Abstract
The insect Toll signaling pathway is activated upon recognition of Gram-positive bacteria and fungi, resulting in the expression of antimicrobial peptides via NF-κB-like transcription factor. This activation is mediated by a serine protease cascade leading to the processing of Spätzle, which generates the functional ligand of the Toll receptor. Recently, we identified three serine proteases mediating Toll pathway activation induced by lysine-type peptidoglycan of Gram-positive bacteria. However, the identities of the downstream serine protease components of Gram-negative-binding protein 3 (GNBP3), a receptor for a major cell wall component β-1,3-glucan of fungi, and their order of activation have not been characterized yet. Here, we identified three serine proteases that are required for Toll activation by β-1,3-glucan in the larvae of a large beetle, Tenebrio molitor. The first one is a modular serine protease functioning immediately downstream of GNBP3 that proteolytically activates the second one, a Spätzle-processing enzyme-activating enzyme that in turn activates the third serine protease, a Spätzle-processing enzyme. The active form of Spätzle-processing enzyme then cleaves Spätzle into the processed Spätzle as Toll ligand. In addition, we show that injection of β-1,3-glucan into Tenebrio larvae induces production of two antimicrobial peptides, Tenecin 1 and Tenecin 2, which are also inducible by injection of the active form of Spätzle-processing enzyme-activating enzyme or processed Spätzle. These results demonstrate a three-step proteolytic cascade essential for the Toll pathway activation by fungal β-1,3-glucan in Tenebrio larvae, which is shared with lysine-type peptidoglycan-induced Toll pathway activation.
Innate immunity is a crucial host defense mechanism against microbial infection in all animals (1–3). A group of germ line-encoded receptors and soluble proteins recognizes infectious microbes by sensing specific molecules called pathogen-associated molecular patterns (PAMPs), such as lipopolysaccharides in Gram-negative bacteria, peptidoglycans (PGs)2 in Gram-positive bacteria, and β-1,3-glucans in fungi (4–7). The Drosophila Toll signaling pathway is activated upon recognition of lysine (Lys)-type PG, which is found in Gram-positive bacteria and β-1,3-glucan, a major component of the fungus cell wall (8, 9). In contrast, the immune deficiency (Imd) pathway is activated primarily after recognition of diaminopimelic acid (DAP)-type PG, a form of PG found in Gram-negative bacteria and Bacillus spp (10). Both pathways lead to the expression of antimicrobial peptides (AMPs) via NF-κB-like transcription factors (11–13).
Elegant genetic studies in Drosophila demonstrated that Lys-type PG is recognized by a complex comprised of the PG recognition protein (PGRP)-SA (14), PGRP-SD (15), and the Gram-negative-binding protein 1 (GNBP1) (16, 17), while β-1,3-glucan from yeast is recognized by GNBP3 (18). Both the PGRP-SA/GNBP1 complex and GNBP3 are believed to mediate the activation of a serine protease cascade that ultimately leads to the cleavage of pro-Spätzle into processed Spätzle (5, 19, 20). Drosophila Spätzle-processing enzyme (SPE), an immune-regulated serine protease with a clip domain, has been identified as the terminal serine protease that cleaves pro-Spätzle (21, 22). Cleaved Spätzle serves as a ligand for the cell membrane receptor Toll, and induces the production of AMPs by the fat body (8, 19, 20). Serine protease cascades play very important roles in conveying and amplifying signals from pattern recognition receptors in the hemolymph (the insect blood) and lead to activation of the intracellular signaling pathway (23, 24). They involve the sequential activation of serine proteases and share similarity to the vertebrate complement system (13, 25). The amplification of these recognition signals represents an efficient host defense strategy in insects, which lack an acquired immune system (1).
Recently, we analyzed the serine protease cascade that regulates the Toll pathway using larvae of the beetle, Tenebrio molitor (26, 27). This large insect enabled us to collect large amount of hemolymph, allowing us to purify several different serine proteases. Our study demonstrated that the recognition of Lys-type PG by the PGRP-SA/GNBP1 complex activates Spätzle through the sequential activation of three different Tenebrio serine proteases: modular serine protease (MSP), Spätzle-processing enzyme-activating enzyme (SAE) and SPE (26).
The activation mechanism of the Toll pathway by β-1,3-glucan is poorly characterized in insects. A recent study (9) showed that Drosophila Grass functioned as a common serine protease, which transferred signals recognizing both Gram-positive bacteria and fungi via the pattern recognition receptors Drosophila PGRP-SA/GNBP1 and GNBP3, respectively. Moreover, these authors showed that the Drosophila Persephone protease, previously shown to be specific for fungal detection in the Toll pathway (18, 28), was also required for the sensing of proteases elicited by bacteria in the hemolymph (9). Even though Drosophila Grass and Persephone proteases are known to be serine proteases that transfer fungal recognition signals to pro-Spätzle during β-1,3-glucan-dependent Drosophila Toll pathway activation, the exact biological functions of these serine proteases have not yet been determined biochemically. Therefore, the purification and biochemical characterization of the immediate downstream molecule(s) of GNBP3 or the upstream serine protease(s) of Spätzle that participates in the fungal-dependent Toll signaling pathway are essential for understanding the host defense against fungal infections and the sensing of fungal virulence factors.
Here, we have identified the serine protease that lies immediately downstream of Tenebrio GNBP3. By performing in vitro reconstitution experiments, we provide biochemical evidence as to how the Tenebrio GNBP3-mediated β-1,3-glucan recognition signal is transferred to Spätzle. We identified three Tenebrio serine proteases, MSP, SAE, and SPE that function sequentially in a cascade linking β-1,3-glucan recognition by GNBP3 to pro-Spätzle activation. Furthermore, we identified two AMPs that were induced upon injection of β-1,3-glucan, activated SAE, or a processed Spätzle into Tenebrio larvae, demonstrating that they constitute bona fide targets of the Toll pathway.
EXPERIMENTAL PROCEDURES
Animals and Collection of Hemolymph
T. molitor larvae (mealworms) were maintained in terraria containing wheat bran. Hemolymph was collected as previously described (29). Briefly, to harvest the hemolymph, a larva was pricked using a 25-gauge needle, and a 10-μl drop of hemolymph was collected in 500 μl of a modified anti-coagulation buffer (136 mm trisodium citrate, 26 mm citric acid, 20 mm EDTA, and 15 mm sodium chloride, pH 5.0). The collected crude hemolymph was centrifuged at 20,000 × g for 15 min at 4 °C. The supernatant was then stored at −80 °C until use.
Purification of Tenebrio GNBP3, MSP, SAE, SPE, Tribolium Spätzle Proteins
The native and recombinant Tenebrio proteins, such as GNBP3, MSP, SAE, and SPE and the recombinant Tribolium castneum Spätzle were obtained as previously described (26).
In Vitro Reconstitution Experiments and Peptide Sequencing
In vitro reconstitution experiments of the proteolytic cascade were performed as previously described (26). After incubation with β-1,3-glucan, the reaction mixtures were electrophoresed using SDS-PAGE under reducing conditions, blotted onto a polyvinylidene difluoride membrane, and then stained with a solution containing 0.1% Coomassie Brilliant Blue (CBB) R-250 and 50% methanol, to map the proteolytic cleavage sites of the serine proteases. The membrane was destained with 50% methanol containing 10% acetic acid (v/v) until the protein bands became visible. The N-terminal amino acid sequences of the cleavage products of the serine proteases or the processed Spätzle, and N-terminal and internal amino acid sequences of the purified Tenecin 2 were identified using a gas phase protein sequencer (Applied Biosystems).
Antibodies and Immunoblotting
Rabbit antisera raised against MSP and Spätzle were used after affinity purification as described (26, 30). Briefly, the polyclonal antibodies of Tenebrio MSP were raised using a keyhole limpet hemocyanin (KLH)-conjugated synthetic peptide. This peptide has the sequence (VNGKPVKKGDYPWQ), which is located at the N terminus of the catalytic serine protease domain of MSP (31). Synthetic KLH-conjugated peptides with more than 95% purity were purchased from Peptron, Inc. (Daejeon, Korea). Rabbits were injected subcutaneously with 500 μg of KLH-conjugated peptides in Freund's complete adjuvant. A booster dose (250 μg in Freund's incomplete adjuvant) was given after 4 weeks, and the animals were bled 2–3 weeks thereafter. The resulting antibodies were affinity-purified as previously described (31). The polyclonal antibodies against the purified recombinant Tribolium Spätzle protein were raised by injecting 10 μg of the purified recombinant Spätzle proteins into each male albino rabbit with complete Freund's adjuvant and giving two booster injections with the same amount of protein 7 and 14 days later. The resulting antibody was also affinity-purified as described above. Western blotting was performed as previously described (31).
Measurement of the β-1,3-Glucan-specific Amidase Activity
To determine the amidase activity in the sample, the commercially available α-thrombin substrate t-butyloxy- carbonyl-benzyl-l-valinyl-l-prolinyl-l-arginine-4-methyl coumaryl-7-amide (Boc-Val-Pro-Arg-MCA) was used. This α-thrombin substrate is specifically cleaved by the active form of Tenebrio SPE (26). The substrate was dissolved in dimethylformamide according to the manufacturer's instructions. A 10-μl sample of crude hemolymph or a fraction from column chromatography was incubated with β-1,3-glucan, in the presence of 490 μl of the reaction solution containing 20 μm substrate in 20 mm Tris-HCl buffer, pH 8.0. After incubating the mixture at 30 °C for 1 h, 500 μl of 17% (v/v) acetic acid was added to terminate the enzyme reaction. The amidase activity was detected by a fluorescence spectrophotometer at λex = 380 nm and λem = 460 nm. One unit of amidase activity was defined as the amount that liberates 1 nmol of 7-amino-4-methylcoumarin per min.
Assay of Antimicrobial Activity
The antimicrobial activity was assayed during the purification of Tenecin 1 and Tenecin 2 essentially as previously described (32). Briefly, the bactericidal activity of the hemolymph that was obtained from insects that were previously injected with either the soluble Lys-type PG, β-1,3-glucan or the active form of SAE or cleaved Spätzle was assayed against Staphylococcus aureus (strain Cowan 1) and Escherichia coli (strain K 12). These bacteria were harvested in the exponential phase of growth and then suspended in 10 mm sodium phosphate buffer containing 130 mm NaCl, pH 6.0 (buffer A). The collected hemolymph that was obtained after injection was diluted serially with buffer A containing 0.2% bovine serum albumin, and a portion (10 μl) of the diluted samples were incubated with 106 S. aureus cells in 200 μl of insect saline (130 mm NaCl, 5 mm KCl, 1 mm CaCl2) for 60 min at 37 °C. Then the mixtures were diluted 2,000-fold with insect saline and aliquots of 50 μl were spread on Difco nutrient agar. The plates were incubated for 18 h at 37 °C, and the colony numbers on test and control plates were compared. Antibacterial susceptibility disc tests against S. aureus, E. coli, or Saccharomyces cerevisiae cells were performed as previously described (26) using lyophilized hemolymph from 100 μl of a whole extract resulting from the injection of insect saline, soluble Lys-type PG, β-1,3-glucan, the active form of SAE, or cleaved Spätzle. The extract was harvested at 36 h after injection. The zones of inhibition were estimated after overnight incubation. Data represent means ± S.E. for three experiments.
Purification and Characterization of Inducible AMPs
To purify inducible AMPs, we collected hemolymph after the injection of soluble Lys-type PG (50 ng/larva), β-1,3-glucan (400 ng), the active form of SAE (50 ng), or cleaved Spätzle (50 ng) and followed the same procedures as previously reported (32, 33). Briefly, Tenebrio hemolymph was collected at 48 h after injection and then heated for 5 min at 100 °C to denature most of the proteins. The clear supernatant was obtained after centrifugation for 15 min at 20,000 × g at 4 °C. The clear solution that was collected was applied to a CAPCELL PAK C18 column (ACR Type 5 μm, 4.6-mm i.d. × 250 mm, Shiseido Fine Chemicals), and then eluted with a 0–100% linear gradient of acetonitrile containing 0.05% (v/v) trifluoroacetic acid over a 60-min period. Active fractions showing antimicrobial activity were concentrated by lyophilization and then loaded onto a Synchropak RP C18 column (4.6 mm i.d. × 250 mm, Synchrom, Inc.), and eluted with a 0–60% linear gradient of acetonitrile containing 0.05% (v/v) trifluoroacetic acid over a 60-min period. For the third column, concentrated active fractions were applied to a TSK gel G2000SWXL size exclusion column (TOSOH) equilibrated with 20 mm Tris/HCl, pH 8.0, and the two active fractions showing antibacterial activities (Tenecin 1 and 2) were separated and collected. For the final purification step, concentrated active fractions were applied to a TSK gel ODS-100S C18 column (TOSOH), and then eluted with a 50% acetonitrile/H2O linear gradient containing 0.05% (v/v) trifluoroacetic acid over a 60-min period. The whole amino acid sequence of the purified Tenecin 2 protein was determined by the assembly of determined partial amino acid sequences, which were obtained from peptides treated with lysyl endopeptidase and endoproteinase Asp-N (supplemental Fig. S2).
Real-time Quantitative PCR (RT-qPCR) Analysis
Total RNA was extracted at the indicated time points with a RNAzol reagent for hemocytes or fat bodies from T. molitor larvae, which had been injected with soluble Lys-type PG, β-1,3-glucan, the active form of SAE or cleaved Spätzle. The first cDNA was synthesized using a first cDNA synthesis kit (Roche Applied Science) according to the manufacturer's instructions. To quantify the AMP gene expression, fluorescence real-time RT-qPCR was performed with the double-stranded DNA dye, SYBR Green (PerkinElmer Life Sciences, Boston, MA). Primer pairs for Tenecin 1 (sense, 5′-ATG AAG CTT ACA ATC TTC GCA-3′; antisense, 5′-TTA TCT GCA AAC GCA GAC CC-3′), Tenecin 2 (sense, 5′-CAG CAA AAC GGA TGG T-3′; antisense, 5′-TGC GTT GAA ATC GTG ATC TTG-3′), and the control ribosomal protein L-27A (RPL27A) (sense, 5′-GCA TGG CAA ACA CAG AAA GCA TC-3′; antisense, 5′-ATG ACA GGT TGG TTA GGC AGG C-3′) were used to detect the target gene transcripts. SYBR Green analysis was performed on an ABI PRISM 7700 system (PE Applied Biosystems) according to the manufacturer's instructions. All samples were analyzed in triplicate, and the levels of the detected mRNAs were normalized to control RPL27A values. The normalized data were used to quantify the relative levels of a given mRNA according to the ΔCt analysis (10).
RESULTS
The Active Form of Tenebrio MSP Is Recruited to the β-1,3-glucan/GNBP3 Complex
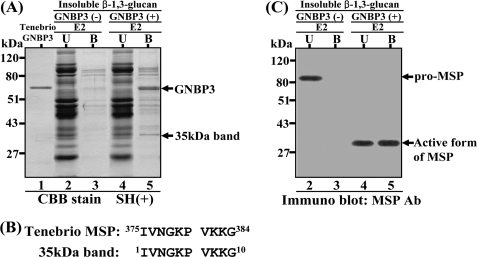
Recently, we demonstrated that Tenebrio crude hemolymph could be fractionated into three fractions (E1, E2, and E3) on a heparin Toyopearl column. We also demonstrated that the E2 and E3 fractions, when incubated with β-1,3 glucan (26), were sufficient to show amidase activity against the α-thrombin substrate, which is specifically cleaved by activated Tenebrio SPE in the hemolymph. This result showed that these two fractions contained all of the components that were essential for the recognition of β-1,3-glucan and processing of pro-Spätzle. To identify the immediate downstream molecule(s) that is recruited by the β-1,3-glucan/GNBP3 complex, insoluble β-1,3-glucan was incubated with the native Tenebrio GNBP3 purified from the E3 fraction (26). The GNBP3/β-1,3-glucan-insoluble fraction was then incubated with the E2 fraction. The molecules binding to the GNBP3/β-1,3-glucan complex were separated on SDS-PAGE under reducing conditions (Fig. 1A). A 35-kDa band (lane 5 in Fig. 1A) was specifically enriched in the GNBP3/β-1,3-glucan insoluble fraction compared with the insoluble β-1,3-glucan fraction lacking GNBP3 (lane 3). The N-terminal amino acid sequence of the 35-kDa protein perfectly matched that of the catalytic serine protease domain of activated Tenebrio MSP (Fig. 1B), suggesting that activated MSP was recruited to the β-1,3-glucan/GNBP3 complex. Tenebrio MSP was initially identified as the apical serine protease activated downstream of PGRP-SA/GNBP1 in response to Lys-type PG (26). To further confirm that the 35-kDa band was generated from Tenebrio pro-MSP in the presence of β-1,3-glucan, we performed a Western blot analysis using an MSP antibody with E2 fraction incubated with β-1,3-glucan and GNBP3 (Fig. 1C). Tenebrio pro-MSP in the E2 fraction was not activated in the absence of GNBP3 (lane 1), but was processed to a 35-kDa active MSP form when both GNBP3 and β-1,3-glucan were added to the E2 fraction (lanes 3 and 4). These results suggest that the pro-MSP is processed into the activated form of 35 kDa in the presence of GNBP3 and β-1,3-glucan and that this active form binds to the β-1,3-glucan/GNBP3 complex. This indicates that MSP is an apical serine protease that functions as the immediate downstream molecule of the β-1,3-glucan/GNBP3 complex.
FIGURE 1.
Identification of a 35-kDa protein binding to β-1,3-glucan/GNBP3 complex. A, lanes were loaded as follows: the purified Tenebrio GNBP3 (lane 1); unbound (U) molecules after incubation of insoluble β-1,3-glucan with the E2 fraction (lane 2); bound (B) molecules after incubation of insoluble β-1,3-glucan with the E2 fraction (lane 3); unbound molecules after incubation of insoluble β-1,3-glucan/GNBP3 complex with the E2 fraction (lane 4); bound molecules after incubation of insoluble β-1,3-glucan/GNBP3 with the E2 fraction (lane 5). The E2 fraction was obtained from Tenebrio hemolymph through heparin-Toyopearl column chromatography as previously described (26). The gel was stained with Coomassie Brilliant Blue R-250 (CBB). B, N-terminal amino acid sequences of the recruited 35-kDa protein and that of the active form of Tenebrio MSP. C, samples described in lanes 2–5 are the same as those of Fig. 1A. Western blot analysis was performed with an affinity-purified anti-MSP antibody.
Pro-MSP Is Processed to the Active Form of MSP after Recognition of the GNBP3/β-1,3-Glucan Complex in the Presence of Ca2+
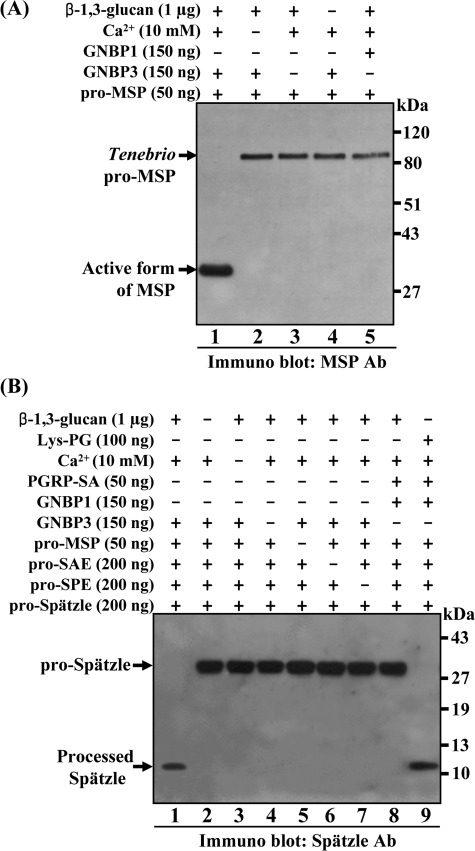
To examine whether the Tenebrio pro-MSP can be converted to the active form of MSP in the presence of Tenebrio GNBP3 and β-1,3-glucan in vitro, we incubated the purified native pro-MSP and GNBP3 in the presence of β-1,3-glucan and Ca2+ (Fig. 2A). As expected, pro-MSP is almost entirely converted to the active form of MSP following a 60-min incubation in the presence of GNBP3, β-1,3-glucan, and Ca2+ (lane 1 in Fig. 2A). This result was confirmed by N-terminal amino acid sequencing (data not shown). However, in the absence of any one of these components, pro-MSP was not cleaved (lanes 2–4 in Fig. 2A). These results clearly demonstrate that pro-MSP was activated after recognition of the β-1,3-glucan/GNBP3 complex in the presence of Ca2+, and again that MSP is an immediate downstream serine protease of the β-1,3-glucan/GNBP3 complex. To further confirm that Tenebrio GNBP3 functions as a distinct β-1,3-glucan recognition protein, pro-MSP was incubated in the presence of β-1,3-glucan, Ca2+ and Tenebrio GNBP1, which is known to be involved in Lys-type PG recognition. As expected, the 35-kDa band (activated MSP) was not generated (lane 5 in Fig. 2A), supporting the hypothesis that GNBP3 is a specific β-1,3-glucan recognition protein.
FIGURE 2.
In vitro reconstitution experiments for the activation of pro-MSP and pro-Spätzle by β-1,3-glucan. A, mixture of purified Tenebrio pro-MSP and GNBP3 in the presence of β-1,3-glucan and Ca2+ was incubated for 60 min and then analyzed by Western blotting with an anti-MSP antibody (lane 1). The 82-kDa pro-MSP and the 35-kDa activated MSP are indicated with arrows. When Tenebrio GNBP1 was added instead of GNBP3, pro-MSP was not cleaved (lane 5). In the absence of any one of the components, such as Ca2+, GNBP3, or β-1,3-glucan, pro-MSP was not converted to its active form (lanes 2–4, respectively). B, mixture of Tenebrio pro-MSP, GNBP3, pro-MSP, pro-SAE, pro-SPE, and Tribolium pro-Spätzle in the presence of β-1,3-glucan and Ca2+ was incubated for 60 min and analyzed by Western blotting with an affinity-purified anti-Spätzle antibody (lane 1). The 30-kDa pro-Spätzle and the 12-kDa-processed Spätzle are indicated with arrows. As a control, when eight components, such as Lys-type PG/PGRP-SA/GNBP1/MSP/SAE/SPE/Spätzle were incubated together, the cleaved 12-kDa Spätzle was generated (lane 9). In the absence of any one of these components, pro-Spätzle was not converted to the processed Spätzle (lanes 2–8).
Five Components Are Sufficient to Transfer the β-1,3-Glucan Recognition Signal to Pro-Spätzle in Vitro
Because Tenebrio pro-MSP is activated in the presence of either the β-1,3-glucan/GNBP3 complex or the Lys-type PGN/PGRP-SA/GNBP1 complex, we assumed that the downstream serine proteases of the β-1,3-glucan/GNBP3 complex would be identical to Tenebrio SAE and SPE that are activated in response to Lys-type PG (26). Therefore, we performed in vitro reconstitution experiments by using five purified proteins: GNBP3, pro-MSP, pro-SAE, pro-SPE, and pro-Spätzle (Fig. 2B). Western blot analysis revealed that processed Spätzle was generated upon incubation of the five proteins, β-1,3-glucan, and Ca2+ (lane 1 in Fig. 2B). Depletion of any of the components resulted in the loss of cleavage of the pro-Spätzle. Under the same conditions, the processing of pro-Spätzle to the cleaved Spätzle was also observed with PGRP-SA, GNBP1, MSP, SAE, SPE, and Spätzle in the presence of Lys-type PG and Ca2+ as recently reported by our group (lane 9 in Fig. 2B) (26). In addition, when the E2 and E3 fractions were incubated with β-1,3-glucan and Ca2+, the activated forms of Tenebrio MSP and SAE were observed (line 7 in supplemental Fig. S1, A and B). These results clearly demonstrate that GNBP3, in the presence of β-1,3-glucan, induces the activation of a three-step proteolytic cascade involving MSP, SAE, and SPE sequentially. This activation leads to the processing of pro-Spätzle into the mature form that functions as a ligand for the Toll receptor. In addition, these data show that the β-1,3-glucan and Lys-type PG recognition signals are sharing a three-step proteolytic cascade to transduce their recognition signals to the Toll receptor.
Injection of Lys-type PG, β-1,3-Glucan, Activated SAE, or Processed Spätzle Induces the Expression of AMPs in the T. molitor Larvae
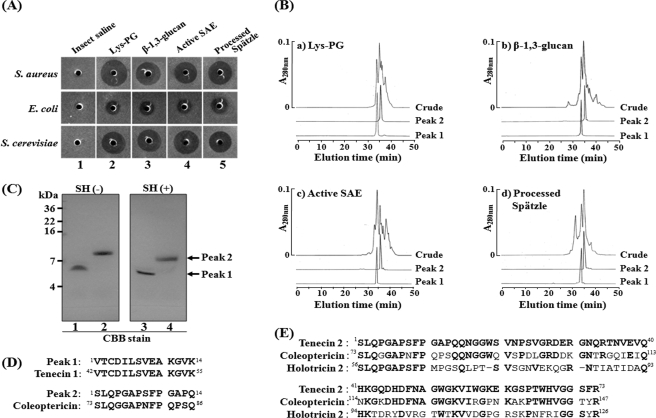
Even though we have provided biochemical evidence for how the β-1,3-glucan recognition signal is transferred to the Toll receptor, we have not demonstrated whether this serine protease cascade is operating in vivo. We hypothesized that if this cascade is working in vivo, the same AMP(s) will be produced in the insect hemolymph when activated molecules of this pathway are injected into Tenebrio larvae. Further, we anticipated that this AMP (these AMPs) will also be induced upon injection of Lys-type PG. To prove this hypothesis, we injected β-1,3-glucan, Lys-type PG, the activated SAE, or the processed Spätzle into Tenebrio larvae. Hemolymph samples collected separately after injection of these four molecules showed strong antimicrobial activities against S. aureus, E. coli, and S. cerevisiae, indicating the presence of AMPs (columns 2–5 in Fig. 3A). We then purified the AMPs from each of the four hemolymph samples. After performing several column chromatography steps, including C18 HPLC and size exclusion chromatography (Fig. 3B), two identical AMPs were purified to homogeneity from each batch of larvae (Fig. 3C). The N-terminal amino acid sequence of peak 1 perfectly matched that of Tenecin 1 (Fig. 3D), an AMP that has bactericidal activity against Gram-positive bacteria, previously identified by our group (32). The amino acid sequence of Tenecin 1 and its disulfide bond arrangement were very similar to Drosophila defensin (35). The N-terminal amino acid sequence of peak 2 showed high homology with an antimicrobial peptide, Coleoptericin (34), which is inducible antibacterial peptide purified from coleopteran insect, Zophobas atratus (Fig. 3D). The entire amino acid sequence of the peak 2 (named Tenecin 2) was determined by the assembly of partial amino acid sequences, which we determined (supplemental Fig. S2). Tenecin 2 shows high sequence identity (65 and 36%) with Coleoptericin and Holotricin 2 (33), respectively. Holotricin 2, previously identified by our group, is also an inducible antibacterial peptide purified from coleopteran insect, Holotrichia diomphalia larvae. Tenecin 2 had bactericidal activity against E. coli and S. cerevisiae. These results demonstrate that Tenecin 1 and Tenecin 2 are induced by the injection of β-1,3-glucan and Lys-type PG, or by the injection of activated SAE, or the processed Spätzle. This strongly supports the notion that β-1,3-glucan and Lys-type PG activate Toll activation using the same three-step proteolytic cascade that results in the production of AMPs.
FIGURE 3.
The purification and characterization of two inducible AMPs following injection of Lys-type PG, β-1,3-glucan, activated SAE, or processed Spätzle. A, antimicrobial activities were measured against S. aureus, E. coli, and S. cerevisiae after injection of insect saline (column 1), Lys-type PG (50 ng, column 2), β-1,3-glucan (400 ng, column 3), activated SAE (50 ng, column 4), or processed Spätzle (50 ng, column 5). B, C18 HPLC elution profiles of peak 1 and peak 2 showing antimicrobial activities. (a) injection of Lys-type PG, (b) β-1,3-glucan, (c) activated SAE, (d) processed Spätzle. C, SDS-PAGE analysis of the purified two Tenebrio AMPs. Pairs of lanes, such as 1 and 3 or 2 and 4, indicate the purified Tenecin 1 (peak 1) and the 7.5-kDa protein (peak 2), respectively. Lanes 1 and 2 and 3 and 4 show the gel mobility of these proteins under reducing and non-reducing conditions, respectively. D, N-terminal amino acid sequence comparison between peak 1 and Tenecin 1 (32), between peak 2 and Coleoptericin (34). E, amino acid sequence comparison between Tenecin 2, Coleoptericin, and Holotricin 2 (33).
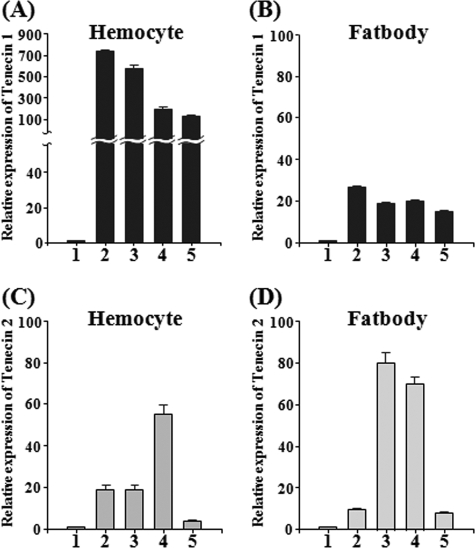
The Expression of Tenecin 1 Gene Is Dramatically Induced in Hemocytes
It is well known that most Drosophila AMP genes are induced in the adult fat body in response to the injection of bacteria (35). To examine the site of expression of Tenecin 1 and Tenecin 2, we carried out RT-qPCR analysis on mRNA fractions that are obtained from the fat body or hemocytes collected 12 h after injection of Lys-type PG, β-1,3-glucan, activated SAE, or cleaved Spätzle. Surprisingly, Tenecin 1 was dramatically induced in Tenebrio hemocytes, but only slightly induced in the fat body (Fig. 4, A and B). Tenecin 2 was moderately induced in the hemocyte and the fat body (Fig. 4, C and D). The equivalent mRNA expression levels of Tenecin 1 were observed at 3, 6, and 18 h after injection of these molecules into hemocytes (supplemental Fig. S3). These results demonstrate that in contrast to Diptera species such as Drosophila, the major site for the production of Tenebrio AMPs during systemic infection is in the hemocytes.
FIGURE 4.
mRNA expression levels of Tenecin 1 and Tenecin 2 in hemocytes and the fat body after injection of Lys-type PG, β-1,3-glucan, activated SAE, or processed Spätzle. A and B represent the mRNA levels of Tenecin 1 in hemocyte and the fat body, respectively. C and D represent the mRNA levels of Tenecin 2 in hemocyte and fat body, respectively. Insect saline (column 1), Lys-type PG (column 2), β-1,3-glucan (column 3), activated SAE (column 4), and processed Spätzle (column 5). The mRNA levels of Tenecin 1 or Tenecin 2 relative to that of insect saline-injected T. molitor larvae at 12 h after injection are shown. T-bars, mean ± S.D. (p < 0.05) of three independent experiments.
DISCUSSION
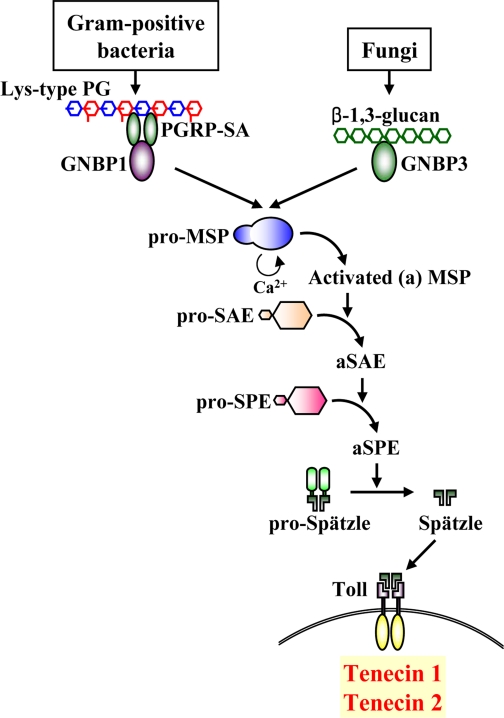
In this study, we report three novel findings regarding the β-1,3-glucan recognition signaling pathway in T. molitor larvae. We show that: 1) Tenebrio MSP functions apically downstream of GNBP3; 2) the β-1,3-glucan recognition signal is transferred via the sequential activation of the three Tenebrio serine proteases, namely MSP, SAE, and SPE, and that this results in the processing of pro-Spätzle to its mature form; and 3) the activation of the GNBP3/MSP/SAE/SPE/Spätzle module cascade regulates the expression of two AMPs, Tenecin 1 and Tenecin 2. Collectively, our results demonstrate that the Tenebrio GNBP3/MSP/SAE/SPE/Spätzle cascade is an essential unit involved in the recognition of fungi and in the activation of Toll-mediated antimicrobial defense (Fig. 5).
FIGURE 5.
A model of the serine proteases cascades regulating the Toll pathway in Tenebrio. When the β-1,3-glucan of fungi is exposed to the insect, Tenebrio GNBP3 binds to β-1,3-glucan and then recruits pro-MSP. In the presence of Ca2+, the β-1,3-glucan/GNBP3 complex induces activation of pro-MSP to activated MSP (aMSP). The aMSP converts pro-SAE to activated SAE (aSAE), which subsequently converts the pro-SPE to activated SPE (aSPE). The aSPE cleaves pro-Spätzle to processed Spätzle leading to the production of AMPs such as Tenecin 1 and Tenecin 2 via the Toll receptor. The Lys-type PG-dependent Tenebrio Toll signaling pathway was previously reported by our group (26).
Our current work, combined with our recent report (26), support a model in which Lys-type PG and β-1,3-glucan activate a common set of three serine protease zymogens sequentially (Fig. 5). This three-step proteolytic cascade-dependent processing of the extracellular pro-Spätzle produces active Spätzle, which then binds to the Toll receptor, resulting in the induction of AMP expression in the Tenebrio larval hemocytes (Fig. 5). Each of the three serine proteases has unique biochemical properties to regulate activation of the Tenebrio Toll pathway. The initial enzyme pro-MSP is an 82-kDa serine protease zymogen with an N-terminal chymotrypsin-like cleavage site and a C-terminal chymotrypsin-like catalytic serine protease domain. Pro-SAE is a 41-kDa serine protease with an N-terminal chymotrypsin cleavage site and a C-terminal trypsin-like catalytic serine protease domain, and pro-SPE is a 44-kDa protein with an N-terminal trypsin-like cleavage site and a C-terminal trypsin-like catalytic serine protease domain. These three different serine protease combinations enhance the specificity of the proteolytic serine protease cascade and prevent nonspecific cleavage of these serine protease zymogens.
Recent genetic studies using either in vivo RNAi strategy or loss-of-function mutation have demonstrated that several Drosophila serine proteases, such as Persephone, Grass, and Spirit, function upstream of the Drosophila SPE during activation of Toll (22). Furthermore, El Chamy et al. (9) demonstrated that Drosophila Grass functions as a signaling component required for both the detection of fungi by GNBP3 and the sensing of Gram-positive bacteria by PGRP-SA. Also, it was suggested that another Drosophila serine protease, Persephone, previously shown to be specific for fungal detection, was required for the sensing of proteases that are released by fungi and bacteria (9). This indicates that Drosophila Persephone defines a parallel proteolytic cascade activated by virulence factors in the hemolymph. However, Drosophila Grass and Persephone genes do not show any homology with Tenebrio MSP, which functions as an apical serine protease common to both Tenebrio GNBP3 and PGRP-SA/GNBP1 complex. These results suggest the existence of significant differences in the proteolytic cascades working upstream of Toll in Tenebrio and Drosophila. In addition to this diversity of serine proteases, the gene expression sites of Tenebrio AMPs are also different from those of Drosophila AMPs. The Tenebrio Tenecin 1 gene was strongly induced in hemocytes of the Tenebrio larvae, while Drosophila AMPs are mainly produced by the fat body. Taken together, the differences in the organization of the serine protease cascade, in the type of AMPs and in the site of AMP synthesis between Drosophila and Tenebrio could represent adaptation mechanisms to fight specific environmental pathogens or may be linked to differences in their physiology. Analyzing differences and similarities in the mechanisms used by insects to fight microbial infection should shed light on the evolution of the immune system in this important phylum.
Recently, we provided clear biochemical evidence that the activated Tenebrio SPE converted both the 79-kDa Tenebrio pro-phenoloxidase and clip-domain serine protease homologue 1 zymogen to their matured forms to generate an active melanization complex (27). This complex, consisting of a 76-kDa Tenebrio-activated phenoloxidase and a 43-kDa serine protease homologue 1, efficiently produced melanin on the surface of bacteria, and this activity had a strong bactericidal effect (27). Because Tenebrio SPE also participates in the activation of the Toll pathway by GNBP3 upon β-1,3-glucan sensing and by PGRP-SA upon Lys-type PG recognition, our results indicate that both the melanization and the Toll pathway immune modules are sharing a common serine protease cascade for the regulation of these two major innate immune responses.
In summary, our biochemical studies shed light on the molecular mechanism regulating the Toll pathway by fungi. Our work supports a model in which bacterial Lys-type PG and β-1,3-glucan recognition activate a common proteolytic cascade involving three different serine protease zymogens that are sequentially processed. This three-step proteolytic cascade leads to the maturation of Spätzle and the activation of the Toll intracellular signaling that control the synthesis of AMPs. A greater understanding of these two cascades could also facilitate the development of novel kits to detect bacteria and fungi in clinical and food products rapidly and sensitively.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PG
- peptidoglycan
- AMP
- antimicrobial peptide
- PGRP
- PG recognition protein
- GNBP
- Gram-negative-binding protein
- SP
- serine protease
- MSP
- modular serine protease
- SPE
- Spätzle-processing enzyme
- SAE
- SPE-activating enzyme
- i.d.
- inner diameter.
REFERENCES
- 1.Hoffmann J. A., Kafatos F. C., Janeway C. A., Ezekowitz R. A. (1999) Science 284, 1313–1318 [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R., Janeway C. A., Jr. (1997) Cell 91, 295–298 [DOI] [PubMed] [Google Scholar]
- 3.Akira S. (2003) Curr. Opin. Immunol. 15, 5–11 [DOI] [PubMed] [Google Scholar]
- 4.Janeway C. A., Jr. (1989) Cold Spring Harb. Symp. Quant. Biol. 54, 1–132700931 [Google Scholar]
- 5.Lemaitre B., Nicolas E., Michaut L., Reichhart J. M., Hoffmann J. A. (1996) Cell 86, 973–983 [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov R., Janeway C. A., Jr. (2002) Science 296, 298–300 [DOI] [PubMed] [Google Scholar]
- 7.Ferrandon D., Imler J. L., Hetru C., Hoffmann J. A. (2007) Nat. Rev. Immunol. 7, 862–874 [DOI] [PubMed] [Google Scholar]
- 8.Levashina E. A., Langley E., Green C., Gubb D., Ashburner M., Hoffmann J. A., Reichhart J. M. (1999) Science 285, 1917–1919 [DOI] [PubMed] [Google Scholar]
- 9.El Chamy L., Leclerc V., Caldelari I., Reichhart J. M. (2008) Nat. Immunol. 9, 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leulier F., Parquet C., Pili-Floury S., Ryu J. H., Caroff M., Lee W. J., Mengin-Lecreulx D., Lemaitre B. (2003) Nat. Immunol. 4, 478–484 [DOI] [PubMed] [Google Scholar]
- 11.Royet J., Reichhart J. M., Hoffmann J. A. (2005) Curr. Opin. Immunol. 17, 11–17 [DOI] [PubMed] [Google Scholar]
- 12.Cherry S., Silverman N. (2006) Nat. Immunol. 7, 911–917 [DOI] [PubMed] [Google Scholar]
- 13.Lemaitre B., Hoffmann J. (2007) Annu. Rev. Immunol. 25, 697–743 [DOI] [PubMed] [Google Scholar]
- 14.Michel T., Reichhart J. M., Hoffmann J. A., Royet J. (2001) Nature 414, 756–759 [DOI] [PubMed] [Google Scholar]
- 15.Bischoff V., Vignal C., Boneca I. G., Michel T., Hoffmann J. A., Royet J. (2004) Nat. Immunol. 5, 1175–1180 [DOI] [PubMed] [Google Scholar]
- 16.Gobert V., Gottar M., Matskevich A. A., Rutschmann S., Royet J., Belvin M., Hoffmann J. A., Ferrandon D. (2003) Science 302, 2126–2130 [DOI] [PubMed] [Google Scholar]
- 17.Pili-Floury S., Leulier F., Takahashi K., Saigo K., Samain E., Ueda R., Lemaitre B. (2004) J. Biol. Chem. 279, 12848–12853 [DOI] [PubMed] [Google Scholar]
- 18.Gottar M., Gobert V., Matskevich A. A., Reichhart J. M., Wang C., Butt T. M., Belvin M., Hoffmann J. A., Ferrandon D. (2006) Cell 127, 1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber A. N., Tauszig-Delamasure S., Hoffmann J. A., Lelièvre E., Gascan H., Ray K. P., Morse M. A., Imler J. L., Gay N. J. (2003) Nat. Immunol. 4, 794–800 [DOI] [PubMed] [Google Scholar]
- 20.Gay N. J., Gangloff M. (2007) Annu. Rev. Biochem. 76, 141–165 [DOI] [PubMed] [Google Scholar]
- 21.Jang I. H., Chosa N., Kim S. H., Nam H. J., Lemaitre B., Ochiai M., Kambris Z., Brun S., Hashimoto C., Ashida M., Brey P. T., Lee W. J. (2006) Dev. Cell 10, 45–55 [DOI] [PubMed] [Google Scholar]
- 22.Kambris Z., Brun S., Jang I. H., Nam H. J., Romeo Y., Takahashi K., Lee W. J., Ueda R., Lemaitre B. (2006) Curr. Biol. 16, 808–813 [DOI] [PubMed] [Google Scholar]
- 23.LeMosy E. K., Hong C. C., Hashimoto C. (1999) Trends Cell Biol. 9, 102–107 [DOI] [PubMed] [Google Scholar]
- 24.Jang I. H., Nam H. J., Lee W. J. (2008) BMB Rep. 41, 102–107 [DOI] [PubMed] [Google Scholar]
- 25.Fujita T. (2002) Nat. Rev. Immunol. 2, 346–353 [DOI] [PubMed] [Google Scholar]
- 26.Kim C. H., Kim S. J., Kan H., Kwon H. M., Roh K. B., Jiang R., Yang Y., Park J. W., Lee H. H., Ha N. C., Kang H. J., Nonaka M., Söderhäll K., Lee B. L. (2008) J. Biol. Chem. 283, 7599–7607 [DOI] [PubMed] [Google Scholar]
- 27.Kan H., Kim C. H., Kwon H. M., Park J. W., Roh K. B., Lee H., Park B. J., Zhang R., Zhang J., Söderhäll K., Ha N. C., Lee B. L. (2008) J. Biol. Chem. 283, 25316–25323 [DOI] [PubMed] [Google Scholar]
- 28.Ligoxygakis P., Pelte N., Hoffmann J. A., Reichhart J. M. (2002) Science 297, 114–116 [DOI] [PubMed] [Google Scholar]
- 29.Zhang R., Cho H. Y., Kim H. S., Ma Y. G., Osaki T., Kawabata S., Söderhäll K., Lee B. L. (2003) J. Biol. Chem. 278, 42072–42079 [DOI] [PubMed] [Google Scholar]
- 30.Park J. W., Je B. R., Piao S., Inamura S., Fujimoto Y., Fukase K., Kusumoto S., Söderhäll K., Ha N. C., Lee B. L. (2006) J. Biol. Chem. 281, 7747–7755 [DOI] [PubMed] [Google Scholar]
- 31.Park J. W., Kim C. H., Kim J. H., Je B. R., Roh K. B., Kim S. J., Lee H. H., Ryu J. H., Lim J. H., Oh B. H., Lee W. J., Ha N. C., Lee B. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6602–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon H. J., Lee S. Y., Kurata S., Natori S., Lee B. L. (1994) J. Biochem. 116, 53–58 [DOI] [PubMed] [Google Scholar]
- 33.Lee S. Y., Moon H. J., Kurata S., Kurama T., Natori S., Lee B. L. (1994) J. Biochem. 115, 82–86 [DOI] [PubMed] [Google Scholar]
- 34.Bulet P., Cociancich S., Dimarcq J. L., Lambert J., Reichhart J. M., Hoffmann D., Hetru C., Hoffmann J. A. (1991) J. Biol. Chem. 266, 24520–24525 [PubMed] [Google Scholar]
- 35.Bulet P., Hetru C., Dimarcq J. L., Hoffmann D. (1999) Dev. Comp. Immunol. 23, 329–344 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.