Abstract
Ubiquitination is essential for the endocytic sorting of various G protein-coupled receptors to lysosomes. Here we identify a distinct function of this covalent modification in controlling the later proteolytic processing of receptors. Mutation of all cytoplasmic lysine residues in the murine δ-opioid receptor blocked receptor ubiquitination without preventing ligand-induced endocytosis of receptors or their subsequent delivery to lysosomes, as verified by proteolysis of extramembrane epitope tags and down-regulation of radioligand binding to the transmembrane helices. Surprisingly, a functional screen revealed that the E3 ubiquitin ligase AIP4 specifically controls down-regulation of wild type receptors measured by radioligand binding without detectably affecting receptor delivery to lysosomes defined both immunochemically and biochemically. This specific AIP4-dependent regulation required direct ubiquitination of receptors and was also regulated by two deubiquitinating enzymes, AMSH and UBPY, which localized to late endosome/lysosome membranes containing internalized δ-opioid receptor. These results identify a distinct function of AIP4-dependent ubiquitination in controlling the later proteolytic processing of G protein-coupled receptors, without detectably affecting their endocytic sorting to lysosomes. We propose that ubiquitination or ubiquitination/deubiquitination cycling specifically regulates later proteolytic processing events required for destruction of the receptor's hydrophobic core.
A fundamental cellular mechanism contributing to homeostatic regulation of receptor-mediated signal transduction involves ligand-induced endocytosis of receptors followed by proteolysis in lysosomes. The importance of such proteolytic down-regulation has been documented extensively for a number of seven-transmembrane or G protein-coupled receptors (GPCRs),3 which comprise the largest known family of signaling receptors expressed in animals, as well as for other important signaling receptors, such as the epidermal growth factor receptor tyrosine kinase (1–5).
One GPCR that is well known to undergo endocytic trafficking to lysosomes is the δ-opioid peptide receptor (DOR or DOP-R) (6). Following endocytosis, DOR traffics efficiently to lysosomes in both neural and heterologous cell models (6–8), whereas many membrane proteins, including various GPCRs, recycle rapidly to the plasma membrane (9–12). Such molecular sorting of internalized receptors between divergent recycling and degradative pathways is thought to play a fundamental role in determining the functional consequences of regulated endocytosis (2, 3, 13, 14). The sorting process that directs internalized DOR to lysosomes is remarkably efficient and appears to occur rapidly (within several min) after receptor endocytosis (11). Nevertheless, biochemical mechanisms that control lysosomal trafficking and proteolysis of DOR remain poorly understood.
A conserved mechanism that promotes lysosomal trafficking of a number of membrane proteins, including various signaling receptors, is mediated by covalent modification of cytoplasmic lysine residues with ubiquitin (4, 15–17). Ubiquitination was first identified as an endocytic sorting determinant in studies of vacuolar trafficking of the yeast GPCR Ste2p (18). Subsequent studies have established numerous examples of lysyl-ubiquitination being required for sorting endocytic cargo to lysosomes and have identified conserved machinery responsible for the targeting of ubiquitinated cargo to lysosomes (3, 17, 19–22).
The CXCR4 chemokine receptor provides a clear example of ubiquitin-dependent lysosomal sorting of a mammalian GPCR. Ubiquitination of the carboxyl-terminal cytoplasmic domain of the CXCR4 receptor, mediated by the E3 ubiquitin ligase AIP4, is specifically required for the HRS- and VPS4-dependent trafficking of internalized receptors to lysosomes. Blocking this ubiquitination event by Lys → Arg mutation of the receptor specifically inhibits trafficking of internalized receptors to lysosomes, resulting in recycling rather than lysosomal proteolysis of receptors after ligand-induced endocytosis (23–25).
Lysosomal trafficking of DOR, in contrast, is not prevented by mutation of cytoplasmic lysine residues (26) and can be regulated by ubiquitination-independent protein interaction(s) (27, 28). Nevertheless, both wild type and lysyl-mutant DORs traffic to lysosomes via a similar pathway as ubiquitin-dependent membrane cargo and require both HRS and active VPS4 to do so (29). These observations indicate that DOR engages the same core endocytic mechanism utilized by ubiquitination-directed membrane cargo but leave unresolved whether ubiquitination of DOR plays any role in this important cellular mechanism of receptor down-regulation.
There is no doubt that DOR can undergo significant ubiquitination in mammalian cells, including HEK293 cells (30–32), where lysosomal trafficking of lysyl-mutant receptors was first observed (26). Ubiquitination was shown previously to promote proteolysis of DOR by proteasomes and to function in degrading misfolded receptors from the biosynthetic pathway (30, 31). A specific role of ubiquitination in promoting proteasome- but not lysosome-mediated proteolysis of DOR has been emphasized (32) and proposed to contribute to proteolytic down-regulation of receptors also from the plasma membrane (33).
To our knowledge, no previous studies have determined if DOR ubiquitination plays any role in controlling receptor proteolysis mediated by lysosomes, although this represents a predominant pathway by which receptors undergo rapid down-regulation following ligand-induced endocytosis in a number of cell types, including HEK293 cells (8). In the present study, we have taken two approaches to addressing this fundamental question. First, we have investigated in greater detail the effects of lysyl-mutation on DOR ubiquitination and trafficking. Second, we have independently investigated the role of ubiquitination in controlling lysosomal proteolysis of wild type DOR. Our results clearly establish the ability of DOR to traffic efficiently to lysosomes in the absence of any detectable ubiquitination. Further, they identify a distinct and unanticipated function of AIP4-dependent ubiquitination in regulating the later proteolytic processing of receptors and show that this distinct ubiquitin-dependent regulatory mechanism operates effectively downstream of the sorting decision that commits internalized receptors for delivery to lysosomes.
EXPERIMENTAL PROCEDURES
Cell Culture, cDNA Constructs, and Transfection
The Myc-tagged AIP4 and the C830A inactive mutant AIP4 have been previously described (24). Nedd4-1, Nedd4-2, WWP1, WWP2, Smurf1, and their corresponding inactive mutant versions were a gift from Laurent Coscoy and Brian Sullivan (University of California, Berkeley). Smurf2, NEDL1, and NEDL2 were a gift of Wes Sundquist (University of Utah School of Medicine) (34). Point mutations of the conserved catalytic cysteine residue were introduced by oligonucleotide-directed site-directed mutagenesis (QuikChange; Stratagene). GFP-AMSH, GFP-AMSH-D348A (D/A), GFP-UBPY, and GFP-UBPY-C786S (C/S) were a gift from Sylvie Urbé (University of Liverpool) and were previously described (35, 36). The FLAG-tagged DOR and the lysine mutant version (DOR-0cK) have been previously described (26). A COOH-terminal HA epitope was added to the F-DOR and F-DOR-0cK using PCR and encoding the HA epitope sequence (YPYDVDDYA) in the reverse primer. The resulting F-DOR-HA and F-DOR-0cK-HA coding sequences were cloned into pcDNA3 (Invitrogen) for generation of stable cell lines. Stably transfected cells expressing epitope-tagged receptors were generated by selection for neomycin resistance using 500 μg/ml G418 (Geneticin; Invitrogen). Resistant colonies were clonally isolated and selected for further study based on comparable levels of receptor expression as assessed by fluorescence microscopy and saturation binding analysis (supplemental Fig. 1). HEK293 cells (ATCC, Manassas, VA) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (University of California, San Francisco, Cell Culture Facility).
For all transient expression of ligases and deubiquitinating enzymes, cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells expressing FLAG-tagged receptors were harvested by washing with EDTA and plated in 60-mm dishes at 80% confluence before transfection with plasmid DNA. Cells were reseeded into polylysine-coated 6-well or 24-well plates and cultured for a further 24 h before experimentation. For knockdown of endogenous AIP4, AMSH, or UBPY levels, the following siRNA duplexes were obtained from Qiagen: AIP4-3 (Hs_ITCH_3), CAAGAGCTATGAGCAACTGAA; AIP4-6 (Hs_ITCH_6), TGCCGCCGACAAATACAAATA; AMSH-7 (Hs_STAMBP_7), ATCACGCTCTTTATTGAGAAA; AMSH-8 (Hs_STAMBP_8), CCGCTCTGGAGTTGAGATTAT; UBPY-1 (Hs_USP8_1), CAGGGTCAATTCAAATCTACA; UBPY-2 (HS_USPB_2), AAGGCTCGTATTCATGCAGAA. They were transfected using Lipofectamine RNAi-max according to the manufacturer's instructions.
Biochemical Detection of Receptor Proteolysis and Protein Levels by Immunblotting
Immunoblotting to assess total cellular receptor levels was carried out as previously described (29). Briefly, cell monolayers were washed three times in ice-cold phosphate-buffered saline (PBS) and lysed in extraction buffer (0.5% Triton X-100, 150 mm NaCl, 25 mm KCl, 25 mm Tris, pH 7.4, 1 mm EDTA) supplemented with a standard protease inhibitor mixture (Roche Applied Science). Extracts were clarified by centrifugation (12,000 × g for 10 min) and then mixed with SDS sample buffer for denaturation. Proteins present in the extracts were resolved by SDS-PAGE using 4–12% BisTris gels (NuPAGE; Invitrogen), transferred to nitrocellulose membranes, and probed for protein by immunoblotting using horseradish peroxidase-conjugated sheep anti-mouse IgG or donkey anti-rabbit IgG (Amersham Biosciences) and SuperSignal detection reagent (Pierce). Apparent molecular mass was estimated using commercial protein standards (SeeBlue Plus2; Invitrogen). Band intensities of unsaturated immunoblots were analyzed and quantified by densitometry using FluorChem 2.0 software (AlphaInnotech Corp.). Antibodies used were anti-FLAG-M1, anti-FLAG-M2-HRP (Sigma), anti-HA-11 (Covance), anti-HA(3F10)-HRP (Roche Applied Science), anti-AIP4/ITCH (BD Biosciences), anti-UBPY (Sigma), and anti-AMSH (a gift from Sylvie Urbé, University of Liverpool).
Biotinylation-Degradation Assay
To specifically label and follow the fate of the surface receptor pool, a previously described cell surface biotinylation assay was used to label FLAG-tagged receptors present in the plasma membrane (26, 29). Briefly, stably transfected HEK293 cells were grown on 60-mm dishes, washed with ice-cold PBS, and incubated with 300 μg/ml sulfo-N-hydroxysuccinimide-biotin (Pierce) in PBS for 30 min at 4 °C to biotinylate surface proteins. Following washing with Tris-buffered saline to remove and quench unreacted biotinylation reagent, cells were returned to 37 °C for incubation in media, in the absence or presence of 10 μm d-Ala-d-Leu-enkephalin (DADLE) for the indicated time period and extracted as described above. Extracts were clarified by centrifugation (12,000 × g for 10 min), and biotinylated proteins were isolated by immobilization on streptavidin-conjugated Sepharose beads (Pierce). Washed beads were eluted with SDS sample buffer before resolving by SDS-PAGE, transferred to nitrocellulose membranes, and probed for FLAG-tagged receptor (M1 antibody; Sigma). Some samples, as indicated, were deglycosylated by the addition of 500 units of peptide N-glycosidase F (New England Biolabs) and incubated for 1 h at 37 °C before the elution with SDS sample buffer.
Biochemical Detection of Receptor Ubiquitination
To ensure the removal of any proteins that might be associated with the receptor, denaturing conditions were used. Cells were transiently transfected with HA-ubiquitin and treated before being lysed in 400 μl of extraction buffer and clarified by centrifugation (12,000 × g for 10 min), mixed with 200 μl of 3× radioimmune precipitation buffer (450 mm NaCl, 150 mm Tris, pH 7.4, 15 mm EDTA, 3% Triton X-100, 1.5% sodium deoxycholate, 30 mm NaF, 30 mm Na2-pyrophosphate, 0.3% SDS), and incubated overnight at 4 °C with 2 μg of M2 anti-FLAG antibody (Sigma). 30 μl of protein A/G-agarose (Pierce) was added for 2 h at 4 °C. Immunoprecipitates were pelleted by centrifugation (3000 rpm, 1 min, 4 °C) and washed three times with 500 μl of radioimmune precipitation buffer before the addition of 20 μl of SDS sample buffer (Invitrogen) supplemented with β-mercaptoethanol and analysis by Western blotting using anti-HA-HRP (Roche Applied Science). Blots were then stripped (Restore Western blot stripping buffer; Pierce) and reprobed with anti-FLAG M2-HRP to verify relative receptor levels.
Analysis of Receptor Levels by Radioligand Binding
Receptor down-regulation was determined by radioligand binding, as previously described (11). Following transfection, HEK293 cells stably expressing FLAG-tagged receptors were replated into 12-well plates. 24 h later, 10 μm DADLE was added to the cells for the indicated time period, cells were washed twice with ice-cold PBS, 300 μl of PBS was added to the cells, and the plates were frozen. Plates were thawed, and cells were resuspended. Binding assays were performed in triplicate in 96-well plates using a 10 nm concentration of the radiolabeled opioid receptor antagonist [3H]diprenorphine (DPN) (88 Ci/mmol; Amersham Biosciences) and incubated for 1 h at room temperature, a saturating concentration that is sufficient to access both surface and internal receptors (11). Incubations were terminated by vacuum filtration through glass fiber filters (Whatman), and unbound radioligand was removed by repeated washes with Tris-buffered saline. Bound radioactivity was determined by liquid scintillation counting of washed filters. Nonspecific binding was determined by carrying out parallel determinations in the presence of excess unlabeled competitive antagonist (10 μm naloxone). Data presented represent the specific binding (total minus nonspecific binding) at each time point, expressed as a percentage of specific binding in similarly transfected but agonist-naive cells.
Fluorescence Microscopy
Colocalization of receptors with late endosome/lysosome markers was visualized using HEK293 cells stably expressing the indicated FLAG-tagged receptor constructs plated on polylysine-coated glass coverslips (Corning Glass). Cells were incubated in the presence of 10 μm DADLE for 2 h before fixation with 4% formaldehyde and permeabilization with 0.1% Triton X-100 in PBS. Cells were labeled using rabbit anti-FLAG (Sigma) and mouse antibodies recognizing LAMP-1 and -2 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), followed by secondary detection using Alexa594-conjugated anti-mouse and Alexa647 anti-rabbit secondary antibodies (Invitrogen). Colocalization of receptors with ubiquitin hydrolases was carried out using an identical procedure but with cells transiently transfected with GFP-ASMH-D348A or GFP-UBPY-C786S. Specimens were imaged by confocal fluorescence microscopy using a Zeiss LSM 510 microscope fitted with a Zeiss ×63, numeric aperture 1.4 objective operated in single photon mode, with standard filter sets verified for lack of detectable cross-channel bleed-through and standard (1 Airy disc) pinhole. Acquired optical sections were analyzed with LSM Image Examiner (Zeiss) and rendered with Adobe Photoshop software.
Statistical Analysis
Quantitative data were averaged across multiple independent experiments, with the number of experiments specified in the corresponding figure legend. Unless indicated otherwise, the error bars represent the S.E. value determined after compiling mean determinations across experiments. The statistical significance of the indicated differences was analyzed using the appropriate variations of one-way ANOVA and post-test and Student's t test, as specified in the figure legends, calculated using Prism 4.0 software (GraphPad Software, Inc.). The relative significance of each of the reported differences is specified by calculated p values that are also listed in the figure legends and annotated graphically in the figures.
RESULTS
Ubiquitination-independent Down-regulation of DOR
Previous findings indicated that mutation of all cytoplasmic lysine residues in the murine DOR does not prevent proteolytic down-regulation of receptors mediated by endosomal sorting complex required for transport (ESCRT)-dependent trafficking of internalized receptors to lysosomes (26, 29). This was unexpected, because lysyl-ubiquitination is known to be essential for lysosomal trafficking of several other GPCRs (3) and because DOR is known to undergo extensive ubiquitination in intact cells (30, 32). Because our previous analysis of receptor proteolysis relied primarily on biochemical detection of a FLAG epitope tag engineered into the NH2-terminal ectodomain of the receptor (F-DOR), we considered the possibility that receptor proteolysis detected in our previous work might reflect limited proteolysis of the proximal NH2-terminal ectodomain, perhaps analogous to proteolytic “shaving” reported for the Ste3p seven-transmembrane receptor in yeast (37). Such limited proteolysis might be insufficient to destroy receptor function, since mutational studies indicate that the proximal NH2 terminus of opioid receptors is not essential for ligand binding (37–39).
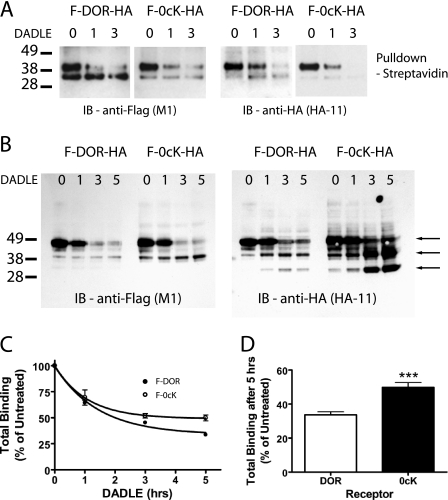
To further evaluate the ubiquitination dependence of receptor proteolysis, we engineered a distinct (HA) epitope tag into the COOH-terminal endodomain of F-DOR and F-DOR-0cK (F-DOR-HA and F-DOR-0cK-HA), to allow monitoring of receptor proteolysis involving both ends of the receptor protein in the primary structure on both sides of the membrane. Stable cell lines generated from these constructs showed expression levels of between 1 and 2 fmol/μg for all constructs used (supplemental Fig. 1), an expression level that is on a similar order as that reported endogenously in the brain (40) and one at which efficient endocytic sorting of receptors occurs in HEK293 cells (11). To specifically follow the fate of the mature surface receptor, HEK293 cells stably expressing F-DOR-HA were labeled by surface biotinylation, and proteolysis of receptors was evaluated by streptavidin affinity purification and immunoblotting after incubating cells for various time periods with an agonist ligand (10 μm concentration of the opioid peptide analogue DADLE) that promotes receptor endocytosis. Anti-FLAG blotting confirmed that the NH2-terminal ectodomains of both the wild type (F-DOR-HA) and lysyl-mutant (F- DOR-0cK-HA) receptors were fully proteolyzed within 2–3 h after ligand-induced endocytosis (Fig. 1A, left), consistent with previous studies using this approach. Significantly, a similarly rapid time course of receptor proteolysis was observed when anti-HA blotting was assessed (Fig. 1A, right). This loss of COOH-terminal immunoreactivity was not simply the result of loss of ectodomain-linked biotin used to label surface receptors, since extensive proteolysis was also evident by immunoblotting of whole cell extracts prior to streptavidin purification (Fig. 1B). Further, this analysis revealed a “ladder” of HA-immunoreactive fragments derived from both F-DOR-HA and F-DOR-0cK-HA, which appeared over a similar time course after DADLE application to cells. Together, these results indicate that both wild type and lysyl-mutant receptors undergo extensive proteolytic fragmentation following ligand-induced endocytosis.
FIGURE 1.
Both DOR and DOR-0cK undergo extensive proteolysis and pharmacological down-regulation after ligand-induced endocytosis. A, HEK293 cells stably expressing F-DOR-HA and F-DOR-0cK-HA were biotinylated (as described under “Experimental Procedures”) before incubation in the presence of 10 μm DADLE for the indicated time period (in hours). Extracts were split in two before pull-down with streptavidin beads, deglycosylation with peptide N-glycosidase F, and SDS-PAGE separation. Shown are representative anti-FLAG blots (left) and anti-HA blots (right) of F-DOR-HA (left) and F-DOR-0cK-HA (right). B, cells stably expressing F-DOR-HA or F-DOR-0cK-HA, as indicated, were incubated for the indicated periods in the presence of 10 μm DADLE before lysis and division into two identical samples. Shown is a representative anti-FLAG (left) and anti-HA (right) immunoblot. The arrows denote major proteolytic cleavage products, indicating that both wild type and lysyl-mutant receptors undergo extensive ligand-induced proteolysis over a similarly rapid time course, as indicated by the generation of multiple proteolytic cleavage events. C, cells stably expressing F-DOR (closed symbols) or F-DOR-0cK (open symbols) were treated for the indicated time with 10 μm DADLE before freeze-thawing and undergoing ligand binding with [3H]DPN. Results shown represent specific binding expressed as a percentage of binding in untreated cells. D, total binding after 5 h of DADLE treatment expressed as a percentage of total binding in untreated cells (***, p < 0.001; Student's t test, n = 10).
As another approach to verify efficient lysosomal proteolysis of lysyl-mutant opioid receptors (DOR-0cK), we assessed proteolytic down-regulation of receptors by loss of receptor-mediated binding of the small molecule radioligand antagonist, [3H]diprenorphine. Mutational studies indicate that diprenorphine interacts primarily with residues located in the transmembrane helical structure of the receptor protein rather than with the receptor ectodomain (39). This intramembrane binding site is also essential for association of physiologically relevant opioid peptides with receptors, although high affinity binding of opioid peptides also requires residues present in extracellular loops (41). Thus, diprenorphine binding represents an independent measure of receptor proteolysis, which is sensitive to the integrity of the receptor's hydrophobic core. Time course analysis indicated that both wild type and lysyl-mutant receptors exhibited pharmacological down-regulation with similarly rapid kinetics (Fig. 1C), which were comparable with (albeit slightly slower than) the kinetics of proteolytic fragmentation estimated biochemically by ecto- and endodomain immunoblotting (Fig. 1A) (see also Refs. 26 and 29). Moreover, as expected, down-regulation of both F-DOR and F-DOR-0cK measured pharmacologically by [3H]diprenorphine binding was inhibited both by the classical inhibitor of lysosomal proteolysis chloroquine and by overexpression of HRS (supplemental Fig. 2). Taken together, these data verify definitively that the DOR-0cK lysyl-mutant receptor does indeed undergo efficient proteolytic degradation following ligand-induced endocytosis, like wild type receptors, and does so via HRS-dependent trafficking to lysosomes.
Although lysyl-mutant receptors were clearly able to undergo extensive proteolysis by lysosomes, careful comparison of the pharmacological results revealed a small reduction in the extent of down-regulation of DOR-0cK (compared with wild type DOR) measured by radioligand binding. Although relatively subtle at all time points, this effect was most noticeable at later time points (>3 h) after endocytosis of receptors. Proteolytic fragmentation of receptors assessed biochemically was already extensive by this time, suggesting that lysyl-mutation affects a relatively late stage in a progressive process of receptor destruction. This quantitative difference in pharmacological down-regulation, although small in absolute magnitude, was statistically significant when evaluated in multiple expression-matched cell clones (Fig. 1D).
The E3 Ligase, AIP4, Specifically Controls Pharmacological Down-regulation of Wild Type DOR
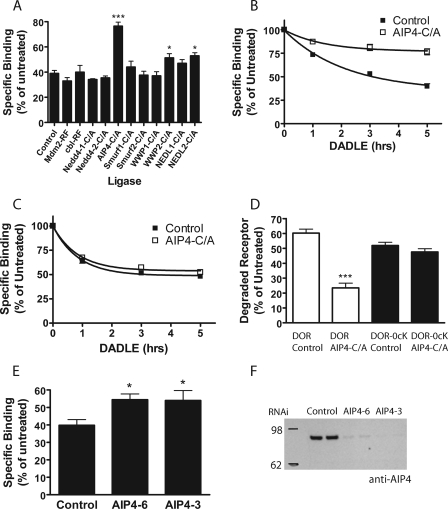
Such kinetic effects could represent a secondary consequence of introducing multiple lysyl-mutations into the receptor, but might also reflect the existence of some previously unappreciated ubiquitin-dependent regulation. To distinguish these possibilities, we focused on wild type receptors and devised a screen to search for ubiquitin ligase(s) that influence ligand-induced down-regulation. A number of E3 ligases have been implicated in lysosomal sorting and/or pharmacological down-regulation of signaling receptors in mammalian cells, specifically two RING finger ligases (c-Cbl and Mdm2) (42–45) and several HECT domain ligases, including Nedd4 and related enzymes (46–50). We cloned catalytically inactive mutant forms of each of these ligases into the same cytomegalovirus-driven vector backbone to facilitate comparable heterologous expression. Radioligand binding was used to test the effect of overexpressing each mutant ligase on down-regulation of DOR measured after 5 h of continuous exposure to agonist. Most of the inactive ligases had little or no effect on ligand-induced down-regulation of DOR. Disrupting the HECT-domain E3 ligase AIP4/Itch (C830A mutation), however, produced a strong inhibition (Fig. 2A).
FIGURE 2.
Effect of ubiquitin ligases on DOR down-regulation. A, F-DOR-expressing HEK293 cells were transiently transfected with the indicated inactive ubiquitin ligase constructs and 24 h later replated in 12-well plates. After 24 h, cells were treated with 10 μm DADLE for 5 h before freeze-thaw and assay by radioligand binding with [3H]DPN. Specific binding was expressed as a percentage of binding in similarly transfected, agonist-naive cells (one-way ANOVA, Bonferroni multiple comparison test; ***, p < 0.001; *, p < 0.05, n ≥ 4). HEK293 cells stably expressing F-DOR (B) or F-DOR-0cK (C) were transiently transfected with AIP4-C/A (open symbols) and 48 h later treated for the indicated time with 10 μm DADLE before freeze-thawing and determining receptor down-regulation by [3H]DPN binding assay. Specific binding detected at the indicated times is expressed as a percentage of that measured in agonist-naive cells. D, the same data expressed as a percentage of receptor degraded after 5 h of DADLE treatment (one-way ANOVA, Bonferroni multiple comparison test; p < 0.001, n = 14). E, F-DOR cells were transfected with siRNA duplexes against AIP4 and, 72 h later, assayed for DADLE-induced down-regulation by [3H]DPN binding. Results shown represent specific binding expressed as a percentage of binding in untreated cells (one-way ANOVA, Bonferroni multiple comparison test; *, p < 0.05, n = 7). F, knockdown was verified by lysing the remaining cells from the down-regulation assay, resolving equal amounts of total cellular protein on NuPAGE 4–12% BisTris gel, and detecting endogenous AIP4 by immunoblotting. A representative Western blot is shown.
Time course analysis confirmed the pronounced inhibitory effect of inactive AIP4 on pharmacological down-regulation of wild type receptors (Fig. 2B). To test for biochemical specificity with respect to ubiquitination of receptors, we next tested effects of the identified ligase on pharmacological down-regulation of lysyl-mutant receptors (F-DOR-0cK). In contrast to its pronounced inhibitory effect on down-regulation of wild type receptors, overexpression of inactive AIP4 (again, with similar expression verified by immunoblotting) did not produce any detectable effect on pharmacological down-regulation of lysyl-mutant receptors (Fig. 2C). This remarkable specificity of AIP4-dependent regulation was verified across multiple experiments and cell clones (Fig. 2D). Depleting endogenous AIP4 by RNA interference also inhibited pharmacological down-regulation of wild type F-DOR (Fig. 2E), and significant inhibition was observed using two independent siRNA duplexes that were verified to produce efficient depletion of endogenous AIP4 protein (Fig. 2F). Together, these results identify an essential function of AIP4-dependent ubiquitination specifically in controlling pharmacological down-regulation of wild type DOR without detectably affecting lysyl-mutant DOR.
The observation that down-regulation of F-DOR-0cK was insensitive to AIP4 disruption indicated that the pronounced inhibition of F-DOR down-regulation did not result from a nonspecific effect of reduced ligase activity and suggested that AIP4 mediates this regulatory effect via ubiquitination of the receptor itself. To test this, we applied an established method to assay incorporation of HA-tagged ubiquitin into F-DOR immunopurified from HEK293 cells. Comparison of control purifications prepared from matched cells not expressing F-DOR verified the specificity of this detection (Fig. 3A, 293 lane). Although a basal level of specific ubiquitin incorporation was clearly observed in F-DOR isolated from cells maintained in the absence of opioid ligand, receptor activation with DADLE produced a transient increase in HA-ubiquitin incorporation. Comparison of wild type (F-DOR; left side of blot) relative to lysyl-mutant (F-DOR-0cK; right side of blot) receptors clearly established that the lysyl-mutations fully prevented detectable ubiquitination of receptors, reducing the HA-ubiquitin signal to control levels even when lanes containing lysyl-mutant receptors were overloaded with immunoisolated receptors (Fig. 3B). We also noted that ubiquitinated F-DOR resolved at considerably higher apparent molecular mass (100–200 kDa; bracket in Fig. 3A) compared with the major immunoreactive receptor species detected by anti-FLAG blot (50–60 kDa; bracket in Fig. 3B).
FIGURE 3.
DOR undergoes AIP4-dependent lysyl-ubiquitination. A, HEK293 cells stably expressing F-DOR or F-DOR-0cK were transfected with HA-tagged ubiquitin and, 24 h later, were stimulated with 10 μm DADLE for the indicated time period before extraction and immunoprecipitation under denaturing conditions with mouse anti-FLAG antibody (M2) and SDS-PAGE separation. Shown is a representative anti-HA (3F10)-HRP blot with untransfected HEK293 cells (no receptor expressed; 293), the F-DOR (left), and F-DOR-0cK (right). B, the same blot stripped and reprobed with anti-FLAG (M2)-HRP to show the relative size and amount of the major receptor species. C, HEK293 cells stably expressing F-DOR were transfected with HA-tagged ubiquitin and either empty pcDNA vector or mycAIP4-C/A construct and 24 h later were stimulated with 10 μm DADLE for 20 min before extraction and immunoprecipitation with mouse anti-FLAG antibody (M2) and SDS-PAGE separation. Shown is a representative anti-HA (3F10)-HRP blot. D, the same blots were stripped and reprobed with anti-FLAG (M2)-HRP to show the relative size and amount of the major receptor species. IP, immunoprecipitation; IB, immunoblot.
Although the major receptor species identified by anti-FLAG blot corresponds to the complex glycosylated receptor monomer, the substantially reduced electrophoretic mobility of the ubiquitinated species suggests that, at steady state, a small population of receptors is modified by extensive multiubiquitination and/or polyubiquitination (rather than monoubiquitination). Overexpression of catalytically inactive AIP4 shifted the distribution of ubiquitinated receptor species toward lower apparent molecular mass but did not fully prevent receptor ubiquitination (Fig. 3C). Nevertheless, we verified in the same cells that mutant AIP4 expression strongly inhibited proteolytic down-regulation of receptors measured by radioligand binding (not shown). Thus, although preventing receptor ubiquitination entirely had little effect on proteolytic down-regulation (F-DOR-0cK; Fig. 1C), partial inhibition of receptor ubiquitination (AIP4 disruption; Figs. 2B and 3C) strongly inhibited this process. These observations are clearly not consistent with the hypothesis that ubiquitin acts simply as a lysosomal sorting signal and suggest, instead, that ubiquitination mediates a distinct regulatory function on the later proteolytic processing of receptors.
AIP4 Does Not Detectably Affect Endocytic Sorting of DOR to Lysosomes
To further investigate this hypothesis, we examined the effect of disrupting AIP4 activity on receptor proteolysis detected biochemically. Despite strongly inhibiting down-regulation of wild type receptors measured by radioligand binding (Fig. 2B), catalytically inactive AIP4 did not detectably affect DOR proteolysis assessed by loss of FLAG immunoreactivity (Fig. 4A shows a representative immunoblot, and Fig. 4B summarizes quantification across multiple experiments). As yet another approach to examine the specificity of the AIP4-dependent regulatory effect, we evaluated the trafficking fate of endocytosed receptors by immunocytochemical localization. Previous studies have established that both wild type F-DOR and lysyl-mutant F-DOR-0cK colocalize with the late endosome/lysosome markers LAMP1 and -2 within ∼2 h after stimulating receptor endocytosis with DADLE (26, 29), consistent with the time course of receptor proteolysis determined biochemically. Overexpression of catalytically inactive mutant AIP4 did not prevent this colocalization, as indicated by the detection in confocal optical sections of numerous endomembrane structures labeled for both DOR (green) and LAMP1/2 (red) immunoreactivity at this time point (Fig. 4C). Colocalized structures were readily apparent when optical cross-sections containing many endocytic structures were examined at higher magnification (insets).
FIGURE 4.
AIP4 is not required for lysosomal delivery of receptors detected biochemically or by immunocytochemical localization. A, F- DOR-expressing HEK293 cells were transfected with GFP or AIP4-C/A and, 48 h later, underwent cell surface biotinylation before incubation for the indicated time periods with 10 μm DADLE. Cells were lysed, and biotinylated protein was pulled down with streptdavidin-agarose beads and separated by SDS-PAGE. A representative anti-FLAG immunoblot is shown. B, blots generated in multiple experiments were scanned to estimate the amount of FLAG-tagged receptor remaining at each time point after incubation in the presence of 10 μm DADLE, expressed as a percentage of that measured in identically transfected cells not exposed to opioid. Results were pooled and averaged across multiple experiments (n = 5). Closed symbols, degradation curve measured in control cells transfected with GFP; open symbols, degradation curve measured in cells transfected with AIP4-C/A-GFP (n = 5 for each set). C, F-DOR cells were transfected with empty pcDNA or AIP4-C/A-GFP (vi), replated onto coverslips, and incubated in the presence of 10 μm DADLE for 2 h before fixation and labeling for anti-FLAG (i and iv) and anti-LAMP1/2 (ii and v). Shown are representative confocal optical sections imaged under nonsaturating conditions and rendered using simple background subtraction and a linear lookup table. Merged images (iii and vii) display DOR and LAMP channels pseudocolored in green and red, respectively. The AIP4 channel has not been included in the merged image displayed, to make receptor localization more easily compared, but this signal can be seen in the grayscale image and pseudocolored inset (vi). IB, immunoblot.
The Endosome-associated Deubiquitinating Enzymes, AMSH and UBPY, Also Specifically Control Pharmacological Down-regulation
The ability of partial inhibition of receptor ubiquitination (by disrupting AIP4 activity) to strongly inhibit late proteolytic processing of receptors, while complete blockade of receptor ubiquitination (by lysyl-mutation) produced little inhibitory effect, suggested that the observed regulation of late proteolytic processing may require both ubiquitination and deubiquitination of receptors. To test this hypothesis, we looked for a potential regulatory effect of receptor deubiquitination by two deubiquitinating enzymes (DUBs), AMSH and UBPY, which are known to associate with endocytic membranes and act on other endocytic cargo (35, 36). The functional screen used to assess ubiquitin ligases was adapted to test these candidate DUBs, again using catalytically inactive mutant versions of each enzyme (AMSH D348A or UBPY C786S). Remarkably, overexpressing inactive versions of either DUB significantly inhibited pharmacological down-regulation of wild type receptors (Fig. 5A). In contrast, down-regulation of lysyl-mutant receptors continued unimpeded in the presence of either mutant DUB (Fig. 5B). Further, neither mutant DUB detectably affected receptor proteolysis assessed biochemically by immunoblot analysis (Fig. 5C shows a representative blot, and Fig. 5D summarizes quantification across multiple experiments). Depletion of endogenous levels of either AMSH or UBPY by RNA interference also inhibited pharmacological down-regulation of wild type F-DOR (Fig. 5E). This was somewhat surprising, since depletion of AMSH has been shown to increase the degradation of epidermal growth factor receptors (35), further suggesting differences in ubiquitin-dependent regulation between DOR and epidermal growth factor receptors. Significant inhibition was again observed using two independent siRNA duplexes for each DUB that were verified to produce efficient depletion of endogenous AMSH or UBPY protein (Fig. 5F). Simultaneous depletion of both DUBs did not have any additional effect on down-regulation as compared with either DUB independently. Finally, we investigated the effect of DUBs on trafficking of F-DOR to the lysosome as visualized by confocal microscopy. Consistent with previous reports (35, 36), both AMSH-D/A and UBPY-C/S were visualized in a largely cytosolic distribution, with increased concentration on enlarged endosomes (Fig. 5G, iii and vii; higher magnification is shown in the inset). Interestingly, these endosomes colocalized with the late endosome/lysosome marker LAMP1/2 (ii and vi). Moreover, F-DOR localized to the same structures following prolonged agonist exposure (i and v). This overlap between internalized receptors and both DUBs in late endosome/lysosome structures is emphasized in the merged color image (iv and viii), particularly when examined at higher magnification (inset).
FIGURE 5.
Endosomal deubiquitinating enzymes specifically affect down-regulation of DOR but not DOR-0cK. HEK293 cells stably expressing F-DOR (A) or F-DOR-0cK (B) were transiently transfected with GFP, GFP-AMSH-D/A, or GFP-UBPY-C/S and, 48 h later, treated for 5 h with 10 μm DADLE before freeze-thawing and radioligand binding assay using [3H]DPN. Data shown represent specific binding expressed as a percentage of binding in untreated cells (one-way ANOVA, Bonferroni multiple comparison test; ***, p < 0.001, n = 5). C, F-DOR expressing HEK293 cells were transfected with GFP, GFP-AMSH-D/A, or GFP-UBPY-C/S and, 48 h later, underwent cell surface biotinylation before incubation of cells for the indicated time period with 10 μm DADLE. Cells were lysed, and biotinylated protein was isolated with streptdavidin-agarose beads and resolved by SDS-PAGE. A representative anti-FLAG blot is shown. D, anti-FLAG blots were scanned to estimate the amount of FLAG-tagged receptor remaining at each time point after DADLE incubation relative to that detected in parallel samples of cells not exposed to opioid agonist. Results were pooled and averaged across multiple experiments (n = 4). Closed squares, results from cells expressing GFP; open squares, results from cells expressing GFP-AMSH-D/A; closed circles, results from cells expressing GFP-UBPY-C/S. E, F-DOR cells were transfected with siRNA duplexes against AMSH or UBPY and, 72 h later, assayed for DADLE-induced down-regulation by [3H]DPN binding. Results shown represent specific binding expressed as a percentage of binding in untreated cells (one-way ANOVA, Dunnett multiple comparison test; *, p < 0.05; **, p < 0.01, n = 7). F, knockdown was verified by lysing the remaining cells, resolving equal protein loads on NuPAGE 4–12% BisTris gel, and detecting endogenous UBPY (top) or AMSH (bottom) by immunoblotting. A representative Western blot is shown for each. G, F-DOR-expressing cells were transfected with GFP-AMSH-D/A (iii) or GFP-UBPY-C/S (vii) and replated onto coverslips and incubated for 2 h with 10 μm DADLE before being fixed and stained with rabbit-anti-FLAG (i and v) antibody and anti-LAMP1/2 (ii and vi). Shown are representative confocal optical sections imaged under nonsaturating conditions and rendered using simple background subtraction and a linear lookup table. Merged images and insets show receptor, LAMP, and the indicated DUB immunoreactivity pseudocolored in green, red, and blue, respectively (iv and viii). IB, immunoblot; RNAi, RNA interference.
DISCUSSION
The present results identify a specific function of AIP4-dependent ubiquitination in regulating the late proteolytic processing of GPCRs, which is clearly distinct from the previously defined function of ubiquitination by this ligase as a sorting determinant required for delivery of internalized receptors to lysosomes. We identified this function by study of a particular member of the GPCR family, DOR, which does not require ubiquitination for endocytic sorting to lysosomes yet traverses a similar endocytic pathway as ubiquitin-directed membrane cargo (26, 29). This feature of DOR trafficking was verified definitively in the present study, using several independent assays of lysosomal delivery and proteolysis and provided an advantageous system for identifying AIP4-dependent regulation of later proteolytic processing.
The specific regulatory effect of AIP4-dependent ubiquitination was manifest primarily by reduced proteolytic down-regulation of receptors detected by loss of binding to the small molecule radioligand [3H]diprenorphine, a ligand that binds to residues present in the transmembrane helices (39). Proteolysis monitored biochemically by epitope tagging, however, is sensitive to extramembrane cleavage and was largely unaffected by disrupting or depleting AIP4, suggesting that ubiquitination by this ligase regulates later proteolytic processing event(s) that mediate destruction of the receptor's hydrophobic core. Moreover, the present data indicate that down-regulation of the receptors indicated by radioligand binding can be almost completely dissociated from lysosomal delivery of receptors assessed by immunocytochemical localization and from extensive proteolytic fragmentation of extramembrane receptor domains by lysosomal proteases observed biochemically.
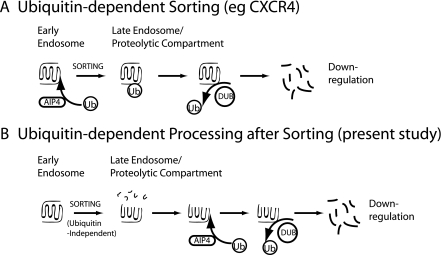
Perhaps the most striking observation from the present study is that ubiquitination of DOR regulates specific step(s) in the proteolytic processing pathway rather than being an absolute requirement for receptor down-regulation. Preventing all detectable ubiquitination of receptors by lysyl-mutation had little or no effect on ligand-induced endocytic trafficking of receptors to lysosomes detected by any of the assays, including down-regulation of radioligand binding (Fig. 1). Remarkably, disrupting AIP4 activity strongly inhibited pharmacological down-regulation of wild type DOR, whereas receptor ubiquitination was only partially reduced. This further supports a distinct regulatory function of DOR ubiquitination and suggests that ubiquitination of receptors is not simply a means to promote proteolysis but, under some conditions, can actually inhibit this process. The similar phenotype of disrupting membrane-associated DUB activities provides even more support for this idea. Interestingly, depleting AMSH or UBPY separately produced comparable effects on receptor down-regulation, and simultaneous knockdown of both DUBs failed to reveal additional effects. These data suggest that AMSH and UBPY are not redundant, and that they probably function at distinct stages in the same pathway of DOR trafficking. Altogether, the present findings support the hypothesis that later proteolytic processing of receptors is controlled by ubiquitination/deubiquitination cycling, involving multiple DUBs and perhaps multiple ligases, a possibility currently being investigated. Fig. 6 summarizes schematically the key features of the ubiquitination-dependent regulatory mechanism identified in the present study of DOR and contrasts this mechanism with the current model of ubiquitin-directed sorting as exemplified by the CXCR4 receptor elucidated previously.
FIGURE 6.
Proposed model for the postsorting function of AIP4 in controlling DOR down-regulation. A, depiction of the current view of AIP4-dependent regulation of GPCR proteolysis, based on previous studies of the CXCR4 receptor (3, 24). Ubiquitination acts as a sorting determinant that is required for delivery of internalized receptors to the late endosome/lysosome pathway, and receptor proteolysis follows. Receptors presumably undergo deubiquitination, as indicated, which is not specifically required for lysosomal sorting of receptors but is thought to occur generally after sorting is complete to prevent depletion of free cytoplasmic ubiquitin (Ub) pools (36). B, the currently proposed function of AIP4-dependent ubiquitination in regulating later proteolytic processing of DOR. Endocytic sorting of the DOR into the late endosome/lysosome pathway does not require receptor ubiquitination, in contrast to that of the CXCR4 receptor, as indicated by the ability of the lysyl-mutant DORs to undergo complete proteolytic destruction at a rate similar to that of wild type receptors. Wild type receptors are also sorted into the late endosome/lysosome pathway irrespective of their ubiquitination state, as indicated by LAMP1/2 colocalization and initial proteolytic fragmentation detected biochemically. Subsequent proteolytic processing required to destroy the hydrophobic core of the receptor is specifically regulated by both ubiquitination/deubiquitination, as indicated by the pronounced inhibition produced by disrupting either ubiquitin ligase (AIP4) or hydrolase (DUB) activity on down-regulation of wild type but not lysyl-mutant receptors detected by radioligand binding. The critical distinctions from model A are 1) that ubiquitination affects proteolytic processing of the wild type DOR clearly after receptor sorting to a lysosomal fate and the occurrence of initial proteolytic cleavage events, and 2) that later proteolytic processing of receptors is specifically regulated by both receptor ubiquitination and deubiquitination.
What physiological significance might ubiquitin-dependent regulation of later proteolytic processing have, considering that this distinct regulatory function is clearly not required for the efficient delivery of endocytosed receptors to lysosomes? One possibility is that lysosomes have a limited capacity to destroy complex polytopic membrane proteins, such as GPCRs, whose hydrophobic core is composed of seven tightly packed helical domains that are buried in the lipid bilayer, whereas extramembrane receptor domains are readily accessible to soluble proteases. Proteolytic destruction of GPCRs might be expected to occur asynchronously, therefore, with destruction of the hydrophobic core occurring relatively slowly. In this case, ubiquitination of receptors or ubiquitination/deubiquitination cycling might function as a biochemical timer to impose order on the sequence of proteolytic events or prevent saturation of slow step(s), thereby optimizing the processing of complex polytopic membrane proteins in lysosomes or preventing overload of the proteolytic potential of individual lysosomes.
Accordingly, important goals for future study are to define more precisely what event(s) in the later proteolytic processing of opioid receptors is controlled by ubiquitination/deubiquitination, to identify specific ubiquitin-binding proteins that function as effectors in regulating late proteolytic processing, and to determine if receptor ubiquitination/deubiquitination serves as a passive biochemical timer or plays a more active “checkpoint” role in sensing the occurrence of specific proteolytic events.
Because the ubiquitination-dependent regulatory function identified in the present study occurs effectively downstream of the critical endocytic sorting step, detecting such regulation of “ubiquitination-dependent” GPCRs would be difficult. Another important question for future study, therefore, is to determine whether the distinct regulatory function of ubiquitination on later proteolytic processing of DOR applies to other receptors. If ubiquitin-dependent regulation of proteolytic processing indeed functions to facilitate destruction of the seven-transmembrane helical bundle or to protect lysosomes from saturation, as we presently hypothesize, one might anticipate this mechanism to affect a wider range of GPCRs and perhaps other endocytic membrane cargo.
In closing, the present results reveal a distinct function of AIP4-mediated ubiquitination in regulating the later proteolytic processing of a mammalian GPCR by lysosomes and establish definitively that this ubiquitin-dependent regulation follows the molecular sorting decision that commits internalized receptors for lysosomal delivery. In addition to providing significant new insight to ubiquitin-dependent regulation of the largest known family of signaling receptors, our findings suggest that ubiquitination/deubiquitination cycling may be relevant more generally to proteolytic processing of diverse polytopic membrane proteins in the endocytic pathway.
Supplementary Material
Acknowledgments
We thank Laurant Coscoy and Brian Sullivan for generously allowing the use of the unpublished Nedd4-1, Nedd4-2, Smurf1, WWP1, and WWP2 and corresponding mutant constructs; Sylvie Urbé for the DUBs and the anti-AMSH antibody; and Wes Sundquist for the Smurf2, NEDL1, and NEDL2 constructs. We also thank Michael Tanowitz for valuable discussion.
This work was supported, in whole or in part, by National Institutes of Health, NIDA, Grants DA012864 and DA010711 (to M. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- GPCR
- G protein-coupled receptor
- DOR
- δ-opioid peptide receptor
- E3
- ubiquitin-protein isopeptide ligase
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- HRP
- horseradish peroxidase
- DADLE
- d-Ala-d-Leu-enkephalin
- DPN
- diprenorphine
- siRNA
- small interfering RNA
- ANOVA
- analysis of variance.
REFERENCES
- 1.Ferguson S. S. (2001) Pharmacol. Rev. 53, 1–24 [PubMed] [Google Scholar]
- 2.Hanyaloglu A. C., von Zastrow M. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 537–568 [DOI] [PubMed] [Google Scholar]
- 3.Marchese A., Paing M. M., Temple B. R., Trejo J. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 601–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shenoy S. K. (2007) Circ. Res. 100, 1142–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorkin A., Goh L. K. (2008) Exp. Cell Res. 314, 3093–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Law P. Y., Wong Y. H., Loh H. H. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 389–430 [DOI] [PubMed] [Google Scholar]
- 7.Law P. Y., Hom D. S., Loh H. H. (1984) J. Biol. Chem. 259, 4096–4104 [PubMed] [Google Scholar]
- 8.Tsao P., von Zastrow M. (2000) Curr. Opin. Neurobiol. 10, 365–369 [DOI] [PubMed] [Google Scholar]
- 9.Maxfield F. R., McGraw T. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 121–132 [DOI] [PubMed] [Google Scholar]
- 10.Vickery R. G., von Zastrow M. (1999) J. Cell Biol. 144, 31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsao P. I., von Zastrow M. (2000) J. Biol. Chem. 275, 11130–11140 [DOI] [PubMed] [Google Scholar]
- 12.Tanowitz M., von Zastrow M. (2003) J. Biol. Chem. 278, 45978–45986 [DOI] [PubMed] [Google Scholar]
- 13.Carman C. V., Benovic J. L. (1998) Curr. Opin. Neurobiol. 8, 335–344 [DOI] [PubMed] [Google Scholar]
- 14.Tsao P. I., von Zastrow M. (2001) Pharmacol. Ther. 89, 139–147 [DOI] [PubMed] [Google Scholar]
- 15.Hicke L. (1999) Trends Cell Biol. 9, 107–112 [DOI] [PubMed] [Google Scholar]
- 16.Urbé S. (2005) Essays Biochem. 41, 81–98 [DOI] [PubMed] [Google Scholar]
- 17.Raiborg C., Rusten T. E., Stenmark H. (2003) Curr. Opin. Cell Biol. 15, 446–455 [DOI] [PubMed] [Google Scholar]
- 18.Hicke L., Riezman H. (1996) Cell 84, 277–287 [DOI] [PubMed] [Google Scholar]
- 19.Katzmann D. J., Babst M., Emr S. D. (2001) Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 20.Katzmann D. J., Odorizzi G., Emr S. D. (2002) Nat. Rev. Mol. Cell Biol. 3, 893–905 [DOI] [PubMed] [Google Scholar]
- 21.Saksena S., Sun J., Chu T., Emr S. D. (2007) Trends Biochem. Sci. 32, 561–573 [DOI] [PubMed] [Google Scholar]
- 22.Russell M. R., Nickerson D. P., Odorizzi G. (2006) Curr. Opin. Cell Biol. 18, 422–428 [DOI] [PubMed] [Google Scholar]
- 23.Marchese A., Benovic J. L. (2001) J. Biol. Chem. 276, 45509–45512 [DOI] [PubMed] [Google Scholar]
- 24.Marchese A., Raiborg C., Santini F., Keen J. H., Stenmark H., Benovic J. L. (2003) Dev. Cell 5, 709–722 [DOI] [PubMed] [Google Scholar]
- 25.Bhandari D., Trejo J., Benovic J. L., Marchese A. (2007) J. Biol. Chem. 282, 36971–36979 [DOI] [PubMed] [Google Scholar]
- 26.Tanowitz M., Von Zastrow M. (2002) J. Biol. Chem. 277, 50219–50222 [DOI] [PubMed] [Google Scholar]
- 27.Whistler J. L., Enquist J., Marley A., Fong J., Gladher F., Tsuruda P., Murray S. R., Von Zastrow M. (2002) Science 297, 615–620 [DOI] [PubMed] [Google Scholar]
- 28.Simonin F., Karcher P., Boeuf J. J., Matifas A., Kieffer B. L. (2004) J. Neurochem. 89, 766–775 [DOI] [PubMed] [Google Scholar]
- 29.Hislop J. N., Marley A., Von Zastrow M. (2004) J. Biol. Chem. 279, 22522–22531 [DOI] [PubMed] [Google Scholar]
- 30.Petaja-Repo U. E., Hogue M., Laperriere A., Bhalla S., Walker P., Bouvier M. (2001) J. Biol. Chem. 276, 4416–4423 [DOI] [PubMed] [Google Scholar]
- 31.Petaja-Repo U. E., Hogue M., Laperriere A., Walker P., Bouvier M. (2000) J. Biol. Chem. 275, 13727–13736 [DOI] [PubMed] [Google Scholar]
- 32.Chaturvedi K., Bandari P., Chinen N., Howells R. D. (2001) J. Biol. Chem. 276, 12345–12355 [DOI] [PubMed] [Google Scholar]
- 33.Yadav P. N., Chaturvedi K., Howells R. D. (2007) J. Pharmacol. Exp. Ther. 320, 1186–1194 [DOI] [PubMed] [Google Scholar]
- 34.Chung H. Y., Morita E., von Schwedler U., Müller B., Kräusslich H. G., Sundquist W. I. (2008) J. Virol. 82, 4884–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCullough J., Clague M. J., Urbé S. (2004) J. Cell Biol. 166, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Row P. E., Prior I. A., McCullough J., Clague M. J., Urbé S. (2006) J. Biol. Chem. 281, 12618–12624 [DOI] [PubMed] [Google Scholar]
- 37.Chen L., Davis N. G. (2002) Traffic 3, 110–123 [DOI] [PubMed] [Google Scholar]
- 38.Befort K., Tabbara L., Bausch S., Chavkin C., Evans C., Kieffer B. (1996) Mol. Pharmacol. 49, 216–223 [PubMed] [Google Scholar]
- 39.Befort K., Tabbara L., Kling D., Maigret B., Kieffer B. L. (1996) J. Biol. Chem. 271, 10161–10168 [DOI] [PubMed] [Google Scholar]
- 40.Scherrer G., Tryoen-Tóth P., Filliol D., Matifas A., Laustriat D., Cao Y. Q., Basbaum A. I., Dierich A., Vonesh J. L., Gavériaux-Ruff C., Kieffer B. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9691–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mosberg H. I., Fowler C. B. (2002) J. Pept. Res. 60, 329–335 [DOI] [PubMed] [Google Scholar]
- 42.Shenoy S. K., McDonald P. H., Kohout T. A., Lefkowitz R. J. (2001) Science 294, 1307–1313 [DOI] [PubMed] [Google Scholar]
- 43.Jacob C., Cottrell G. S., Gehringer D., Schmidlin F., Grady E. F., Bunnett N. W. (2005) J. Biol. Chem. 280, 16076–16087 [DOI] [PubMed] [Google Scholar]
- 44.Duan L., Miura Y., Dimri M., Majumder B., Dodge I. L., Reddi A. L., Ghosh A., Fernandes N., Zhou P., Mullane-Robinson K., Rao N., Donoghue S., Rogers R. A., Bowtell D., Naramura M., Gu H., Band V., Band H. (2003) J. Biol. Chem. 278, 28950–28960 [DOI] [PubMed] [Google Scholar]
- 45.Levkowitz G., Waterman H., Ettenberg S. A., Katz M., Tsygankov A. Y., Alroy I., Lavi S., Iwai K., Reiss Y., Ciechanover A., Lipkowitz S., Yarden Y. (1999) Mol. Cell 4, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 46.Medina G., Pincetic A., Ehrlich L. S., Zhang Y., Tang Y., Leis J., Carter C. A. (2008) Virology 377, 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McNatt M. W., McKittrick I., West M., Odorizzi G. (2007) Mol. Biol. Cell 18, 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin-Serrano J., Perez-Caballero D., Bieniasz P. D. (2004) J. Virol. 78, 5554–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shenoy S. K., Xiao K., Venkataramanan V., Snyder P. M., Freedman N. J., Weissman A. M. (2008) J. Biol. Chem. 283, 22166–22176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staub O., Rotin D. (2006) Physiol. Rev. 86, 669–707 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.