Abstract
Rpb9 is a small non-essential subunit of yeast RNA polymerase II located on the surface on the enzyme. Deletion of the RPB9 gene shows synthetic lethality with the low fidelity rpb1-E1103G mutation localized in the trigger loop, a mobile element of the catalytic Rpb1 subunit, which has been shown to control transcription fidelity. Similar to the rpb1-E1103G mutation, the RPB9 deletion substantially enhances NTP misincorporation and increases the rate of mismatch extension with the next cognate NTP in vitro. Using pre-steady state kinetic analysis, we show that RPB9 deletion promotes sequestration of NTPs in the polymerase active center just prior to the phosphodiester bond formation. We propose a model in which the Rpb9 subunit controls transcription fidelity by delaying the closure of the trigger loop on the incoming NTP via interaction between the C-terminal domain of Rpb9 and the trigger loop. Our findings reveal a mechanism for regulation of transcription fidelity by protein factors located at a large distance from the active center of RNA polymerase II.
The fidelity of transcription is essential for maintenance of genetic information content and for accurate gene expression. Given the structural and mechanistic similarities shared among all classes of multisubunit nucleic acid polymerases, the kinetic and mechanistic basis of fidelity is likely to be conserved in transcription. Nucleotide incorporation involves at least five steps: binding of NTP, isomerization into a catalytically competent complex, phosphoryl transfer, a second conformational change associated with release of pyrophosphate, and translocation (1–4). Although any of these steps can serve as fidelity checkpoints, early pre-steady state analyses suggested that the fidelity of DNA polymerases might be controlled at the isomerization step following the dNTP binding (3–6). The prevailing hypothesis holds that isomerization corresponds to the open-to-closed conformation transition in the protein after the initial binding of dNTP (7, 8). Arnold and co-workers (1, 9, 10) established that a conformational change preceding catalysis is a key fidelity checkpoint for the poliovirus RNA-dependent RNA polymerase RdRp (3Dpol). Importantly, they demonstrated that a mutation G64S, located far from the catalytic center of 3Dpol, affects isomerization of the active site (11).
It has recently been shown that the fidelity of RNA polymerase II (pol II)2 is similarly controlled at the isomerization step mediated by the trigger loop (TL), a mobile structural element within the largest Rpb1 subunit (12–14). The TL connects the ends of two α-helices forming a helical hairpin (15). This hairpin is located beneath another long helix (the bridge helix), which is proposed to participate in the catalysis and pol II translocation (16). The TL moves toward the active center upon binding of the complementary NTP. This transition involves a long range motion of the TL from the enzyme surface to the active center referred to as the open-to-closed TL transition (14). Previous studies provided evidence that the TL closing properly positions the NTP for catalysis and prevents spontaneous NTP release from the active center (13, 14). The TL adopts an open configuration in pol II elongation complexes co-crystallized with a non-complementary substrate (14). Several mutations in the TL increase misincorporation (12). Recently, we have characterized a mutation in the distal part of the helical hairpin located at a significant distance from the active center, which rendered pol II error-prone (13, 17). This mutation, rpb1-E1103G, was proposed to destabilize the open conformation of the TL, causing overly efficient trapping of non-complementary NTPs and misincorporation (13). Therefore, the dynamics of the active site isomerization mediated by the TL movement appear to control transcription fidelity.
In addition to Rpb1, it has been suggested that Rpb9, a small subunit of pol II, plays a role in transcription fidelity (18). Rpb9 is a non-essential subunit located on the surface of the enzyme (19, 20). Rpb9 comprises two zinc ribbon domains joined by a conserved linker: an N-terminal domain (Zn1) forming a part of the primary DNA-binding channel of pol II and a C-terminal domain (Zn2) interacting with the secondary channel, a special pore connecting the active center with the surface of the enzyme (see Fig. 6). Based on pre-steady state kinetic analysis of bond formation by human pol II, it has been proposed that NTP substrates enter the enzyme through the primary DNA-binding channel (main channel) by prehybridization to the template DNA strand 2–3 nucleotides ahead of the active center (21). The N-terminal domain of Rpb9 constitutes a significant part of the primary channel, which suggests that Rpb9 deletion may affect fidelity by altering the structure of the putative NTP preloading site. Alternatively, the secondary channel has been proposed to serve for the NTP entry to the active center and release of pyrophosphate after bond formation (22, 23). The secondary channel also provides access to the active center for transcript cleavage factor TFIIS (22), which is involved in proofreading transcription errors (24) and rescuing arrested complexes of pol II (25). Interestingly, the C-terminal domain of Rpb9 is structurally similar to the zinc ribbon domain of TFIIS (25, 26). The Rpb9 subunit is highly conserved among eukaryotes and shows significant sequence similarity with the C11 subunit in pol III and the A12.2 in pol I, both required for the intrinsic cleavage activity of their related polymerases (27–29). Although Rpb9 has never been demonstrated to contribute to the intrinsic cleavage activity of pol II, it is nevertheless required for optimal function of transcript cleavage factor TFIIS (26, 30). Yeast cells harboring the RPB9 deletion confer a slow growth phenotype with disruption of DST1, the gene encoding elongation factor TFIIS in certain genetic backgrounds (31). RPB9 deletion in yeast causes hypersensitivity to the nucleotide-depleting drug 6-azauracil (31), a phenotype frequently associated with transcription elongation defects (32). Recently, Rpb9 has been implicated in maintenance of transcription fidelity in vivo based on the increased frequency of transcriptional frameshift and base substitution events in an RPB9 deletion strain (18, 33). The fidelity checkpoint, affected by the RPB9 deletion, remains to be identified. The synergistic phenotype of RPB9 deletion with DST1 deletion (31) and altered TFIIS-dependent transcript cleavage pattern in ternary elongation complexes (TECs) lacking the Rpb9 subunit (30) suggest that Rpb9 could be involved in TFIIS-mediated error correction (24, 33). However, the reports regarding the effect of TFIIS on transcription fidelity in vivo remain controversial. Koyama et al. (34) showed that TFIIS contributes to maintenance of transcription fidelity and that Rpb9 also controls fidelity in a TFIIS-independent manner (33). Therefore, Rpb9 might also be involved in fidelity control at another step of the transcription elongation cycle, distinct from the TFIIS-dependent error correction step. In contrast, several other genetic tests failed to detect a significant effect of TFIIS in transcription fidelity in vivo (18, 35).
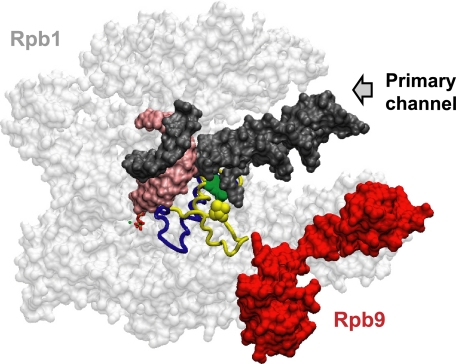
FIGURE 6.
Model describing the effect of RPB9 deletion on transcription fidelity. TEC structures of Saccharomyces cerevisiae pol II with the closed trigger loop (blue) and the open trigger loop (yellow) (PDB: 2E2H (14) and PDB: 1Y1V (22)) were aligned using the VMD software (59). Rpb1 (gray) is made semitransparent to visualize the location of the TL inside the pol II structure, and Rpb9 is shown in red. The NTP in the active center of pol II is shown on the left side of the TL. The DNA backbone and RNA are shown in dark gray and pink, respectively. The primary DNA-binding channel in pol II is indicated by the leftward arrow. The secondary channel is located at the opposite side of the structure. The Rpb9 subunit stabilizes the open conformation of the TL by interaction between the C-terminal domain of Rpb9 with the tip of the TL (yellow), similar to the proposed interaction between Rpb1-Glu1103 (green) with Rpb1-Thr1095 (yellow) (13).
In this work, we established a genetic interaction between the Rpb9 subunit and the TL by showing that deletion of the RPB9 gene is synthetic lethal with the rpb1-E1103G mutation. We also showed that Rpb9-deficient pol II confers a significant decrease in transcription fidelity in vitro. Pre-steady state kinetic analysis revealed that the absence of Rpb9 promoted NTP sequestration in the polymerase active center before catalysis, similar to that observed for the rpb1-E1103G mutation (13). Our findings support a crucial role for active site isomerization in transcription fidelity and establish a paradigm for catalytic control of pol II by cis-acting regulatory elements like Rpb9 and the TL or trans-acting elongation factors like TFIIS.
EXPERIMENTAL PROCEDURES
Media, Yeast Manipulations, and Strains
Media, growth conditions, and yeast manipulations were as described previously (Malagon et al. (17)). Briefly, GRY3041 (MATα, can1, his-3Δ200, leu2-Δ1, trp1, ura3-52, pep4::HIS3, prb1-Δ1.6, RPB3::6xHisBio, rpb9::kanMX4, GAL+) was made by one step gene disruption of the RPB9 gene with the kanMX4 cassette (from the Open Biosystems deletion collection) and is related to BJ5464 (ATTC Yeast Genetic Stock Center). To determine whether rpb9::Nat and rpb1-E1103G show a synthetic interaction, the strains GRY3185 (MATα, his3-Δ1, lys2-Δ0, leu2-Δ0, met15-Δ0, trp-, URA3-CMV-tTA, KanMX-Ptet-RPB1, rpb9::Nat) and GRY3028 (MATa, his3-Δ1, leu2-Δ0, lys2-Δ0, met15-Δ0, trp1::hisG, URA3-CMV-tTA, rpb1-E1103G) were crossed and subjected to tetrad analysis. The KanMX-Ptet-RPB1 allele and the CMV-tTA transactivator were introduced by crosses with the strain YSC1180-7428981 (Open Biosystems (36); see Ref. 17). All yeast strain genotypes are described in supplemental Table 1.
Enzymes, Oligonucleotides, and Reagents
DNA and RNA oligonucleotides (supplemental Table 2) were purchased from IDT (San Diego, CA). NTPs were purchased from Amersham Biosciences, and dNTPs were from Invitrogen. The NTPs used for misincorporation assays were additionally purified as described previously (13). Exonuclease III was from New England Biolabs (Beverly, MA). WT pol II and pol IIΔ9 were purified as described previously (13). Subunit composition of pol II and pol IIΔ9 was analyzed by gel electrophoresis on a 4–12% polyacrylamide gel in denaturing conditions, and proteins were visualized by silver staining.
TEC Assembly and in Vitro Transcription Assays
Transcription elongation complexes were assembled on synthetic DNA and RNA oligonucleotides and immobilized on Ni2+-NTA agarose beads (Qiagen, Valencia, CA) as described previously (37, 38). Briefly, the 5′ radioactively labeled RNA (RNA7, RNA7–20, or RNA9) and the template DNA oligonucleotide (TDS45G, TDS45C, TDS50, or TDS65) were incubated with the immobilized pol II followed by the subsequent addition of the non-template DNA oligonucleotide (NDS45G, NDS45C, NDS50, or NDS65). When indicated, the RNA was extended to different positions subsequent to incubation with NTP subsets (5 μm, final concentration). The TECs were eluted by 100 mm imidazole, filtered using Ultrafree-MC 0.45-μm centrifugal filters (Millipore, Billerica, MA), and diluted with bovine serum albumin-containing transcription buffer (20 mm Tris-HCl, pH 7.9, 5 mm MgCl2, 1 mm 2-mercaptoethanol, 40 mm KCl, 0.1 mm mg/ml bovine serum albumin, 12% glycerol). The reactions with the correct NTP were performed on the model RFQ-3 and RFQ-4 chemical quench flow instruments (KinTek Corp., Austin, TX). TECs were incubated with NTPs for 0.002–0.5 s at 25 °C followed by stopping the reaction by quenching with 0.5 m EDTA or 1 m HCl. The misincorporation reactions were stopped manually by quenching with the gel-loading buffer (5 m urea; 25 mm EDTA final concentrations). The RNA products were resolved in 20% denaturing polyacrylamide gel, visualized with a Typhoon 8600 PhosphorImager, and quantified using ImageQuant software (GE Healthcare). The analyses of the experimental data were performed using the OriginPro 7.5 software (OriginLab Corp., Northampton, MA).
Exonuclease III Footprinting
Elongation complexes were assembled with the wild-type enzyme and pol IIΔ9 with RNA9 on TDS65CCC with the fully complementary NDS65CCC (13). Both the transcript and the template DNA strand were 32P-labeled at the 5′ end using [γ32P]ATP and T4 polynucleotide kinase. The non-labeled DNA strand in the scaffold contained a Biotin linkage at the 3′ end position to inhibit cleavage of that strand by exonuclease III (Exo III). The TECs were eluted from the Ni2+-NTA agarose beads as described above. The footprinting reactions were initiated by mixing 15 μl of the TECs with 15 μl of transcription buffer containing 100 units of Exo III. To examine the effect of the incoming NTP on translocation, either the next complementary NTP (CTP) or a non-complementary NTP (UTP) was mixed with Exo III followed by adding TEC to the reaction. The NTP concentration was 1 mm final. Reactions were stopped by the addition of an equal volume of 2× gel loading buffer. Products of the reactions were resolved by 12% PAGE in the presence of 7 m urea.
RESULTS
Synthetic Lethal Interaction of rpb1-E1103G and rpb9-Δ
Identification and characterization of the rpb1-E1103G mutation (17) that decreases transcription fidelity (13) provided a tool for the identification of other mutations affecting fidelity in vivo. pol II carrying this substitution has an elevated level of base substitutions in the RNA it produces. In vivo, the rpb1-E1103G mutation results in a dependence on the transcription factor TFIIS consistent with the editing role of TFIIS in removing misincorporated bases. Because it has been reported that rpb9 mutants have elevated transcription errors (18), we tested for a genetic interaction between rpb1-E1103G and an open reading frame deletion allele of rpb9. A strain (GRY3185) carrying a deletion allele of rpb9 marked with nourseothricin resistance (rpb9::nat) and an allele of RPB1 marked with G418 resistance (RPB1::kanMX) was crossed to a yeast strain (GRY3028) carrying rpb1-E1103G. Tetrads from this strain showed a clear synthetic lethal segregation pattern indicating that the double mutant rpb1-E1103G, rpb9::nat is inviable. Of the 290 meiotic segregants carrying the RPB1::kanMX allele, 56% carried the rpb9::nat allele, whereas none of the 162 rpb1-E1103G segregants that survived carried the rpb9::nat allele. The lethal phenotype of the double mutant could reflect a direct role in avoiding errors, which has a synergistic lethal consequence with rpb1-E1103G when Rpb9 is absent, or an essential role for Rpb9 in correcting errors elevated in rpb1-E1103G cells. We assessed the in vitro catalytic properties and pre-incorporation transcription fidelity of pol II lacking the Rpb9 subunit (pol IIΔ9).
Loss of Rpb9 Does Not Significantly Affect the Catalytic Properties of pol II
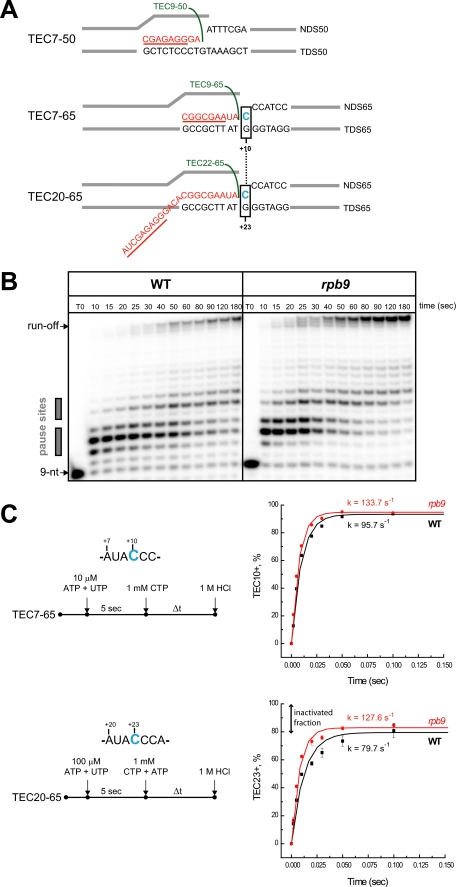
It was previously reported that the Rpb9 subunit is involved in the control of pol II elongation rate in vitro (26, 30). First, we compared the elongation properties of the WT pol II and pol IIΔ9. We utilized a previously published technique for assembly of the functional TECs of the yeast pol II in the absence of promoter sequence and initiation factors (38). This method employs the core enzyme of pol II assembled on synthetic DNA and short RNA primers (7–9 nt). For the bulk transcription elongation assay, the TECs were assembled on the 50-nt template and non-template DNA oligonucleotides with 7-nt RNA (TEC7) primer and immobilized on Ni2+-NTA agarose beads (Fig. 1A). TEC7 was extended to TEC9 with ATP and GTP, washed with transcription buffer, eluted from the beads (see “Experimental Procedures”), and chased with all four NTPs. Incubation with NTPs resulted in the accumulation of a 28-nt runoff product, which occurred 2–3 times faster with pol IIΔ9 than with the WT enzyme (Fig. 1B). This increased rate observed for pol IIΔ9 may predominantly derive from the decreased pausing at the intrinsic +11/+12 pause sites in the template (Fig. 1B). These results are consistent with previously reported properties of yeast pol II lacking the Rpb9 subunit (26, 30).
FIGURE 1.
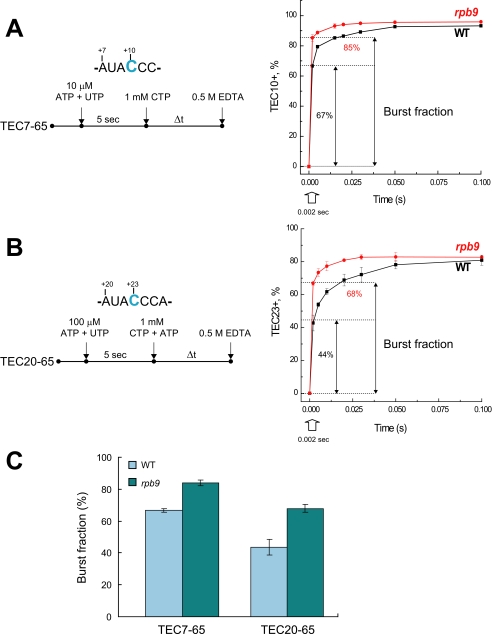
RPB9 deletion has a minor effect on the catalytic properties of pol II. A, design of the assembled TEC7–50, TEC7–65, and TEC20–65. RNA is colored in red, and the sequences of the RNA primers for the TEC assembly are underlined and described in supplemental Table 2. B, bulk elongation assays. TEC7–50 was walked to position +9 with 5 μm each of GTP and ATP (TEC9–50, indicated in green), washed, and incubated with 2 μm of all four NTPs for various time intervals. The pause sites are indicated by gray boxes. C, time course of CTP incorporation at positions +10 and +23 of the transcript by TEC7–65 and TEC20–65, respectively. Black symbols and lines show the data set for WT pol II, and red symbols and lines represent pol IIΔ9. The curves represent a single exponential fit of the experimental data and the apparent rates (k) are shown for each reaction. Note that for the experiments described in the legends for Figs. 1C and 5, TEC9–65 and TEC22–65 were chased with CTP or with a mixture of CTP and ATP, respectively. The presence of ATP in the chase mixture does not affect CTP incorporation kinetics by TEC22–65.
Bulk assays do not measure the dynamics of elongation at a single nucleotide level but provide information solely on the average rate of elongation, mostly defined by rare pausing of pol II on a few DNA sequences. Therefore, we performed pre-steady state kinetic analysis of a single NTP addition at a non-pause site with TEC7–65 and TEC20–65 (Fig. 1A). These two TECs share the same sequences near the 3′ end of the RNA but carry transcripts of different lengths. Moreover, TEC20–65 contains a long 20-nt transcript and is capable of backtracking along DNA. Backtracking involves extrusion of the 3′ end of the nascent transcript to the secondary channel, placing it in close proximity to the C-terminal domain of the Rpb9 subunit (19). For the pre-steady state kinetic analysis, both TECs were incubated with ATP and UTP for 5 s, allowing extension of TEC7 to TEC9 and TEC20 to TEC22, and chased with 1 mm CTP (TEC9) or with CTP and ATP (TEC22). The reactions were quenched after various times with 1 m HCl (Fig. 1C, right-hand panel), which stops the reaction instantly and provides information on the rate of the bond formation (21). By fitting the time-dependent formation of C10 or C23 transcripts to a single exponential equation, we determined the pseudo-first-order rate constants of CTP incorporation for TECs formed with pol II and pol IIΔ9 (Fig. 1C). We found that deletion of the Rpb9 subunit either did not affect (Table 1) or moderately increased (Figs. 1C and 3A) the rate for the correct CTP incorporation depending on the local sequence context. This 1.3–1.5-fold rate increase was similar to that reported for the TL rpb1-E1103G mutation (13). These data indicate that the Rpb9 subunit is not involved in the bond synthesis per se, which is fully consistent with its location far from the catalytic site of pol II.
TABLE 1.
Deletion of the Rpb9 subunit confers a 5-fold decrease in fidelity of transcription in vitro
TEC9 was assembled on the template 65 and chased with 0.01–1 mm CTP or 0.1–2 mm UTP. The kpol and KD parameters were obtained from the hyperbolic fit as described in the supplemental text. The fidelity parameter in the last column (bold) for the WT and pol IIΔ9 is determined by dividing kpol/KD for the correct CTP and kpol/KD for the incorrect UTP.
| pol II variant | NTP | kpol | KD | kpol/KD | Fidelity |
|---|---|---|---|---|---|
| s−1 | μm | s−1μm−1 | |||
| WT | CTP (correct) | 75 ± 4 | 68 ± 7 | 1.1 ± 0.2 | |
| UTP (incorrect) | (7.8 ± 0.9) × 10−3 | 3531 ± 556 | (2.2 ± 0.6) × 10−6 | (5 ± 2.1) × 105 | |
| rpb9 | CTP (correct) | 60.5 ± 4.6 | 76.1 ± 19.1 | 0.8 ± 0.2 | |
| UTP (incorrect) | (20 ± 3.9) × 10−3 | 2860 ± 792 | (6.9 ± 3) × 10−6 | (1.1 ± 0.6) × 105 |
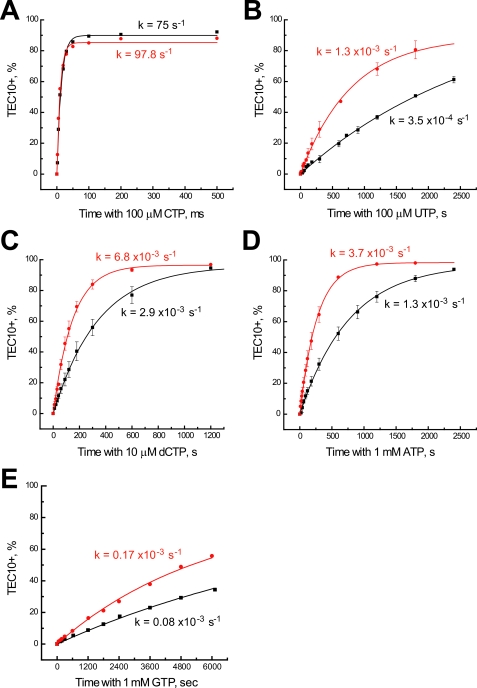
FIGURE 3.
RPB9 deletion causes a uniform 3-fold increase of different types of misincorporation. WT or pol IIΔ9 TEC8 obtained from TEC7 on template 45C was incubated with 10 μm ATP (TEC9) for 30 s, after which 0.1 mm CTP (A), 0.1 mm UTP (B), 10 μm dCTP (C), 1 mm ATP (D), or 1 mm GTP (E) was added. The reaction with CTP was stopped with 1 m HCl. The experimental data set obtained for the WT (black) and pol IIΔ9 (red) are represented as in Fig. 1C. Error bars indicate S.E. Panel E shows GTP misincorporation.
A comparison of the CTP incorporation dynamics by TEC9 and TEC22 revealed that although nearly 100% of the pol II that starts elongation from TEC9 continues to incorporate through position +10, only about 80% of the TEC22 continues to elongate to TEC23. We interpreted these 20% inactive TEC22 complexes as stalled by backtracking (Fig. 1C, bottom graph). This result indicated that TEC22 was subject to the catalytic inactivation by backtracking along DNA and that deletion of the Rpb9 subunit did not affect the backtracking.
Rpb9 Deletion Promotes NTP Misincorporation
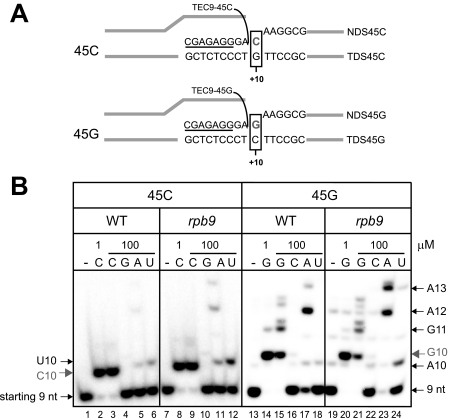
To test the role of the Rpb9 subunit in transcription accuracy, we compared the transcription error pattern of pol II and pol IIΔ9 in a single NTP misincorporation assay in vitro. The templates 45C and 45G are identical except at the +10 position, where CTP or GTP is incorporated into the RNA as a cognate substrate to TEC9 (Fig. 2A). WT and pol IIΔ9 TECs were assembled on the 45-nt template using the 7-nt RNA (TEC7), extended 2 bases to TEC9, washed, and incubated for 10 min with highly purified NTP substrates. The distinct mobility of the products carrying different misincorporated NMPs (Fig. 2B, compare lanes 3, 5, and 6 for templates 45C and lanes 15, 17, and 18 for templates 45G) eliminated the possibility that the RNA extension in this assay derived from contamination of NTP preparations with trace amounts of cognate NTPs. Notably, Rpb9 deletion led to a moderate increase in “transition-type” misincorporation, which resulted in purine-to-purine or pyrimidine-to-pyrimidine substitution (Fig. 2B, compare lanes 6 and 12 or lanes 17 and 23). Second, the deletion also promoted some transversion-type events, such as incorporation of ATP for CTP (lanes 5 and 11) and incorporation of UTP for GTP (lanes 18 and 24). The Rpb9 deletion also appeared to promote extension of a misincorporated base with the next cognate NTP more efficiently than the WT (lanes 5 and 11; lanes 17 and 23), which we addressed in more detail in the supplemental data and supplemental Figs. S1 and S2.
FIGURE 2.
RPB9 deletion promotes NTP misincorporation by pol II in vitro. A, TECs were assembled with fully complementary 45-nt template and non-template DNA strands and 7-nt RNA oligonucleotide (underlined). The TECs were then walked to TEC9 with 5 μm each of ATP and GTP, washed, and incubated with purified NTPs for 10 min. B, the resulting 10-nt products are named according to the base at the 3′ end (G10 incorporated GMP at the 3′ end, etc.). Gray and black arrows indicate normal incorporation products and misincorporation, respectively. The longer RNAs G11, A12, and A13 derived from misincorporation events followed by mismatch extension.
To establish the quantitative effect of Rpb9 deletion on fidelity, we compared the incorporation rates for the correct (CTP) and incorrect (dCTP, ATP, UTP, and GTP) nucleotides by the WT and pol IIΔ9 at a given template position (Fig. 3). The correct CTP incorporation proceeded rapidly with the WT and pol IIΔ9, reaching completion by 50 ms at 100 μm concentration of the cognate substrate (Fig. 3A). In this sequence context, the absence of the Rpb9 subunit conferred only a 30% increase in the rate of the correct NTP incorporation (Fig. 3A). In contrast, the effect of Rpb9 deletion on misincorporation was substantially more pronounced (Fig. 4, B–E). pol IIΔ9 incorporated the non-complementary NTPs (ATP, UTP, and GTP) and dCTP 2-to-3-fold faster than the WT pol II. As a result, the accuracy of NTP and dCTP incorporation by pol IIΔ9 is decreased when compared with the WT enzyme.
FIGURE 4.
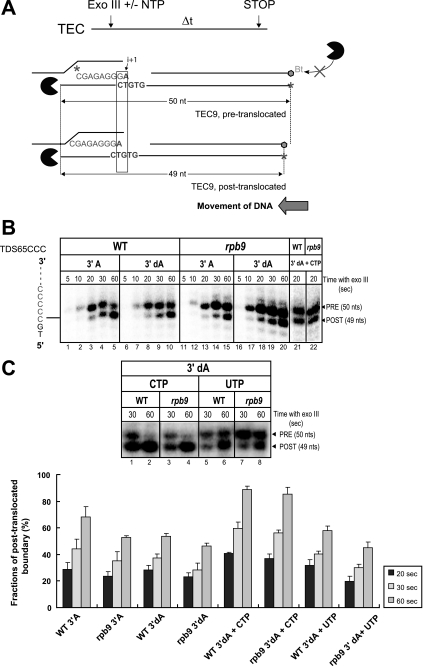
Mapping of the translocation state of TECs using exonuclease III footprinting of the rear of pol II boundary. A, design of the Exo III experiment. The figure illustrates the pre- and post-translocated states of TEC9. The length of the template DNA strand protected from the digestion differs by one nucleotide between the pre-translocated (top) and post-translocated (bottom) state of TEC9. A biotin group (in black) at the 3′ end of the non-template strand prevents its degradation by Exo III (in gray). B, TECs by WT and pol IIΔ9 were assembled on templates NDS65CCC with the 5′-labeled RNA7 followed by walking to TEC8 by incubation with 5 μm GTP. TEC9 was obtained by a 5-min incubation of TEC8 with 10 μm ATP or 3′-dATP and was incubated for short time intervals with Exo III. C, forward translocation of 3′-dA-terminated TEC9 is stimulated by the correct CTP added at 1 mm simultaneously with Exo III. The bottom panel depicts dynamics of the appearance of the post-translocated footprint of TEC9. Error bars indicate S.E.
The kinetics of nucleotide incorporation at a given substrate concentration is a function of the NTP binding to the enzyme and the maximal rate of nucleotide incorporation. Comparison of incorporation and misincorporation rates at physiological concentrations of NTP (Fig. 3) provides an estimation of the effect of Rpb9 deletion on transcription fidelity. Quantitatively, replication and transcription fidelity is calculated as the ratio of (kpol/KD) parameters for the correct and the incorrect NTP (1, 4, 13). We determined these parameters for incorporation of the correct (CTP) and the incorrect (UTP) nucleotide at position +10 of the transcript by WT pol II and pol IIΔ9. The reaction rates were determined over a 0.01–1 mm range of CTP and a 0.1–2 mm range of UTP (supplemental Fig. S3) and plotted against the substrate concentration (supplemental Fig. S4). The maximum polymerization rate and the dissociation constant were obtained from the resulting data by a non-linear fit using the following equation k = (kpol × [NTP])/(KD + [NTP]). The results for incorporation and misincorporation are shown in Table 1. The dissociation constant (KD, app) for the non-cognate NTP was slightly lower for pol IIΔ9 when compared with the WT enzyme (Table 1). The absence of the Rpb9 subunit caused a 3-fold increase (from 0.0078 to 0.02 s−1) of the maximal rate constant for the incorrect nucleotide incorporation (kpol for UTP). As a result, the fidelity of pol IIΔ9 is almost five times lower than the fidelity of the WT pol II for UTP misincorporation.
Rpb9 Subunit and Translocation Properties of pol II
Exo III footprinting is widely used for evaluation of the translocation state of a bacterial TEC stalled on a double-stranded DNA template (15, 39–41). Recently, we optimized the Exo III footprinting approach (supplemental data) to analyze the pre- and post-translocated equilibrium of pol II (13). The pre- and post-translocated states are defined with regard to the position of the 3′ end of the transcript relative to the pol II active site. The pre-translocated TEC contains the 3′ end of the RNA, occluding the NTP-binding pocket in the active center, preventing loading of the next substrate. The post-translocated TEC originates from the pre-translocated complex by one-base pair forward movement of the enzyme along the RNA and the DNA, leading to the clearance of the NTP-binding pocket for binding the next substrate. In the absence of NTPs, the TEC appears to be in equilibrium between these two states at each template position. The translocation register can be detected using Exo III footprinting as described below and in the supplemental data. We used TECs carrying a 9-nt RNA (Fig. 4A), which resist backtracking due to the short transcript length; backtracking could interfere with the identification of the correct front and rear boundaries of TECs (40). We analyzed a time course of template digestion because the equilibrium between the pre- and post-translocation states in the TEC is detected at shorter time points. Only two upstream rear boundaries appear on the TEC9 footprints with the WT and pol IIΔ9 enzymes: (i) the boundary located 50-nt from the labeled 5′ end of the template DNA (and 16 bp upstream from the RNA 3′ end), which was detected at the shortest incubation time with Exo III, and (ii) the boundary located 49 nt from the labeled end, which was evident upon longer incubation with Exo III. We assigned the first boundary to the pre-translocated TEC9 and the second boundary to the post-translocated state of the complex (Fig. 4A).
For the WT TEC9, the pre-translocated boundary appeared after 10 s of incubation with Exo III followed by its slow conversion to the post-translocated boundary (Fig. 4B, lanes 1–5), leading to an equal distribution between those two at ∼30-s incubation with Exo III (lane 4). Thus, WT TEC9 was predominantly dwelling in the pre-translocated state. Notably, the distribution of the pre/post boundaries in pol IIΔ9 TEC9 showed that the TEC is more inclined to reside in the pre-translocated state (Fig. 4B, lanes 11–15 and lanes 5 and 15), which was similar to the translocation distribution earlier reported for the rpb1-E1103G pol II mutant (13). In summary, pol IIΔ9 exhibits a minor increase of the elongation rate and a significant increase in NTP misincorporation, which is associated with the preferential dwelling of the mutant TEC in the pre-translocated state (13). These results challenge the commonly accepted correlation between the post-translocated state of the TEC as a source of rapid bond formation (15, 42) and also as a cause of decreased fidelity (40).
The binding of a complementary NTP to pol II in conditions prohibiting formation of the phosphodiester bond induces a characteristic shift of the equilibrium in the TEC to the post-translocated state (13, 15, 40, 43). This effect is attributed to the stabilization of the post-translocated state by the cognate substrate bound in the active center. We modified TEC9 by incorporating a 3′-dAMP to prevent the addition of the next NTP (43) and tested whether deletion of the Rpb9 subunit affected the NTP-induced lateral stabilization. We found that incorporation of the terminating analogue increased the pre-translocated fraction of the TEC (Fig. 4B, compare lanes 1–5 and lanes 6–10) and practically eliminated the difference in the translocation equilibrium between the WT and pol IIΔ9 (Fig. 4B, compare lanes 6–10 and lanes 16–20). Binding of the complementary CTP equally shifted the equilibrium in TEC9 toward the post-translocated state for both enzyme variants, whereas the non-cognate UTP produced no effect (Fig. 4B, lanes 21–22, and 4C). We concluded that absence of the Rpb9 subunit does not significantly affect the stabilization of the post-translocated conformation by the incoming NTP.
Rpb9 Deletion Promotes Sequestration of the NTP in the Active Center of pol II
A pre-steady state kinetic analysis of yeast and human pol II revealed that the complementary NTP rapidly binds to the enzyme followed by a reversible isomerization of the complex into the catalytically competent closed conformation poised for bond formation (13, 21). NTP binding can be distinguished from the subsequent chemical step by employing HCl or EDTA to quench the reaction (13, 21, 44). The instant inactivation of the enzyme by acid provides information on the rate of bond synthesis (chemistry), which fits to a single exponential curve (21). In contrast, the kinetics of NTP incorporation obtained with EDTA quench appears biphasic (13, 21): (i) a rapid incorporation (burst) phase, with a rate exceeding 1000 s−1, followed by (ii) a slower phase with a rate of 50–100 s−1. The addition of EDTA depletes the free NTP-Mg2+ pool without affecting the enzyme-bound NTP-Mg2+ and the subsequent phosphodiester bond formation (13). Therefore, the burst fraction corresponds to the TEC-NTP-Mg2+ complex that completes the phosphodiester bond formation before NTP-Mg2+ dissociation. Furthermore, kinetic modeling revealed that the biphasic nature of the EDTA quench might be explained by a reversible closing (isomerization) of the active site, which prevents NTP-Mg2+ dissociation (sequesters NTP-Mg2+). According to this model, the burst amplitude is proportional to the dwell time of the closed complex; an increase of the burst reflects a delay in transition to the open complex. We previously observed that the rpb1-E1103G mutation increased the burst fraction in the EDTA quench and concluded that the mutation rendered NTP sequestration less reversible (13).
We compared pre-steady state kinetics using EDTA quench in TECs by the WT and pol IIΔ9 assembled on the 65-bp DNA template (Fig. 5). The assembled complexes were incubated with ATP and UTP at the indicated concentration for 5 s and chased with the addition of CTP at 1 mm, allowing extension from TEC9 to TEC10 (Fig. 5A), or by the addition of ATP and CTP to extend TEC22 to TEC23 and beyond (Fig. 5B). The reactions were quenched with 0.5 m EDTA after a 0.002–0.5-s incubation with NTPs. As expected, the time dependence of C10 or C23 (reported as TEC10+ or TEC23+ plus longer transcripts) synthesis is described by biphasic kinetics. Fig. 5A illustrates that pol IIΔ9, despite being in the pre-translocated state, sequestered the incoming complementary CTP, as judged by the EDTA quench, more efficiently than the WT pol II, as revealed by the increase of the burst fraction at the 2-ms quench time (13). The increase was from 67% (WT) to 85% (pol IIΔ9) in the TEC with the short transcript and from 44% (WT) to 68% (pol IIΔ9) in the TEC with the longer transcript (Fig. 5C). This result suggests that the Rpb9 subunit favors the opening of the pol II active center. A similar increase in the apparent NTP incorporation burst (from 70 to 90%) has been observed for rpb1-E1103G pol II, in which the equilibrium between the closed and open TL is shifted toward the closed TL (13). Such a phenomenon has also been reported for the poliovirus RNA-dependent RNA polymerase 3Dpol (9). These authors observed the burst of a single bond formation in 3Dpol in the presence of manganese ion substituting for magnesium in the reaction quenched with EDTA. This result was interpreted as a more efficient sequestration of the incoming NTP by the enzyme in the presence of Mn2+ in the active center. Similarly, we observed that replacement of Mg2+ by Mn2+ promotes NTP sequestration for both pol II variants (supplemental data and supplemental Fig. S5B). Moreover, Mn2+ dramatically promotes misincorporation for both the WT and the pol IIΔ9 (supplemental Fig. S5A). We concluded that Mn2+, deletion of the Rpb9 subunit, and the rpb1-E1103G mutation decreased the fidelity of yeast pol II by a similar mechanism, which involves the increased isomerization of TEC into the closed conformation mediated by premature closing of the trigger loop.
FIGURE 5.
RPB9 deletion promotes NTP sequestration in the active center of pol II. A and B, time course of CTP incorporation monitored in TEC7–65 (A) and TEC20–65 (B) at positions +10 and +23, respectively, in reactions quenched with 0.5 m EDTA for the WT (black) and pol IIΔ9 (red). The double-headed arrows indicate the burst fraction observed at 2 ms of quench time. C, quantification of the results of panel B showing the size of the apparent burst of CTP incorporation within the first 2 ms. Error bars indicate S.E.
DISCUSSION
Our present work identifies a novel transcription fidelity mechanism involving the peripheral Rpb9 subunit of yeast pol II. In vivo results obtained by Koyama et al. (33) imply that transcription fidelity is controlled at the pre-incorporation and post-incorporation (error correction) levels; the pre-incorporation step is mediated by Rpb9, whereas the post-incorporation step involves both Rpb9 and TFIIS. The pre-incorporation fidelity checkpoint depends on the function of the TL, a mobile element of the catalytic Rpb1 subunit (14). TL closure promotes the phosphodiester bond formation with the correctly paired substrate via hydrogen bonding of the Rpb1-His1085 residue with β- and γ-phosphates of the incoming NTP (14). Mutations of His1085 compromise the fidelity of pol II (12). Additionally, substitution distal to the NTP-interacting part of the TL decreases pol II selectivity as well (12, 13). Based on characterization of one of these mutants, rpb1-E1103G, we proposed that dissociation of any non-cognate NTP substrate from the enzyme is facilitated by a delay in the TL closure caused by direct interaction of the Rpb1-Glu1103 residue with the Rpb1-Thr1095 residue localized to the mobile part of the TL (13). Therefore, the TL is hypothesized to play a dual role in fidelity: (i) to delay catalysis of inappropriate substrate NTP and (ii) to render NTP binding more reversible.
We show that the rpb1-E1103G mutation exhibits synthetic lethality with RPB9 gene deletion. Both RPB9 deletion and the rpb1-E1103G mutation suppress transient pausing of pol II. Strikingly, pol II lacking the Rpb9 subunit possesses the same fidelity defects as rpb1-E1103G pol II, including promotion of misincorporation errors and impaired discrimination between dNTP and rNTP. Both pol II variants sequester the incoming NTP in the active center of the enzyme with higher efficiency than the wild-type pol II. Collectively, the genetic interaction and the close biochemical similarity between the E1103G mutation and the RPB9 deletion support the idea that deletion of RPB9 affects the same fidelity checkpoint as the one impaired by the rpb1-E1103G TL mutation.
How does the Rpb9 subunit, located at the perimeter of the enzyme and at a significant distance from the active center (19, 20), affect TL closure? Multiple conformations of the TL have been revealed in different crystals with positions in various proximities to the active center (14, 22, 45). Upon close examination of the structure, we noted that in the open loop conformation, the C-terminal domain of Rpb9 localizes to the close proximity of the tip of the TL (Fig. 6 and supplemental Fig. S6). This spatial vicinity may delay the TL closure, similar to that proposed for Thr1095 residue in the flexible part of the TL with Glu1103 (13). In addition, further opening of the TL toward the Rpb9 subunit is not constrained by the surrounding protein in all crystals of pol II TEC that we examined, further supporting the possibility of their direct interaction. This contact becomes broken upon closure of the TL. One attractive model, based on available structural data, suggests that the TL-Rpb9 interaction might involve a series of charged residues (90QRRKDTS96) in the C-terminal domain of Rpb9 pointing toward the tip of the open TL (Fig. 6 and supplemental Fig. S6). Previous in vivo studies provided further evidence that this acidic domain is involved in the TFIIS-independent fidelity control mediated by Rpb9 (33). To test this model, we investigated one of the possibilities by site-directed mutagenesis targeting the Lys93 residue and assessed the effect of the Rpb9-K93A mutation on fidelity in vitro. We found that, contrary to our expectations, the mutant Rpb9-K93A did not promote misincorporation and NTP sequestration (supplemental text and supplemental Fig. S7). Nevertheless, stabilization of the open conformation of the TL by Rpb9 may be mediated by the adjacent charged residues in the patch.
Stabilization of the open conformation of TL by Rpb9 may be consistent with the synthetic lethality of the rpb1-E1103G mutation and RPB9 deletion. Indeed, persistence of the loop in the closed configuration caused by the double mutation may decrease transcription fidelity or alter the elongation properties of pol II beyond a level that can be tolerated by yeast cells. This model explains the suppression of transient pausing during transcription by the pol II lacking the Rpb9 subunit (Fig. 1B) because an intermediate positioning of the TL between the open and closed state was shown to contribute to pausing of Escherichia coli RNA polymerase (15). The similar connections between a remote site and the active center have also been reported for the viral RNA-dependent RNA polymerase 3Dpol G64S (1, 11). Similarly to our present results, the G64S mutation that increased the fidelity of 3Dpol also decreased the sequestration of the correct substrate (11).
In the co-crystal structure of yeast pol II with transcription factor TFIIS, Rpb9 contacts the primary DNA-binding channel via its N-terminal domain (22). The downstream DNA interaction within the main channel has been shown to affect transcription initiation, pausing, arrest, and termination, suggesting that protein-DNA interactions in the main channel of pol II may contribute to the conformation of the active center (46–49). Recently, kinetic studies have suggested that NTP substrates enter pol II through the main channel (2, 50). This model posits that the NTPs prehybridize to the downstream template two and three nucleotides ahead of the active center, where screening for the correct substrates may occur prior to their loading to the active center, therefore enhancing fidelity (2). Although Rpb9 might also affect fidelity through an alternative pathway involving contacts within the main channel, our data provide evidence for regulation through the secondary channel.
The Rpb9 subunit occupies a position in the enzyme that is similar to that of a large 193-amino-acid segment (GNCD (region G non-conserved domain)) in the β′-subunit of E. coli RNA polymerase, referred to as the conserved region G-trigger loop (51), but is missing in eukaryotes, archaea, and some bacteria. In the bacterial polymerase, this segment is inserted at the tip of the TL and protrudes into the DNA-binding channel of RNA polymerase (supplemental Fig. S8). Therefore, the Rpb9 subunit and the GNCD segment may possess a similar role of stabilizing the TL in the open position. In addition, the Rpb9-like subunits in pol I (A12.2) and pol III (C11) showed significant sequence similarity in both the N-terminal and the C-terminal domains and occupied similar positions in their cognate polymerases, suggesting that Rpb9-like subunits are structurally and functionally homologous proteins (27, 28, 52–54).
The Rpb9 subunit was linked to a post-incorporation error-correcting mechanism (33), where removal of transcription errors by endonucleolytic cleavage of the RNA is stimulated by the transcription factor TFIIS (24). The hypothetical involvement of Rpb9 in TFIIS-mediated proofreading has been proposed based on the slow growth phenotype of RPB9 and DST1 double deletion mutant and on the altered TFIIS-stimulated transcript cleavage pattern observed in purified pol II lacking the Rpb9 subunit (26, 30). A similar post-incorporational proofreading mechanism was suggested for the archaeal RNA polymerase and eukaryotic pol III (55, 56). In archaea, a small protein TFS, closely related to both Rpb9 and TFIIS, has been shown to improve transcription fidelity in vitro by stimulating the intrinsic cleavage activity of RNA polymerase (25, 57). In contrast, C11, the Rpb9 counterpart for pol III, mediates the cleavage activity of pol III specifically and independently of any accessory factor. Notably, mutations abolishing the C11-dependent nuclease activity are lethal (27), similar to a double deletion of RPB9 and DST1 (31). Our findings indicate that Rpb9 controls transcription fidelity by a mechanism involving selection of the correct substrate NTP. Based on these phenotypic and structural similarities, it is conceivable that Rpb9 structural analogues might also control the accuracy of transcription at the pre-incorporation level. Interestingly, E. coli RNA polymerase deleted for the β′-GNCD (39, 58), as well as yeast pol III lacking the C11 subunit (27), were shown to respond inefficiently to normal transcriptional pause sites in vitro, similar to pol II lacking the Rpb9 subunit.
It has recently been reported that Rpb9 plays a role in preventing base substitutions and frameshift transcription errors in vivo by a TFIIS-independent mechanism (18). Sequencing of cDNA revealed two major observations: (i) a higher frequency of base substitutions (mostly transitions) over base insertions and deletions in the wild-type strains and (ii) a moderate 1.6-fold increase in base substitutions and a 5-fold increase in base insertion frequency in rpb9Δ strains. Consistent with these findings, our in vitro system indicates that RPB9 deletion induces a substantial increase in misincorporation events; however, our assay was not sensitive enough to detect base insertions or deletion events for both the wild-type pol II and pol IIΔ9 (supplemental Figs. S1 and S2). The apparently higher effect of RPB9 deletion on base insertions when compared with base substitutions observed in vivo might be explained by the presence of the TFIIS-mediated error correction factor. It is conceivable that cells may be less efficient in correcting base insertions than misincorporation errors.
In summary, our findings represent an example of pre-incorporation transcription fidelity control involving a long distance communication between distal subunits and the active center of multisubunit RNA polymerases. The wiring of the deeply buried active center with the surface of pol II may be a part of transcription regulation by extrinsic protein factors that may require intermediary contacts with different parts of the trigger loop.
Supplementary Material
Acknowledgments
We thank Brenda Schafer for technical help, Donald Court and Zachary Burton for helpful discussion and Donald Court, Zachary Burton, and Dwight Nissley for critical reading of the manuscript.
This work was supported, in whole or in part, by the Intramural Research Program of the National Institutes of Health through the NCI. The contents of this publication do not necessarily reveal the views of the policy of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement of the U.S. government.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” supplemental Figs. S1–S8, and supplemental Tables S1–S2.
- pol II
- RNA polymerase II
- Exo III
- exonuclease III
- TL
- trigger loop
- WT
- wild type
- Ni2+-NTA
- nickel-nitrilotriacetic acid
- TEC
- ternary elongation complex
- nt
- nucleotide(s)
- GNCD
- region G non-conserved domain.
REFERENCES
- 1.Arnold J. J., Cameron C. E. (2004) Biochemistry 43, 5126–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton Z. F., Feig M., Gong X. Q., Zhang C., Nedialkov Y. A., Xiong Y. (2005) Biochem. Cell Biol. 83, 486–496 [DOI] [PubMed] [Google Scholar]
- 3.Patel S. S., Wong I., Johnson K. A. (1991) Biochemistry 30, 511–525 [DOI] [PubMed] [Google Scholar]
- 4.Wong I., Patel S. S., Johnson K. A. (1991) Biochemistry 30, 526–537 [DOI] [PubMed] [Google Scholar]
- 5.Kuchta R. D., Mizrahi V., Benkovic P. A., Johnson K. A., Benkovic S. J. (1987) Biochemistry 26, 8410–8417 [DOI] [PubMed] [Google Scholar]
- 6.Mizrahi V., Henrie R. N., Marlier J. F., Johnson K. A., Benkovic S. J. (1985) Biochemistry 24, 4010–4018 [DOI] [PubMed] [Google Scholar]
- 7.Johnson K. A. (1993) Annu. Rev. Biochem. 62, 685–713 [DOI] [PubMed] [Google Scholar]
- 8.Joyce C. M., Benkovic S. J. (2004) Biochemistry 43, 14317–14324 [DOI] [PubMed] [Google Scholar]
- 9.Arnold J. J., Gohara D. W., Cameron C. E. (2004) Biochemistry 43, 5138–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro C., Arnold J. J., Cameron C. E. (2005) Virus Res. 107, 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold J. J., Vignuzzi M., Stone J. K., Andino R., Cameron C. E. (2005) J. Biol. Chem. 280, 25706–25716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan C. D., Larsson K. M., Kornberg R. D. (2008) Mol. Cell 30, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kireeva M. L., Nedialkov Y. A., Cremona G. H., Purtov Y. A., Lubkowska L., Malagon F., Burton Z. F., Strathern J. N., Kashlev M. (2008) Mol. Cell 30, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D., Bushnell D. A., Westover K. D., Kaplan C. D., Kornberg R. D. (2006) Cell 127, 941–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toulokhonov I., Zhang J., Palangat M., Landick R. (2007) Mol. Cell 27, 406–419 [DOI] [PubMed] [Google Scholar]
- 16.Brueckner F., Cramer P. (2008) Nat. Struct. Mol. Biol. 15, 811–818 [DOI] [PubMed] [Google Scholar]
- 17.Malagon F., Kireeva M. L., Shafer B. K., Lubkowska L., Kashlev M., Strathern J. N. (2006) Genetics 172, 2201–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nesser N. K., Peterson D. O., Hawley D. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3268–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cramer P., Bushnell D. A., Kornberg R. D. (2001) Science 292, 1863–1876 [DOI] [PubMed] [Google Scholar]
- 20.Gnatt A. L., Cramer P., Fu J., Bushnell D. A., Kornberg R. D. (2001) Science 292, 1876–1882 [DOI] [PubMed] [Google Scholar]
- 21.Zhang C., Burton Z. F. (2004) J. Mol. Biol. 342, 1085–1099 [DOI] [PubMed] [Google Scholar]
- 22.Kettenberger H., Armache K. J., Cramer P. (2003) Cell 114, 347–357 [DOI] [PubMed] [Google Scholar]
- 23.Westover K. D., Bushnell D. A., Kornberg R. D. (2004) Science 303, 1014–1016 [DOI] [PubMed] [Google Scholar]
- 24.Jeon C., Agarwal K. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13677–13682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fish R. N., Kane C. M. (2002) Biochim. Biophys. Acta 1577, 287–307 [DOI] [PubMed] [Google Scholar]
- 26.Hemming S. A., Edwards A. M. (2000) J. Biol. Chem. 275, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 27.Chédin S., Riva M., Schultz P., Sentenac A., Carles C. (1998) Genes Dev. 12, 3857–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn C. D., Geiger S. R., Baumli S., Gartmann M., Gerber J., Jennebach S., Mielke T., Tschochner H., Beckmann R., Cramer P. (2007) Cell 131, 1260–1272 [DOI] [PubMed] [Google Scholar]
- 29.Whitehall S. K., Bardeleben C., Kassavetis G. A. (1994) J. Biol. Chem. 269, 2299–2306 [PubMed] [Google Scholar]
- 30.Awrey D. E., Weilbaecher R. G., Hemming S. A., Orlicky S. M., Kane C. M., Edwards A. M. (1997) J. Biol. Chem. 272, 14747–14754 [DOI] [PubMed] [Google Scholar]
- 31.Hemming S. A., Jansma D. B., Macgregor P. F., Goryachev A., Friesen J. D., Edwards A. M. (2000) J. Biol. Chem. 275, 35506–35511 [DOI] [PubMed] [Google Scholar]
- 32.Exinger F., Lacroute F. (1992) Curr. Genet. 22, 9–11 [DOI] [PubMed] [Google Scholar]
- 33.Koyama H., Ito T., Nakanishi T., Sekimizu K. (2007) Genes Cells 12, 547–559 [DOI] [PubMed] [Google Scholar]
- 34.Koyama H., Ito T., Nakanishi T., Kawamura N., Sekimizu K. (2003) Genes Cells 8, 779–788 [DOI] [PubMed] [Google Scholar]
- 35.Shaw R. J., Bonawitz N. D., Reines D. (2002) J. Biol. Chem. 277, 24420–24426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mnaimneh S., Davierwala A. P., Haynes J., Moffat J., Peng W. T., Zhang W., Yang X., Pootoolal J., Chua G., Lopez A., Trochesset M., Morse D., Krogan N. J., Hiley S. L., Li Z., Morris Q., Grigull J., Mitsakakis N., Roberts C. J., Greenblatt J. F., Boone C., Kaiser C. A., Andrews B. J., Hughes T. R. (2004) Cell 118, 31–44 [DOI] [PubMed] [Google Scholar]
- 37.Kireeva M. L., Lubkowska L., Komissarova N., Kashlev M. (2003) Methods Enzymol. 370, 138–155 [DOI] [PubMed] [Google Scholar]
- 38.Komissarova N., Kireeva M. L., Becker J., Sidorenkov I., Kashlev M. (2003) Methods Enzymol. 371, 233–251 [DOI] [PubMed] [Google Scholar]
- 39.Artsimovitch I., Svetlov V., Murakami K. S., Landick R. (2003) J. Biol. Chem. 278, 12344–12355 [DOI] [PubMed] [Google Scholar]
- 40.Bar-Nahum G., Epshtein V., Ruckenstein A. E., Rafikov R., Mustaev A., Nudler E. (2005) Cell 120, 183–193 [DOI] [PubMed] [Google Scholar]
- 41.Kashkina E., Anikin M., Brueckner F., Pomerantz R. T., McAllister W. T., Cramer P., Temiakov D. (2006) Mol. Cell 24, 257–266 [DOI] [PubMed] [Google Scholar]
- 42.Nedialkov Y. A., Gong X. Q., Hovde S. L., Yamaguchi Y., Handa H., Geiger J. H., Yan H., Burton Z. F. (2003) J. Biol. Chem. 278, 18303–18312 [DOI] [PubMed] [Google Scholar]
- 43.Tuske S., Sarafianos S. G., Wang X., Hudson B., Sineva E., Mukhopadhyay J., Birktoft J. J., Leroy O., Ismail S., Clark A. D., Jr., Dharia C., Napoli A., Laptenko O., Lee J., Borukhov S., Ebright R. H., Arnold E. (2005) Cell 122, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C., Zobeck K. L., Burton Z. F. (2005) Mol. Cell Biol. 25, 3583–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westover K. D., Bushnell D. A., Kornberg R. D. (2004) Cell 119, 481–489 [DOI] [PubMed] [Google Scholar]
- 46.Ederth J., Artsimovitch I., Isaksson L. A., Landick R. (2002) J. Biol. Chem. 277, 37456–37463 [DOI] [PubMed] [Google Scholar]
- 47.Keene R. G., Mueller A., Landick R., London L. (1999) Nucleic Acids Res. 27, 3173–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerppola T. K., Kane C. M. (1990) Biochemistry 29, 269–278 [DOI] [PubMed] [Google Scholar]
- 49.Telesnitsky A., Chamberlin M. J. (1989) Biochemistry 28, 5210–5218 [DOI] [PubMed] [Google Scholar]
- 50.Xiong Y., Burton Z. F. (2007) J. Biol. Chem. 282, 36582–36592 [DOI] [PubMed] [Google Scholar]
- 51.Chlenov M., Masuda S., Murakami K. S., Nikiforov V., Darst S. A., Mustaev A. (2005) J. Mol. Biol. 353, 138–154 [DOI] [PubMed] [Google Scholar]
- 52.Bischler N., Brino L., Carles C., Riva M., Tschochner H., Mallouh V., Schultz P. (2002) EMBO J. 21, 4136–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lorenzen K., Vannini A., Cramer P., Heck A. J. (2007) Structure 15, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 54.Van Mullem V., Landrieux E., Vandenhaute J., Thuriaux P. (2002) Mol. Microbiol. 43, 1105–1113 [DOI] [PubMed] [Google Scholar]
- 55.Alic N., Ayoub N., Landrieux E., Favry E., Baudouin-Cornu P., Riva M., Carles C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10400–10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lange U., Hausner W. (2004) Mol. Microbiol. 52, 1133–1143 [DOI] [PubMed] [Google Scholar]
- 57.Hausner W., Lange U., Musfeldt M. (2000) J. Biol. Chem. 275, 12393–12399 [DOI] [PubMed] [Google Scholar]
- 58.Artsimovitch I., Svetlov V., Anthony L., Burgess R. R., Landick R. (2000) J. Bacteriol. 182, 6027–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 27–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.