Abstract
Skeletal myogenesis is potently regulated by the extracellular milieu of growth factors and cytokines. We observed that cardiotrophin-1 (CT-1), a member of the interleukin-6 (IL-6) family of cytokines, is a potent regulator of skeletal muscle differentiation. The normal up-regulation of myogenic marker genes, myosin heavy chain (MyHC), myogenic regulatory factors (MRFs), and myocyte enhancer factor 2s (MEF2s) were inhibited by CT-1 treatment. CT-1 also represses myogenin (MyoG) promoter activation. CT-1 activated two signaling pathways: signal transducer and activator of transcription 3 (STAT3), and mitogen-activated protein kinase kinase (MEK), a component of the extracellular signal-regulated MAPK (ERK) pathway. In view of the known connection between CT-1 and STAT3 activation, we surprisingly found that pharmacological blockade of STAT3 activity had no effect on the inhibition of myogenesis by CT-1 suggesting that STAT3 signaling is dispensable for myogenic repression. Conversely, MEK inhibition potently reversed the inhibition of myotube formation and attenuated the repression of MRF transcriptional activity mediated by CT-1. Taken together, these data indicate that CT-1 represses skeletal myogenesis through interference with MRF activity by activation of MEK/ERK signaling. In agreement with these in vitro observations, exogenous systemic expression of CT-1 mediated by adenoviral vector delivery increased the number of myonuclei in normal post-natal mouse skeletal muscle and also delayed skeletal muscle regeneration induced by cardiotoxin injection. The expression pattern of CT-1 in embryonic and post-natal skeletal muscle and in vivo effects of CT-1 on myogenesis implicate CT-1 in the maintenance of the undifferentiated state in muscle progenitor cells.
Terminal differentiation of skeletal myogenic cells, termed myogenesis, consists of a series of well characterized highly regulated steps that has become a paradigm for lineage acquisition and cellular differentiation. Initially, pluripotent mesodermal stem cells commit to become myogenic precursor cells. Commitment to the myogenic lineage then results in the binary state of either maintenance of proliferative potential and pluripotency, or, on appropriate cues, withdrawal from the cell cycle, activation of a battery of structural, contractile, and metabolic genes constituting the differentiation program and ultimately formation of multinucleated myotubes (1). The field of myogenesis has benefited from the use of well established in vitro cell-culture systems, which faithfully recapitulate the in vivo differentiation program. During myogenesis, a group of basic helix-loop-helix transcription factors, myogenic differentiation-1 (MyoD),2 myogenic factor-5 (Myf5), myogenin (MyoG), and myogenic regulatory factor-4, collectively termed the myogenic regulatory factors (MRFs), play essential roles in differentiation (2–4). Most promoter-enhancer regions of muscle-specific genes contain the cognate binding site, E-box (CANNTG), for the MRFs, and the E-box is often essential for the induction of these genes during differentiation (5, 6). For example, early and late muscle-specific genes, MyoG, and muscle-specific myosin heavy chain (MyHC), respectively, are transcriptionally regulated by MyoD and other MRFs through E-boxes in their proximal promoter regions (4, 7). The molecular and genetic requirement for the MRFs during myogenesis has been confirmed in many studies both in vitro and in vivo (2, 8, 9). The MRFs also cooperate with another class of myogenic transcription factors, comprising the myocyte enhancer factor two family (MEF2) (10, 11). MEF2 genes are taxonomically part of the MADS-box gene superfamily that encode DNA-binding proteins involved in yeast mating type decisions (mini chromosome maintenance-1), plant development (Agamous and Deficiens), and serum responsivity of mammalian cells (serum response factor) (12–15).
As well as the detailed knowledge of core transcriptional regulatory circuits mediated by myogenic transcription factors and their accessory factors, much work has contributed to the identification of a number of growth factor and cytokine-mediated signaling pathways that positively and negatively impact myogenesis (16–19). In some cases, these pathways regulate the decision to differentiate or not, a critical regulatory point, because differentiation in muscle is terminal and absolutely required for viability of all metazoan life. Moreover, negative regulation of differentiation is equally important, because it underpins the maintenance of the proliferative state and pluripotency.
A number of growth factors and cytokines, such as insulin-like growth factors, insulin, transforming growth factor-β, fibroblast growth factor, and epidermal growth factor, that influence myogenesis has been identified (17–20), however, a detailed understanding of their corresponding signal transduction pathways and transcriptional network targets is still rudimentary. One group of cellular signaling cascades that are known to affect myogenesis in a complex manner consists of the MAPK pathways. For example, p38 MAPK, a member of one of the MAPK pathways, directly phosphorylates and activates E47, which forms a productive dimer with MyoD (21, 22). p38 MAPK also regulates MEF2 (23, 24) transcription factors as well as being involved in the recruitment of ATP-dependent chromatin remodeling factors to myogenic loci (25–27). Conversely the ERK-MAPK cascade plays a bi-phasic role in myogenic cells, being inhibitory in the initial phases of the differentiation program while being required for later stage events, such as cell fusion (28).
CT-1 is a member of the IL-6 family, which is composed of IL-11, leukemia inhibitory factor (LIF), ciliary neurotrophic factor, and oncostatin M. These cytokines are structurally related and form a variety of oligomeric ligand-receptor complexes. IL-6 and IL-11 form a complex with a homodimer of the glycoprotein-130 (Gp130) receptor or heterodimers of gp130 and leukemia inhibitory factor receptor-β (LIFRβ). Gp130/LIFRβ also recognizes LIF, CT-1, ciliary neurotrophic factor, oncostatin M, and cardiotrophin-like cytokine. Oncostatin M binds to the Gp130 and oncostatin M receptor. Upon formation of the requisite complex with the respective cytokine, the preponderant view is that the oligomeric receptor complex transduces its signal through the Janus kinase (JAK)-STAT signaling pathway (29).
CT-1 was originally identified in conditioned medium from embryoid bodies (30). In developing embryos, CT-1 is expressed in heart, skeletal muscle, liver, and dorsal root ganglia (31). In adults, human CT-1 mRNA is detected in the heart, skeletal muscle, ovary, colon, prostate, and testis and in fetal kidney and lung (32). The functions of CT-1 in the cardiovascular system have been extensively researched. Patients with ischemic and valvular heart disease have elevated levels of CT-1 in their sera (33). Further study of the role of CT-1 in the heart indicated that it has a cardioprotective role by reducing apoptosis (31, 34) and may be involved in regeneration of cardiac muscle after infarction (35). Exogenously administered CT-1 also induces cardiac hypertrophy in vitro (31). Although the modulation of cardiomyocyte phenotype by CT-1 has been well documented, the underlying signaling pathways are still unclear, and the role of CT-1 in skeletal muscle has not, thus far, been characterized.
In this report, we demonstrate that CT-1 is a potent inhibitor of skeletal muscle differentiation. In C2C12 cells, CT-1 represses molecular markers of muscle differentiation and phenotypic myogenesis. Also, the transcriptional networks involved in the induction of key myogenic genes such as the MyoG and MCK genes are suppressed by CT-1 signaling. Surprisingly, small chemical inhibitors of MEK, PD98059 and U0126, reversed these repressive effects on skeletal myogenesis by CT-1, whereas inhibition of STAT3 activation was without effect. Collectively, these data show that CT-1 interferes with the transcriptional network required for muscle differentiation through the activation of the MEK-MAPK signaling module. Furthermore, in vivo, adenovirus-mediated expression of CT-1 increases satellite cell number and delays regeneration of damaged muscle by cardiotoxin injection. These observations indicate that CT-1 represses myogenesis and serves to maintain myogenic progenitors in their proliferative, multipotent state in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Plasmids
MRF expression plasmids were constructed in pEMSV as described elsewhere (36). An activated (ΔN3 S218D/S222E) human MEK1 expression construct was a kind gift from A. Natalie (37). The reporter construct pMCK-eGFP was a gift from A. Ferrer-Martinez (Universitat de Barcelona, Spain). Transcription reporter constructs, pMCK-luc (38) and pCMV-β-galactosidase, were described elsewhere (39). The myogenin promoter region was excised from pMyoG-luc by SacI/BglII digestion. The resultant 1152-bp fragment was inserted at the SacI/BglII sites of pGL4–10 vector (Promega, Madison, WI). The dsRed2-N1 expression construct was purchased from Clontech Laboratories.
Antibodies
The primary antibodies used in this study obtained from Santa Cruz Biotechnology (Santa Cruz, CA) were MyoD (C-20), Myf5 (C-20), actin (I-19), and ERK1 (C-16). MEF2D (610775) was from BD Biosciences. Stat3 (9132), phospho-Stat3 (Tyr-705, 58E12, 9135), phospho-Stat3 (Ser-727, 6E4, 9136), MEK1/2 (9122), phospho-MEK1/2 (Ser-217/221, 9121), and phospho-p44/42 MAPK (Thr-202/Tyr-204, E10, 9106) were from Cell Signaling Technology. Myogenin (F5D) and MyHC (MF20) were from the Developmental Studies Hybridoma Bank. MyoD1 (clone 5.8A, M3512) was from DakoCytomation. Polyclonal antibody for MEF2A was prepared as described previously (23). Normal mouse IgG (sc-2025) was from Santa Cruz Biotechnology.
Cell Culture
C2C12 myoblasts were obtained from American Type Culture Collection (CLR-1772) and cultured in growth medium (GM) consisting of 10% fetal bovine serum (HyClone) in high-glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 1% penicillin-streptomycin (Invitrogen) at 37 °C and 5% CO2. Myotube formation was induced by replacing GM with differentiation medium (DM), which consisted of 2% horse serum (Atlanta Biologicals) in Dulbecco's modified Eagle's medium supplemented with 1% penicillin-streptomycin. For CT-1 treatment, recombinant mouse CT-1 (R&D system, 438-CT) was resuspended with solvent (4 mm HCl, 0.1% bovine serum albumin) and supplemented into the media. For myotube formation assays, DM with CT-1 (10 ng/ml) was replenished every 2 days. Inhibitors (PD98059 (Cell Signaling Technology, 9900), U0126 (Cell Signaling Technology, 9903), and P6 (2-(1,1-dimethylethyl)-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinolin-7-one, Pyridone 6, Calbiochem, 420097)) were resuspended with DMSO and added into the cell culture media for 30 min prior to adding CT-1.
Sacromeric Myosin Heavy Chain Detection
C2C12 cells were washed with Phosphate-buffered saline (PBS, pH 7.4) and fixed with 90% methanol at −20 °C for 10 min. After fixation, the cells were incubated in 5% milk in PBS for 30 min at 37 °C for blocking. Cells were incubated at room temperature with MF-20 (primary antibody) diluted in blocking buffer (5% milk PBS) for 1 h. After incubation, the cells were washed three times with PBS and incubated for 60 min at room temperature with an horseradish peroxidase-conjugated α-mouse secondary antibody. The cells were again washed three times with PBS and incubated in developer (0.6 mg/ml DAB, 0.1% H2O2 in PBS) to detect MyHC by immunocytochemistry. The nuclei were counter-stained with hematoxylin. Images were recorded with a microscope (Axiovert 35; Carl Zeiss MicroImaging) with either 4X NA 0.10 or 10X NA 0.25 Achrostigmat objective lenses with a digital camera (Canon, EOS D60).
Proliferation Assay
After 72 h in DM and in the presence of CT-1 (10 ng/ml) (or solvent), cells were incubated with 100 μm of bromodeoxyuridine (Sigma) for 1 h at 37 °C. Cells were washed with cold 1x PBS then fixed with 70% ethanol for 1 h at 4 °C. The cells were then washed with 1x PBS and incubated with 2N HCl for 1 h at 37 °C to denature the DNA. The cells were blocked in 10% goat serum (Sigma) diluted in 1x PBS for 2 h at room temperature with shaking and then incubated with bromodeoxyuridine primary antibody (G3G4: Developmental hybridoma bank, Iowa) diluted in 1.5% goat serum (Sigma) for 1.5 h at room temperature with shaking. Cells were washed with 1x PBS-T (0.5% Tween20) and incubated with anti-mouse secondary antibody conjugated to FITC (Sigma) diluted in 1.5% goat serum (Sigma) for 2 h at room temperature with shaking. Cells were washed with 1x PBS-T (0.5% Tween20).
Microscopy and Fluorescence
Fluorescence and phase contrast pictures were obtained using an epifluoresence microscope (Axiovert 35; Carl Zeiss MicroImaging), with appropriate phase and filter settings, and either 4X NA 0.10 or 10X NA 0.25 Achrostigmat objective lenses. Images were recorded with a digital camera (Canon, EOS D60).
Western Blotting Analysis
Total cellular protein extracts were prepared in Nonidet P-40 lysis buffer (0.1% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, 50 mm Tris-HCl pH 8.0, 1 mm sodium vanadate, 1 mm PMSF, supplemented with a protease inhibitor mixture (Sigma, P-8340)). Protein concentrations were determined by a standard Bradford assay (Bio-Rad). Equivalent amounts of protein were resolved by SDS-PAGE gels, followed by electrophoretic transfer to an Immobilon-P membrane (Millipore) as directed by the manufacturer (Millipore). Blots were incubated with the indicated primary antibody in 5% milk in PBS or Tris buffered saline (TBS)-T (10 mm Tris-HCl pH8.0, 150 mm NaCl, 0.1% Tween-20) or 5% Bovine serum albumin (bovine serum albumin) in TBS-T according to the manufacturer's protocol at 4 °C overnight with gentle agitation. After washing briefly, the blots were incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies in 5% milk in PBS or TBS-T at room temperature according to the manufacturer's protocols (Santa Cruz Biotechnology and Cell Signaling Technology). After washed three times with 1× PBS or 1× TBS (depending on the primary antibody) at room temperature, the blots were treated with the enhanced chemiluminescence reagent (Amersham Biosciences) to detect immunoreactive proteins. The blots were exposed to BioMax film (Kodak) for visual representation.
Transcription Reporter Gene Assays
C2C12 myoblasts were transfected by a standard calcium phosphate-DNA precipitation method with the indicated reporter gene and expression constructs and pCMV-β-galactosidase to monitor transfection efficiency. After transfection, the cells were washed with PBS and maintained in GM and then treated as indicated. Total cellular protein was extracted with luciferase lysis buffer (20 mm Tris-HCl, pH 7.4, 0.1% Triton X-100). Luciferase and β-galactosidase enzyme assays were performed according to the manufacturer's protocol (Promega). Luciferase activity was quantified using a luminometer (Berthold Lumat, 9501) and standardized according to the β-galactosidase activity. Relative luciferase units normalized for the β-galactosidase activity (relative luciferase units) were determined and plotted as an average of triplicate determinations, and error bars represent the standard deviations of the triplicate values. Each experiment was repeated at least three times.
Semi-quantitative Reverse Transcription-PCR Analysis
Total RNA was extracted from cells with TRIzol (Invitrogen) according to the manufacturer's protocol. cDNA was generated from the isolated total RNA (1 μg) with SuperScript III (Invitrogen) and oligo-dT16 primer (Sigma) by the protocol provided by the manufacturer. To amplify a target transcript, a pair of primers was designed that flanked an intron based on the mouse gene sequences. The target transcripts were amplified by Taq DNA polymerase (New England Biolabs) with gene-specific primers. An amplified DNA was separated in an agarose gel and visualized by ethidium bromide (Sigma) staining and UV exposure. Detailed information about the primers is in the supplemental material.
Co-immunoprecipitation Analysis
An equal amount of total cellular protein (250 μg) was diluted with Nonidet P-40 lysis buffer to a final concentration of 1 μg/μl. Protein complexes were immunoprecipitated with the indicated antibody and 25 μl of protein G-Plus Sepharose beads (50% slurry, Santa Cruz Biotechnology) by incubation at 4 °C overnight on a rotating platform. The beads were washed with three changes of NETN wash buffer (0.1% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, and 50 mm Tris-HCl, pH 8.0). Beads were boiled in SDS sample buffer, and protein complexes were resolved by SDS-PAGE and immunoblotted as described above.
CT-1 Adenovirus
The CT-1 adenovirus was previously described (40). Briefly, full-length murine CT-1 cDNA was isolated by PCR and the CT-1 reading frame was fused with a 60-br pre-nerve growth factor leader sequence to promote secretion of the CT-1 protein. The CT-1 cDNA was cloned in-frame with the long terminal repeat of the Rous sarcoma virus (40). A LacZ-containing adenovirus (CTRL) was used as a control for all injection experiments. This adenovirus was kindly provided by Dr. Robin Park at the Ottawa Health Research Institute, Ottawa, Canada.
In Vivo Administration of CT-1: Muscle Injury
To test CT-1 in vivo, B6C3F1 mice were subjected to systemic delivery of the CT-1 adenovirus. Briefly, animals were anesthetized with halothane. The injections were administered via intra-cardiac chamber delivery using a 29-gauge insulin needle (VWR) with 50 μl of Ad-CT-1 at a concentration of 3.0 × 108 plaque forming units/ml (n = 3). A control group of B6C3F1 mice were injected with 50 μl of Ad-CTRL at a similar concentration (n = 3). In a separate group of animals, cardiotoxin was used to induce muscle injury immediately prior to AdCT-1 and Ad-CTRL injection (n = 3 for each group). 25 μl of 10 μm cardiotoxin (Latoxan) was injected directly into the tibialis anterior (TA) muscle using a 29G1/2 insulin syringe in halothane-anesthetized mice (41). Post-recovery, mice were monitored closely for weight loss, dehydration, and cardiac distress. All injections were administered by a trained animal care technician according to the standards of the Animal Care Committee at the University of Ottawa, Ottawa, Canada.
Immunohistology
At 7 days post-injection, skeletal muscle was excised and rinsed in cold 1× PBS. The muscle was fixed in 4% Pefabloc A in PBS for 2days then embedded in paraffin, sectioned at 10 μm, and counterstained with hematoxylin and eosin to visualize the nuclei and cytoplasm. Sections were dehydrated in a graded ethanol series ending in CitriSolv (Fisher Scientific). For immunohistology, sections were treated with antigen-unmasking solution (Vector Labs), blocked with 5% bovine serum albumin, incubated overnight at 4 °C with a primary antibody, then incubated in donkey-anti-goat CY3 antibody (Chemicon) and finally counterstained with 4′,6-diamidino-2-phenylindole (Sigma). Five fields of view per section and five sections per TA muscle were analyzed. The micrographs presented are representative views.
Stem Cell/Progenitor Cell Isolation
Side population (muscle progenitor cells) were collected as previously described (42). Contralateral TA muscle was collected from Ad-CT-1 and Ad-CTRL mice, and all visible connective tissue and blood vessels were removed by dissection. Muscle was digested in collagenase B (10 mg/ml, Roche Applied Science) plus dispase II (2 units/ml, Roche Applied Science) for and the resulting single ell suspensions were then stained with Hoechst dye 33342 (5 μg/ml, Sigma-Aldrich) at 37 °C for 90 min. As an SP control, the drug verapamil (50 μm, Sigma-Aldrich) was added to an aliquot of cells simultaneously stained with Hoechst 33342. Cells were finally re-suspended in 500 μl of Hanks' balanced salt solution with 2% fetal bovine serum and 10 mm Hepes. The cells were filtered through a 50 μm Cell Tric® (disposable filters made of monofil nylon material, Partec GmbH) and remained on ice until fluorescence-activated cell sorting analysis (42). Cell sorting was performed using a MoFlo high speed cell sorter (DakoCytomation) (42). Forward and side scatter was measured at 488 nm (Spectraphysic argon laser). The Hoechst dye was excited at 359 nm (I90C laser from Coherent). Blue emission was measured at 424 nm (424/44 band pass filter), and red emission was above 675 nm (675 AGLP long pass filter). All data were collected and analyzed with SummitTM data acquisition and analysis software (DakoCytomation).
Methylcellulose Stem Cell/Progenitor Cell Culture
Side population cells (2 × 104) were re-suspended in 2.5 ml of Methocult media GF3434 (Stem Cell Technologies) using a 5-ml syringe and a 12-gauge needle (42). Cells were then plated on 2-cm plastic Petri dishes and incubated in humidity chambers at 37 °C and 5% CO2 for 14 days. At 14 days post plating, colonies were counted using a Zeiss inverted microscope.
Statistical Analysis
Differences between Ad-CT-1- and Ad-CTRL-injected samples were evaluated for statistical significance using one tailed, unpaired Student's t test. Differences were considered statistically significant at a p value <0.05.
RESULTS
CT-1 Represses Myogenic Differentiation
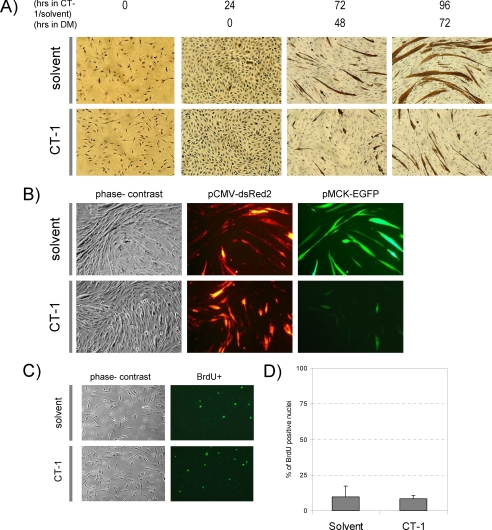
Major sites of CT-1 expression during embryonic development are heart and skeletal muscle (31). Although the role of CT-1 in the cardiovascular system is being defined (43), its role in skeletal muscle is not characterized. To begin to elucidate CT-1 function in skeletal muscle, we initially treated C2C12 cells chronically with CT-1 (10 ng/ml, 0.5 nm) and assessed muscle differentiation by the formation of multinucleated myotubes and accumulation of a skeletal muscle differentiation marker protein, MyHC. Solvent-treated C2C12 cells began to exhibit multinucleated myotubes after 48 h in DM. Thereafter, the control, solvent-treated C2C12 cells developed MyHC-positive myotubes with large numbers of nuclei at later time points (Fig. 1A). In contrast, C2C12 cells in the CT-1-containing DM failed to form multinucleated myotubes at 48 h. At later time points, some myogenesis occurred, although the number and caliber of MyHC-positive myotubes were greatly reduced in the presence of CT-1 compared with the corresponding controls (Fig. 1A). In addition, the MCK promoter activity was strongly inhibited by CT-1 as indicated by the transfection of an MCK promoter-reporter gene fused to enhanced signal green fluorescent protein (EGFP) (pMCK-EGFP) (Fig. 1B). We also observed that CT-1 did not affect the cellular proliferation rate of differentiating myoblasts in DM assessed by bromodeoxyuridine incorporation rate (Fig. 1C&D). Therefore, these data document that CT-1 represses the skeletal muscle differentiation program without affecting proliferation rate.
FIGURE 1.
CT-1 represses myogenic differentiation. A, C2C12 cells were seeded onto cell culture plates at equal density and maintained in CT-1 (10 ng/ml) or solvent containing growth medium (GM) or differentiation medium (DM) for the indicated time period. The cells were fixed and stained for muscle myosin heavy chain (MyHC) detection by immunochemistry. The photomicrographs are representative fields in each condition. B, C2C12 cells were plated at equal density and transfected with pCMV-deRed2 and pMCK-eGFP constructs. The transfected cells were maintained in CT-1 (10 ng/ml) or solvent containing DM for 72 h to induce myotube formation. The cell morphology was recorded by phase-contrast microscopy and transfected cells were monitored by the red fluorescence signal. MCK promoter activity was assessed by the green fluorescence signal. C, C2C12 cells were maintained in DM for 72 h and CT-1 (10 ng/ml) or solvent was added every 24 h. After 72 h in low serum conditions cells were incubated with 100 μm of bromodeoxyuridine for 1 h. Cells were then fixed with 70% ethanol and then incubated with 2N HCl to denature the DNA. The cells were then blocked with 10% goat serum prior to incubation with bromodeoxyuridine primary antibody. The cells were then washed with 1x PBS-T and incubated with secondary antibody conjugated to FITC. The cells were washed with 1x PBS-T and mounted using fluorescence mounting media and viewed under a fluorescence microscope. D, the average of percentage of bromodeoxyuridine positive nuclei over total nuclei in 12 individual fields per condition was calculated and graphed. (error = standard variation).
CT-1 Represses the Expression of Pro-differentiation Transcriptional Regulators (MyoG and MEF2A/D)
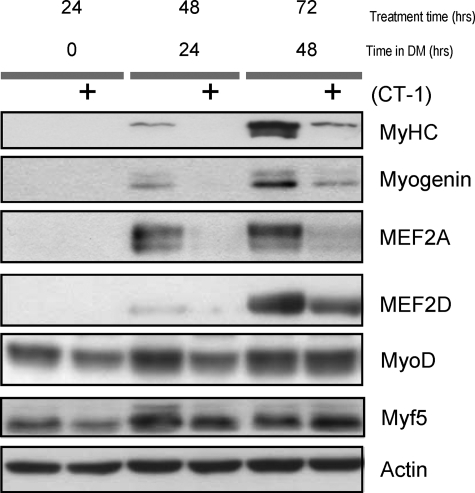
To generate multinucleated myotubes from mono-nucleated myoblasts, the MRFs and MEF2s play an essential synergistic role at various stages of the differentiation program (44). Therefore, we postulated that CT-1 might interfere with muscle differentiation through the MRFs and/or MEF2. First, to establish that the repression of myogenesis by CT-1 was observed in this analysis the levels of MyHC, a structural marker of muscle differentiation were assessed. As we expected that MyHC accumulated in the solvent-treated C2C12 cells at late time points. Conversely, this accumulation of MyHC was largely attenuated in C2C12 cells treated with CT-1 (Fig. 2). Having determined that myogenesis was repressed by CT-1 at the molecular level, we next assessed the levels of various muscle transcription factors. Under these conditions, the expression of MyoG, a key MRF required for differentiation (45), was repressed by CT-1 compared with the solvent-treated cells, in which it was strongly induced (Fig. 2). In addition, MEF2A and MEF2D were also lower in the cells treated with CT-1 (Fig. 2). These data indicate that CT-1 inhibits myogenic differentiation by interfering with the up-regulation of MyoG and MEF2 factors. Interestingly, MyoD and Myf5 protein levels were relatively unaffected by CT-1 suggesting that the lesion in the hierarchical differentiation program lies between the MRFs required for lineage commitment (MyoD and Myf5) and the pro-differentiation transcriptional regulators (MyoG and MEF2A and MEF2D).
FIGURE 2.
CT-1 represses the expression of pro-differentiation transcriptional regulators (MyoG and MEF2A/D). C2C12 cells were induced to differentiate in DM with CT-1 (10 ng/ml) or solvent. The cells were maintained in the indicated conditions for specific time periods. Total protein samples were extracted from the cells and equal amounts of total protein (20 μg) were subjected to Western blotting analysis. The levels of indicated proteins were assessed by a standard immuno-blotting technique with a specific primary antibody. Actin indicates equal amounts of protein loading into each lane.
Transcriptional Induction of the MyoG Promoter by MyoD Is Repressed by CT-1 Signaling
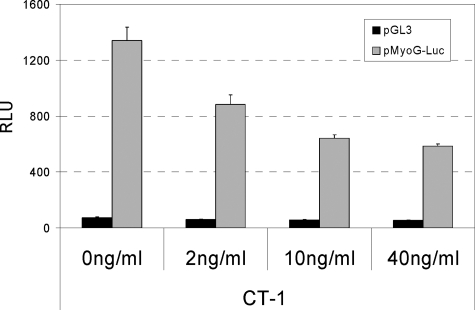
Because MyoD, along with Myf5, plays an early “commitment” role in the myogenic cascade and also plays an important role in the induction of the myoG gene (26), we hypothesized that CT-1 might interfere with the trans-activation properties of MyoD and therefore its ability to activate myoG transcription. To begin to address this hypothesis, we initially measured myoG promoter activity using reporter gene assays. In the absence of CT-1, the myoG promoter was activated in differentiating C2C12 cells in DM (Fig. 3). In the presence of CT-1, the activation of the myoG promoter was markedly inhibited in a dose-dependent manner (Fig. 3). These data indicate that reduced MyoG levels observed with CT-1 (Fig. 2) result from a loss of transcriptional induction of the myoG locus.
FIGURE 3.
Transcriptional induction of the myoG promoter by MyoD is repressed by CT-1. C2C12 cells were transfected with either pGL3 (empty control) or a Myogenin promoter-luciferase reporter gene construct (pMyoG-Luc), and to monitor transfection efficiency, pCMV-β-gal construct was included in each condition. The transfected cells were maintained for 16 h in the indicated concentration of CT-1 or its solvent in DM. Total protein samples were harvested with a luciferase lysis buffer. Luciferase activity in each condition was measured independently and normalized according to β-Galactosidase activity.
trans-Activation Properties of the MRFs Are Repressed by CT-1
Based on our observation that myoG gene transcription was attenuated by CT-1, we next focused on whether MyoD trans-activation properties might be altered by CT-1, because MyoD expression levels remained unaffected with CT-1 treatment (Fig. 2).
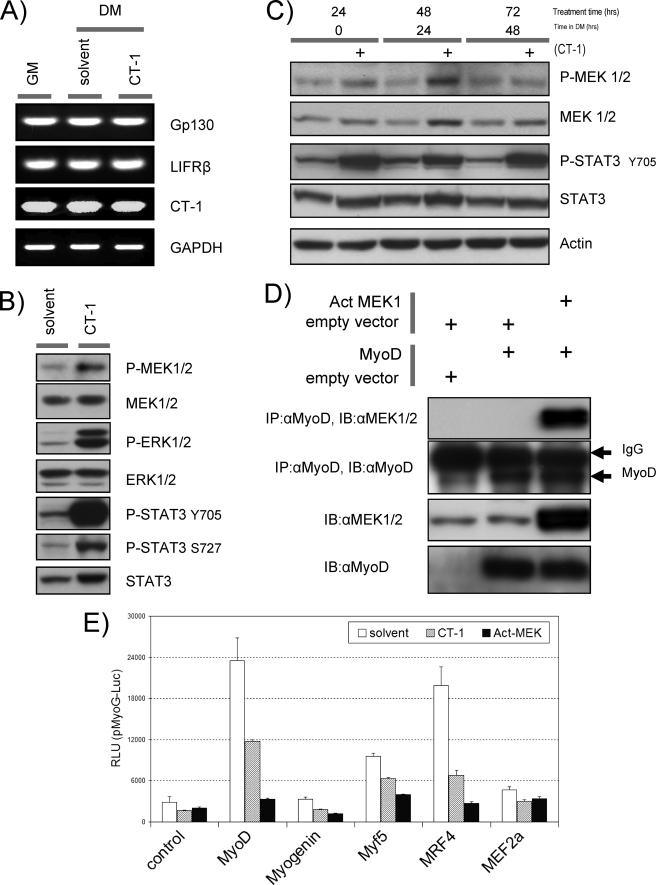
The trans-activation capacity of MyoD has already been documented to be a heavily regulated aspect of its function, both positively and negatively, by a variety of mechanisms (46–49). Bioinformatic analysis of MyoD-interacting proteins revealed that MEK1 (50) and STAT3 (51) also share the property that they are known to be activated by phosphorylation in the presence of IL-6 family cytokines in different cell types. Therefore, we first confirmed the expression of CT-1 and its signal transduction receptors, Gp130 and LIFRβ, in C2C12. Endogenous CT-1 and its receptor expression were confirmed by semi-quantitative reverse transcription-PCR analysis in the presence or absence of recombinant exogenous CT-1 in the media. We detected these transcripts in C2C12 cells, and their expression levels were not affected in the presence of CT-1 (Fig. 4A). We then surveyed these signaling molecules first by determining the phosphorylation levels of MEK1 and STAT3 in C2C12 cells acutely (Fig. 4B) or chronically (Fig. 4C) treated with CT-1 by Western blotting analysis. Indeed, levels of phosphorylated MEK-1 and STAT3 proteins in C2C12 cells were elevated in the presence of CT-1 compared with those in solvent control cells (Fig. 4, B and C). A previous study indicated that MyoD transcriptional activation properties can be inhibited by a direct interaction with MEK1 (50). Therefore, we sought to test this interaction by co-immunoprecipitation analysis. These experiments revealed that exogenous expression of an activated form of MEK1 (Act-MEK: MEK1 R4F) and MyoD resulted in co-purification of the two molecules in the same complex, suggesting the possibility that this interaction can occur (Fig. 4D). In addition we observed that the typical activation of the myoG promoter (Fig. 4E) and MCK promoter (supplemental Fig. S1) by exogenously expressed MRFs was repressed by CT- 1 signaling (recombinant CT-1 or Act-MEK1) (Fig. 4E). These results further support the idea that MyoD trans-activation properties are repressed by CT-1 and that MEK activation is a key component of that repression.
FIGURE 4.
Trans-activation properties of the MRFs are repressed by CT-1. A, total RNA was isolated from C2C12 cells in GM (lane 1), DM with solvent (48 h) (lane 2), and DM with CT-1 (10 ng/ml) (48 h) (lane 3) and subjected to semi-quantitative reverse transcription-RCP analysis with indicated gene specific primer pairs. reverse transcription-PCR amplified DNA was separated in a TAE/agarose-gel, and ethidium bromide stained DNA was visualized by UV irradiation. GAPDH serves as an internal loading control. B, C2C12 cells were plated at equal density and kept in DM for 16 h. CT-1 (10 ng/ml) or equal volume of the solvent was added to the media. The cells were harvested after 20min of CT-1/solvent addition. Total protein samples were subjected to Western blotting analysis to estimate the levels of indicated proteins. C, Western blotting analysis was performed as described above. However, the cells were maintained in DM with CT-1 or solvent for indicated time periods. D, C3H10T1/2 cells were transfected with combinations of the indicated constructs. Total protein samples were extracted from the cells maintained in DM. Exogenous-expression of MyoD and an activated form of MEK1 was confirmed by immuno-blotting (IB) (10 μg loading) with the specific antibodies. An immuno-precipitation (IP) analysis was performed with the total protein extract (250 μg) with MyoD antibody (mouse) and proteinG conjugated beads. Precipitated immuno-complex were eluted off the proteinG beads and subjected for an immunoblotting with MEK antibody (Rabbit). Equal amount of IgG loading was monitored with MyoD immuno-blotting with MyoD specific antibody (Rabbit). E, C2C12 cells were transfected with the indicated expression constructs or its empty vector (1 μg) and myoG promoter- (pMyoG-Luc) promoter-Luciferase reporter construct (0.5 μg). In addition, an activated form of MEK1 expression vector (1 μg) or its empty vector (for CT-1 and solvent) was included. To monitor transfection efficiency, a pCMV-β-Gal construct was also included (0.3 μg). After transfection, the cells were maintained in DM containing CT-1 (10 ng/ml) or its solvent for 16 h. The cells were harvested and subjected to luciferase assay and β-Galactosidase assay.
CT-1 Inhibits the Transcriptional Properties of the MRFs through Activation of MEK Signaling
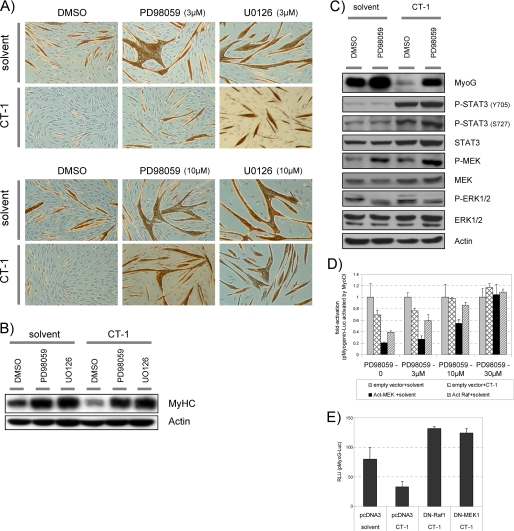
To directly test the idea that CT-1 activation of MEK is responsible for MyoD trans-repression, we utilized MEK-specific inhibitors, PD98056 and U0126. First, we reasoned that if MEK activation is absolutely required for CT-1 repression of myogenesis, then we should abrogate CT-1 effects on myogenesis by repression of MEK. In the absence of CT-1 (solvent), C2C12 cells formed multinucleated myotubes, and they accumulated MyHC proteins (brown color) after 2 days in DM (Fig. 5A). These morphological changes were not observed in the presence of CT-1. However, addition of MEK inhibitors neutralized the inhibitory effect of CT-1 on both myotube formation and MyHC accumulation in a dose-dependent manner (3 μm versus 10 μm) assessed by immunochemistry (Fig. 5A). Western blotting analysis of MyHC levels further confirmed the above observations (Fig. 5B). In agreement with this, a more detailed Western blotting analysis showed that MyoG protein levels were lower in the presence of CT-1, and this inhibitory effect was reversed by MEK inhibition (PD98059), which prevented CT-1-mediated induction of phosphorylation of ERK (an MEK activity indicator). It was noted that, as previously reported in different systems (52, 53), the MEK inhibitor caused hyperphosphorylation of MEK. However, in the presence of PD98059 up-regulation of phospho-ERK by CT-1 was clearly inhibited (Fig. 5C). Therefore, this MEK inhibitor prevents CT-1-mediated activation of MEK. We also noticed that the MEK inhibitor reversed these CT-1 effects without affecting the phosphorylation levels of STAT3 (Fig. 5C, see below). Furthermore, luciferase reporter gene assays also showed that myoG promoter activity driven by exogenously expressed MyoD was repressed by CT-1. Furthermore, exogenous expression of an activated form of MEK1 or Raf (components of the MAPK signaling pathway), also repressed myoG activation, and these effects were reversed in a dose-dependent manner by MEK inhibition (Fig. 5D), and by expression of dominant negative form of MEK1 or Raf1 (Fig. 5E). Therefore, these data indicate that MEK inhibition “rescues” muscle differentiation from the inhibitory effect of CT-1, both morphologically and biochemically; and repression of MyoD trans-activation properties by CT-1 is also reversed by MEK inhibition. Taken together, CT-1 represses skeletal myogenic differentiation through interference of the transcriptional activity of MyoD by the activation of MEK signaling.
FIGURE 5.
CT-1 inhibits the transcriptional activity of the MRFs through activation of MEK signaling. A, C2C12 cells were plated at equal density and induced differentiation transferred into DM upon about reaching confluence. The cells were maintained in indicated concentration of MEK inhibitor (PD98059, U0126, or DMSO; 3 μm or 10 μm) with without CT-1 (10 ng/ml). After 2days in the indicated conditions, the cells were fixed and stained for MyHC detection by immunochemistry with MF-20 mouse monoclonal antibody. MyHC protein accumulation was indicated by brown color. The photomicrographs are representative fields. B and C, C2C12 cells were maintained in DM with CT-1 (10 ng/ml) and or PD98059 (10 μm), or their solvents for 2days (C) or 3days (B) to induce myotube formation. Total cellular proteins were extracted from the cells in each condition. The total protein lysate samples (20 μg) were subjected to Western blotting analysis. Actin levels indicate loading of an equal amount of the total protein into each lane. D, C2C12 cells were transfected with a pMyoG-Luc (0.5 μg), a MyoD expression vector (1 μg), a pCMV-β-Gal (0.3 μg), and also the indicated kinase expression vector (act.MEK1, act.Raf) or an empty vector (1 μg). The transfected cells were maintained in DM containing CT-1 (10 ng/ml) or solvent, and the indicated concentration of PD98059 MEK inhibitors for 16 h. The cells were harvested and subjected to Luciferase assay and β-Gal assay. Luciferase activity was normalized according to the β-galactosidase activity from a co-transfected pCMV-β-Gal expression construct by calculating the Relative Luciferase Unit (relative luciferase unit) for each individual condition, and the fold-activation was calculated with respect to the average relative luciferase unit of the “empty vector + solvent” at the corresponding concentration of PD98059. E, C2C12 cells were transfected with a pMyoG-Luc (0.5 μg), a pCMV-β-Gal (0.3 μg), and also the indicated kinase expression vector (DN-MEK1, DN-Raf1) or an empty vector (1 μg). The transfected cells were maintained in DM containing CT-1 (10 ng/ml) or solvent for 16 h. The cells were harvested and subjected to Luciferase assay and β-Gal assay.
STAT3 Activation by CT-1 Is Not Sufficient for Inhibition of Myogenesis
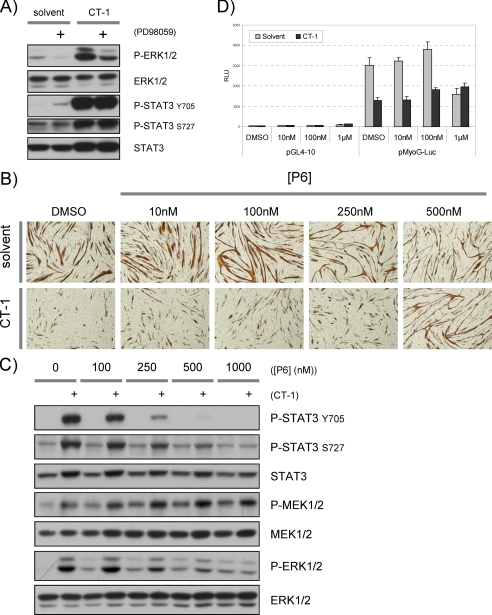
We documented that STAT3 is highly phosphorylated at tyrosine 705 (Tyr-705) and serine 727 (Ser-727) in response to CT-1 treatment (Fig. 4, A and B). The Tyr-705 phosphorylation is required for STAT3 dimer formation, nuclear translocation, and transcriptional regulatory activity of STAT3 (54–56). Since a previous study showed that activated STAT3 can inhibit the transcriptional properties of MyoD (51), we postulated that STAT3 might also be involved in the repression of MyoD by CT-1 signaling. Western blotting analysis showed that the MEK inhibitor inhibited phospho-ERK1/2 (an indicator of MEK activity) activation by CT-1. However, MEK inhibition had no apparent effect on the phosphorylation levels of STAT3 at Tyr-705 or Ser-727 by acute or chronic CT-1 treatment (Figs. 5C and 6A). Because MEK inhibition rescues myogenic repression but does not alter STAT3 phosphorylation by CT-1, this indicates that STAT3 activation is not sufficient to inhibit myogenesis. To further address this issue, we next used a pan-JAK kinase inhibitor, P6 (57), because STAT3 is phosphorylated by the Gp130/LIFRβ-associated JAK kinases. As previously observed, CT-1 inhibited myotube formation and MyHC accumulation in DM compared with controls (Fig. 6B). In assessing the dose dependence of the P6, we observed no effect on CT-1-mediated myogenic repression up to a concentration at 250 nm. However, at 500 nm, P6 clearly neutralized the inhibitory effect of CT-1. Because P6 inhibits tyrosine kinase activity of other kinases at high levels (58), we assessed the inhibitory effect of P6 on the phosphorylation levels of, STAT3, MEK1/2, and ERK1/2 by Western blotting analysis. As is claimed for this inhibitor, increased phosphorylation of STAT3 (Tyr-705 and Ser-727) by CT-1 was inhibited by P6 in a dose-dependent manner (Fig. 6C). However, at a high concentration (500 nm), P6 also repressed phosphorylation of ERK1/2 in the presence of CT-1. Because significant repression of phosphorylation of STAT3 was seen with the P6 inhibitor at low concentrations (up to 250 nm), but such concentrations had no effect in reversing CT-1 effects on myogenesis, we conclude that STAT3 activation by CT-1 is not sufficient to inhibit myogenesis. In agreement with the above results, MyoD-driven myoG promoter activity was clearly inhibited in the presence of CT-1 (Fig. 6D). However, at any concentration tested, P6 had little effect on the inhibitory effect of CT-1. In addition, exogenous expression of constitutively active (A662C and N664C) (54), phospho-mimetic mutant (Y705D and S727D), or dominant negative forms of STAT3 (S705F and S727A) (59) had no apparent effect on myogenesis phenotypically and biochemically in the presence of CT-1 (data not shown). Therefore, these results indicate that inhibition of MEK1/2 activity but not JAK activity is required for reversing the inhibitory effect of CT-1 on myogenesis. Taken together, we conclude that CT-1 inhibits skeletal muscle differentiation primarily through activation of MEK and, surprisingly, does not require STAT3 activation.
FIGURE 6.
STAT3 activation by CT-1 is not sufficient for inhibition of myogenesis. A, C2C12 cells were plated at equal density maintained in DM. A MEK inhibitor (PD98059 (10 μm)) or DMSO was added 30min before the addition of CT-1 (10 ng/ml) or its solvent. After 20 min of CT-1 or solvent treatment, the cells were harvested, and total protein samples were extracted for each condition. The protein samples (20 μg) were subjected to Western blotting analysis. B, an equal number of C2C12 cells were plated and maintained in DM containing CT-1 (10 ng/ml) or its solvent, in addition, the indicated concentration of pan-JAK kinase inhibitor, P6, was included in the DM. The cells were fixed after maintained in the DM for 3days, and accumulation of MyHC was visualized by immunochemistry. The brown color indicates MyHC accumulation in the cells. The photomicrographs are representative fields of each condition. C, C2C12 cells were plated at equal density and maintained in DM for 16 h. Thirty min before adding CT-1 (10 ng/ml) or its solvent, the cells were treated with indicated concentration of P6 (pan-JAK kinase inhibitor). After 20 min of CT-1 or solvent addition to the media, the cells were harvested. Total protein samples were extracted from the cells in each condition, and equal amounts of the protein (20 μg) was subjected for Western blotting analysis. D, C2C12 cells were transfected with an either pMyoG-Luc or pGL4–10 (0.5 μg), and a MyoD expression vector (1 μg), a pCMV-β-Gal (0.3 μg). The transfected cells were maintained in DM containing CT-1 (10 ng/ml) or solvent, and the indicated concentration of P6 pan-JAK inhibitor for 16 h. The cells were harvested and subjected to Luciferase assay and β-Gal assay.
CT-1 Increases the Number of Muscle Precursor Cells and Delays Regeneration of Damaged Muscle in Vivo
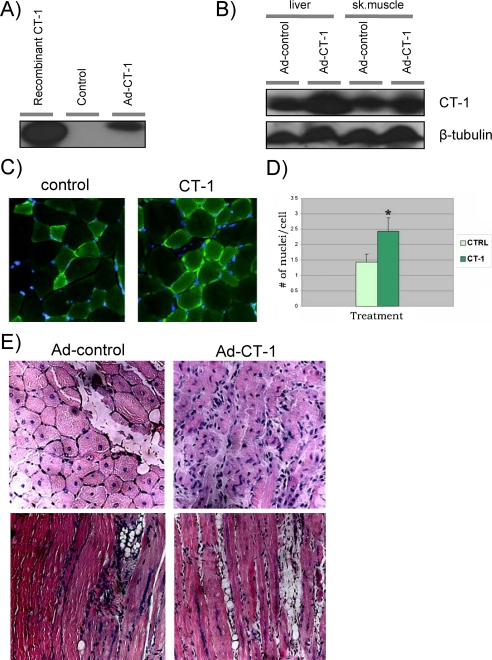
To test the effect of CT-1 on in vivo skeletal muscle function, we utilized systemic delivery of a CT-1-expressing adenovirus, AdCT-1 (40). AdCT-1 infection causes accumulation of CT-1 protein in cell-culture medium (Fig. 7A), and AdCT-1 injection leads to accumulation of CT-1 in liver and skeletal muscle (Fig. 7B). Although it did not lead to gross morphologic alterations in skeletal muscle (Fig. 7C), we noted a significant increase in the number of 4′,6-diamidino-2-phenylindole positive nuclei per myofiber following exposure to AdCT-1 compared with control-injected animals (Fig. 7, C and D, p < 0.05). This observation suggested that CT-1 exposure represses differentiation leading to an increase in the number of undifferentiated myogenic precursors in vivo, similar to the effect elicited in C2C12 myoblast cell cultures. To test the possibility that CT-1 elicited an expansion of the myoblast/muscle precursor cell population, we also investigated the impact of CT-1 administration on the endogenous skeletal muscle progenitor pool. Skeletal muscle contains a population of cells that retain stem cell/progenitor like characteristics, and these cells can be isolated based on Hoechst dye exclusion, referred to as side population (SP) cells (41, 60, 61). Skeletal muscle-derived muscle progenitor cells from CT-1-injected animals were substantially increased compared with the number of progenitor cell colonies derived from control-injected animals (10.6 versus 1.0, p < 0.05, n = 7). Based on our in vitro observations, we postulated that CT-1 exposure might also limit the differentiation of myoblasts in vivo. To test this supposition we induced muscle regeneration via cardiotoxin injection in animals that received either AdCT-1 or the control adenovirus. Cardiotoxin injury elicits a well defined response in which the myofibers are damaged, followed by expansion and differentiation of myogenic precursors to renew or replace the lost myofibers. Interestingly, CT-1-injected animals displayed a limited regeneration, exemplified by a marked reduction in the number of myofibers with centrally located nuclei and an expansion of mononucleated cells associated with regenerating myofibers compared with controls (Fig. 7E). These results suggest that CT-1 targets myoblasts/muscle progenitor cells in vivo and actively represses the differentiation program. Taken together, our results implicate a role for CT-1 in the maintenance of the undifferentiated state in muscle progenitor cells.
FIGURE 7.
CT-1 delays regeneration of damaged skeletal muscle in vivo. A, immunoblotting was used to verify the efficacy of Adenovirus CT-1 production. Recombinant CT-1 protein (100 ng) was used as a positive control as well as media from CT-1 adenovirus infected myocytes. At 72 h post-infection, the media from the treated and untreated cells was collected and subjected to Western blotting analysis with a CT-1 antibody. B, at 7 days post-injection, skeletal muscle (sk.muscle) and liver samples were excised from adenovirus-injected mice. Frozen tissue was homogenized and a total of 300 μg of protein was electrophoresed on a 15% SDS-PAGE. An equal protein loading was verified by Western blotting analysis using β-tubulin specific antibody. C, at 7days post-injection, skeletal muscle was excised, fixed then embedded in paraffin, and sectioned at 10 μm. These sections were counterstained with hematoxylin and eosin to visualize the nuclei and cytoplasm. For immuno-histological detection of α-actinin, the sections were incubated with α-actinin antibody (Abcam), then incubated in donkey-anti-goat CY3 antibody (Chemicon) and finally counterstained with 4′,6-diamidino-2-phenylindole (Sigma). The micrographs were representative fields. D, 5 fields of view per section and 5 sections per TA muscle were analyzed. Differences between Ad-CT-1 and Ad-CTRL-injected samples were evaluated for statistical significance using one tailed, unpaired Student's t test. Differences were considered statistically significant at a p value less than 0.05. (n = 3). E, B6C3F1 mice were subject to systemic delivery of the CT-1 adenovirus. The injections were administered via intra-cardiac chamber delivery with 50 μl of Ad-CT-1 at a concentration of 3.0 × 108 plaque forming units/ml (n = 3). A control group of B6C3F1 mice were injected with 50 μl Ad-CTRL at a similar concentration (n = 3). In a separate group of animals, cardiotoxin was used to induce muscle injury immediately prior to AdCT-1 and Ad-CTRL injection (n = 3 for each group). 25 μl of 10 μm cardiotoxin (Latoxan) was injected directly into the TA muscle. 5 fields of view per section and 5 sections per TA muscle were analyzed. The micrographs presented are representative views. During post-recovery, mice were monitored closely for weight loss, dehydration, and cardiac distress.
DISCUSSION
In this study, we have characterized CT-1 as a potent inhibitory cytokine for the skeletal muscle differentiation program. We document that CT-1 activates MEK, which functionally abrogates the transcriptional activation properties of MyoD, a master regulator of myogenesis. Repression of this core muscle transcriptional network extinguishes induction of the myoG gene, an essential downstream regulator of the muscle differentiation program. Inhibition of muscle differentiation by CT-1 is MEK-dependent, because well established MEK-specific inhibitors, PD98059 and U0126, reverse the inhibitory effects of CT-1 on myogenesis both biochemically and phenotypically. Conversely, even though STAT3 is highly phosphorylated in the presence of CT-1, our experiments indicate that the phosphorylated STAT3 at Tyr-705 and Ser-727 is not sufficient to inhibit myogenesis. Thus, we conclude that CT-1-mediated inhibition of myogenesis requires MEK activation, which subsequently interferes with the trans-activation properties of MyoD. This repression is independent of JAK-STAT signaling, because pharmacological blockade of this pathway has no effect on the repression of myogenesis by CT-1.
Is MEK-ERK Signaling a Convergent Regulatory Nexus for Cytokine-mediated Myogenic Repression?
Several cytokines and growth factors such as fibroblast growth factor and EGF inhibit myogenesis through activation of MEK-ERK signaling. There are, however, some exceptions, such as insulin-like growth factor and insulin, which activate MEK-ERK but paradoxically enhance muscle differentiation under some conditions. Because insulin-like growth factors and insulin also activate the phosphatidylinositol 3-kinase-Akt pathway, and inhibition of phosphatidylinositol 3-kinase or Akt neutralizes their effect on myogenesis (62), it is likely that inhibition of differentiation is a “ground state” that can be overcome by pro-myogenic signals such as those mediated by Akt (27). This is essentially the sequence of events during ontogeny in which the muscle progenitor cells are held in an undifferentiated state until appropriate cues and conditions for differentiation are established. Thus, the dominance of pro-myogenic over inhibitory signals is a prerequisite for differentiation to occur. There is now substantial evidence suggesting that MEK activation is a point of convergence for several growth factors in repressing myogenesis (63–65). Evidence to date indicates that an activated nuclear MEK interacts with the MRFs and inhibits their transcriptional activation properties (50). The MRFs have consensus MAPK phosphorylation sites. However, MEK is capable of inhibiting the activity of a mutated form of Myf5, which does not have intact ERK phospho-acceptor sites. Therefore, the phosphorylation of the MRFs by MEK is not necessarily required for the repression (66). Recently, transcriptional regulators have been found to recruit kinases in a stable manner to target promoters to phosphorylate other components at the transcriptional machinery (48). Therefore, it is possible that the recruitment of kinases to muscle promoters is required for the inhibitory effects on differentiation. This is consistent with our data, which indicate that the physical association of MyoD with MEK is crucial for the anti-myogenic activity of CT-1.
Interestingly, another member of the IL-6 cytokine family, LIF, was shown to inhibit skeletal myogenesis in vitro (67). In agreement with our observations, LIF-mediated repression was also correlated with MEK-ERK pathway activation (67). LIF and CT-1 transduce signals in a similar manner through β-receptors such as Gp130 and LIFRβ. Prior to binding to the β-receptors, at least some of the IL-6 family cytokines bind to ligand-specific α-receptors, and expression levels of the α-receptor in some cell types is known to regulate the sensitivity of the responsiveness to the specific ligand. Although LIF appears to bind β-receptors directly, CT-1 forms a complex with an α-receptor (29). However, this receptor has so far not been fully characterized, so a tissue-specific role of this receptor has yet to be determined. In C2C12 skeletal muscle cells, we have confirmed that Gp130 and LIFRβ are expressed, and further characterization of the CT-1 α-receptor will delineate the precise receptor system. The convergence of LIF and CT-1 on MEK-ERK signaling suggests that this is a common nodal point for Gp130-linked cytokines.
CT-1 was originally isolated as a hypertrophic factor for cardiomyocytes in vitro (30). Chronic administration of CT-1 into the mouse, indeed, causes hypertrophic hearts and also increases the size of liver, kidney, and spleen. This is, at least partially, the result of induction of the vascular endothelial growth factor gene in cardiac myocytes through activation of the Gp130-JAK-STAT3 pathway (68). In addition, CT-1 activates MAPK pathways and the Akt-PI3 kinase pathway and protects cardiomyocytes from apoptosis (34, 43, 69). One of the target genes of CT-1 in this cardioprotective role is the small proline-rich repeat protein-1A (SPRR1A) gene. CT-1 induces SPRR1A expression transcriptionally through activation of MEK-AP-1 and CCAAT/enhancer-binding protein-β pathways. This SPRR1A gene induction by CT-1 is independent of STAT3 activity but blunted by small chemical inhibitors of MEK activity, PD98059, and U0126 (70). Therefore, in other systems, CT-1 activates MEK kinases and regulates their downstream transcription factors. In C2C12 cells, we observed that the SPRR1A promoter was also up-regulated by CT-1 or an activated form of MEK, and this induction was dependent on MEK activation. However, SPRR1A overexpression does not inhibit myogenesis, suggesting that this CT-1 target gene is not responsible for myogenic repression (data not shown). Our observations are discordant with a previous study, in which it was shown that activated STAT3 and MyoD physically interact and functionally antagonize each other by competing for limited amounts of co-activators, such as P300 and PCAF (51). We document that the pan-JAK inhibitor, P6, reduced the phosphorylation levels of STAT3 (Tyr-705 and Ser-727) by CT-1 but had little effect on myotube formation or the transcriptional activity of MyoD at the concentration at which P6 inhibits phosphorylation of STAT3. Therefore, although we do not completely rule out the possibility that the inhibition of MyoD activity may be partly mediated by STAT3, we conclude that activation of MEK but not STAT3 is the primary molecular event responsible for CT-1's inhibitory effect on myogenesis. Further support for this idea was recently provided by the observation that STAT3 and JAK2 were shown to be required for muscle differentiation C2C12 (71). Thus, the notion that STAT3 also functions in an inhibitory manner is unlikely. In addition, we observed that a well established JAK2 inhibitor, AG490, inhibited muscle differentiation in a dose-dependent manner as previously reported (supplemental Fig. S2A) (71). However, this JAK2 inhibitor surprisingly had no effect on phosphorylation of STAT3 by CT-1 (supplemental Fig. S2B). Therefore, the JAK/STAT pathway does not appear to play a repressive role during myogenesis.
Given the temporal and spatial patterns of CT-1 expression during myogenesis, a pervasive consideration is whether CT-1 plays a role in the maintenance of the undifferentiated state or even pluripotency of progenitor cells in an autocrine or paracrine manner. Because CT-1 is expressed in skeletal muscle at key times during embryogenesis, and, as we observed, has a potent role in which it can reversibly repress myogenesis in vitro and delay regeneration in vivo. The observed in vitro and in vivo role of CT-1 in skeletal muscle cells defines it as a potential target of therapeutic interventions in which small molecule cell permeable inhibitors can be used to manipulate pro- and anti-differentiation pathways. Moreover, knowledge of these pathways could be instrumental in ex vivo programming of progenitor cells, which may have critical implications for a variety of cellular-based muscle therapies.
In summary, we have documented that the CT-1 cytokine has a potent repressive effect on skeletal myogenesis in vivo and in vitro. This effect, which is reversible, requires MEK-ERK signaling and, surprisingly, does not require STAT3 activation. The expression patterns of CT-1 and its in vivo and in vitro properties described here make it a viable candidate to play a role in the maintenance of the undifferentiated muscle progenitor cell state in embryonic and postnatal skeletal muscle.
Supplementary Material
Acknowledgments
We thank Joseph Chan for technical assistance. We also thank Dr. Robert L. Perry for providing MyHC, Myogenin, and Myc antibodies and valuable suggestions.
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes of Health Research (to J. C. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- MyoD
- myogenic differentiation-1
- Myf5
- myogenic factor-5
- MyoG
- myogenin
- MRF
- myogenic regulatory factor
- MyHC
- myosin heavy chain
- MEF2
- myocyte enhancer factor 2
- EGF
- epidermal growth factor
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MEK
- MAPK/ERK kinase
- CT-1
- cardiotrophin-1
- LIF
- leukemia inhibitory factor
- IL-6
- interleukin-6
- Gp130
- glycoprotein-130
- LIFRβ
- leukemia inhibitory factor receptor-β
- JAK
- Janus kinase
- STAT3
- signal transducer and activator of transcription 3
- CMV
- cytomegalovirus
- GM
- growth medium
- DM
- differentiation medium
- PBS
- phosphate-buffered saline
- CTRL
- LacZ-containing adenovirus
- SPRR1A
- small proline-rich repeat protein-1A
- P6
- 2-(1,1-dimethylethyl)-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinolin-7-one, Pyridone 6
- TA
- tibialis anterior.
REFERENCES
- 1.Perry R. L., Rudnick M. A. (2000) Front. Biosci. 5, D750–767 [DOI] [PubMed] [Google Scholar]
- 2.Kassar-Duchossoy L., Gayraud-Morel B., Gomès D., Rocancourt D., Buckingham M., Shinin V., Tajbakhsh S. (2004) Nature 431, 466–471 [DOI] [PubMed] [Google Scholar]
- 3.Pownall M. E., Gustafsson M. K., Emerson C. P., Jr. (2002) Annu. Rev. Cell Dev. Biol. 18, 747–783 [DOI] [PubMed] [Google Scholar]
- 4.Rudnicki M. A., Jaenisch R. (1995) Bioessays 17, 203–209 [DOI] [PubMed] [Google Scholar]
- 5.Tapscott S. J. (2005) Development 132, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 6.Walsh K., Gualberto A. (1992) J. Biol. Chem. 267, 13714–13718 [PubMed] [Google Scholar]
- 7.Penn B. H., Bergstrom D. A., Dilworth F. J., Bengal E., Tapscott S. J. (2004) Genes Dev. 18, 2348–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myer A., Olson E. N., Klein W. H. (2001) Dev. Biol. 229, 340–350 [DOI] [PubMed] [Google Scholar]
- 9.Edmondson D. G., Olson E. N. (1989) Genes Dev. 3, 628–640 [DOI] [PubMed] [Google Scholar]
- 10.Naya F. J., Olson E. (1999) Curr Opin Cell Biol. 11, 683–688 [DOI] [PubMed] [Google Scholar]
- 11.Olson E. N., Perry M., Schulz R. A. (1995) Dev. Biol. 172, 2–14 [DOI] [PubMed] [Google Scholar]
- 12.Jarvis E. E., Clark K. L., Sprague G. F., Jr. (1989) Genes Dev. 3, 936–945 [DOI] [PubMed] [Google Scholar]
- 13.Sommer H., Beltrán J. P., Huijser P., Pape H., Lönnig W. E., Saedler H., Schwarz-Sommer Z. (1990) EMBO J. 9, 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldmann K. A., Meyerowitz E. M. (1990) Nature 346, 35–39 [DOI] [PubMed] [Google Scholar]
- 15.Norman C., Runswick M., Pollock R., Treisman R. (1988) Cell 55, 989–1003 [DOI] [PubMed] [Google Scholar]
- 16.Ridgeway A. G., Petropoulos H., Wilton S., Skerjanc I. S. (2000) J. Biol. Chem. 275, 32398–32405 [DOI] [PubMed] [Google Scholar]
- 17.Engert J. C., Berglund E. B., Rosenthal N. (1996) J. Cell Biol. 135, 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olwin B. B., Hauschka S. D. (1988) J. Cell Biol. 107, 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Templeton T. J., Hauschka S. D. (1992) Dev. Biol. 154, 169–181 [DOI] [PubMed] [Google Scholar]
- 20.Allen R. E., Boxhorn L. K. (1987) J. Cell. Physiol. 133, 567–572 [DOI] [PubMed] [Google Scholar]
- 21.Lluís F., Perdiguero E., Nebreda A. R., Muñoz-Cánoves P. (2006) Trends Cell Biol. 16, 36–44 [DOI] [PubMed] [Google Scholar]
- 22.Lluís F., Ballestar E., Suelves M., Esteller M., Muñoz-Cánoves P. (2005) EMBO J. 24, 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox D. M., Du M., Marback M., Yang E. C., Chan J., Siu K. W., McDermott J. C. (2003) J. Biol. Chem. 278, 15297–15303 [DOI] [PubMed] [Google Scholar]
- 24.Zetser A., Gredinger E., Bengal E. (1999) J. Biol. Chem. 274, 5193–5200 [DOI] [PubMed] [Google Scholar]
- 25.McKinsey T. A., Zhang C. L., Olson E. N. (2002) Curr. Opin. Cell Biol. 14, 763–772 [DOI] [PubMed] [Google Scholar]
- 26.de la Serna I. L., Ohkawa Y., Berkes C. A., Bergstrom D. A., Dacwag C. S., Tapscott S. J., Imbalzano A. N. (2005) Mol. Cell. Biol. 25, 3997–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serra C., Palacios D., Mozzetta C., Forcales S. V., Morantte I., Ripani M., Jones D. R., Du K., Jhala U. S., Simone C., Puri P. L. (2007) Mol. Cell 28, 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett A. M., Tonks N. K. (1997) Science 278, 1288–1291 [DOI] [PubMed] [Google Scholar]
- 29.Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., Schaper F. (2003) Biochem. J. 374, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pennica D., King K. L., Shaw K. J., Luis E., Rullamas J., Luoh S. M., Darbonne W. C., Knutzon D. S., Yen R., Chien K. R., et al. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1142–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheng Z., Pennica D., Wood W. I., Chien K. R. (1996) Development 122, 419–428 [DOI] [PubMed] [Google Scholar]
- 32.Pennica D., Swanson T. A., Shaw K. J., Kuang W. J., Gray C. L., Beatty B. G., Wood W. I. (1996) Cytokine 8, 183–189 [DOI] [PubMed] [Google Scholar]
- 33.Freed D. H., Moon M. C., Borowiec A. M., Jones S. C., Zahradka P., Dixon I. M. (2003) Mol. Cell. Biochem. 254, 247–256 [DOI] [PubMed] [Google Scholar]
- 34.Brar B. K., Stephanou A., Pennica D., Latchman D. S. (2001) Cytokine 16, 93–96 [DOI] [PubMed] [Google Scholar]
- 35.Freed D. H., Cunnington R. H., Dangerfield A. L., Sutton J. S., Dixon I. M. (2005) Cardiovasc. Res. 65, 782–792 [DOI] [PubMed] [Google Scholar]
- 36.Davis R. L., Weintraub H., Lassar A. B. (1987) Cell 51, 987–1000 [DOI] [PubMed] [Google Scholar]
- 37.Mansour S. J., Resing K. A., Candi J. M., Hermann A. S., Gloor J. W., Herskind K. R., Wartmann M., Davis R. J., Ahn N. G. (1994) J. Biochem. 116, 304–314 [DOI] [PubMed] [Google Scholar]
- 38.Donoviel D. B., Shield M. A., Buskin J. N., Haugen H. S., Clegg C. H., Hauschka S. D. (1996) Mol. Cell. Biol. 16, 1649–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kollias H. D., Perry R. L., Miyake T., Aziz A., McDermott J. C. (2006) Mol. Cell. Biol. 26, 6248–6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bordet T., Schmalbruch H., Pettmann B., Hagege A., Castelnau-Ptakhine L., Kahn A., Haase G. (1999) J. Clin. Invest. 104, 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asakura A., Seale P., Girgis-Gabardo A., Rudnicki M. A. (2002) J. Cell Biol. 159, 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hierlihy A. M., Seale P., Lobe C. G., Rudnicki M. A., Megeney L. A. (2002) FEBS Lett. 530, 239–243 [DOI] [PubMed] [Google Scholar]
- 43.Sheng Z., Knowlton K., Chen J., Hoshijima M., Brown J. H., Chien K. R. (1997) J. Biol. Chem. 272, 5783–5791 [DOI] [PubMed] [Google Scholar]
- 44.Kaushal S., Schneider J. W., Nadal-Ginard B., Mahdavi V. (1994) Science 266, 1236–1240 [DOI] [PubMed] [Google Scholar]
- 45.Myer A., Wagner D. S., Vivian J. L., Olson E. N., Klein W. H. (1997) Dev. Biol. 185, 127–138 [DOI] [PubMed] [Google Scholar]
- 46.Kim C. H., Neiswender H., Baik E. J., Xiong W. C., Mei L. (2008) Mol. Cell. Biol. 28, 2941–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polesskaya A., Naguibneva I., Duquet A., Bengal E., Robin P., Harel-Bellan A. (2001) Mol. Cell. Biol. 21, 5312–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puri P. L., Iezzi S., Stiegler P., Chen T. T., Schiltz R. L., Muscat G. E., Giordano A., Kedes L., Wang J. Y., Sartorelli V. (2001) Mol. Cell 8, 885–897 [DOI] [PubMed] [Google Scholar]
- 49.Reynaud E. G., Leibovitch M. P., Tintignac L. A., Pelpel K., Guillier M., Leibovitch S. A. (2000) J. Biol. Chem. 275, 18767–18776 [DOI] [PubMed] [Google Scholar]
- 50.Perry R. L., Parker M. H., Rudnicki M. A. (2001) Mol. Cell 8, 291–301 [DOI] [PubMed] [Google Scholar]
- 51.Kataoka Y., Matsumura I., Ezoe S., Nakata S., Takigawa E., Sato Y., Kawasaki A., Yokota T., Nakajima K., Felsani A., Kanakura Y. (2003) J. Biol. Chem. 278, 44178–44187 [DOI] [PubMed] [Google Scholar]
- 52.Chen C., Sytkowski A. J. (2004) Blood 104, 73–80 [DOI] [PubMed] [Google Scholar]
- 53.Yip-Schneider M. T., Klein P. J., Wentz S. C., Zeni A., Menze A., Schmidt C. M. (2009) J. Pharmacol. Exp. Ther. 329, 1063–1070 [DOI] [PubMed] [Google Scholar]
- 54.Bromberg J. F., Wrzeszczynska M. H., Devgan G., Zhao Y., Pestell R. G., Albanese C., Darnell J. E., Jr. (1999) Cell 98, 295–303 [DOI] [PubMed] [Google Scholar]
- 55.Wen Z., Zhong Z., Darnell J. E., Jr. (1995) Cell 82, 241–250 [DOI] [PubMed] [Google Scholar]
- 56.Yu C. L., Meyer D. J., Campbell G. S., Larner A. C., Carter-Su C., Schwartz J., Jove R. (1995) Science 269, 81–83 [DOI] [PubMed] [Google Scholar]
- 57.Pedranzini L., Dechow T., Berishaj M., Comenzo R., Zhou P., Azare J., Bornmann W., Bromberg J. (2006) Cancer Res. 66, 9714–9721 [DOI] [PubMed] [Google Scholar]
- 58.Thompson J. E., Cubbon R. M., Cummings R. T., Wicker L. S., Frankshun R., Cunningham B. R., Cameron P. M., Meinke P. T., Liverton N., Weng Y., DeMartino J. A. (2002) Bioorg. Med. Chem. Lett. 12, 1219–1223 [DOI] [PubMed] [Google Scholar]
- 59.Kaptein A., Paillard V., Saunders M. (1996) J. Biol. Chem. 271, 5961–5964 [DOI] [PubMed] [Google Scholar]
- 60.Muskiewicz K. R., Frank N. Y., Flint A. F., Gussoni E. (2005) J. Histochem. Cytochem. 53, 861–873 [DOI] [PubMed] [Google Scholar]
- 61.Jackson J. D., Zhou G., Kuszynski C. A., Cai J., Fox I. J. (2002) Cell Transplant. 11, 779–785 [PubMed] [Google Scholar]
- 62.Xu Q., Wu Z. (2000) J. Biol. Chem. 275, 36750–36757 [DOI] [PubMed] [Google Scholar]
- 63.Rommel C., Clarke B. A., Zimmermann S., Nuñez L., Rossman R., Reid K., Moelling K., Yancopoulos G. D., Glass D. J. (1999) Science 286, 1738–1741 [DOI] [PubMed] [Google Scholar]
- 64.Ramocki M. B., Johnson S. E., White M. A., Ashendel C. L., Konieczny S. F., Taparowsky E. J. (1997) Mol. Cell. Biol. 17, 3547–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Page J. L., Wang X., Sordillo L. M., Johnson S. E. (2004) J. Biol. Chem. 279, 30966–30972 [DOI] [PubMed] [Google Scholar]
- 66.Winter B., Arnold H. H. (2000) J. Cell Sci. 113, 4211–4220 [DOI] [PubMed] [Google Scholar]
- 67.Jo C., Kim H., Jo I., Choi I., Jung S. C., Kim J., Kim S. S., Jo S. A. (2005) Biochim. Biophys. Acta 1743, 187–197 [DOI] [PubMed] [Google Scholar]
- 68.Jin H., Yang R., Keller G. A., Ryan A., Ko A., Finkle D., Swanson T. A., Li W., Pennica D., Wood W. I., Paoni N. F. (1996) Cytokine 8, 920–926 [DOI] [PubMed] [Google Scholar]
- 69.Liao Z., Brar B. K., Cai Q., Stephanou A., O'Leary R. M., Pennica D., Yellon D. M., Latchman D. S. (2002) Cardiovasc. Res. 53, 902–910 [DOI] [PubMed] [Google Scholar]
- 70.Pradervand S., Yasukawa H., Muller O. G., Kjekshus H., Nakamura T., St Amand T. R., Yajima T., Matsumura K., Duplain H., Iwatate M., Woodard S., Pedrazzini T., Ross J., Firsov D., Rossier B. C., Hoshijima M., Chien K. R. (2004) EMBO J. 23, 4517–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang K., Wang C., Xiao F., Wang H., Wu Z. (2008) J. Biol. Chem. 283, 34029–34036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.