Abstract
The flavoprotein iodotyrosine deiodinase (IYD) salvages iodide from mono- and diiodotyrosine formed during the biosynthesis of the thyroid hormone thyroxine. Expression of a soluble domain of this membrane-bound enzyme provided sufficient material for crystallization and characterization by x-ray diffraction. The structures of IYD and two co-crystals containing substrates, mono- and diiodotyrosine, alternatively, were solved at resolutions of 2.0, 2.45, and 2.6 Å, respectively. The structure of IYD is homologous to others in the NADH oxidase/flavin reductase superfamily, but the position of the active site lid in IYD defines a new subfamily within this group that includes BluB, an enzyme associated with vitamin B12 biosynthesis. IYD and BluB also share key interactions involving their bound flavin mononucleotide that suggest a unique catalytic behavior within the superfamily. Substrate coordination to IYD induces formation of an additional helix and coil that act as an active site lid to shield the resulting substrate·flavin complex from solvent. This complex is stabilized by aromatic stacking and extensive hydrogen bonding between the substrate and flavin. The carbon-iodine bond of the substrate is positioned directly over the C-4a/N-5 region of the flavin to promote electron transfer. These structures now also provide a molecular basis for understanding thyroid disease based on mutations of IYD.
The micronutrient iodide is essential for the biosynthesis of thyroxine (3,3′,5,5′-tetraiodothyronine), a hormone used by a wide range of organisms as a master control of metabolic rate. In mammals, iodide homeostasis in the thyroid gland is critical for generating thyroxine and is achieved by sequestering and salvaging iodide. Both of these functions are critical for human health, and congenital defects in either may lead to hypothyroidism (1, 2). Sequestration of iodide from the circulatory system is accomplished by a Na+/I− symporter located in the plasma membrane of thyroid follicular cells (2). Salvage of iodide is accomplished by iodotyrosine deiodinase (IYD)3 located in the apical plasma membrane surrounding the thyroid colloid in which thyroglobulin is stored and processed (3). Proteolysis of mature thyroglobulin releases thyroxine as well as mono- and diiodotyrosine (MIT and DIT, respectively). IYD catalyzes a reductive deiodination of MIT and DIT selectively to prevent loss of iodide that would otherwise occur by excretion of these amino acids. The gene encoding IYD has recently been identified (3, 4) and has provided an initial basis for correlating its mutation with hypothyroidism and goiter observed in certain patients (1). The crystal structure described in this work now supersedes the previous structural models.
IYD represents one of only two enzymes known to promote reductive dehalogenation in mammals (Fig. 1). The other enzyme, iodothyronine deiodinase, acts alternatively to activate and deactivate thyroxine by deiodinating the outer or inner ring, respectively (5). Interestingly two distinct strategies based on two distinct protein architectures have been recruited for catalyzing essentially the same deiodination. Iodothyronine deiodinase is a member of the thioredoxin structural superfamily and utilizes an active site selenocysteine for its catalysis. In contrast, IYD is a member of the NADH oxidase/flavin reductase superfamily (3, 4) and requires neither selenocysteine nor cysteine for catalysis (6). Instead a bound flavin mononucleotide (FMN) is required (7). Flavin is integral to many catalytic functions and even certain biological halogenation reactions (8–11). However, its role in reductive dehalogenation is quite unusual and not yet characterized. Flavin has also been implicated in just one bacterial reductive dehalogenation, although little information is available on this additional system other than its use of flavin-adenine dinucleotide (FAD) rather than FMN (12).
FIGURE 1.
Reductive deiodination for iodide salvage catalyzed by IYD (A) and metabolism of the hormone thyroxine by iodothyronine deiodinase (ID) (B).
Structural characterization of IYD is critical for identifying the active site properties that support its unique catalytic ability and substrate selectivity. Moreover this information contributes a molecular understanding of how mutations in IYD can disrupt function to cause iodide deficiency and hypothyroidism. Such mutants are particularly tragic because their detection often occurs only after developmental damage to patients has occurred (1). Studies on IYD to date have been limited by its membrane association and transient expression in mammalian cells (3, 4). However, deletion of its N-terminal membrane anchor yields a soluble and active enzyme (6) that has been expressed in Sf9 cells in sufficient quantities to support the crystallographic analysis reported here.
EXPERIMENTAL PROCEDURES
Gene Construction of a Soluble and Affinity-tagged IYD
Wild-type Mus musculus IYD cDNA (I.M.A.G.E. clone 5061638) was obtained from ATCC (Manassas, VA) and amplified with primers 5′-AAGCTTAAGCTTGGATCCGCCACCATGGCTCAAGTTCAGCCC-3′ and 5′-CTCGAGCTCGAGCTAATGGTGATGGTGATGGTGTACTGTCACCATGATC-3′ to generate a derivative lacking codons for amino acids 2–33 and gaining codons for a C-terminal His6 tag. The amplified product and pFASTBAC1 (Invitrogen) were digested with BamHI and XhoI, ligated together, and transformed into One Shot Top10 cells. DNA was isolated from colonies resistant to ampicillin and sequenced to verify the desired gene sequence (IYD(ΔTM)His6). Bacmid containing IYD(ΔTM)His6 was generated through transposition of pFASTBAC1-IYD(ΔTM)His6 into the baculovirus shuttle vector bMON14272 according to the Bac-to-Bac baculovirus expression system protocols provided by Invitrogen.
Sf9 cells adapted to SF-900 II SFM (Invitrogen) were transfected with the pFASTBAC1-IYD(ΔTM)His6 recombinant bacmid DNA and Cellfectin reagent (Invitrogen). The resulting recombinant baculovirus was collected from the growth medium 72 h after transfection. The virus was then amplified with a multiplicity of infection of 0.05 (plaque-forming unit/cell) as directed by the Bac-to-Bac protocols, and virus concentration was determined by standard end point dilution analysis (13).
Expression and Purification of Protein
Sf9 cells were infected by pFASTBAC1-IYD(ΔTM)His6 recombinant baculovirus stock in SF-900 II SFM with a multiplicity of infection of 1 (plaque-forming unit/cell). Cells were incubated at 27 °C for 72 h and then harvested by centrifugation at 500 × g for 5 min (room temperature). The cell pellet was resuspended in 500 mm NaCl, 50 mm sodium phosphate, 10 mm imidazole, pH 8.0 and lysed by three freezing and thawing cycles followed by three passages through a 20-gauge needle. Lysates were centrifuged at 20,000 × g for 1 h (4 °C). The supernatant was filtered through a 0.22-μm membrane and then loaded onto a HisTrap HP column (1 ml) with chelated Ni2+. The column was washed with 500 mm NaCl, 50 mm sodium phosphate, 20 mm imidazole, pH 8.0 (5 ml) and then eluted with a linear gradient of increasing imidazole (20–300 mm; 20 ml). Fractions containing the desired protein were identified by SDS-PAGE, pooled, and dialyzed against 10 mm potassium phosphate, pH 7.4 (4 °C). Protein solutions were stored at 4 °C without loss of catalytic activity for at least 4 weeks.
Crystallization
Initial crystallization was explored with sparse matrix screening (WizardTM I, II, and III (Emerald Biosciences); PEGSuiteTM and CryoSuiteTM (Qiagen); and NatrixTM and IndexTM (Hampton Research)) using an Art Robbins Phoenix high throughput liquid handler. Ultimately IYD was crystallized at 20 °C in three different crystal forms by the hanging drop vapor diffusion method using two parts IYD (16 mg/ml; 10 mm potassium phosphate, pH 7.4) to one part reservoir solution. IYD crystals were obtained using a reservoir solution containing 20% (w/v) polyethylene glycol 3000, and 0.1 m sodium acetate, pH 4.5. IYD·MIT co-crystals were obtained by supplementing the enzyme solution with 2 mm MIT prior to addition of the reservoir solution containing 0.2 m magnesium chloride, 20% (w/v) polyethylene glycol 3350, 10 mm potassium phosphate, pH 7.4. IYD·DIT co-crystals were obtained by supplementing the enzyme solution with 2 mm DIT prior to addition of the reservoir solution containing 0.2 m ammonium acetate, 45% 2-methyl-2,4-pentanediol, 0.1 m BisTris, pH 5.5. In each case, yellow crystals appeared within 24 h.
Data Collection
Initial crystals were screened for diffraction and cryo conditions using a Bruker Microstar H2 generator with Proteum Pt135 CCD detector at 100 K. Diffraction data sets were then collected at the Northeastern Collaborative Access Team Beamline 24-ID, Advanced Photon Source, Argonne National Laboratories. Diffraction data for IYD(ΔTM)His6 was collected at a wavelength of 1.653 Å for single anomalous diffraction phasing using sulfur atoms. Diffraction data of co-crystals containing MIT and DIT alternatively were collected at 0.9795 Å. All collected data were integrated and scaled using HKL2000 (14).
Phasing and Refinement
All nine sulfur positions within IYD(ΔTM)His6 were found using HKL2MAP (15). Heavy atom positions were refined, and initial phases were determined using the AUTOSHARP program set (16). Initial model building was performed by WARP within AUTOSHARP. Because of the complexity of the monomeric fold, nearly half of the residues were not built and required manual building. Several rounds of model building and refinement were needed for IYD(ΔTM)His6 using Refmac5 with TLS refinement and COOT for molecular visualization and rebuilding (17, 18). A monomer of IYD(ΔTM)His6 was used as a model for molecular replacement to solve the structure of the co-crystal containing MIT. One dimer of this co-crystal was used in turn as the model for molecular replacement to solve the co-crystal containing DIT. Molecular replacement was performed by PHASER within the CCP4 program suite (19, 20). These structures were also rebuilt and refined by iterations of Refmac5 and COOT. Refinement statistics are provided in Table 1.
TABLE 1.
Data collection and refinement statistics
| IYD | IYD·MIT | IYD·DIT | |
|---|---|---|---|
| Protein Data Bank code | 3GB5 | 3GFD | 3GH8 |
| Data collection | |||
| Space group | P3121 | P3221 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 87.8, 87.8, 62.7 | 105.3, 105.3, 162.2 | 50.61, 112.56, 189.25 |
| α, β, γ (°) | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 | 90.00, 89.99, 90.00 |
| Molecules/asymmetric unit | 1 | 2 | 8 |
| Wavelength (Å) | 1.653 | 0.9795 | 0.9795 |
| Resolution (Å) | 50-2.0 | 50-2.45 | 50-2.6 |
| Rsym | 0.057 (0.581)b | 0.081 (0.439)b | 0.086 (0.295)b |
| I/σI | 63.7 (4.3)b | 20.0 (3.7)b | 9.7 (2.4)b |
| Completeness (%) | 99.8 (99.0)b | 99.2 (99.3)b | 97.1 (92.9)b |
| Redundancy | 14.0 (8.9)b | 4.7 (4.7)b | 2.6 (2.5)b |
| Refinement | |||
| Resolution (Å) | 30-2.0 | 30-2.45 | 30-2.6 |
| Rwork/Rfree (%) | 16.7/18.9 | 14.3/17.8 | 18.1/26.4 |
| No. protein residues in monomers A/B/C/D/E/F/G/H | 220 | 221/220 | 220/219/220/219/220/219/220/219 |
| No. non-protein atoms | |||
| Ligand | 43 | 102 | 370 |
| Solvent | 132 | 540 | 300 |
| Mean B-factors (Å2) | 32.6 | 38.8 | 35.5 |
| r.m.s.a deviations | |||
| Bond lengths (Å) | 0.020 | 0.019 | 0.019 |
| Bond angles (°) | 1.569 | 2.016 | 1.926 |
| Ramachandran plot | |||
| Most favorable (%) | 92.5 | 92.2 | 88.3 |
| Additionally allowed (%) | 7.5 | 7.3 | 10.9 |
| Generously allowed (%) | 0.0 | 0.5 | 0.3 |
| Disallowed (%) | 0.0 | 0.0 | 0.4 |
a Root mean square.
b Statistics for last shell.
RESULTS
Overall Structure of the Soluble Domain of IYD
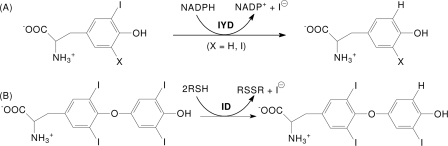
In the absence of the N-terminal membrane anchor, IYD becomes a soluble enzyme (6). A suitably truncated form of the IYD gene (M. musculus) encoding residues 33–285 (IYD(ΔTM)His6) was expressed in Sf9 insect cells and purified by nickel-based affinity chromatography. The resulting protein was screened for crystallization, and crystals were obtained for IYD alone and IYD containing its substrates MIT and DIT alternatively. In the absence of substrate, IYD crystallized with one monomer per asymmetric unit. However, analysis of the crystal packing interactions revealed an extensive interface with a symmetry-related molecule (2490 Å2 per monomer) indicating that the enzyme forms a domain-swapped dimer as observed in related proteins within the same structural superfamily (4, 21). The IYD·MIT complex crystallized with two monomers per asymmetric unit, and the IYD·DIT complex crystallized with eight monomers per asymmetric unit. All three structures were solved from distinct crystal forms (Table 1). Electron density for the first 34 amino acids of the truncated enzyme IYD(ΔTM)His6 was not observed for any of the three crystals. These residues in the native protein likely provide a flexible linker between the soluble domain and the N-terminal anchor embedded in the membrane.
IYD contains the characteristic α-β fold that is common to all proteins of the NADH oxidase/flavin reductase superfamily (Fig. 2). The closest structural neighbor to IYD as determined by DaliLite (22) is BluB (Protein Data Bank code 2ISK) (23) with an root mean square deviation of 3.1 Å for 198 structurally equivalent residues of 219 possible residues. Despite the structural similarities, their sequence identity is quite low (19%). The net catalytic turnover of BluB appears to be very different from IYD, and BluB has been recently identified as the source of the lower ligand of vitamin B12 by sacrificing its bound FMN cofactor to oxidation (23). Still IYD and BluB form similar dimer interfaces at their core with criss-crossing helices and similar domain swaps with their extended N- and C-terminal fingers. Neither enzyme conforms well to the existing subcategories within their superfamily (21), although they do retain many protein-FMN interactions common to the superfamily and additionally share a few unique interactions as discussed below.
FIGURE 2.
IYD structure. A, an overall view of the native homodimer of IYD crystallized in the absence of substrate. Each monomer is distinguished by color. Disordered regions consisting of residues 156–177 and 195–208 connect to the structure as indicated by ★ and ☆, respectively. B, native homodimer of IYD crystallized in the presence of its substrate, MIT. Only the structure induced upon substrate binding is highlighted in the colors of the monomers shown in A. The surface properties of IYD (C) and its complex with monoiodotyrosine (D) were calculated using vacuum electrostatics in PyMOL (47). Blue indicates positive charge, and red indicates negative charge. E, ionic interactions and hydrogen bonding stabilize the FMN·monoiodotyrosine complex formed by IYD. F, the interaction between FMN and MIT in the active site of IYD. An Fo − Fc electron density map calculated after refinement in the absence of FMN and MIT is shown contoured at 3σ.
Interface Structure of the Homodimer
The dimer interface is highly conserved among the NADH oxidase/flavin reductase superfamily. One helix of the α-β fold of each monomer assembles together to form the central interface of IYD. Domain swapping within IYD is also consistent with the superfamily. For IYD, N- and C-terminal extensions of each polypeptide wrap around the other. These extensions span distances of greater than 37 Å and comprise a minimum of 26 amino acids near the N terminus and 18 amino acids at the C-terminus. Two equivalent active sites are located within the dimer interface, and each active site is comprised of residues from both subunits. Accordingly subunit association is essential for FMN binding and catalytic activity. Because the IYD structure was solved as a monomer per asymmetric unit, its dimer was generated by crystallographic symmetry, and consequently no differences between the two monomers were observed. Differences observed between the monomeric units of IYD·MIT and IYD·DIT were minimal as evident from their root mean square deviation values of 0.196 and 0.254 Å, respectively.
Substrate-induced Conformational Changes in the Active Site
Two unstructured regions were identified within substrate-free IYD by the lack of electron density corresponding to residues 156–177 and 195–208. Even in the substrate-bound co-crystals, the regions containing residues 195–208 exhibit relatively weak electron density compared with other regions of the molecule. Only portions of this region, some with higher than average B-factors, could be built for IYD bound to substrate.
In contrast, residues 156–177 gain detectable structure based on the additional electron density observed for the co-crystals IYD·MIT and IYD·DIT. Substrate binding appears to induce the extension of one helix (αC) and the formation of another short helix (αD) in this region to cover the active site and protect substrate and flavin from solvent (Fig. 2, B and D). This active site lid is stabilized in part by numerous interactions with the bound substrate that is in turn anchored by aromatic stacking with the isoalloxazine ring of FMN as well as by polar contacts with FMN and Ala-126 (described below) (Fig. 2E). Eight residues of the lid assemble within 4 Å of the substrate, and three of these form polar contacts (Glu-153, Tyr-157, and Lys-178).
Haloaromatic Recognition
Noncovalent recognition of halogen substituents has recently been featured in a review describing halogen bonds in analogy to hydrogen bonds (24). Such bonding typically involves electron donation from a lone pair of electrons contributed by an oxygen, nitrogen, or sulfur atom to a halogen and, in particular, to highly polarizable iodine. The environment surrounding the iodine of MIT by 4 Å is primarily nonpolar and consists of the β-carbon of Tyr-207, α-carbon of Gly-125, and two aromatic carbons of Tyr-208. The aromatic ring of Tyr-208 interacts edge-on with the iodine and precludes interaction with its aromatic π-electrons (25). In contrast, the π-system of the isoalloxazine ring defines one surface of the substrate binding pocket. Potential donors for a halogen bond are present but remain slightly out of range (farther than 3.5 Å) and not optimally oriented for the most favorable C–I···N/O angle of 130–180° (26, 27). The amide nitrogen of Ala-126 and indole nitrogen of Trp-165 are 3.57 and 3.99 Å from the iodine, respectively, and form angles with the C–I bond of 93° and 81°. Still the distances are at least consistent with those observed by crystallography to surround the iodine substituent of a thyroxine mimic bound to transthyretin (28). The next closest polar atoms, the N-5 of the isoalloxazine ring and the amide nitrogen of Tyr-208, are even farther from the iodine at 4.10 and 5.82 Å, respectively. Perhaps most importantly, the C-4a of the isoalloxazine ring closely approaches the carbon bearing the iodine (3.65 Å; Fig. 2F) and provides a likely path for electron transfer during reductive deiodination.
Substrate Coordination to FMN
Many of the contacts between the phosphoribose component of FMN and IYD are relatively common to flavoproteins in general and especially representative of the NADH oxidase/flavin reductase superfamily (29). For example, the phosphate group is coordinated within the protein interior by multiple arginines, which in the case of IYD are Arg-96, Arg-97, and Arg-275. Additionally a variety of serine hydroxyl groups provide hydrogen bonding to the ribityl hydroxyl groups as evident with Ser-98 and Ser-124 of IYD. The pyrimidine ring of the isoalloxazine system typically associates with a number of hydrogen bond donors and acceptors of the protein active site. Such interactions are crucial for modulating the chemistry of the cofactor and optimizing catalysis (30–32).
The role of substrate coordination to the pyrimidine ring of FMN is unique to IYD. Both the α-ammonium and carboxylate groups of MIT interact with the N-3 and O4 of isoalloxazine (Fig. 2E). The zwitterionic nature of the substrate is thus essential for controlling the chemistry of the flavin cofactor as well as securing the active site lid. Close association between the phenolic hydroxyl group of MIT and a ribityl hydroxyl group of FMN (2.55 Å; Fig. 2F) adds an additional contact to the considerable interplay between substrate and cofactor and is also likely essential for catalysis as presented in the discussion section. The phenolic hydroxyl group additionally interacts with the backbone nitrogen of Ala-126 providing yet another substrate-mediated bridge between protein and FMN.
DISCUSSION
Relationship of IYD within Its Structural Superfamily
Flavoproteins collectively catalyze a highly diverse set of reactions. Even within structural subfamilies, the diversity of catalysis can be broad. Flavoproteins of the NADH oxidase/flavin reductase superfamily are characterized by an α-β fold and subunit dimerization stabilized by an extensive protein-protein interface and domain swapping (Fig. 2, A and B). Most representatives of this superfamily promote oxidation of NAD(P)H (29, 33–38). The ultimate acceptors of the reducing equivalents are far more varied and in many cases not yet defined in vivo. Some representatives such as flavin reductase P (FRP) generate reduced flavin required for bioluminescence (33). Other representatives reduce nitroaromatic compounds and quinones that may play a critical role in activation and deactivation of drugs and pollutants (29, 34). These nitroreductases are oxygen-insensitive suggesting an obligate two-electron transfer from the reduced flavoprotein to the nitroaromatic substrate.
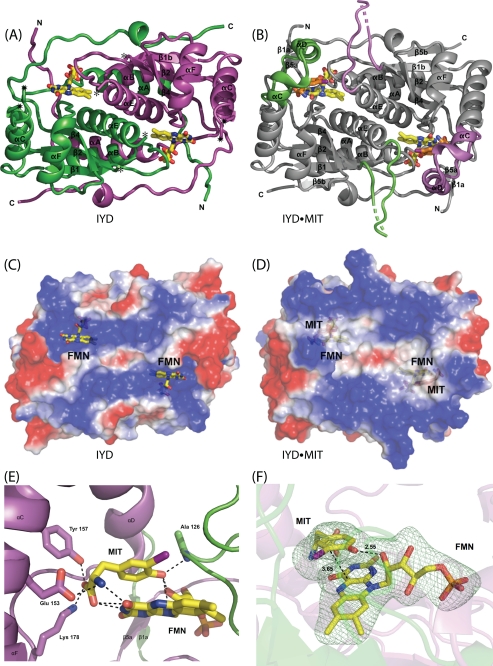
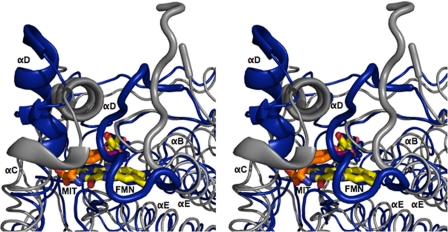
Until recently, all representatives of this structural superfamily belonged to one of two subclasses represented here by NADH oxidase from Thermus thermophilus (35) and FRP from Vibrio harveyi (33) (Fig. 3). Overall the organization of α-helices and β-sheets is very similar and notably different only in the region used to create their active site lid. For NADH oxidase, the lid (Fig. 3, boxed area) is derived from a central region of its polypeptide. For FRP, the lid is derived from a C-terminal extension. For IYD, the lid is derived from elongation of a central α-helix common to the superfamily (αC) and insertion of a loop and an additional α-helix (αD) not present in NADH oxidase or FRP. BluB, one of the most recent additions to the NADH oxidase/flavin reductase superfamily, also contains a lid in a position equivalent to that of IYD (Fig. 3). The chemistry promoted by this latter enzyme is likely quite different from the oxygen-insensitive nitroreductases because its biological function appears to involve degradation of its bound flavin by molecular oxygen (23). Thus, it likely activates, rather than suppresses, one-electron processes. The catalytic mechanism of IYD, like its structure, may also share more similarities with BluB than with the other members of the superfamily. Structural analysis alone argues for establishing a third subclass within the superfamily that is represented by BluB and IYD and is defined by the position of their active site lids.
FIGURE 3.
Alignment of secondary structure for representatives of the NADH oxidase/flavin reductase superfamily. IYD and BluB now define a third subclass of the NADH oxidase/flavin reductase superfamily. NADH oxidase (NOX) and FRP illustrate the α-β fold for the original two subclasses of this superfamily. The boxed regions indicate the sequences that form the active site lids. The dotted lines indicate spacing inserted for alignment. Structural assignments were derived from crystallographic data (Protein Data Bank codes 3GFD, 2ISL, 1NOX, and 2BKJ, respectively).
Substrate Binding in the NADH Oxidase/Flavin Reductase Superfamily
Considerable effort has focused on defining the mode of binding of nicotinamide to this superfamily because no standard nucleotide binding motif is present despite the use of reduced nicotinamides during catalysis (29, 33–38). A co-crystal of nitroreductase NfsB (Escherichia coli) and nicotinic acid demonstrates the potential for NAD+ to stack over the isoalloxazine ring in an orientation optimal for hydride transfer from the C-4 position of the nicotinamide ring to the N-5 of the isoalloxazine ring (38). A variety of dinitrobenzamide prodrugs and dicoumarol also bind to NfsB in an equivalent mode by stacking over the active site isoalloxazine ring (39). Similar binding orientations are evident for dicoumarol in a co-crystal of flavin reductase (Vibrio fischeri) (36) and benzoate in a co-crystal of nitroreductase (Enterobacter cloacae) (34). Both enzymes share membership in the same subfamily as NsfB. Only one structure from the entire superfamily has been solved with nicotinamide dinucleotide (NAD+) bound in its active site, and this was derived from FRP, a representative of a subfamily distinct from that above (40). FRP does not associate with the nicotinamide ring by stacking onto the isoalloxazine of FMN. Instead the nicotinamide and adenine rings of NAD+ associate intramolecularly as they do in solution, and the pyrophosphate of NAD+ stacks above the isoalloxazine ring system.
Nicotinamides are not expected to associate with either IYD or BluB in contrast to the proteins of the two subclasses above. Although NADPH is thought to donate its reducing equivalents for net deiodination in vivo, this process most likely is indirect and involves at least one intervening oxidoreductase (6, 41). Even less is known about the physiological reduction of BluB (23). The site equivalent to that occupied by nicotinic acid in NfsB is used by IYD to bind the iodophenol portion of its substrates MIT and DIT (Figs. 2F and 4A). The aromatic phenol stacks over the isoalloxazine ring, and a hydrogen bond between the substrate phenol and the 2′-hydroxyl ribityl group of FMN is formed that is equivalent to that within the co-crystal of nicotinic acid and NsfB (Fig. 2F) (38).
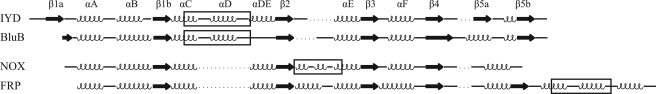
FIGURE 4.
Conformational changes in the IYD active site to accommodate MIT and DIT. A, the surface characteristics of the active site of IYD for the IYD·MIT co-crystal crystal calculated using vacuum electrostatics in PyMOL (47). Blue indicates positive charge, and red indicates negative charge. B, alignment of active site structures of IYD bound with MIT (orange) and DIT (cyan) illustrates the minor conformational change required to accommodate the larger substrate.
Once the active site lid of IYD is formed, the resulting pocket surrounding the substrate and FMN has little or no access to solvent (Figs. 2D and 4A). The electrostatics of this pocket are complementary to both the substrate and FMN as expected. The ribityl phosphate group is proximal to the most electropositive surface, and the α-amino group of the substrate is proximal to the most electronegative surface. The active site structure induced by MIT is not sufficient to accommodate the larger DIT. However, only minor shifts of Leu-169, Thr-174, and Leu-172 are necessary for DIT to align over the isoalloxazine equivalently to MIT (Fig. 4B).
Recognition of the zwitterion of the substrates remains constant for both MIT and DIT. The active site residues Glu-153, Tyr-157, and Lys-178 are primarily responsible for these interactions that are likely essential for stabilizing the active site lid as well as indirectly influencing the redox characteristics of the FMN (Fig. 2E). This influence is transmitted through the zwitterionic portion of the substrate that coordinates to the isoalloxazine ring. In a complementary manner, substrate selectivity is likely controlled by the steric constraints imposed by the lid and the chelation established cooperatively by the protein and the isoalloxazine ring. Neither thyroxine nor 3,3′,5-triiodothyronine are processed by IYD (42), and neither are likely capable of binding to IYD in a productive orientation because of their extended size. Such selectivity is especially beneficial because deiodination of thyroglobulin and its hormone products within the thyroid would be counterproductive.
FMN Coordination in IYD and Related Enzymes
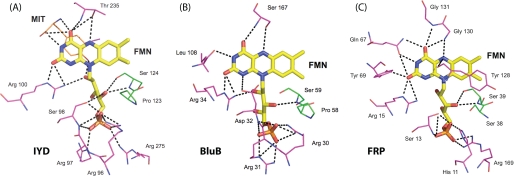
Proteins generally coordinate and control the reactivity of flavin by establishing a network of hydrogen bonds and electrostatic interactions particularly to its pyrimidine ring. The NADH oxidase/flavin reductase superfamily is no exception (29, 33–38). For example, Asn-67 of FRP hydrogen bonds to the N-3 and O4 of the isoalloxazine (Fig. 5). Concurrently Tyr-69 and Arg-15 coordinate to its O2, and Arg-15 also coordinates to its N-1 position. Other members of this family maintain a similar array of coordination. As illustrated in Fig. 5, IYD, BluB, and FRP all maintain contact between an arginine and the O2, if not also the N-1, of the isoalloxazine. Such an interaction is considered particularly important for stabilizing the reduced form of the cofactor (30). These three proteins also stabilize the ribityl phosphate moiety by the common presence of multiple serine and arginine residues.
FIGURE 5.
Polar contacts between bound FMN and IYD (A), BluB (B), and FRP (C). Coordination of FMN by protein is highlighted for residues within 4 Å of the flavin in the crystal structures for Protein Data Bank codes 3GDF, 2ISJ, and 2BKJ, respectively.
Interactions between the pyrimidine ring of the isoalloxazine system and the BluB protein are more sparse than typical. The N-3 and O4 are only stabilized by backbone interactions. IYD offers even less direct coordination to this same region of its isoalloxazine. However, as mentioned above, contacts are instead provided by the amino acid portion of the substrate, and hence the catalytic properties of the bound FMN are likely to depend strongly on the presence of substrate. The substrate-FMN interactions presented by the co-crystals of IYD·MIT and IYD·DIT are considerably more extensive than those typical of flavoproteins.
The nature of the hydrogen bonding between IYD and the N-5 of the isoalloxazine ring is also distinct within its structural superfamily and flavoproteins in general. Hydrogen bonding to this site is very common but is usually derived from a backbone or side chain N–H bond (30). The NADH oxidase/flavin reductase superfamily follows this same trend as illustrated in FRP by hydrogen bonding between the backbone of Gly-130 and the N-5 of the isoalloxazine ring (Fig. 5). In contrast, a comparable hydrogen bond in IYD is created by the side chain hydroxyl group of Thr-235, and similarly, BluB uses an equivalent side chain of Ser-167 that has already been shown to be essential for activity (Fig. 5) (23). This type of hydrogen bonding is also evident in a family of electron transfer flavoproteins containing FAD that are quite distinct from IYD and BluB (43). For these transferases, mutagenesis of the participating serine suggested that its hydrogen bond to the N-5 of the isoalloxazine ring helps to stabilize its anionic semiquinone form (43). The role of the similar hydrogen bond in IYD and BluB may be comparable or at least allow for single electron chemistry within a structural framework that previously was known only to promote two-electron processes (34, 36).
The mechanistic significance of hydrogen bonding between the phenolic hydroxyl group of MIT and the ribityl 2′-hydroxyl group of FMN bound to IYD is not as clear as that for the contacts above. This type of bonding is also common to other enzymes within the same superfamily as well as enzymes of other families that, for example, include the FAD-containing acyl-CoA dehydrogenases (44, 45). For these later enzymes, coordination by the 2′-hydroxyl group appears to stabilize the enolate form of the acyl-CoA substrate and hence increase the acidity of the α-C–H bond required for deprotonation during turnover. The substrates of IYD, MIT and DIT, already exist in their enol(ate) forms under ambient conditions and, in contrast, have been proposed to undergo ketonization prior to deiodination (46). Still coordination by the hydroxyl group of FMN could increase the electrophilicity of the substrate in either case to promote its reduction. Equivalent coordination is evident for the molecular oxygen that binds within the active site of BluB (23) and likely facilitates single electron transfer.
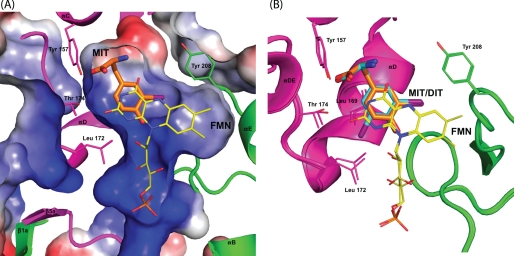
Active Site Structures of IYD and BluB
Similarities between IYD and BluB extend beyond their coordination of FMN. Their overall structures and most interestingly their active site morphology are also closely related. FMN is nearly superimposable upon the alignment of IYD·MIT and BluB using PyMOL (47) (Fig. 6). The presence of MIT induces formation of a helix/turn and a coil that close the active site of IYD and control substrate specificity. BluB forms a comparable helix/turn to define one side of the active site and a related coil appropriate for selecting its substrate, molecular oxygen. In contrast to the coil of IYD, the coil of BluB folds into the active site cavity to protect the substrate·FMN complex and exclude anything larger than molecular oxygen. Interestingly this substrate of BluB aligns closely with the phenolic hydroxyl of MIT/DIT in IYD, and both hydrogen bond to the 2′-ribityl hydroxyl of their FMN.
FIGURE 6.
Structural overlay of IYD and BluB. Structural differences between IYD·MIT (gray) and BluB (blue) are highlighted using an overlaid stereoimage. Structural changes near the active site are depicted with the schematic representation. The full-view overlay can be seen as a stereoimage in the supplemental information.
Structural Basis for Deficiency of IYD in Humans
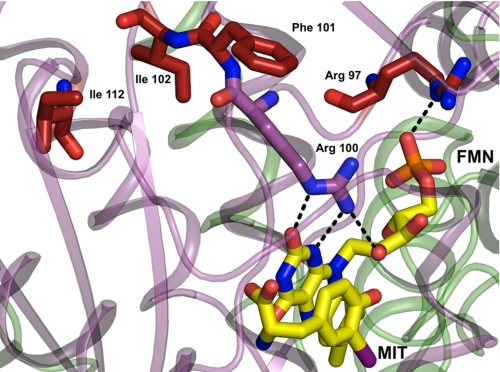
Mutations of IYD and the resulting deficiency in retaining iodide equivalents may not be detected during standard tests for thyroid function (1). Only very recently has a sensitive assay been developed to measure elevated concentrations of MIT and DIT in urine as expected from a deficiency of IYD (48). The phenotypes of all four human mutations of IYD described to date are well rationalized by the current structure of the IYD from mouse in part because of the high identity (91%) between their NADH oxidase/flavin reductase domains responsible for catalysis. The most severe consequences were observed for mutations in the human gene that are equivalent to R97W and a combined I102L and Phe-101 deletion in the mouse gene (Fig. 7). These and related mutants retain only minimal deiodinase activity and are unresponsive to addition of exogenous FMN (1). This is consistent with the loss of FMN binding because Arg-97 directly contacts the phosphate of FMN, and residues 101 and 102 are necessary to orient Arg-100 for direct contact with the 2′-ribityl hydroxyl group as well as the N-1 and O2 of FMN. Mutation at residue 112 (human residue 116) is more distal to FMN than those above. This in turn causes less severe depression of IYD activity and a milder phenotype (1). As suggested previously, the human mutation corresponding to A216T in mouse may destabilize the dimer interface (48) and likely leads to premature degradation of IYD in vivo. The crystal structures presented in this work now provide a predictive understanding of other mutations that would compromise our ability to conserve iodide effectively. The insensitivity of IYD to C217A and C239A as reported earlier (6) could also have been predicted from these structures. The native cysteine residues are far from the active site in contrast to previous expectations based on a model derived from the structure of NfsB, another member of the NAD oxidase/flavin reductase superfamily.
FIGURE 7.
Mapping human mutations onto the structure of IYD. Native residues of IYD (M. musculus) highlighted in red correspond to sites associated with human mutations identified clinically to cause hypothyroidism (1). Other color coding is consistent with the previous illustrations (see legends) and distinguishes the two subunits within the dimer and the FMN.
Supplementary Material
Acknowledgments
We thank Chiwei Hung and Prof. Bentley for help with protein expression in Sf9 cells. This work is based upon research conducted in part at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source supported by Award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the United States Department of Energy, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. We are grateful to the excellent staff at the Northeastern Collaborative Access Team for assistance with data collection and analysis.
This work was supported by a Herman Kraybill Biochemistry fellowship (to P. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3GB5, 3GFD and 3GH8) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IYD
- iodotyrosine deiodinase
- MIT
- monoiodotyrosine
- DIT
- diiodotyrosine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- FRP
- flavin reductase P.
REFERENCES
- 1.Moreno J. C., Klootwijk W., van Toor H., Pinto G., D'Alessandro M., Lèger A., Goudie D., Polak M., Grüters A., Visser T. J. (2008) N. Engl. J. Med. 358, 1811–1818 [DOI] [PubMed] [Google Scholar]
- 2.Dohán O., De la Vieja A., Paroder V., Riedel C., Artani M., Reed M., Ginter C. S., Carrasco N. (2003) Endocr. Rev. 24, 48–77 [DOI] [PubMed] [Google Scholar]
- 3.Gnidehou S., Caillou B., Talbot M., Ohayon R., Kaniewski J., Noël-Hudson M. S., Morand S., Agnangji D., Sezan A., Courtin F., Virion A., Dupuy C. (2004) FASEB J. 18, 1574–1576 [DOI] [PubMed] [Google Scholar]
- 4.Friedman J. E., Watson J. A., Jr., Lam D. W., Rokita S. E. (2006) J. Biol. Chem. 281, 2812–2819 [DOI] [PubMed] [Google Scholar]
- 5.Bianco A. C., Salvatore D., Gereben B., Berry M. J., Larsen P. R. (2002) Endocr. Rev. 23, 38–89 [DOI] [PubMed] [Google Scholar]
- 6.Watson J. A., Jr., McTamney P. M., Adler J. M., Rokita S. E. (2008) ChemBioChem 9, 504–506 [DOI] [PubMed] [Google Scholar]
- 7.Goswami A., Rosenberg I. N. (1979) J. Biol. Chem. 254, 12326–12330 [PubMed] [Google Scholar]
- 8.De Colibus L., Mattevi A. (2006) Curr. Opin. Struct. Biol. 16, 722–728 [DOI] [PubMed] [Google Scholar]
- 9.Joosten V., van Berkel W. J. (2007) Curr. Opin. Chem. Biol. 11, 195–202 [DOI] [PubMed] [Google Scholar]
- 10.Mansoorabadi S. O., Thibodeaux C. J., Liu H. W. (2007) J. Org. Chem. 72, 6329–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Pée K. H., Patallo E. P. (2006) Appl. Microbiol. Biotechnol. 70, 631–641 [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T., Kasia N. (2003) Trends Glycosci. Glycotechnol. 15, 329–349 [Google Scholar]
- 13.O'Reilly D. R., Miller L. K., Luckow V. A. (1994) Baculovirus Expression Vectors: a Laboratory Manual, pp. 130–134, Oxford University Press, Inc., New York [Google Scholar]
- 14.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 15.Pape T., Schneider T. R. (2004) J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 16.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 17.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 18.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 19.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collaborative (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 21.Tu S. C. (2001) Antioxid. Redox Signal. 3, 881–897 [DOI] [PubMed] [Google Scholar]
- 22.Holm L., Park J. (2000) Bioinformatics 16, 566–567 [DOI] [PubMed] [Google Scholar]
- 23.Taga M. E., Larsen N. A., Howard-Jones A. R., Walsh C. T., Walker G. C. (2007) Nature 446, 449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metrangolo P., Meyer F., Pilati T., Resnati G., Terraneo G. (2008) Angew. Chem. Int. Ed. Engl. 47, 6114–6127 [DOI] [PubMed] [Google Scholar]
- 25.Matter H., Nazaré M., Güssregen S., Will D. W., Schreuder H., Bauer A., Urmann M., Ritter K., Wagner M., Wehner V. (2009) Angew. Chem. Int. Ed. Engl. 48, 2911–2916 [DOI] [PubMed] [Google Scholar]
- 26.Auffinger P., Hays F. A., Westhof E., Ho P. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16789–16794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cody V., Murray-Rust P. (1984) J. Mol. Struct. 112, 189–199 [Google Scholar]
- 28.Gales L., Macedo-Ribeiro S., Arsequell G., Valencia G., Saraiva M. J., Damas A. M. (2005) Biochem. J. 388, 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkinson G. N., Skelly J. V., Neidle S. (2000) J. Med. Chem. 43, 3624–3631 [DOI] [PubMed] [Google Scholar]
- 30.Fraaije M. W., Mattevi A. (2000) Trends Biochem. Sci. 25, 126–132 [DOI] [PubMed] [Google Scholar]
- 31.Legrand Y. M., Gray M., Cooke G., Rotello V. M. (2003) J. Am. Chem. Soc. 125, 15789–15795 [DOI] [PubMed] [Google Scholar]
- 32.Lyubimov A. Y., Lario P. I., Moustafa I., Vrielink A. (2006) Nat. Chem. Biol. 2, 259–264 [DOI] [PubMed] [Google Scholar]
- 33.Tanner J. J., Lei B., Tu S. C., Krause K. L. (1996) Biochemistry 35, 13531–13539 [DOI] [PubMed] [Google Scholar]
- 34.Haynes C. A., Koder R. L., Miller A. F., Rodgers D. W. (2002) J. Biol. Chem. 277, 11513–11520 [DOI] [PubMed] [Google Scholar]
- 35.Hecht H. J., Erdmann H., Park H. J., Sprinzl M., Schmid R. D. (1995) Nat. Struct. Biol. 2, 1109–1114 [DOI] [PubMed] [Google Scholar]
- 36.Koike H., Sasaki H., Kobori T., Zenno S., Saigo K., Murphy M. E., Adman E. T., Tanokura M. (1998) J. Mol. Biol. 280, 259–273 [DOI] [PubMed] [Google Scholar]
- 37.Kobori T., Sasaki H., Lee W. C., Zenno S., Saigo K., Murphy M. E., Tanokura M. (2001) J. Biol. Chem. 276, 2816–2823 [DOI] [PubMed] [Google Scholar]
- 38.Lovering A. L., Hyde E. I., Searle P. F., White S. A. (2001) J. Mol. Biol. 309, 203–213 [DOI] [PubMed] [Google Scholar]
- 39.Johansson E., Parkinson G. N., Denny W. A., Neidle S. (2003) J. Med. Chem. 46, 4009–4020 [DOI] [PubMed] [Google Scholar]
- 40.Tanner J. J., Tu S. C., Barbour L. J., Barnes C. L., Krause K. L. (1999) Protein Sci. 8, 1725–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goswami A., Rosenberg I. N. (1977) Endocrinology 101, 331–341 [DOI] [PubMed] [Google Scholar]
- 42.Solís-S. J. C., Villalobos P., Orozco A., Valverde-R. C. (2004) J. Endocrinol. 181, 385–392 [DOI] [PubMed] [Google Scholar]
- 43.Yang K. Y., Swenson R. P. (2007) Biochemistry 46, 2289–2297 [DOI] [PubMed] [Google Scholar]
- 44.Thorpe C., Kim J. J. (1995) FASEB J. 9, 718–725 [DOI] [PubMed] [Google Scholar]
- 45.Vock P., Engst S., Eder M., Ghisla S. (1998) Biochemistry 37, 1848–1860 [DOI] [PubMed] [Google Scholar]
- 46.Kunishima M., Friedman J. E., Rokita S. E. (1999) J. Am. Chem. Soc. 121, 4722–4723 [Google Scholar]
- 47.Delano W. (2002) PyMOL, Delano Scientific, San Carlos, CA [Google Scholar]
- 48.Afink G., Kulik W., Overmars H., de Randamie J., Veenboer T., van Cruchten A., Craen M., Ris-Stalpers C. (2008) J. Clin. Endocrinol. Metab. 93, 4894–4901 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.