Abstract
The Sulfolobus solfataricus protein acetyltransferase (PAT) acetylates ALBA, an abundant nonspecific DNA-binding protein, on Lys16 to reduce its DNA affinity, and the Sir2 deacetylase reverses the modification to cause transcriptional repression. This represents a “primitive” model for chromatin regulation analogous to histone modification in eukaryotes. We report the 1.84-Å crystal structure of PAT in complex with coenzyme A. The structure reveals homology to both prokaryotic GNAT acetyltransferases and eukaryotic histone acetyltransferases (HATs), with an additional “bent helix” proximal to the substrate binding site that might play an autoregulatory function. Investigation of active site mutants suggests that PAT does not use a single general base or acid residue for substrate deprotonation and product reprotonation, respectively, and that a diffusional step, such as substrate binding, may be rate-limiting. The catalytic efficiency of PAT toward ALBA is low relative to other acetyltransferases, suggesting that there may be better, unidentified substrates for PAT. The structural similarity of PAT to eukaryotic HATs combined with its conserved role in chromatin regulation suggests that PAT is evolutionarily related to the eukaryotic HATs.
Sulfolobus solfataricus, a thermoacidophile, is a member of the archaeal domain of life, and is likely to have diverged from bacteria and eukaryotes early during evolution. Despite its lack of a nucleus or other organelles, archaeal DNA replication and chromatin regulation seem to more closely resemble eukaryotes than bacteria (1, 2). Sulfolobus belongs to the phylum Crenarchaeota, which lacks histones, and instead uses two analogous chromatin proteins: Sul7d and ALBA3 (acetylation lowers binding affinity). Both proteins have been shown to undergo post-translational modification in Sulfolobus. Sul7d is monomethylated (3) and ALBA is acetylated (4, 5). The acetylation of ALBA by protein acetyltransferase (PAT) on Lys16 has been shown to reduce DNA-binding affinity, and deacetylation of ALBA by archaeal Sir2 deacetylase has been shown to repress transcription in what appears to be a primitive form of chromatin regulation by reversible post-translational modification (4, 5). PAT is also likely to regulate other proteins in Sulfolobus. Based on its homology to PAT from Salmonella enterica, PAT from Sulfolobus may also play a role in metabolism by regulating the activity of acetyl-coenzyme A synthetase (6).
There are at least four families of histone acetyltransferases (HATs) in eukaryotes: the Gcn5/PCAF family that also shows sequence and structural homology to the GNAT (Gcn5-related acetyltransferase) superfamily, which includes many small molecule acetyltransferases such as antibiotic acetyltransferases (aminoglycoside N-acetyltransferases) and serotonin N-acetyltransferase; the MYST family, named from the founding members of MOZ, Ybf2/Sas3, Sas2, and Tip60; the metazoan-specific transcriptional coactivators p300 and CREB-binding protein; and the recently characterized fungal-specific Rtt109 (regulator of Ty1 transposition gene product 109). Recent structures of p300 and Rtt109 reveal that these seemingly unrelated HAT families share considerable structural homology throughout the HAT domain, even in the absence of sequence homology (7, 8). Strikingly, each of the HAT families contains a homologous acetyl-CoA binding core segment. This raises the possibility that the eukaryotic acetyltransferase families may have evolved from a common ancestral or “primordial” scaffold.
Efforts to characterize the diversity in structure, mechanism, and substrate selectivity derived from a conserved acetyl-CoA binding scaffold by the different families of HATs are ongoing and the topic is covered in depth in recent reviews (9–11). Kinetic analysis combined with structural information has revealed significant diversity in the mechanism and catalytic residues employed in the reaction. For some HATs the rate-determining step is the deprotonation of the incoming substrate lysine to activate it for direct nucleophilic attack on the acetyl-CoA in an Ordered Bi Bi ternary complex mechanism. It has been demonstrated for Gcn5 that a mutation of the conserved general base glutamate residue (173 in Saccharomyces cerevisiae) to glutamine leads to a 320-fold reduction in activity (12). Other acetyltransferases do not appear to depend on a single general base residue. An investigation of the catalytic residues in serotonin acetyltransferase found that His120 and His122 had redundant roles as the general base (13). Some ambiguity remains about whether all acetyltransferases use a ternary complex mechanism. A ping-pong mechanism, in which the acetyl group is temporarily transferred to a nucleophilic side chain in the active site to form an acetyl-enzyme intermediate has been proposed for yeast Esa1 (14); however, a more recent investigation of Esa1 activity in complex with other subunits reports data that supports a ternary complex mechanism (15), suggesting that other associated protein factors might influence HAT activity. Structural and enzymatic studies on the p300/CBP HAT reveals that it employs a conserved tryptophan and tyrosine for catalysis with the tyrosine likely functioning as a general acid with no key general base residue and a Theorell-Chance Bi Bi ternary complex mechanism (7). Finally, recent enzymatic studies on the Rtt109 histone acetyltransferase reveals a more complex reaction mechanism that does not follow Michaelis-Menten kinetics and key general acid and base residues have not yet been identified (8). Together, the picture that emerges is that the four histone acetyltransferase families have evolved to use a conserved templating structural scaffold to mediate acetyl transfer, albeit through different chemical strategies.
The regulation of chromatin through acetylation and deacetylation of ALBA in Sulfolobus is a valuable paradigm for understanding the origins of gene regulation by HATs in eukaryotes. The structure of ALBA from Sulfolobus has been reported (16). Structures and biochemical characterization of the archaeal Sir2 deacetylase have also been carried out (17, 18). Here we report the structure of PAT from S. solfataricus to complete the structural basis for this model system. We then report the results of mutational analysis to probe the mechanism and substrate selectivity of PAT, and we discuss the similarities and differences between PAT and the more complex eukaryotic HATs.
EXPERIMENTAL PROCEDURES
PAT Cloning, Expression, and Purification
The gene encoding PAT from S. solfataricus, amino acids 1–160, was cloned into a pET-28a vector using NdeI and XhoI restriction sites. Overnight expression at 18 °C in BL21(DE3) (Novagen) yields PAT with a thrombin cleavable His6 tag. Following lysis and nickel-nitrilotriacetic acid purification, the histidine tag was removed by incubation with thrombin protease at 4 °C overnight. The cleavage product was then further purified by Superdex-200 size exclusion chromatography in 25 mm Tris-HCl, pH 7.5, 200 mm NaCl buffer. The protein was finally concentrated to 35 mg/ml and stored at 4 °C until use. A plasmid expressing glutathione S-transferase-tagged ALBA protein was a generous gift of Professor Malcolm White and ALBA was expressed and purified as described previously (19). Protein mutants were generated by site-directed mutagenesis based on the QuikChange protocol from Stratagene (20). The point mutants were purified as described above. A selenomethionine derivative of PAT was prepared by expressing PAT in a methionine auxotroph strain of Escherichia coli, B834 (Novagen), in minimal medium (Molecular Dimensions, Ltd.) containing 50 μg/liter of selenomethionine (Sigma) and was purified as described above.
Crystallization and Data Collection
A PAT-coenzyme A complex for crystallization was prepared by mixing concentrated PAT, with concentrated coenzyme A, in a 1:2 molar ratio. The final concentration of PAT for crystallization was 30 mg/ml. Crystals of PAT-CoA were grown by hanging drop vapor diffusion in 20 days at 20 °C using a well solution containing 0.1 m MES, pH 6.5, and 12% PEG 20,000. Crystals were cryoprotected using well solution supplemented with 30% glycerol. A three wavelength MAD dataset was collected at beamline 23ID-D of the Advanced Photon Source. Data were processed using HKL2000 (21).
Structure Determination and Refinement
The three wavelength anomalous signal (MAD) was used to find three selenium sites in the asymmetric unit using Solve and Resolve (22, 23). Model building was carried out with Coot (24) and refinement done with CNS (25). The model was initially refined with simulated annealing, energy minimization and group B-factor refinement. For later stages of refinement, solvent molecules were added to the model and individual atomic B-factors were refined. The final model was checked for errors against a simulated annealing omit map. Refinement of the structure resulted in a model with excellent statistics and geometries (Table 1). Figures were prepared using PyMOL (DeLano Scientific, Palo Alto, CA) and CCP4mg (26).
TABLE 1.
Data collection, phasing, and refinement statistics for the PAT·CoA complex
Values in parentheses are for the highest resolution shell. Rfree was calculated using 10% of the reflection data chosen randomly and omitted at the start of refinement.
| Data statistics | Se Peak | Inflection | Remote |
|---|---|---|---|
| Space group | P212121 | ||
| Cell a (α) | 44.701 (90) | ||
| Cell b (β) | 46.750 (90) | ||
| Cell c (γ) | 68.624 (90) | ||
| Wavelength (Å) | 0.97932 | 0.97945 | 0.94932 |
| Resolution (Å) | 50-1.84 | 50-1.84 | 50-1.84 |
| Unique reflections | 23,578 | 23,316 | 23,521 |
| Completeness (%) | 98.4 (86.4) | 97.9 (81.8) | 98.6 (89.2) |
| Multiplicity | 6.1 | 6.1 | 5.9 |
| I/σ | 25.0 (2.7) | 22.6 (2.2) | 23.1 (2.5) |
| Rmerge (%) | 6.4 (39.4) | 6.8 (45.8) | 6.8 (42.3) |
| Phasing (solve) | |||
| Figure of merit | 0.62 | ||
| Z-score | 14.0 | ||
| Refinement (CNS) | |||
| Resolution (Å) | 1.84 | ||
| Rfree | 22.1 | ||
| Rwork | 20.2 | ||
| Bond length root mean square deviation (Å) | 0.006 | ||
| Bond angle root mean square deviation (°) | 1.15 | ||
| Average B-factor (Å2) | |||
| All atoms | 27.5 | ||
| Protein | 26.6 | ||
| Coenzyme A | 27.8 | ||
| Water | 40.3 | ||
| Ramachandran plot | |||
| Most favored | 94.4% | ||
| Allowed | 4.6% | ||
| Generously allowed | 0.0% | ||
| Disallowed | 0.9% | ||
PAT Acetyltransfer Assays
PAT assays were carried out with 1 μm PAT at 75 °C for 1 h in 25 mm Tris-HCl, pH 8.0, 50 mm NaCl, and 0.01% bovine serum albumin buffer. For comparison of wild type and mutant PAT activities, concentrations of 833 μm acetyl-CoA and 3 mm ALBA 11-mer peptide, with the sequence VLIGKKPVMNY, were used. Radiolabled [14C]acetyl-coenzyme A (4 mCi/mmol) from PerkinElmer Life Sciences was used to measure the formation of acetylated product. Following 75 °C incubation, 30-μl reaction mixtures were cooled to 4 °C to quench the reaction, 4 μl of 1 m HEPES pH 7.5 was added to adjust the pH, and 20 μl of the reaction mixture was bound to P81 paper (Whatman). The paper disks were washed three times for 5 min for each wash, with 10 mm HEPES pH 7.5 to remove unreacted acetyl-CoA, and then dried with acetone. Scintillation fluid was added and signal was measured using a Packard Tri-Carb 1500 liquid scintillation analyzer. The values derived from a reaction lacking PAT and from a reaction lacking substrate were subtracted from the counts to account for both background acetyl transfer in the absence of enzyme and possible autoacetylation of PAT. Substrate Km values were determined by titrating substrate with acetyl-CoA concentration fixed at a saturating concentration of 500 μm. Counts were converted to rate using a standard curve, and data were fit to a one-site binding equation in Graph Pad Prism software. Acetyl-CoA Km values were determined by titrating acetyl-CoA with peptide substrate fixed at a saturating concentration of 3000 μm. All experiments were done at least in duplicate. The pH rate profile was determined using a three-component buffer as described previously (12). The pH values were corrected to account for the change in pH at 75 °C. The peptides used in this study were C-terminal amidated and were greater than 95% pure. Peptides were obtained from GenScript (Piscataway, NJ), except for the histone H4 peptide (sequence: GKGGAKRHRKI), which was provided by Santosh Hodawadekar.
RESULTS
Overall Structure
Full-length PAT from S. solfataricus, amino acids 1–160, was initially crystallized following 3 weeks of incubation by hanging drop vapor diffusion. Initial attempts to reproduce the crystals failed and it was eventually found that the crystallized form of PAT had degraded into two chains with sizes of ∼5 and 11 kDa, as confirmed by SDS-PAGE (supplemental Fig. S1). It is unclear whether the degradation resulted from contamination or occurred spontaneously; however, we found that limited proteolysis with V8 protease from Staphylococcus aureus accelerated formation of the identical crystal form to 3 days. The structure of PAT was determined by MAD from crystals of selenomethionine-derivatized protein, and most of the protein could be readily traced into the experimental electron density map. There was no observable density corresponding to internal residues 42–52 and C-terminal residues 147–160 in the structure. The two protein chains resulting from these interruptions in the sequence are consistent with the 5- and 11-kDa fragments observed in the washed crystals. Repeated attempts to obtain crystals of full-length PAT or a PAT-(1–146) construct were unsuccessful, suggesting that residues 42–52 and 147–160 are flexible, susceptible to proteolysis, and inhibit crystallization.
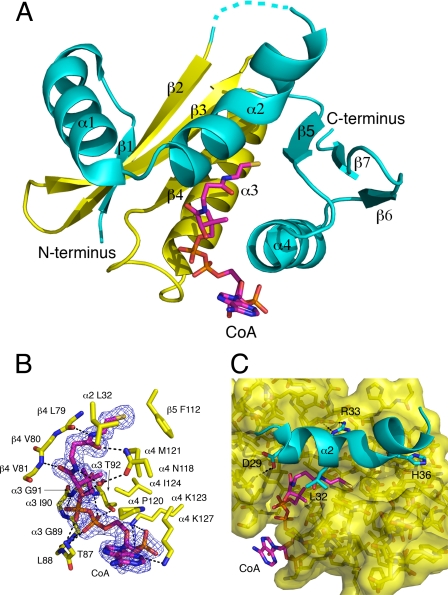
The 1.84-Å structure of PAT (Table 1 and Fig. 1A) reveals amino acids 5–41 and 53–146 making up a mixed α/β-fold, with a structurally conserved acetyl-CoA binding core region made up of three β-strands (β2–4) and one α-helix (α4) that is characteristic of GNATs and other HATs. The pantetheine group of the CoA makes β-strand mimicking hydrogen bonds with β4, and the N terminus of helix α4 is capped by a P-loop motif found in many nucleotide-binding proteins, which binds the pyrophosphate moiety of the CoA using backbone hydrogen bonds from Thr87, Leu88, Gly89, Gly91, and Thr92 (Fig. 1B). In addition to interactions with the conserved acetyl-CoA binding core region, the side chain of Asn118 makes a hydrogen bond to O5 of the pantetheine group, Lys127 forms a salt bridge with the 3′ phosphate of CoA, and Lys123 hydrogen bonds to N3 of the adenine moiety of CoA (Fig. 1B). This last interaction combined with hydrophobic stacking against Pro120 likely accounts for the observation that the adenine moiety is well ordered in the crystal structure. Many previously reported acetyltransferase structures, including the Gcn5/PCAF and Esa1 HATs, show poor density for the adenine due to its conformational flexibility.
FIGURE 1.
Structure of the PAT·CoA complex. A, overall structure of the PAT·CoA complex. The conserved acetyl-CoA binding core region is in yellow, less conserved segments are colored in cyan, and CoA is colored by element. B, PAT-CoA interactions. The electron density is from a simulated annealing omit map contoured at 1.0 σ around the CoA. C, representation of the bent helix (α2) in cyan and the interactions that anchor it proximal to the active site of PAT. CoA is colored by element.
An unusual feature of the PAT structure is the presence of a “bent helix,” from residues 32–41 (α2), which sits in a position that is proximal to where protein substrate is likely to bind (Fig. 1C). The α2 helix is held in place by a series of salt bridges between Asp29 and Arg83/Arg86, Arg33 and Glu68, and His36 and Glu76, and van der Waals interactions between Leu32 and Leu115. Although it is possible that the proteolysis of residues 42–52 may have led to the helix assuming a non-native conformation, we postulate that the α2 helix may play some dynamic autoregulatory function in PAT activity. To investigate this possibility we mutated side chains that formed salt bridges to hold the helix in its observed conformation and compared their activity to wild type PAT (see below).
Structural Relationship to Other Acetyltransferases
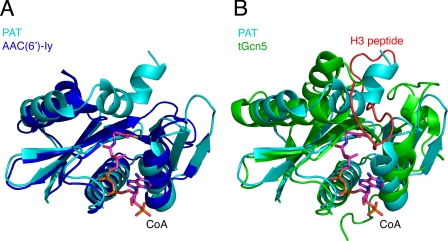
We found that roughly 100 residues of PAT aligned well with both aminoglycoside N-acetyltransferases (AAC) (Fig. 2A and Table 2), serotonin acetyltransferase, and Gcn5/PCAF (Fig. 2B and Table 2) HAT structures, in addition to showing significant but lower structural similarity to the larger eukaryotic Esa1, p300, and Rtt109 acetyltransferases (Table 2). Specifically, the root mean square deviation of Cα atoms ranged from 2 to 3 Å for each of these enzymes. Interestingly, the sequence identity of more than 20% is greatest for the AAC(6′)-Iy and Gcn5 acetyltransferases, whereas sequence identity with the other acetyltransferases ranges from 9 to 18%. This structural similarity in the face of limited sequence identity highlights the evolutionary conservation of the acetyltransferase enzyme fold that mediates gene regulation and small molecule metabolism. This observation also raises the possibility that PAT serves as both a small molecule and protein acetyltransferase for Sulfolobus. Dual activity of a GNAT toward both aminoglycoside antibiotics and histones has been reported for aminoglycoside 6′-N-acetyltransferase (AAC(6′)-Iy) from S. enterica; however, the physiological substrates of AAC(6′)-Iy are not known (27).
FIGURE 2.
Structural comparison of PAT to other acetyltransferases. A, PAT·CoA complex aligned with S. enterica AAC(6′)-Iy (Protein Data Bank code 1S5K). B, PAT·CoA complex aligned with a tetrahymena Gcn5 ternary complex (PDB code 1PUA) illustrating overlap between the Gcn5 substrate peptide (red) and helix α2 in the active site of PAT.
TABLE 2.
Structure and sequence alignment for the acetyl-CoA binding core region and the full-length acetyltransferase domain of PAT with other acetyltransferases
Aminoglycoside 6′-N-acetyltransferase from S. enterica (AAC(6′)-Iy, PDB code 1S5K), aminoglycoside 2′-N-acetyltransferase from Mycobacterium tuberculosis (AAC(2′)-Ic, PDB code 1M4I), serotonin N-acetyltransferase from Ovis aries (sheep) (AANAT, PDB code 1CJW), Gcn5 from Tetrahymena (PDB code 1PUA), Esa1 from S. cerevisiae (PDB code 1FY7), human p300 (PDB code 3BIY), and Rtt109 from S. cerevisiae (PDB code 3D35).
| Comparison | Acetyl-CoA binding core |
Full acetyltransferase domain |
||||
|---|---|---|---|---|---|---|
| Residues aligned | Cα root mean square deviation | Sequence identity | Residues aligned | Cα root mean square deviation | Sequence identity | |
| Å | % | Å | % | |||
| PAT versus AAC(6′)-Iy | 52/56 | 1.0 | 19.2 | 101/153 | 1.9 | 20.8 |
| PAT versus AAC(2′)-Ic | 52/65 | 1.1 | 13.5 | 112/181 | 1.9 | 10.7 |
| PAT versus AANAT | 51/53 | 1.3 | 21.6 | 112/166 | 2.1 | 17.9 |
| PAT versus Gcn5 | 51/58 | 1.4 | 23.5 | 110/162 | 2.4 | 21.8 |
| PAT versus Esa1 | 50/61 | 1.7 | 22.0 | 92/273 | 2.5 | 15.2 |
| PAT versus p300 | 50/77 | 2.2 | 10.0 | 103/317 | 2.6 | 12.6 |
| PAT versus Rtt109 | 37/70 | 2.5 | 8.1 | 108/355 | 3.0 | 9.3 |
Active Site Mutants and Insights into Catalysis
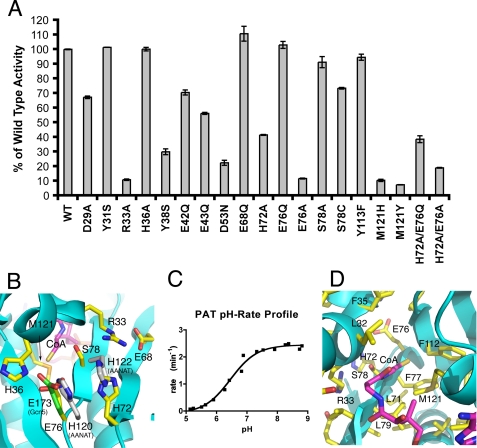
To investigate the catalytic properties of PAT, a total of 19 mutants were made and their steady-state activity at 75 °C was measured using a 14C-based assay (Fig. 3A). The mutants fall into five categories. The D29A, R33A, and H36A mutants were made to disrupt salt bridges that hold the putative α2 autoregulatory helix within the substrate binding site. The Y31S, Y38S, E42Q, E43Q, D53N, E68Q, H72A, E76Q, Y113F, H72A/E76Q, and H72A/E76A mutants were made to try to identify one or more general base residues required for deprotonation of the incoming substrate lysine. The S78A and S78C mutants were made to explore what role the hydroxyl of Ser78 might play in substrate binding or if it could form an acetylserine/acetylcysteine intermediate indicative of a ping-pong mechanism. The M121H and M121Y mutants were made to investigate the effect of having a non-polar versus polar side chain in the active site. Last, the E76A mutant was made to compare with E76Q and investigate the role of Glu76 in hydrogen bonding with incoming substrate rather than acting as a general base.
FIGURE 3.
The PAT active site. A, steady-state kinetic analysis of PAT mutants. Assays were done in duplicate with 833 μm [14C]acetyl-coenzyme A and 3 mm ALBA peptide. Bars represent the activity of each mutant measured in counts and plotted as a percent of wild type PAT counts. Error bars represent the range for the two measurements. B, the putative substrate binding face of PAT showing some of the residues mutated. The position of the general bases of yeast Gcn5 (green, Glu173 from PDB code 1YGH) and AANAT (gray, His120 and His122 from PDB code 1CJW) are shown following superposition of the structures with the PAT·CoA complex. C, pH-rate profile for PAT. Rates represent the mean of two measurements. The pH of each assay buffer was measured at 75 °C to account for the shift in pH at high temperature. D, the active site of PAT highlighting hydrophobic residues Leu79, Phe112, and Met121 around the sulfur of CoA.
As summarized in Fig. 3A, the α2 mutant H36A shows activity similar to the wild type enzyme, whereas the α2 mutants D29A and R33A show a reduction in activity by about 2-fold for D29A and a reduction to near background levels for R33A. A more detailed kinetic analysis of the R33A mutant reveals that the Km for Ac-CoA is similar to wild type and elevated by about 2-fold for protein substrate, whereas the overall kcat is reduced about 5-fold relative to the wild type protein (Table 3). Taken together, this data are consistent with a role of the α2 helix in facilitating protein substrate acetylation, and inconsistent with a role in autoinhibition, as might be predicted from its position in the structure, although it is possible that the α2 helix might have an autoinhibitory role for non-cognate substrates. The α2 helix therefore likely plays a dynamic role in facilitating PAT activity on cognate substrates.
TABLE 3.
Bisubstrate kinetic analysis of wild type PAT compared with the R33A, E76A, H72A/E76Q, M121Y, and M121H mutants
Values represent the mean of two measurements. Km values are rounded to 2 significant figures. The values for each of the two measurements are shown in parentheses.
| PAT mutants | kcat | ALBA 11-mer Km | Acetyl-CoA Km | kcat/Km peptide |
|---|---|---|---|---|
| min−1 | μm | m−1min−1 | ||
| Wild type | 2.31 (2.36, 2.25) | 580 (592, 565) | 48 (44, 52) | 4.0 × 103 |
| R33A | 0.50 (0.51, 0.50) | 1200 (1194, 1266) | 37 (36, 38) | 4.2 × 102 |
| E76A | 0.40 (0.40, 0.39) | 940 (1062, 825) | 59 (54, 64) | 4.3 × 102 |
| H72A/E76Q | 1.32 (1.25, 1.39) | 910 (800, 1022) | 39 (40, 38) | 1.5 × 103 |
| M121Y | 0.40 (0.44, 0.36) | 1300 (1525, 1057) | 50 (49, 50) | 3.1 × 102 |
| M121H | 0.55 (0.57, 0.54) | 1200 (1253, 1071) | 48 (50, 45) | 4.6 × 102 |
If PAT relies on a single general base residue for catalysis, we would expect that mutation of that residue to have a significant effect on PAT activity, as seen with mutation of the glutamate general base residue in Gcn5 (12) and Esa1 (14). As seen in Fig. 3A, mutation of Glu76 to Gln, corresponding in three-dimensional space to the general base Glu173 residue of yeast Gcn5 (Fig. 3B), had no effect on activity. We also carried out LC-MS/MS of trypsin-digested PAT to confirm the identity and stability of the E76Q mutant to heating during enzymatic analysis. Also, mutation of His72 to Ala, corresponding in three-dimensional space to one of two histidine residues in AANAT that function as a general base (Fig. 3B), had about a 2-fold effect on PAT activity suggesting that it is not a required catalytic residue. Mutants of other polar residues near the active site that could play a catalytic role (His36, Tyr38, Glu42, Glu43, Asp53, Glu68, Ser78, and Tyr113) still exhibited significant acetyltransferase activity on ALBA peptide substrate. Together, these mutational studies suggest that PAT does not use a single general base side chain for substrate deprotonation. To investigate if His72 and Glu76 have a redundant role in substrate deprotonation, similar to two histidines in AANAT (Fig. 3B), the H72A/E76Q mutant was tested and still found to have significant activity. These findings suggest that PAT either relies on another strategy for deprotonating the substrate lysine, or substrate deprotonation may not be the rate-determining step. The modest reduction in activity for the Y38S, E42Q, E43Q, D53N, and H72A mutants suggests that they may be acting as a proton wire to shuttle protons out of the active site.
Ser78 is the nearest polar residue to the sulfhydryl of CoA in the structure and is the only candidate nucleophile to form an acetyl-enzyme intermediate if PAT utilizes a ping-pong mechanism for catalysis. To investigate if Ser78 is important for catalysis, S78A and S78C mutants were made. The activity for both mutants was close to wild type PAT (Fig. 3A) suggesting that Ser78 is not important for catalysis and ruling out the possibility that PAT might use a ping-pong mechanism for catalysis.
A pH/rate profile was carried out to identify the optimal pH for catalysis and perhaps identify a titratable residue that is utilized in the mechanism. An inflection was observed at pH 6.5 in the profile with the rate reaching a plateau at pH 7.5 (Fig. 3C). Because none of the general base candidate residues were found to be essential for activity, this pH inflection could represent the sum of contributions from several residues. Alternatively, the inflection could represent the pKa of the incoming substrate lysine, which has been significantly reduced from its typical pKa of 10.5 due to the significant hydrophobicity of the active site (Fig. 3D). The pocket surrounding the sulfur atom of acetyl-CoA is comprised of the hydrophobic side chains of Leu79, Phe112, and Met121. It has been demonstrated that the energetic barrier to putting a charged residue, such as a substrate lysine, in a hydrophobic pocket can significantly alter the pKa of the residue (28, 29). As an interesting alternative to using a general base for catalysis, this hydrophobic pocket of PAT may lower the pKa of the incoming substrate lysine or select for deprotonated substrate that is capable of nucleophilic attack on the acetyl-CoA. Consistent with this hypothesis, mutation of Met121 to His or Tyr has the most significant effect on catalysis by PAT (Fig. 3A) resulting in about a 2-fold increase in protein substrate Km and about a 5-fold decrease in overall kcat, despite little change in the Km for acetyl-CoA (Table 3).
Protein Substrate Binding and Selectivity by PAT
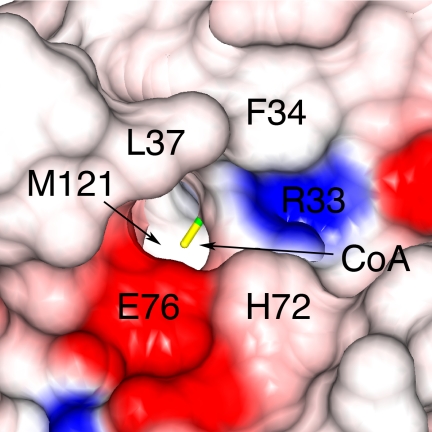
An analysis of the electrostatic surface of PAT around the active site reveals what appears to be a substrate binding surface with a small tunnel leading to the CoA (Fig. 4). This suggested that the lowered activity for the H72A and R33A mutants may be due to steric changes to the substrate binding surface or elimination of substrate orienting hydrogen bonds rather than removal of a catalytic residue. To further investigate this putative substrate-binding surface, E76A and H72A/E76A mutants were made and activity was found to be reduced to near background levels (Fig. 3A and Table 3). The observation that E76Q had wild type activity suggests that Glu76 may have an important role in substrate binding rather than performing a chemical step in catalysis. Moreover, the correlation between lowered kcat and increased substrate Km observed for the debilitating PAT mutants (Table 3) suggests that a diffusional step such as substrate binding or CoA product dissociation, not substrate deprotonation, may be the rate-determining step under these conditions.
FIGURE 4.
The putative substrate binding surface of PAT. Access to the active site is shown in a surface representation with the electrostatic surface prepared using CCP4mg. Red represents negative charge and blue represents positive charge at physiological pH.
To investigate whether PAT shows a preference for ALBA substrate, and if specific residues adjacent to Lys16 of ALBA form a substrate recognition sequence, kinetic analysis was done to compare ALBA peptide substrate with full-length ALBA protein. PAT showed an ∼5-fold lower Km for full-length substrate versus ALBA 11-mer peptide (Table 4). This suggests that surfaces or specific side chains distal to Lys16 of ALBA may be recognized by PAT. It has been proposed that P(X)4–7GK may be a substrate recognition sequence for PAT (5). To test this hypothesis we prepared P6A, P8A, and G15A mutants of ALBA for comparison with wild type ALBA protein (Table 4). These ALBA mutants were found to have Km values similar to wild type ALBA (Table 4), thus revealing that these residues do not play a significant role of PAT acetylation of ALBA. Taken together, these findings suggest a modest selectivity for full-length ALBA, possibly due to a preference for the conformation of Lys16 in the folded substrate. A substrate Km of 107 μm suggests weak binding compared with the Gcn5 Km of 28 μm for histones, or the p300 Km of 12.5 μm for histone H4 peptide (7, 12).
TABLE 4.
Kinetic analysis of PAT substrates
Values represent the mean of two measurements. Km values are rounded to 2 significant figures. The values for each of the two measurements are shown in parentheses.
| Substrate | kcat | Substrate Km | kcat/Km |
|---|---|---|---|
| min−1 | μm | m−1min−1 | |
| Full-length Alba SSO | 2.31 (2.36, 2.25) | 110 (116, 99) | 2.1 × 104 |
| Full-length P6A ALBA SSO | 2.41 (2.37, 2.45) | 160 (161, 155) | 1.5 × 104 |
| Full-length P8A ALBA SSO | 2.81 (2.84, 2.78) | 100 (96, 104) | 2.8 × 104 |
| Full-length G15A ALBA SSO | 2.43 (2.44, 2.42) | 86 (87, 85) | 2.8 × 104 |
| ALBA SSO 11-mer | 2.31 (2.36, 2.25) | 580 (592, 565) | 4.0 × 103 |
| ALBA SSO K16R 11-mer | 0.78 (0.77, 0.78) | 1300 (1243, 1412) | 6.0 × 102 |
| ALBA SSO 21-mer | NDa | >1500 | ND |
| Histone H4 peptide | 4.31 (4.30, 4.32) | 220 (213, 218) | 2.0 × 104 |
a ND, not determined.
The observation that PAT achieves a very modest kcat/Km of just 2.1 × 104 m−1 min−1 toward full-length ALBA substrate at 75 °C also suggests that it is a relatively inefficient enzyme, consistent with the absence of key catalytic residues that mediate catalysis. It may also be possible that better PAT substrates may exist that have not yet been identified, or that PAT may associate with activating subunits to achieve increased turnover or greater substrate selectivity similar to recent reports that yeast Rtt109 requires a histone chaperone protein, Vps75 or Asf1, for activity (30–33).
DISCUSSION
The finding that post-translational histone modifications effect chromatin structure and gene expression has lead to remarkable advances in our understanding of eukaryotic gene expression and how misregulation of gene expression can cause disease. Less is understood about the analogous mechanisms used by lower organisms to regulate chromatin structure and there seems to be an evolutionary gap in our understanding of the origins of chromatin regulation. PAT from S. solfataricus is a GNAT acetyltransferase from a lower organism that is reported to regulate chromatin-like structures through reversible acetylation of Lys16 of ALBA, and a better understanding of the structure and activity of PAT provides new insights into protein acetylation as a mechanism for regulation of gene expression.
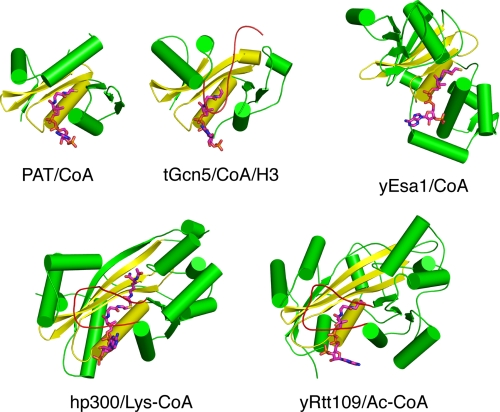
A comparison of PAT with eukaryotic histone acetyltransferases suggests that these proteins have in common a structurally conserved core region that provides functional conservation for acetyl-CoA binding and a structural framework for catalysis (Fig. 5). Based on this structural similarity, it is tempting to speculate that PAT represents a minimal scaffold similar to the original HAT; however, there is no direct evidence of this and considerable evolution has likely occurred since archaea and eukaryotes branched from a common ancestor. Its similarity to other GNATs reported to acetylate small molecules, such as aminoglycosides and serotonin, suggests that PAT could also have unidentified small molecule substrates in Sulfolobus.
FIGURE 5.
Comparison of PAT with eukaryotic HATs. The HATs Gcn5, Esa1, p300, and Rtt109 are shown. Their shared CoA binding core is highlighted in yellow with the surrounding structures shown in green.
Despite the structural conservation between histone acetyltransferases, these enzymes have evolved different chemical strategies for mediating acetylation. This is likely facilitated by the relative simplicity of carrying out an acetyltransferase reaction thus leading to several different chemical strategies, probably tailored to the different substrate and biological requirements of the particular enzyme. PAT appears to be the least efficient acetyltransferase, catalyzing only about 5-fold faster acetyl transfer compared with the uncatalyzed reaction at 75 °C and PAT is nearly completely inactive at room temperature (data not shown). PAT is likely to achieve a higher turnover at even higher temperatures consistent with the environment of Sulfolobus. It is also possible that better, yet unidentified PAT substrates exists. It could also suggest that PAT represents a primitive acetyltransferase scaffold upon which additional chemical groups evolved to stimulate the reaction rate. The poor turnover is consistent with the finding that PAT does not appear to use essential catalytic residues for acetylation. The putative role of the α2 helix of PAT in autoregulating PAT activity might also represent an evolutionary connection to the autoregulation of the fungal Rtt109 and metazoan p300/CBP proteins by acetylation. Another interesting connection between PAT and p300 is that the PAT Lys127 salt bridge with the 3′-phosphate of CoA that orders the adenine base of the cofactor is analogous to a similar CoA contact mediated by Arg1410 of p300. Taken together, the PAT-CoA structure reported here reveals a small and inefficient protein acetyltransferase scaffold with distinct similarities and differences to the larger and more selective eukaryotic acetyltransferases.
Supplementary Material
Acknowledgments
We thank Xin Liu, Yong Tang, Dario Segura, Santosh Hodawadekar, Chris Lanci, Mary Fitzgerald, Jasna Maksimoska, Brandi Sanders, and Cheng Luo for advice on the structure solution, development of the acetyltransfer assay, and help with making figures. We thank Kaye Speicher, Tom Beer, and David Speicher of the Wistar Institute Proteomics Core Facility for in-gel digestion, LC-MS/MS, data base searching, and analysis of the unheated and heat-treated wild type and E76Q PAT mutant. We also thank the staff of the GM/CA-CAT beamline 23ID-D of the Advanced Photon Source for beam time and assistance with data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant GM060293 (to R. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (code 3F8K) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ALBA
- acetylation lowers binding affinity
- PAT
- protein acetyltransferase
- HAT
- histone acetyltransferase
- GNAT
- Gcn5-related N-acetyltransferase
- CoA
- coenzyme A
- MAD
- multiple anomalous dispersion
- CREB
- cAMP-response element-binding protein
- MES
- 4-morpholineethanesulfonic acid
- AAC
- aminoglycoside N-acetyltransferase
- LC-MS
- light chromatography-mass spectrometry.
REFERENCES
- 1.Barry E. R., Bell S. D. (2006) Microbiol. Mol. Biol. Rev. 70, 876–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandman K., Reeve J. N. (2005) Curr. Opin. Microbiol. 8, 656–661 [DOI] [PubMed] [Google Scholar]
- 3.Edmondson S. P., Shriver J. W. (2001) Methods Enzymol. 334, 129–145 [DOI] [PubMed] [Google Scholar]
- 4.Bell S. D., Botting C. H., Wardleworth B. N., Jackson S. P., White M. F. (2002) Science 296, 148–151 [DOI] [PubMed] [Google Scholar]
- 5.Marsh V. L., Peak-Chew S. Y., Bell S. D. (2005) J. Biol. Chem. 280, 21122–21128 [DOI] [PubMed] [Google Scholar]
- 6.Starai V. J., Escalante-Semerena J. C. (2004) J. Mol. Biol. 340, 1005–1012 [DOI] [PubMed] [Google Scholar]
- 7.Liu X., Wang L., Zhao K., Thompson P. R., Hwang Y., Marmorstein R., Cole P. A. (2008) Nature 451, 846–850 [DOI] [PubMed] [Google Scholar]
- 8.Tang Y., Holbert M. A., Wurtele H., Meeth K., Rocha W., Gharib M., Jiang E., Thibault P., Verreault A., Verrault A., Cole P. A., Marmorstein R. (2008) Nat. Struct. Mol. Biol. 15, 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodawadekar S. C., Marmorstein R. (2007) Oncogene 26, 5528–5540 [DOI] [PubMed] [Google Scholar]
- 10.Vetting M. W. S., de Carvalho L. P., Yu M., Hegde S. S., Magnet S., Roderick S. L., Blanchard J. S. (2005) Arch. Biochem. Biophys. 433, 212–226 [DOI] [PubMed] [Google Scholar]
- 11.Wang L., Tang Y., Cole P. A., Marmorstein R. (2008) Curr. Opin. Struct. Biol. 18, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanner K. G., Trievel R. C., Kuo M. H., Howard R. M., Berger S. L., Allis C. D., Marmorstein R., Denu J. M. (1999) J. Biol. Chem. 274, 18157–18160 [DOI] [PubMed] [Google Scholar]
- 13.Scheibner K. A., De Angelis J., Burley S. K., Cole P. A. (2002) J. Biol. Chem. 277, 18118–18126 [DOI] [PubMed] [Google Scholar]
- 14.Yan Y., Harper S., Speicher D. W., Marmorstein R. (2002) Nat. Struct. Biol. 9, 862–869 [DOI] [PubMed] [Google Scholar]
- 15.Berndsen C. E., Albaugh B. N., Tan S., Denu J. M. (2007) Biochemistry 46, 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wardleworth B. N., Russell R. J., Bell S. D., Taylor G. L., White M. F. (2002) EMBO J. 21, 4654–4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min J., Landry J., Sternglanz R., Xu R. M. (2001) Cell 105, 269–279 [DOI] [PubMed] [Google Scholar]
- 18.Avalos J. L., Celic I., Muhammad S., Cosgrove M. S., Boeke J. D., Wolberger C. (2002) Mol. Cell 10, 523–535 [DOI] [PubMed] [Google Scholar]
- 19.Wardleworth B. N., Russell R. J., White M. F., Taylor G. L. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1893–1894 [DOI] [PubMed] [Google Scholar]
- 20.Braman J., Papworth C., Greener A. (1996) Methods Mol. Biol. 57, 31–44 [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 22.Terwilliger T. C. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 26.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 27.Vetting M. W., Magnet S., Nieves E., Roderick S. L., Blanchard J. S. (2004) Chem. Biol. 11, 565–573 [DOI] [PubMed] [Google Scholar]
- 28.Mehler E. L., Fuxreiter M., Simon I., Garcia-Moreno E. B. (2002) Proteins 48, 283–292 [DOI] [PubMed] [Google Scholar]
- 29.Harris T. K., Turner G. J. (2002) IUBMB Life 53, 85–98 [DOI] [PubMed] [Google Scholar]
- 30.Driscoll R., Hudson A., Jackson S. P. (2007) Science 315, 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fillingham J., Recht J., Silva A. C., Suter B., Emili A., Stagljar I., Krogan N. J., Allis C. D., Keogh M. C., Greenblatt J. F. (2008) Mol. Cell. Biol. 28, 4342–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han J., Zhou H., Li Z., Xu R. M., Zhang Z. (2007) J. Biol. Chem. 282, 14158–14164 [DOI] [PubMed] [Google Scholar]
- 33.Tsubota T., Berndsen C. E., Erkmann J. A., Smith C. L., Yang L., Freitas M. A., Denu J. M., Kaufman P. D. (2007) Mol. Cell 25, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.