Abstract
The pneumococcal surface protein C (PspC) is a major adhesin of Streptococcus pneumoniae, the cause of lobar pneumonia and invasive diseases. PspC interacts in a human-specific manner with the ectodomain of the human polymeric immunoglobulin receptor (pIgR) produced by respiratory epithelial cells. By adopting the retrograde machinery of human pIgR, this protein-protein interaction promotes colonization and transcytosis across the epithelial layer. Here, we explored the role of Rho family guanosine triphosphatases (GTPases), phosphatidylinositol 3-kinase (PI3K) and protein kinase B (Akt) for ingestion of pneumococci via the human pIgR. Inhibition experiments suggested that the host-cell actin microfilaments and microtubules are essential for this pneumococcal uptake mechanism. By using specific GTPase-modifying toxins, inhibitors, and GTPase expression constructs we demonstrate that Cdc42, but not Rac1 and RhoA are involved in PspC-mediated invasion of pneumococci into host cells. Accordingly, Cdc42 is time-dependently activated during ingestion of pneumococci. In addition, PI3K and Akt are essential for ingestion of pneumococci by respiratory epithelial cells via the PspC-pIgR interaction. The subunit p85α of PI3K and Akt was activated during the infection process. Moreover, Akt activation upon pneumococcal invasion depends on PI3K. In conclusion, our results illustrate for the first time key signaling molecules of host cells that are required for PspC-pIgR-mediated invasion of pneumococci into epithelial cells. This unique and specific bacterial entry process is dependent on the cooperation and activation of Rho family GTPase Cdc42, PI3K, and Akt.
Streptococcus pneumoniae (pneumococci) is (are) the etiologic agent of community-acquired pneumonia and life-threatening invasive diseases such as septicemia and bacterial meningitis (1). Pneumococci use several strategies to colonize the respiratory tract, which is considered to be the initial and essential step prior to their transmigration into the lungs and bloodstream. Adherence of pneumococci to host cells is facilitated by serum or matrix proteins such as Factor H, thrombospondin-1, and vitronectin (2–4). More importantly, pneumococci produce adhesins, which interact directly with cellular receptors and, consequently, these interactions promote bacterial adherence to and invasion into host cells (5). The pneumococcal surface protein C (PspC),3 also referred to as CbpA or SpsA, is a multifunctional choline-binding protein and a major adhesin of pneumococci residing on mucosal respiratory surfaces. PspC interacts directly and in a human-specific manner with the ectodomain of the polymeric immunoglobulin receptor (pIgR), which is also known as the secretory component (SC) (6). The PspC-hpIgR interaction has been characterized in detail on the molecular level and also on the structural level with regard to the PspC protein. A hexameric peptide within the N-terminal repeat domains (termed R1 or R2) of PspC recognizes human-specific amino acids in ectodomains D3 and D4 of pIgR (6–9). After binding to pIgR, pneumococci are ingested and transcytosed across epithelial cells by adopting the pIgR retrograde transcytosis machinery (7, 10). Additionally, the N terminus of PspC interacts in a human-specific manner with the innate immune regulator Factor H, and this interaction mediates immune evasion and adherence to host cells (2, 11–13).
The pIgR, which is broadly expressed by epithelial cells of the respiratory tract, mediates the transport of polymeric IgA (dIgA) or pIgM across the mucosal epithelial barriers from the basolateral to apical surface (14). Although unloaded pIgR undergoes constitutive transcytosis, binding of dIgA stimulates the receptor transcytosis in in vitro and in vivo situations (15, 16). The model of pIgR-dIgA transcytosis from the basolateral to the apical cell surface is based largely on studies using Madin-Darby canine kidney (MDCK) cells expressing exogenous rabbit or rat pIgR (15–17). The studies provided important insights into receptor sorting, intracellular compartments involved in transcytosis, and regulation of the endocytic pathways (14). After endocytosis in clathrin-coated vesicles at the basolateral surface, pIgR is delivered in an actin- and microtubule-dependent manner to the common recycling endosomes. At the apical surface unloaded receptor can be recycled and transported in retrograde. The dIgA-stimulated pIgR transcytosis is regulated by Rho family GTPases, phosphatidylinositol-3-kinase (PI3K), and requires the production of secondary messengers, including inositol 1,4,5-triphosphate and free intracellular calcium (17–23). In addition, the activation of these signaling molecules depends on the Src family protein tyrosine kinase p62yes and may stimulate a network of downstream pathways (24). Although it has become clear that pneumococci can adopt the pIgR-transcytosis machinery for invasion, the induced signal transduction cascades have not yet been explored. The goal of this study was, therefore, to assess the induced intracellular signaling pathways during PspC-hpIgR-mediated pneumococcal invasion into host cells. We asked whether this process depends on the dynamics of the actin cytoskeleton as suggested by earlier observations by electron microscopy (5) and which member(s) of the Rho family of small GTPases are the key players in this uptake mechanism. In addition, we have analyzed the role of the PI3K and of protein kinase B (Akt; also known as PKB). Akt is phosphorylated during activation, and phosphorylation at Ser-473 depends on PI3K activity (25, 26). By using GTPase-modifying toxins, pharmacological inhibitors, GTPase constructs, and GTPase activation assays we demonstrate for the first time that pneumococcal invasion via the PspC-hpIgR interaction requires the small GTPase member Cdc42, PI3K, and Akt activity.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
S. pneumoniae (NCTC10319; serotype 35A) were cultured in Todd-Hewitt broth (Oxoid, Basingstoke, UK) supplemented with 0.5% yeast extract to mid-log phase or grown on blood agar plates (Oxoid). The use of this strain for PspC-pIgR-mediated adherence and invasion and construction of the isogenic pspC mutant was described earlier (2, 7).
Cell Culture
MDCK (ATCC CCL-34) epithelial cells that were stably transfected with the hpIgR cDNA in pCB6 (MDCK-hpIgR) (27) and pIgR-expressing human lung epithelial cell line Calu-3 (ATCC HTB-55) were cultured in Eagle's minimum essential medium supplemented with 10% fetal bovine serum, 2 mm glutamine, penicillin G (100 IU ml−1), and streptomycin (100 μg ml−1) (all from PAA Laboratories) at 37 °C under 5% CO2. The medium for Calu-3 cells was further supplemented with 1 mm sodium pyruvate and 0.1 mm non essential amino acids (PAA Laboratories). Primary human bronchial epithelial cells previously used to investigate production of SC (28) were purchased from PromoCell (Heidelberg, Germany) and cultured according to the instructions of the manufacturer.
Reagents and Antibodies
Cytochalasin D was purchased from MP Biomedicals and nocodazole was obtained from Sigma. Latrunculin B, jasplakinolide, Y27632, NSC23766, wortmannin, LY294002, and Akt inhibitor VIII (Akti-1/2) were purchased from Calbiochem. NSC23766 is a specific inhibitor of Rac1, whereas Y27632 is an inhibitor of Rho-associated protein kinase (ROCK) and targets p160ROCK of Rho protein kinases. The inhibitors were reconstituted in DMSO and stored according to the manufacturer's instructions. Clostridium difficile toxins TcdB1470 and TcdB10463 were kindly provided by Klaus Aktorius and Gudula Schmidt, Institute of Experimental and Clinical Pharmacology and Toxicology University of Freiburg, Germany (29). Secramine A, a specific inhibitor of Cdc42, was kindly provided by Tom Kirchhausen, Immune Disease Institute, Harvard Medical School, Boston, MA (30, 31). The following antibodies were used: rabbit anti-Rac1, rabbit anti-Cdc42, rabbit anti-RhoA, rabbit anti-phospho-Akt (Ser-437), rabbit anti-Akt (all from Cell Signaling Technology), and goat phospho-PI3K p85α (Tyr-508, from Santa Cruz Biotechnology), horseradish peroxidase-conjugated goat anti-rabbit, and horseradish peroxidase-conjugated anti-goat IgG (Dako, Hamburg, Germany). Rabbit antiserum to human free SC was a kind gift from Jean-Pierre Vaerman (Brussels, Belgium) and described earlier (32).
Transfection Experiments
Transfection of dominant-negative GTPase constructs containing cDNAs of mutated GTPases (Rac1-T17N, Cdc42-T17N, and RhoA-T19N) were performed using Fugene6 transfection reagent (Roche Applied Science) according to the manufacturer's instruction. Transfection of constitutively active (ca) Cdc42-Q61L (33) was performed using Lipofectamine LTX reagent (Invitrogen) according to the manufacturer's instruction. The Cdc42-Q61L construct was purchased from Addgene. Uptake of pneumococci by transiently transfected MDCK-hpIgR was evaluated by infecting the host cells for 1 h with pneumococci. All dominant-negative constructs were expressed as N-terminal c-Myc fusion proteins and were kindly provided by Christof Hauck, University of Konstanz, Germany (34).
Infection Experiments and Inhibitor Studies
For the infection assays host cells were seeded on glass coverslips (diameter, 12 mm) or directly in the wells of a 24-well plate (Cellstar, Greiner, Germany) at a density of 5 × 104 cells per well and cultivated to give cell monolayers with ∼2 × 105 cells per well. The cells were washed three times with Dulbecco's modified Eagle's medium containing HEPES (Dulbecco's modified Eagle's medium-HEPES, PAA Laboratories, Coelbe, Germany) supplemented with 1.0% fetal bovine serum and then infected with pneumococci. In a standardized assay we used a multiplicity of infection of 50 bacteria per host cell, and the bacteria were centrifuged onto the host cells using 120 × g for 3 min. Infections were carried out for 1 h at 37 °C and 5.0% CO2. Thereafter, unbound bacteria were removed by rinsing the cells three times with Dulbecco's modified Eagle's medium-HEPES. The infection dose (colony forming units) per well was controlled by serial plating of the bacteria on blood agar plates. The pharmacological inhibitors used to study the impact of the cytoskeleton and the signaling molecules were solved in DMSO, and the cells were preincubated for 1 h at 4 °C and 30 min at 37 °C for nocodazole and for 30 min with the other inhibitors prior to the infection. Cells were preincubated with TcdB1470 and TcdB10463 for 4 h and 1 h respectively, whereas secramine A was used 10–15 min prior to infections. Infection assays were performed in the presence of the inhibitors. As a control, the cells were incubated with DMSO alone and infected as indicated. The amount of DMSO used in our assay had no influence on cell viability, cell morphology, and pneumococcal adherence as determined by immunofluorescence microscopy, trypan blue staining, and electron microscopy (data not shown).
Quantification of Pneumococcal Adherence and Invasion
After the infection cells were thoroughly washed and subjected to saponin-mediated lysis (1.0% w/v). The total number of attached and invasive bacteria was evaluated after serial plating of bacteria on blood agar plates. Antibiotic protection assays were performed to quantify the total number of internalized and recovered pneumococci after the infection experiments. The bacteria were briefly centrifuged over the cells at 120 × g for 3 min, and infections were performed as described above. Thereafter, the host cell layers were washed thoroughly to remove unbound bacteria. To kill extracellular, adherent pneumococci, host cells were incubated for 1 h with Dulbecco's modified Eagle's medium-HEPES containing 100 μg of gentamicin and 100 units of penicillin G at 37 °C and 5.0% CO2. Intracellular pneumococci were released by a saponin-mediated host cell lysis (1.0% w/v), and the total number of invasive and recovered pneumococci was monitored after plating sample aliquots on blood agar plates, followed by colony formation and enumeration. Each experiment was repeated at least three times, and results are expressed as mean ± S.D.
Immunofluorescence
For immunofluorescence microscopy the infected host cells were fixed on the glass coverslips with 3.0% paraformaldehyde. The immunofluorescence staining of pneumococci attached to the host cells and in some case also the differentiation between extracellular and intracellular bacteria was carried out as described recently (3). Briefly, extracellular pneumococci were stained using a polyclonal anti-pneumococcal antiserum and secondary goat anti rabbit IgG coupled to Cy5 (blue, Dianova, Germany) or AlexaFluor-488 (green, Invitrogen). Intracellular pneumococci were stained with AlexaFluor-568 (red, Invitrogen). The host cell actin-cytoskeleton was stained with phalloidin AlexaFluor-488 (Invitrogen). The samples were viewed with a confocal laser scanning microscope (Leica TCS SP5 AOBS or Zeiss LSM510Meta), and the appropriate software (LAS AF SP5 or LSM510) was used for image acquisition.
Cell Lysis and Western Blotting
At various time points of infection, cells were washed once with ice-cold phosphate-buffered saline and lysed with lysis buffer (10 mm Tris, 100 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm NaF, 20 mm Na4P2O7, 2 mm, Na3VO4, 0.1% SDS, 1.0% Triton X-100, 10% glycerol, 0.5% deoxycholate) containing a complete protease inhibitor mixture tablet (Roche Applied Science). The amount of protein in the samples was determined using the Bradford protein quantification method (Sigma), and equal amounts of protein lysates were separated by SDS-PAGE and Western blotting was performed as described (3). The membranes were blocked with 5% skim milk (Roth) prior to incubations with specific antibodies at 4 °C overnight. Membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Following washing, antibody binding was detected using enhanced chemiluminescence (ECL, Amersham Biosciences). To confirm equal protein amounts in each sample, blots were stripped and reprobed with antibodies specific for the non-phosphorylated form/total protein of the analyzed proteins.
Pulldown Assay for Activated GTPase
For pulldown assays, 100 μg of whole cell lysates containing equivalent amounts of protein were mixed with p21 binding domain of PAK1 fused to glutathione S-transferase (GST-PAK) or the Rho binding domain fused to GST (GST-mDia) (35, 36) conjugated to Sepharose beads for 1 h at 4 °C. Thereafter, the beads were collected by centrifugation and washed twice with radioimmune precipitation assay buffer (50 nm Tris HCl, pH 7.7, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS). Activated Rac and Cdc42 were then visualized by immunoblotting as described above. To confirm equal amounts of protein for each sample, aliquots of the lysate from different time points were also analyzed by immunoblotting.
Statistical Evaluation
The infection experiments were performed at least three times, each in duplicate, and the data were expressed as mean ± S.D. Differences in adherence and internalization of pneumococci were analyzed by the two-tailed unpaired Student's t test. In all analyzes, p values of <0.05 were considered statistically significant.
RESULTS
PspC-hpIgR Interaction Is Essential for Pneumococcal Internalization into Host Epithelial Cells
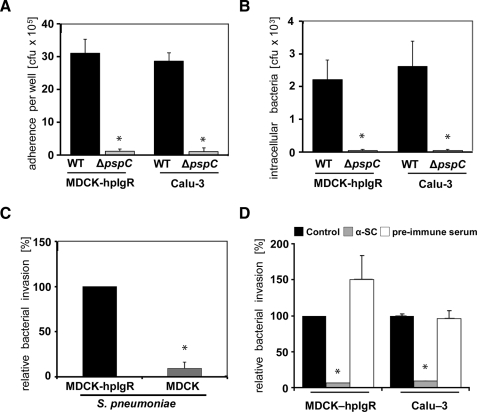
Pneumococcal adherence to and internalization into human pIgR-producing host cells were determined by cell culture infection experiments. Human epithelial cells Calu-3, which produce endogenously human pIgR, and Madin-Darby canine kidney cells stably transfected with hpIgR (MDCK-hpIgR) were infected for 1 h with S. pneumoniae wild-type bacteria or its isogenic pspC-mutant (ΔpspC). Although wild-type bacteria showed significant levels of adherence to and invasion into these host cells, PspC-deficient bacteria demonstrated a massive reduction in bacterial adherence and invasion (Fig. 1, A and B). The requirement of pIgR for efficient pneumococcal internalization was confirmed using the wild-type and non-transfected MDCK cells as a control (Fig. 1C). To confirm the requirement of a functional hpIgR for pneumococcal invasion in this setup, inhibition assays were performed by using polyclonal antibodies recognizing specifically the ectodomain SC of hpIgR. The results revealed a significant reduction in the number of host cell attached bacteria (data not shown). Consequently, the number of ingested wild-type pneumococci recovered from the intracellular compartment of anti-SC pretreated MDCK-hpIgR and Calu-3 cells, respectively, was significantly decreased (Fig. 1D).
FIGURE 1.
PspC-hpIgR-mediated adherence and invasion of MDCK-hpIgR and Calu-3 cells by wild-type pneumococci and its isogenic pspC-mutant. A, adherence of pneumococci was determined by counting the cfu (colony forming unit) per well obtained from sample aliquots plated onto blood agar plates after 1 h of infection. *, p < 0.02 relative to infections carried out with the wild-type strain. B and C, invasion and intracellular survival of pneumococci were determined by the antibiotic protection assay. *, p < 0.02 relative to infections carried out with the wild-type strain or relative to MDCK-hpIgR. D, the invasion and intracellular survival of pneumococci in host cells were determined in the presence of antibodies recognizing the secretory component of hpIgR (α-SC, 8 μg/well), pre-immune serum, or absence of antibody using the antibiotic protection assay. Invasion of S. pneumoniae in the absence of α-SC was set to 100%. *, p < 0.001 relative to infections carried out in absence of antibodies.
Cytoskeletal Dynamics Are Essential for PspC-hpIgR-mediated Internalization of Pneumococci
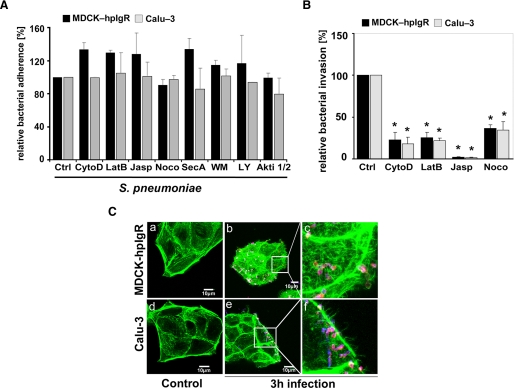
The rabbit-pIgR-pIgA transcytosis across eukaryotic cells is regulated by both microtubules and actin microfilaments (37). Therefore, the impact of these host cytoskeleton components on uptake of pneumococci by host cells via the PspC-hpIgR-mediated pathway was investigated. Infections of our host cells were conducted in the presence of pharmacological inhibitors cytochalasin D, latrunculin B, jasplakinolide, or nocodazole. Cytochalasin D and latrunculin B inhibit actin polymerization, whereas jasplakinolide stabilizes actin filaments. Nocodazole inhibits polymerization of microtubules. The presence of actin cytoskeleton inhibitors and nocodazole significantly blocked pneumococcal invasion in both MDCK-hpIgR and Calu-3 cells, as determined by enumeration of recovered intracellular pneumococci (Fig. 2B). However, no significant differences were observed for pneumococcal adherence to inhibitor treated cells when compared with untreated host cells (Fig. 2A). Moreover, changes in the actin cytoskeleton of pIgR-expressing host cells after infections with pneumococci were visualized by staining f-actin with AlexaFluor-488 phalloidin (Fig. 2C). In conclusion, these results clearly demonstrated the involvement of the host cell cytoskeleton dynamics in pneumococcal ingestion by respiratory host epithelial cells via the human pIgR mechanism.
FIGURE 2.
Invasion of pneumococci via pIgR requires the dynamics of the host cell actin cytoskeleton and microtubuli. A, pneumococcal adherence to MDCK-hpIgR and Calu-3 cells was determined in the absence (control, Ctrl) or presence of various inhibitors. Adherence of S. pneumoniae in the absence of an inhibitor was set to 100%. B, invasion of MDCK-hpIgR or Calu-3 cells with pneumococci was followed in the absence (control) or presence of inhibitors of actin filaments and microtubules, including cytochalasin D (CytoD, 125 nm), latrunculin B (LatB, 50 nm), jasplakinolide (Jasp, 100 nm), and nocodazole (Noco, 10 μm), by the antibiotic protection assay. Invasion of S. pneumoniae in the absence of an inhibitor was set to 100%. *, p < 0.001 relative to infections carried out in absence of an inhibitor. C, immunofluorescence microscopy illustrating changes of the actin cytoskeleton after infecting host cells for 3 h with pneumococci. F-actin was stained with AlexaFluor-488 phalloidin, and intracellular pneumococci were stained with AlexaFluor-568 (red), whereas adherent bacteria were stained with Cy5 and hence, bacteria appear pink (blue/red stain). Uninfected host cells (a and d) and infected host cells (b and e), respectively. Higher magnifications (c and e) illustrate changes of the actin cytoskeleton during pIgR mediated infection of MDCK-hpIgR or Calu-3 cells.
Inactivation of Cdc42, but Not Rac1 and RhoA, Inhibits PspC-hpIgR-mediated Internalization of Pneumococci
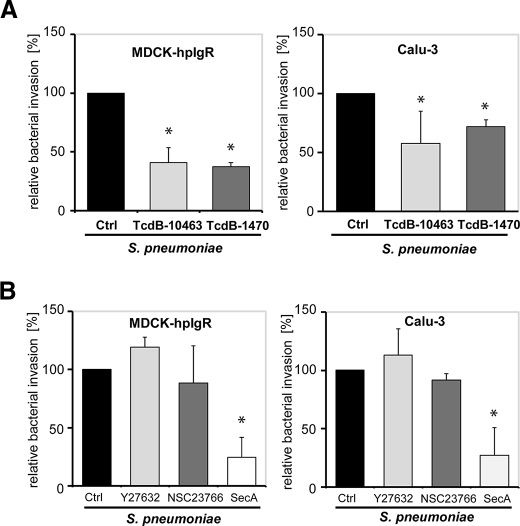
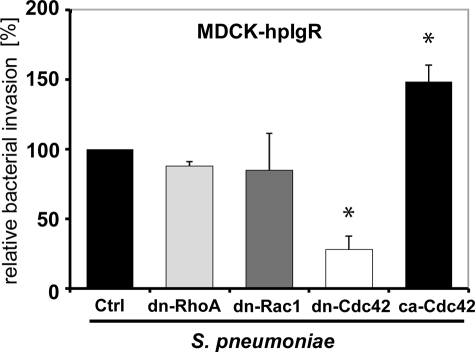
To elucidate the impact of the Rho GTPase members on PspC-hpIgR-mediated pneumococcal internalization into host cell, C. difficile toxin B, TcdB-10463 and a variant of toxin B from C. difficile strain 1470, namely TcdB1470, were employed in infection experiments. TcdB-10463 glucosylates the Rho family of small GTPases Rho (A/B/C), Rac1, and Cdc42 (29). Glucosylation of Rho proteins causes their functional inactivation due to an impaired coupling to effector and regulatory proteins (38, 39). Toxin TcdB1470 glucosylates Rac1 and Cdc42 but not Rho (A/B/C) (29). Pretreatment of MDCK-hpIgR and Calu-3 cells with TcdB10463 significantly reduced the number of internalized pneumococci (Fig. 3A). Similarly, TcdB1470 pretreatment of host cells also significantly reduced the number of internalized pneumococci (Fig. 3A) suggesting that Rac1 and Cdc42 but not RhoA are involved in the PspC-hpIgR-mediated internalization of pneumococci by mucosal epithelial cells. Importantly, immunofluorescence microscopy of host cell attached pneumococci indicated that the presence of these toxins did not alter bacterial adherence to pIgR-expressing host cells (data not shown). To corroborate these results, infection studies were performed in the presence of Y27632, a specific Rho-associated protein kinase inhibitor, NSC23766, a specific Rac1 inhibitor, or secramine A, which is a potent inhibitor of Cdc42 activation. Secramine A stabilizes the association of Cdc42 with Rho GDP dissociation inhibitor 1, thereby decreasing the availability of Cdc42 for activation and downstream signaling (30). Treatment of MDCK-hpIgR and Calu-3 cells with Y27632 and NSC23766 did not affect pneumococcal internalization, whereas pretreatment of host cells with secramine A significantly reduced invasion of pneumococci (Fig. 3B). The essential role of Cdc42 was confirmed with MDCK-hpIgR cells that were transiently transfected with dominant-negative (dn) alleles of Rac1 (Rac1-T17N), Cdc42 (Cdc42-T17N), Rho (Rho-T19N), or constitutively active (ca) allele of Cdc42 (Cdc42-Q61L). The antibiotic protection assay performed after 1-h post infection demonstrated that expression of dn-Rac1 (Rac1-T17N) or dn-Rho (Rho-T19N) did not influence pneumococcal internalization, whereas expression of dn-Cdc42 (Cdc42-T17N) significantly reduced pneumococcal uptake. Moreover, expression of ca-Cdc42 significantly increased pneumococcal uptake by these host cells (Fig. 4). Taken together, these data demonstrated the essential role of Cdc42 for pneumococcal internalization into host epithelial cells expressing human pIgR.
FIGURE 3.
Effect of bacterial protein toxins and pharmacological inhibitors targeting small Rho GTPases on PspC-hpIgR-mediated host cell internalization of pneumococci. Pneumococcal invasion in MDCK-hpIgR and Calu-3 cells was determined in the absence (Ctrl) or presence of (A) C. difficile toxin B, TcdB-10463 (30 ng ml−1), TcdB-1470 (100 ng ml−1), and (B) specific individual inhibitors of Rho family GTPases such as Y27632 (50 μm), Rac1 inhibitor NSC23766 (50 μm), or Cdc42 inhibitor secramine A (10 μm) by the antibiotic protection assay. Invasion of S. pneumoniae in the absence of toxin or inhibitor was set to 100%. *, p < 0.05 relative to infections carried out in absence of toxins or inhibitor.
FIGURE 4.
Activity of small GTPases Cdc42 is essential for PspC-hpIgR-mediated invasion of MDCK-hpIgR cells by pneumococci. The effect of expression of dominant-negative (dn) forms of Rho GTPases Rho-T19N, Rac1-T17N, Cdc42-T17N, or constitutively active (ca) Cdc42-Q61L on pneumococcal invasion was assessed by the antibiotic protection assay after the constructs were transiently transfected into MDCK-hpIgR cells. Invasion by S. pneumoniae into vector-transfected host cells (Ctrl) was set to 100%. *, p < 0.001 for dominant-negative Cdc42, and p < 0.03 for constitutively active Cdc42 relative to infections performed with vector-transfected cells.
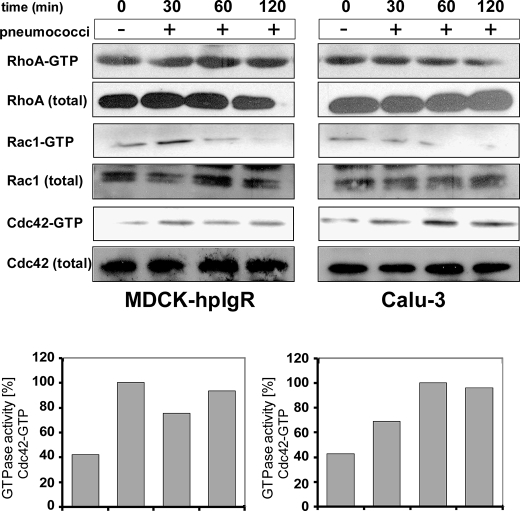
Activation of Endogenous Cdc42 in Response to Pneumococcal Uptake by Host Cells
To assess the activation of Rho GTPase members during pneumococcal invasion into pIgR-expressing host cells, pulldown assays were performed. In particular, GTP loading onto Cdc42 and/or Rac1 was determined by specific binding of active GTPases to the p21 binding domain of PAK1 (GST-PAK), whereas activated RhoA was detected with the Rho binding domain of mDia (GST-mDia) that associates preferentially with GTP-bound RhoA. Host cells were infected with pneumococci for different time periods and by using the prepared host cell lysates in conjunction with the specific binding domains, GTP-loaded Rac1, Cdc42, or RhoA were separately precipitated in a pulldown assay. As a control, uninfected host cell lysates were used. The immunoblot analysis revealed a time-dependent activation of Cdc42 in infected MDCK-hpIgR and Calu-3 cells, whereas no activation was detected for Rac1 and RhoA (Fig. 5). A decrease in Rac1 activation was observed that reduced to undetectable levels between 60 and 120 min post-infection. In conclusion, these results demonstrate the critical role of Cdc42 for pneumococcal uptake into mucosal host epithelial cells and suggest that the deterioration of Cdc42 activation cannot be rescued by other members of the Rho family of GTPases.
FIGURE 5.
Time-dependent activation of Cdc42 GTPase during pIgR-mediated pneumococcal invasion. Host cell lysates of MDCK-hpIgR and Calu-3 cells, respectively, were prepared after infection with pneumococci for indicated time points and were employed in pulldown assays of small GTPases (upper panels). GST-PAK was used for Rac1 and Cdc42, whereas GST-mDia was employed for RhoA. Precipitates were separated by 14% SDS-PAGE and analyzed using GTPase-specific antibodies. The pulldown assays from lysates that were prepared from uninfected host cells were used as controls (0 min). Total amounts of GTPases Rac1, Cdc42, or RhoA were analyzed using sample aliquots of lysates (lower panels). Quantification of GTPase activity of Cdc42: 100% of activity corresponds to the highest amount of detected GTPase-GTP levels.
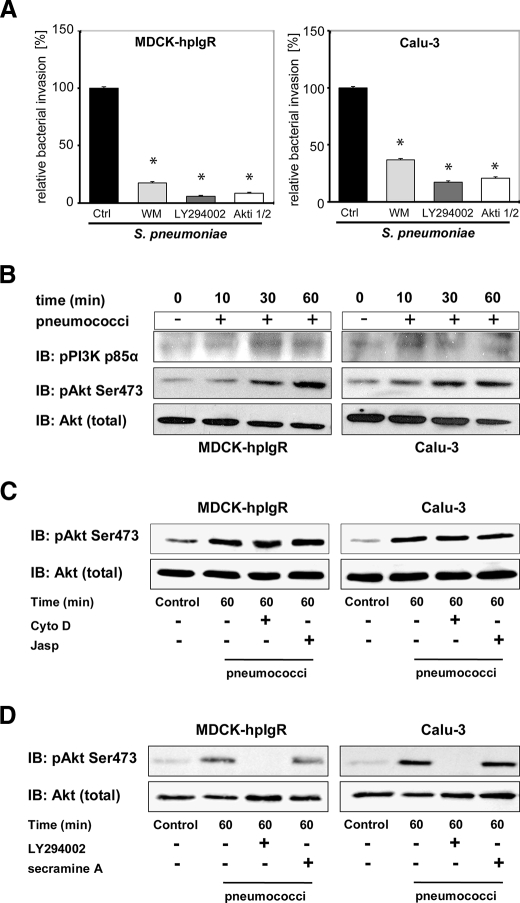
PspC-hpIgR-mediated Internalization of Pneumococci Requires Activity of PI3K and Akt
PI3K is a key regulatory protein and modulates many cytoskeleton-based processes, including phagocytosis. The activity of PI3K is essential for invasion of several pathogenic bacteria such as Listeria monocytogenes, Helicobacter pylori, and Escherichia coli (40–42). To explore the role of PI3K in PspC-hpIgR-mediated pneumococcal ingestion the invasion of pneumococci into MDCK-hpIgR and Calu-3 cells was determined in the presence of PI3K-specific inhibitors. Wortmannin as well as LY294002 significantly reduced PspC-hpIgR-mediated pneumococcal ingestion of pneumococci by pIgR (Fig. 6A). However, immunofluorescence microscopy demonstrated no significant changes in pneumococcal adhesion to inhibitor treated host cell compared with untreated cells (Fig. 2A). One of the main target molecules downstream of PI3K is Akt, which is phosphorylated at Thr-308 through the 3-phosphoinositide-dependent kinase, whereas phosphorylation at Ser-473 was shown to depend on PI3K activity and mammalian target of rapamycin (25, 26, 43–45). Therefore, we have also assessed the impact of Akt on pneumococcal internalization via pIgR. Inhibition of Akt by a specific Akt inhibitor (Akti1/2 VIII) significantly reduced, similar to wortmannin, the number of internalized pneumococci (Fig. 6A). Again, the presence of the inhibitor did not alter pneumococcal adherence to host cells expressing pIgR as determined by immunofluorescence staining and quantification of pneumococcal adherence (Fig. 2A). In addition, immunoblot analysis indicated that the p85α subunit of PI3K and Akt were time dependently phosphorylated after infecting pIgR-expressing host cells with pneumococci (Fig. 6B). Remarkably, inhibition of the actin cytoskeleton dynamics with cytochalasin D or jasplakinolide, both preventing pneumococcal uptake by host cells (Fig. 2B) but not adherence, influenced activation of Akt only to a minor degree (Fig. 6C). Finally, activation of Akt in pIgR-expressing host cells infected with pneumococci was completely abolished in the presence of PI3K inhibitor LY294002 (Fig. 6D), whereas inhibition of Cdc42 by secramine A had no influence on Akt phosphorylation during the infection (Fig. 6D).
FIGURE 6.
PI3K and Akt are activated during pneumococcal invasion via pIgR. A, invasion and intracellular survival of the bacteria in MDCK-hpIgR and Calu-3 cells were monitored in the absence (Ctrl) or presence of PI3K inhibitors wortmannin (50 nm) or LY294002 (50 μm) and in the presence of Akt Inhibitor VIII (Akti, 10 μm) by the antibiotic protection assay. Pneumococcal invasion in the absence of the inhibitor was set to 100%. *, p < 0.001 relative to infections carried out in the absence of inhibitor. B, pneumococcal infection of MDCK-hpIgR and Calu-3 cells induces phosphorylation of PI3K subunit p85α and Akt. Host cell lysates of MDCK-hpIgR and Calu-3 cells prepared after pneumococcal infections and separated by 10% SDS-PAGE. Activation of kinases were analyzed using antibodies against phosphorylated forms of PI3K subunit p85α (upper panel) or Akt (pAkt) (middle panel). The membrane was stripped and reprobed with total Akt antibody and used as loading control (lower panel). C, activation of Akt is independent of pneumococcal invasion. Pneumococcal invasion was blocked by inhibition of the actin cytoskeleton dynamics with Cytochalasin D (CytoD) or jasplakinolide (Jasp) and activation of Akt followed. Total Akt served as loading control (lower panel). D, Akt is activated in a PI3K-dependent but Cdc42-independent manner. Activation of Akt was followed in the presence of PI3K inhibitor LY294002 (50 μm) or Cdc42 inhibitor secramine A (10 μm). Total Akt served as loading control (lower panel).
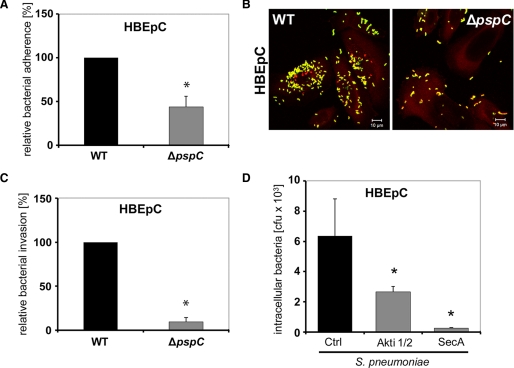
Pneumococcal Internalization into Primary Respiratory Epithelial Cells Depends on Cdc42 and Akt
To assess the biological significance of these interactions, primary human airway epithelial cells were infected for 1 h with S. pneumoniae wild-type bacteria or its isogenic pspC mutant. Although wild-type bacteria showed significant levels of adherence to and invasion into primary epithelial cells, PspC-deficient bacteria demonstrated a significant reduction in bacterial adherence (Figs. 7, A–C). In addition, we have explored the impact of small GTPase Cdc42 and Akt on pneumococcal internalization into primary host cells. Inhibition of Cdc42 using secramine A or inhibition of Akt by its specific Akt inhibitor (Akti1/2 VIII) significantly reduced invasion of pneumococci (Fig. 7D). Taken together, these strengthen the role of small GTPase Cdc42 and Akt for PspC-mediated pneumococcal uptake into host cells.
FIGURE 7.
Akt and Cdc42 are essential for PspC-mediated pneumococcal uptake by primary respiratory host cells. A, adherence of pneumococci was determined by counting the cfu per well obtained from sample aliquots plated onto blood agar plates after 1 h of host cell infection. *, p < 0.02 relative to infections carried out with the wild-type strain. B, immunofluorescence microscopy of S. pneumoniae wild type (WT) and its isogenic PspC mutant attached to primary human respiratory epithelial cells. C, invasion and intracellular survival of pneumococci were determined by the antibiotic protection assay. *, p < 0.02 relative to infections carried out with the wild-type strain. D, invasion and intracellular survival of the bacteria in primary cells human bronchial epithelial cell cells were determined in the absence (Ctrl) or presence of Akt Inhibitor VIII (Akti 1/2, 10 μm) and the presence of Cdc42 inhibitor secramine A (10 μm) by the antibiotic protection assay. *, p < 0.02 relative to infections carried out in the absence of inhibitor.
DISCUSSION
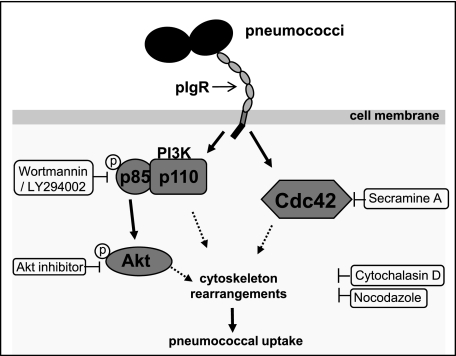
The polymeric immunoglobulin receptor is involved in the transport of immunoglobulins (IgA and IgM) across the mucosal epithelial barriers (14). However, some pathogens and viruses exploit the pIgR for their invasion into the epithelium. This includes the infection by type 2 herpes simplex virus or Epstein-Barr virus, where the virus-specific pIgA antibodies act as a bridging molecule to connect the pathogen with the pIgR-expressing epithelial cells thereby facilitating internalization of viruses (46–48). The study of Zhang et al. (10) hypothesized that S. pneumoniae exploit the apical recycling pathway of human pIgR, i.e. the transport in the retrograde fashion to basolateral surface, for bacterial translocation across human epithelial barriers. The pIgR is recognized by PspC, which is one of the choline-binding proteins produced by pneumococci and considered as the major pneumococcal adhesin on respiratory epithelial cells (5). However, the exact mechanism by which this PspC-human pIgR interaction promotes pneumococcal uptake by epithelial cells and the induced host cell signaling pathways have not yet been explored. In this study we demonstrated for the first time one of the induced signal cascades during pneumococcal ingestion by respiratory epithelial cells via the PspC-pIgR interaction (Fig. 8). Our data clearly show that the PspC-pIgR-mediated pneumococcal internalization of host epithelial cells requires the dynamics of the cytoskeleton and, consequently, inhibition of the host cytoskeleton resulted in a significant reduction in the pneumococcal uptake by pIgR-expressing host epithelial cells. In the presence of inhibitors of actin polymerization such as cytochalasin D and latrunculin B, pneumococcal uptake by pIgR-expressing MDCK-hpIgR and Calu-3 cells was significantly reduced. Similarly, pretreatment of these cell lines with jasplakinolide, a potent inducer of actin polymerization, also reduced uptake of pneumococci. In addition, inhibition of polymerization of microtubules by using nocodazole significantly blocked the PspC-hpIgR-mediated pneumococcal ingestion by host epithelial cells. Taken together, the inhibition experiments suggested that the host cell cytoskeleton dynamics plays a key role during pneumococcal ingestion by host epithelial cells via their binding to the human pIgR.
FIGURE 8.
Schematic model of pneumococci-induced signaling molecules. Solid arrows depict activated signaling molecules due to invasion of respiratory epithelial cells by pneumococci via the PspC-pIgR interaction. The dotted arrows point to the known implication of these signaling molecules toward modulation of host cell cytoskeleton.
Several bacterial pathogens can modify, as part of their virulence mechanisms, central host cellular functions. The strategy to subvert host cellular functions is often beneficial for the pathogen and facilitates invasion or survival. Typically this involves rearrangements of the host cell cytoskeleton by modulating the activity of key regulators such as Rho family of small GTPases (49). Rho GTPases cycle between an active, GTP-bound, and an inactive, GDP-bound, state. Rho-GTPases interact with their effectors mostly in their GTP-bound states, thereby transducing incoming signals to downstream signaling pathways. In mammalian cells, several Rho subfamily proteins have been identified. However, RhoA, Rac1, and Cdc42, which are the most extensively studied GTPases, play a crucial role in actin cytoskeleton regulation (50). Rho subtype proteins are involved in formation of stress fibers and focal adhesion complexes, whereas Rac1 induces lamellipodia formation and membrane ruffling. Cdc42 has been shown to induce formation of filopodia or microspikes. However, in some cell types these GTPases act on the actin cytoskeleton in a cascade-like manner (51). Rho GTPases are particularly attractive targets for pathogens and play a crucial role in host cell invasion of many pathogenic bacteria (52, 53). For example, it has been demonstrated that RhoA is important for uptake of Mycobacterium avium and Pseudomonas aeruginosa (54, 55), whereas Rac1 and Cdc42 play a crucial role in host cell invasion of Salmonella enterica, Shigella flexneri, and Campylobacter jejuni (56–58). In addition, Rho family GTPases RhoA, Rac1, and Cdc42 are required for efficient invasion of HeLa cells by group B streptococci (59). Although the Rho family GTPases were shown to be involved in the regulation of rabbit-pIgR-dimeric IgA transcytosis across mucosal epithelium (20–22), the involvement of Rho family GTPases in human pIgR-mediated pneumococcal infections of host epithelial cells has not yet been explored. Here, we have identified Cdc42 as a key GTPase regulating PspC-hpIgR-mediated pneumococcal invasion of epithelial cells, including primary human respiratory cells. Inhibition of endogenous Rho family members by C. difficile toxin TcdB-10463 or TcdB-1470 or inhibition of Cdc42 using its specific pharmacological inhibitor secramine A significantly reduced pneumococcal ingestion by pIgR-expressing epithelial cells. In contrast, specific inhibition of Rac1 using NSC23766 or blocking of Rho-associated protein kinase using the inhibitory substance Y27632 had no effect on pneumococcal uptake by pIgR-expressing host epithelial cells. Moreover, the transient transfection of MDCK-hpIgR cells with dn-Cdc42 (Cdc42-T17N) effectively reduced pneumococcal invasion. In contrast, constitutively active expression of Cdc42 enhanced pneumococcal uptake by host cells. Similar to our inhibition experiments transient transfections of epithelial cells with dn alleles of Rac1 (Rac1-T17N) or RhoA (Rho-T19N) did not reduce significantly pneumococcal internalization into MDCK-hpIgR cells. Finally, precipitation of GTP-bound Cdc42 following pneumococcal infections of pIgR-expressing epithelial cells demonstrated the activation of Cdc42 during pneumococcal uptake by host cells. These assays also revealed no changes in the level of RhoA activation following pneumococcal infections. In contrast, activation of Rac1 demonstrated a gradual decrease during the course of infection with pneumococci. Hence, these data confirmed that Cdc42 is a major player regulating host cell internalization of S. pneumoniae via the PspC-hpIgR mechanism. Rho-GTPases were also considered as part of the TLR2- and TLR4-signaling pathways (60, 61), and pneumococci were shown to activate Rac1 in bronchial epithelial cells via TLR1/2 (62). However, this host cell modulation was independent of PspC and pIgR, because the used cell lines BEAS-2B and HEK-293 do not produce pIgR. Recent data also revealed that sublytic amounts of the pneumococcal toxin pneumolysin, which is a cholesterol-binding cytolysin and intracellularly produced by pneumococci, activate RhoA and Rac1 GTPases (63). Although we have used pneumolysin-producing pneumococci, these GTPases were not activated in our experiments. It can be hypothesized that our infection time did not result in sublytic amounts of released pneumolysin sufficient to activate RhoA and/or Rac1.
Another interesting finding in this study is the implication of the PI3K-Akt pathway in human pIgR-mediated pneumococcal uptake by host epithelial cells. The PI3K signaling pathway is implicated in a variety of cellular functions, including regulation of the actin cytoskeleton and vesicle trafficking (64). The importance of PI3K and phosphoinositide metabolism for bacterial pathogenesis has also been shown. Several pathogens have been shown to require PI3K activity for invasion of host cells such as group B streptococci (59), group A streptococci (65), P. aeruginosa (66), H. pylori (41), Chlamydia pneumoniae (67), E. coli K1 (42), and Listeria monocytogenes (40). Inhibition of PI3K activity with specific inhibitors abolished PspC-pIgR-mediated pneumococcal invasion. A key downstream effector of PI3K is the serine-threonine kinase Akt, which is in response to PI3K activation phosphorylated and in turn regulates the activity of a number of other target molecules, including kinases, transcription factors, and other regulatory molecules (68). Inhibition of PI3K in MDCK-hpIgR and Calu-3 cells or inhibition of Akt caused a significant reduction of pneumococcal invasiveness. Similar results for Akt were obtained with primary host cells. In addition, kinetic infections demonstrated phosphorylation of PI3K subunit p85α and Akt. Activation of Akt was dependent on PI3K activity but independent of Cdc42 as demonstrated in inhibition assays with LY294002 and secramine A, respectively, followed by immunoblot analysis. Strikingly, vitronectin-αVβ3 integrin-mediated pneumococcal invasion of host epithelial cells was recently shown to depend on integrin-linked kinase activity and also on the PI3K-Akt pathway (4). Although inhibition of actin cytoskeleton dynamics prevents pneumococcal invasion, activation of Akt was only slightly affected, suggesting that pneumococcal adherence to host cells is sufficient to initiate activation of signaling molecules such as Akt.
In conclusion, these results demonstrate that pneumococcal invasion via PspC-pIgR is a highly complex process and involves the concerted role of host cell cytoskeleton and signaling pathways as depicted in the schematic model in Fig. 8. We have clearly shown the key role of the small GTPase Cdc42, PI3K, and Akt pathway during this specific uptake mechanism employed by pneumococci. The activation of these molecules contributes to cell membrane dynamics and, hence, promotes pneumococcal ingestion by host cells. The involvement of other host cell signaling pathways and the process of pneumococcal endocytosis via pIgR must be studied in the future.
Acknowledgments
We are grateful to Tomas Kirchhausen (Havard Medical School, Boston, MA) and Bo Xu and Gerald B. Hammond (University of Louisville, KY) for providing secramine A and to Gudula Schmidt and Klaus Aktories (University of Freiburg, Germany) for kindly providing toxins TcdB-1470 and TcdB-10463.
This work was supported in part by the Deutsche Forschungsgemeinschaft (DFG, Sonderforschungsbereich 479, Teilprojekt A7 (to S. H.) and DFG Ha 3125/4-1).
- PspC
- pneumococcal surface protein C
- pIgR
- polymeric immunoglobulin receptor
- SC
- secretory component
- Akt
- protein kinase B
- PI3K
- phosphatidylinositol 3-kinase
- MDCK
- Madin-Darby canine kidney cell
- GST
- glutathione S-transferase
- dn
- dominant-negative
- ca
- constitutively active
- dIgA
- polymeric IgA (or pIgM)
- PAK
- p21-activated kinase.
REFERENCES
- 1.Cartwright K. (2002) Eur. J. Pediatr. 161, 188–195 [DOI] [PubMed] [Google Scholar]
- 2.Hammerschmidt S., Agarwal V., Kunert A., Haelbich S., Skerka C., Zipfel P. F. (2007) J. Immunol. 178, 5848–5858 [DOI] [PubMed] [Google Scholar]
- 3.Rennemeier C., Hammerschmidt S., Niemann S., Inamura S., Zähringer U., Kehrel B. E. (2007) FASEB J. 21, 3118–3132 [DOI] [PubMed] [Google Scholar]
- 4.Bergmann S., Lang A., Rohde M., Agarwal V., Rennemeier C., Grashoff C., Preissner K. T., Hammerschmidt S. (2009) J. Cell Sci. 122, 256–267 [DOI] [PubMed] [Google Scholar]
- 5.Hammerschmidt S. (2006) Curr. Opin. Microbiol. 9, 12–20 [DOI] [PubMed] [Google Scholar]
- 6.Hammerschmidt S., Tillig M. P., Wolff S., Vaerman J. P., Chhatwal G. S. (2000) Mol. Microbiol. 36, 726–736 [DOI] [PubMed] [Google Scholar]
- 7.Elm C., Braathen R., Bergmann S., Frank R., Vaerman J. P., Kaetzel C. S., Chhatwal G. S., Johansen F. E., Hammerschmidt S. (2004) J. Biol. Chem. 279, 6296–6304 [DOI] [PubMed] [Google Scholar]
- 8.Lu L., Lamm M. E., Li H., Corthesy B., Zhang J. R. (2003) J. Biol. Chem. 278, 48178–48187 [DOI] [PubMed] [Google Scholar]
- 9.Luo R., Mann B., Lewis W. S., Rowe A., Heath R., Stewart M. L., Hamburger A. E., Sivakolundu S., Lacy E. R., Bjorkman P. J., Tuomanen E., Kriwacki R. W. (2005) EMBO J. 24, 34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J. R., Mostov K. E., Lamm M. E., Nanno M., Shimida S., Ohwaki M., Tuomanen E. (2000) Cell 102, 827–837 [DOI] [PubMed] [Google Scholar]
- 11.Dave S., Carmicle S., Hammerschmidt S., Pangburn M. K., McDaniel L. S. (2004) J. Immunol. 173, 471–477 [DOI] [PubMed] [Google Scholar]
- 12.Lu L., Ma Y., Zhang J. R. (2006) J. Biol. Chem. 281, 15464–15474 [DOI] [PubMed] [Google Scholar]
- 13.Lu L., Ma Z., Jokiranta T. S., Whitney A. R., DeLeo F. R., Zhang J. R. (2008) J. Immunol. 181, 7138–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rojas R., Apodaca G. (2002) Nat. Rev. Mol. Cell Biol. 3, 944–955 [DOI] [PubMed] [Google Scholar]
- 15.Giffroy D., Langendries A., Maurice M., Daniel F., Lardeux B., Courtoy P. J., Vaerman J. P. (1998) Int. Immunol. 10, 347–354 [DOI] [PubMed] [Google Scholar]
- 16.Song W., Bomsel M., Casanova J., Vaerman J. P., Mostov K. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 163–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardone M. H., Smith B. L., Mennitt P. A., Mochly-Rosen D., Silver R. B., Mostov K. E. (1996) J. Cell Biol. 133, 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luton F., Cardone M. H., Zhang M., Mostov K. E. (1998) Mol. Biol. Cell 9, 1787–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luton F., Mostov K. E. (1999) Mol. Biol. Cell 10, 1409–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung S. M., Rojas R., Maples C., Flynn C., Ruiz W. G., Jou T. S., Apodaca G. (1999) Mol. Biol. Cell 10, 4369–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jou T. S., Leung S. M., Fung L. M., Ruiz W. G., Nelson W. J., Apodaca G. (2000) Mol. Biol. Cell 11, 287–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas R., Ruiz W. G., Leung S. M., Jou T. S., Apodaca G. (2001) Mol. Biol. Cell 12, 2257–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen S. H., Olsson A., Casanova J. E. (1995) J. Biol. Chem. 270, 28425–28432 [DOI] [PubMed] [Google Scholar]
- 24.Luton F., Vergés M., Vaerman J. P., Sudol M., Mostov K. E. (1999) Mol Cell 4, 627–632 [DOI] [PubMed] [Google Scholar]
- 25.Delcommenne M., Tan C., Gray V., Rue L., Woodgett J., Dedhar S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11211–11216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawlor M. A., Alessi D. R. (2001) J. Cell Sci. 114, 2903–2910 [DOI] [PubMed] [Google Scholar]
- 27.Tamer C. M., Lamm M. E., Robinson J. K., Piskurich J. F., Kaetzel C. S. (1995) J. Immunol. 155, 707–714 [PubMed] [Google Scholar]
- 28.Godding V., Sibille Y., Massion P. P., Delos M., Sibille C., Thurion P., Giffroy D., Langendries A., Vaerman J. P. (1998) Eur. Respir. J. 11, 1043–1052 [DOI] [PubMed] [Google Scholar]
- 29.Genth H., Dreger S. C., Huelsenbeck J., Just I. (2008) Int. J. Biochem. Cell Biol. 40, 592–597 [DOI] [PubMed] [Google Scholar]
- 30.Pelish H. E., Peterson J. R., Salvarezza S. B., Rodriguez-Boulan E., Chen J. L., Stamnes M., Macia E., Feng Y., Shair M. D., Kirchhausen T. (2006) Nat. Chem. Biol. 2, 39–46 [DOI] [PubMed] [Google Scholar]
- 31.Xu B., Pelish H., Kirchhausen T., Hammond G. B. (2006) Org. Biomol. Chem. 4, 4149–4157 [DOI] [PubMed] [Google Scholar]
- 32.Giffroy D., Courtoy P. J., Vaerman J. P. (2001) Scand. J. Immunol. 53, 56–64 [DOI] [PubMed] [Google Scholar]
- 33.Nalbant P., Hodgson L., Kraynov V., Toutchkine A., Hahn K. M. (2004) Science 305, 1615–1619 [DOI] [PubMed] [Google Scholar]
- 34.Nobes C. D., Hall A. (1999) J. Cell Biol. 144, 1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benard V., Bohl B. P., Bokoch G. M. (1999) J. Biol. Chem. 274, 13198–13204 [DOI] [PubMed] [Google Scholar]
- 36.Kimura K., Tsuji T., Takada Y., Miki T., Narumiya S. (2000) J. Biol. Chem. 275, 17233–17236 [DOI] [PubMed] [Google Scholar]
- 37.Maples C. J., Ruiz W. G., Apodaca G. (1997) J. Biol. Chem. 272, 6741–6751 [DOI] [PubMed] [Google Scholar]
- 38.Aktories K., Just I. (2005) Curr. Top. Microbiol. Immunol. 291, 113–145 [DOI] [PubMed] [Google Scholar]
- 39.Just I., Gerhard R. (2004) Rev. Physiol. Biochem. Pharmacol. 152, 23–47 [DOI] [PubMed] [Google Scholar]
- 40.Ireton K., Payrastre B., Chap H., Ogawa W., Sakaue H., Kasuga M., Cossart P. (1996) Science 274, 780–782 [DOI] [PubMed] [Google Scholar]
- 41.Kwok T., Backert S., Schwarz H., Berger J., Meyer T. F. (2002) Infect. Immun. 70, 2108–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy M. A., Prasadarao N. V., Wass C. A., Kim K. S. (2000) J. Biol. Chem. 275, 36769–36774 [DOI] [PubMed] [Google Scholar]
- 43.Boudeau J., Miranda-Saavedra D., Barton G. J., Alessi D. R. (2006) Trends Cell Biol. 16, 443–452 [DOI] [PubMed] [Google Scholar]
- 44.Lynch D. K., Ellis C. A., Edwards P. A., Hiles I. D. (1999) Oncogene 18, 8024–8032 [DOI] [PubMed] [Google Scholar]
- 45.Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 46.Gan Y. J., Chodosh J., Morgan A., Sixbey J. W. (1997) J. Virol. 71, 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin C. T., Lin C. R., Tan G. K., Chen W., Dee A. N., Chan W. Y. (1997) Am. J. Pathol. 150, 1745–1756 [PMC free article] [PubMed] [Google Scholar]
- 48.Sixbey J. W., Yao Q. Y. (1992) Science 255, 1578–1580 [DOI] [PubMed] [Google Scholar]
- 49.Finlay B. B. (2005) Curr. Top. Microbiol. Immunol. 291, 1–10 [DOI] [PubMed] [Google Scholar]
- 50.Caron E., Hall A. (1998) Science 282, 1717–1721 [DOI] [PubMed] [Google Scholar]
- 51.Nobes C. D., Hall A. (1995) Cell 81, 53–62 [DOI] [PubMed] [Google Scholar]
- 52.Cossart P., Sansonetti P. J. (2004) Science 304, 242–248 [DOI] [PubMed] [Google Scholar]
- 53.Rottner K., Lommel S., Wehland J., Stradal T. E. (2004) J. Pathol. 204, 396–406 [DOI] [PubMed] [Google Scholar]
- 54.Kazmierczak B. I., Jou T. S., Mostov K., Engel J. N. (2001) Cell. Microbiol. 3, 85–98 [DOI] [PubMed] [Google Scholar]
- 55.Sangari F. J., Goodman J., Bermudez. L. E. (2000) Cell. Microbiol. 2, 561–568 [DOI] [PubMed] [Google Scholar]
- 56.Hardt W. D., Chen L. M., Schuebel K. E., Bustelo X. R., Galán J. E. (1998) Cell 93, 815–826 [DOI] [PubMed] [Google Scholar]
- 57.Krause-Gruszczynska M., Rohde M., Hartig R., Genth H., Schmidt G., Keo T., König W., Miller W. G., Konkel M. E., Backert S. (2007) Cell. Microbiol. 9, 2431–2444 [DOI] [PubMed] [Google Scholar]
- 58.Tran Van Nhieu G., Caron E., Hall A., Sansonetti P. J. (1999) EMBO J. 18, 3249–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burnham C. A., Shokoples S. E., Tyrrell G. J. (2007) FEMS Microbiol. Lett. 272, 8–14 [DOI] [PubMed] [Google Scholar]
- 60.Arbibe L., Mira J. P., Teusch N., Kline L., Guha M., Mackman N., Godowski P. J., Ulevitch R. J., Knaus U. G. (2000) Nat. Immunol. 1, 533–540 [DOI] [PubMed] [Google Scholar]
- 61.Hippenstiel S., Soeth S., Kellas B., Fuhrmann O., Seybold J., Krüll M., Eichel-Streiber C., Goebeler M., Ludwig S., Suttorp N. (2000) Blood 95, 3044–3051 [PubMed] [Google Scholar]
- 62.Schmeck B., Moog K., Zahlten J., van Laak V., N'Guessan P. D., Opitz B., Rosseau S., Suttorp N., Hippenstiel S. (2006) Respir. Res. 7, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iliev A. I., Djannatian J. R., Nau R., Mitchell T. J., Wouters F. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2897–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stokoe D. (2005) Expert Rev. Mol. Med. 7, 1–22 [DOI] [PubMed] [Google Scholar]
- 65.Purushothaman S. S., Wang B., Cleary P. P. (2003) Infect. Immun. 71, 5823–5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kierbel A., Gassama-Diagne A., Mostov K., Engel J. N. (2005) Mol. Biol. Cell 16, 2577–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coombes B. K., Mahony J. B. (2002) Cell. Microbiol. 4, 447–460 [DOI] [PubMed] [Google Scholar]
- 68.Scheid M. P., Woodgett J. R. (2003) FEBS Lett. 546, 108–112 [DOI] [PubMed] [Google Scholar]