Immunol. 2, 346–353 [DOI] [PubMed] [Google Scholar]
- 9.Thielens N. M., Aude C. A., Lacroix M. B., Gagnon J., Arlaud G. J. (1990) J. Biol. Chem. 265, 14469–14475 [PubMed] [Google Scholar]
- 10.Busby T. F., Ingham K. C. (1990) Biochemistry 29, 4613–4618 [DOI] [PubMed] [Google Scholar]
- 11.Tsai S. W., Poon P. H., Schumaker V. N. (1997) Mol. Immunol. 34, 1273–1280 [DOI] [PubMed] [Google Scholar]
- 12.Thielens N. M., Enrie K., Lacroix M., Jaquinod M., Hernandez J. F., Esser A. F., Arlaud G. J. (1999) J. Biol. Chem. 274, 9149–9159 [DOI] [PubMed] [Google Scholar]
- 13.Thielens N. M., Cseh S., Thiel S., Vorup-Jensen T., Rossi V., Jensenius J. C., Arlaud G. J. (2001) J. Immunol. 166, 5068–5077 [DOI] [PubMed] [Google Scholar]
- 14.Zundel S., Cseh S., Lacroix M., Dahl M. R., Matsushita M., Andrieu J. P., Schwaeble W. J., Jensenius J. C., Fujita T., Arlaud G. J., Thielens N. M. (2004) J. Immunol. 172, 4342–4350 [DOI] [PubMed] [Google Scholar]
- 15.Chen C. B., Wallis R. (2001) J. Biol. Chem. 276, 25894–25902 [DOI] [PubMed] [Google Scholar]
- 16.Gregory L. A., Thielens N. M., Arlaud G. J., Fontecilla-Camps J. C., Gaboriaud C. (2003) J. Biol. Chem. 278, 32157–32164 [DOI] [PubMed] [Google Scholar]
- 17.Gregory L. A., Thielens N. M., Matsushita M., Sorensen R., Arlaud G. J., Fontecilla-Camps J. C., Gaboriaud C. (2004) J. Biol. Chem. 279, 29391–29397 [DOI] [PubMed] [Google Scholar]
- 18.Teillet F., Gaboriaud C., Lacroix M., Martin L., Arlaud G. J., Thielens N. M. (2008) J. Biol. Chem. 283, 25715–25724 [DOI] [PubMed] [Google Scholar]
- 19.Feinberg H., Uitdehaag J. C., Davies J. M., Wallis R., Drickamer K., Weis W. I. (2003) EMBO J. 22, 2348–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero A., Romão M. J., Varela P. F., Kölln I., Dias J. M., Carvalho A. L., Sanz L., Töpfer-Petersen E., Calvete J. J. (1997) Nat. Struct. Biol. 4, 783–788 [DOI] [PubMed] [Google Scholar]
- 21.Teillet F., Lacroix M., Thiel S., Weiguny D., Agger T., Arlaud G. J., Thielens N. M. (2007) J. Immunol. 178, 5710–5716 [DOI] [PubMed] [Google Scholar]
- 22.Lacroix M., Dumestre-Pérard C., Schoehn G., Houen G., Cesbron J. Y., Arlaud G. J., Thielens N. M. (2009) J. Immunol. 182, 456–465 [DOI] [PubMed] [Google Scholar]
- 23.Gaboriaud C., Thielens N. M., Gregory L. A., Rossi V., Fontecilla-Camps J. C., Arlaud G. J. (2004) Trends Immunol. 25, 368–373 [DOI] [PubMed] [Google Scholar]
- 24.Arlaud G. J., Sim R. B., Duplaa A. M., Colomb M. G. (1979) Mol. Immunol. 16, 445–450 [DOI] [PubMed] [Google Scholar]
- 25.Journet A., Tosi M. (1986) Biochem. J. 240, 783–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo C., Thielens N. M., Gagnon J., Gal P., Sarvari M., Tseng Y., Tosi M., Zavodszky P., Arlaud G. J., Schumaker V. N. (1992) Biochemistry 31, 4254–4262 [DOI] [PubMed] [Google Scholar]
- 27.Lacroix M., Rossi V., Gaboriaud C., Chevallier S., Jaquinod M., Thielens N. M., Gagnon J., Arlaud G. J. (1997) Biochemistry 36, 6270–6282 [DOI] [PubMed] [Google Scholar]
- 28.Bally I., Rossi V., Thielens N. M., Gaboriaud C., Arlaud G. J. (2005) J. Immunol. 175, 4536–4542 [DOI] [PubMed] [Google Scholar]
- 29.King L. A., Possee R. D. (1992) in The Baculovirus Expression System: A Laboratory Guide, pp. 111–114, Chapman and Hall, Ltd., London [Google Scholar]
- 30.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 31.Cseh S., Vera L., Matsushita M., Fujita T., Arlaud G. J., Thielens N. M. (2002) J. Immunol. 169, 5735–5743 [DOI] [PubMed] [Google Scholar]
- 32.Bersch B., Hernandez J. F., Marion D., Arlaud G. J. (1998) Biochemistry 37, 1204–1214 [DOI] [PubMed] [Google Scholar]
- 33.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 34.Gál P., Sárvári M., Szilágyi K., Závodszky P., Schumaker V. N. (1989) Complement Inflamm. 6, 433–441 [DOI] [PubMed] [Google Scholar]
- 35.Arlaud G. J., Gaboriaud C., Thielens N. M., Budayova-Spano M., Rossi V., Fontecilla-Camps J. C. (2002) Mol. Immunol. 39, 383–394 [DOI] [PubMed] [Google Scholar]
- 36.Thielens N. M., Illy C., Bally I. M., Arlaud G. J. (1994) Biochem. J. 301, 378–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strang C. J., Siegel R. C., Phillips M. L., Poon P. H., Schumaker V. N. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perkins S. J., Villiers C. L., Arlaud G. J., Boyd J., Burton D. R., Colomb M. G., Dwek R. A. (1984) J. Mol. Biol. 179, 547–557 [DOI] [PubMed] [Google Scholar]
- 39.Budayova-Spano M., Lacroix M., Thielens N. M., Arlaud G. J., Fontecilla-Camps J. C., Gaboriaud C. (2002) EMBO J. 21, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]