Abstract
Serine proteinases in insect plasma have been implicated in two types of immune responses; that is, activation of prophenoloxidase (proPO) and activation of cytokine-like proteins. We have identified more than 20 serine proteinases in hemolymph of the tobacco hornworm, Manduca sexta, but functions are known for only a few of them. We report here functions of two additional M. sexta proteinases, hemolymph proteinases 6 and 8 (HP6 and HP8). HP6 and HP8 are each composed of an amino-terminal clip domain and a carboxyl-terminal proteinase domain. HP6 is an apparent ortholog of Drosophila Persephone, whereas HP8 is most similar to Drosophila and Tenebrio spätzle-activating enzymes, all of which activate the Toll pathway. proHP6 and proHP8 are expressed constitutively in fat body and hemocytes and secreted into plasma, where they are activated by proteolytic cleavage in response to infection. To investigate activation and biological activity of HP6 and HP8, we purified recombinant proHP8, proHP6, and mutants of proHP6 in which the catalytic serine was replaced with alanine, and/or the activation site was changed to permit activation by bovine factor Xa. HP6 was found to activate proPO-activating proteinase (proPAP1) in vitro and induce proPO activation in plasma. HP6 was also determined to activate proHP8. Active HP6 or HP8 injected into larvae induced expression of antimicrobial peptides and proteins, including attacin, cecropin, gloverin, moricin, and lysozyme. Our results suggest that proHP6 becomes activated in response to microbial infection and participates in two immune pathways; activation of PAP1, which leads to proPO activation and melanin synthesis, and activation of HP8, which stimulates a Toll-like pathway.
Innate immune systems of mammals and arthropods include extracellular serine proteinase cascade pathways, which rapidly amplify responses to infection and stimulate killing of pathogens. These proteinase-driven processes include the complement system of vertebrates (1, 2) and pathways in arthropods involving proteinases containing amino-terminal clip domains (3). Clip domain proteinases function in blood coagulation (4, 5), activation of prophenoloxidase (proPO) that leads to melanin synthesis (6–9), and stimulation of the Toll pathway to promote synthesis of antimicrobial peptides/proteins (AMPs)2 secreted into the hemolymph (10, 11).
The serine proteinase systems best characterized in arthropods are the horseshoe crab hemolymph coagulation pathway and the cascade leading to activation of the Toll pathway in dorsal-ventral development in Drosophila (12–14). Recent research also has led to better characterization of the proPO activation pathway in Manduca sexta (7, 15, 16) and the Toll-signaling pathway in the Drosophila immune response (17, 18) and to both the proPO and Toll pathways in the beetle Tenebrio molitor (11, 19).
In the proPO activation pathway, soluble pattern recognition proteins initially recognize pathogen-associated molecular patterns such as bacterial peptidoglycan or fungal β-1,3-glucan (20–22). This interaction stimulates the sequential activation of a series of serine proteinases in hemolymph, leading to the activation of proPO-activating proteinase (PAP), also known as proPO activating enzyme (7, 23). Activated PAP converts inactive proPO to PO. PO catalyzes the hydroxylation of monophenols to o-diphenols and the oxidation of o-diphenols to quinones that are involved in microbial killing, melanin synthesis, sequestration of parasites or pathogens, and wound healing (24, 25). Other proteins required for proPO activation are clip-domain serine proteinase homologs (SPHs), whose catalytic serine is replaced with glycine and, therefore, lack proteolytic activity (26, 27). Serine proteinase inhibitors, including members of the serpin superfamily, regulate the activation of proPO by inhibiting the activating proteinases (28, 29).
Drosophila clip-domain serine proteinases Persephone, Grass, Spirit, and spätzle-processing enzyme (SPE) participate in the activation of Toll pathway, stimulating synthesis of antimicrobial peptides as an innate immune response (18, 30–32). Although genetic evidence indicates that Persephone and Spirit are upstream of SPE in the cascade, the substrate(s) of Persephone and Spirit have not been identified, and which proteinase directly activates SPE is unknown. Neither is it clear whether these enzymes may be related to the melanization pathway, which involves clip-domain proteinases MP2 and MP1 (33).
Here we report the functional characterization of M. sexta HP6 and HP8, probable orthologs of Drosophila Persephone and SPE, respectively. We developed methods to activate purified recombinant proHP6 and proHP8 and discovered that HP6 participates in proPO activation by activating proPAP1 and that both HP6 and HP8 function in a pathway that stimulates the synthesis of AMPs in M. sexta.
EXPERIMENTAL PROCEDURES
Insect Rearing
M. sexta eggs were originally purchased from Carolina Biological Supplies. The larvae were reared on an artificial diet (34).
Sequence Analysis
Sequence comparisons and phylogenetic analyses were performed using MEGA Version 4 software (35). Sequences were aligned using the ClustalW program in MEGA (see supplemental Fig. S1 for the alignment). Trees were constructed by the neighbor-joining method, with statistical analysis by the bootstrap method using 1000 repetitions. The sequences (with GenBankTM accession number) used for the analyses were: M. sexta HP6 (AAV91004), HP8 (AAV91006), HP21 (AAV91019), PAP1 (AAX18636), PAP2 (AAL76085), PAP3 (AAO74570); Bombyx mori BAEEase (ABB58762), proPO-activating enzyme (NP_001036832); Drosophila melanogaster Easter (NP_524362), Grass (NP_733197), Persephone (NP_573297), SPE (NP_651168), Spirit (NP_727276), Snake (NP_524338); Holotrichia diomphalia PPAF1 (BAA34642); Limulus polyphemus proclotting enzyme (AAA30094); T. molitor SPE-activating enzyme (AB363979), SPE (AB363980).
Reverse Transcriptase (RT)-PCR
Fifth-instar-day 2 larvae were injected with 50 μl of sterile water containing formalin-killed Escherichia coli XL1-Blue (Stratagene, 1 × 107 cells/ml), dried Micrococcus luteus ATCC 4698 (Sigma, 10 μg/μl), or curdlan from Alcaligenes faecalis (Sigma, 10 μg/μl) or with water alone as a control (n = 3 larvae for each treatment). After 24 h, total RNA samples were prepared using TRizol Reagent (Invitrogen) from fat body and hemocytes. First-strand cDNA was synthesized from an oligo(dT) primer following the instructions for BD SprintTM PowerSriptTM PrePrimed Single Shots kit (Clontech). M. sexta ribosomal protein S3 (rpS3) cDNA was used as an internal standard to adjust the template amounts in a preliminary PCR experiment. The primers for amplifying HP6, HP8, and rpS3 were: HP6-RTf (5′-TGGTTTCTGATTGTCCAGCAG-3′) and HP6-RTr (5′-CCGCATTTGTCACTTGGAAC-3′), HP8-RTf (5′-CTTCTGCCCAACAGGCGTG-3′) and HP8-RTr (5′-CGCAAGTCCTCAGTTGTCG-3′), and rpS3f (5′-GCCGTTCTTGCCCTGTT-3′) and rpS3r (5′-CGCGAGTTGACTTCGGT-3′). The thermal cycling conditions were 25 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s followed by incubation at 72 °C for 5 min. The PCR products were separated by electrophoresis on a 1.0% agarose gel.
Immunoblot Analysis
Cell-free hemolymph samples were collected as described previously (36), and these plasma samples or purified proteins were separated by 10% SDS-PAGE (37). Immunoblot analysis was performed using rabbit polyclonal antiserum against HP6 or HP8 as the primary antibody (diluted 1:2000) and goat anti-rabbit IgG-alkaline phosphatase conjugate (Bio-Rad, diluted 1:3000) as the secondary antibody.
Production of Recombinant proHP6, proHP8, and proHP6 Mutants
The entire proHP6 or proHP8 coding region, including the signal peptide, was amplified by PCR using specific primers (proHP6f, 5′-ACGGTACCATGTGGTTAATGGTGA-3′, nucleotides 253–268, the KpnI site is underlined; proHP6r, 5′-CCGGAATTCTTAATTAGGCCAAACA-3′, reverse complement of nucleotides 1311–1326, EcoRI site is underlined; proHP8f, 5′-GCGGTACCATGAATACTATACGTG-3′, nucleotides 64–79, the KpnI site is underlined; proHP8r, 5′-CCGATATCCTAAGGTCGTAACTTTG-3′, reverse complement of nucleotides 1163–1179, the EcoRV site is underlined). The KpnI-EcoRI fragment for proHP6 or KpnI-EcoRV fragment for proHP8 were recovered and inserted into the same restriction sites in the vector pMT/V5-His A (Invitrogen). The resulting proHP6 plasmid, after sequence confirmation, was used as the template to produce mutant plasmids according to the instructions of QuikChange multisite-directed mutagenesis kit (Stratagene). A mutation was introduced to change the codon for active site Ser287 (TCT) to Ala (GCG) by using the mutagenic oligonucleotide primer (5′-CACGTGTCAGGGCGACGCGGGCGGGCCTCTTCAGC-3′). This construct, producing an inactive proteinase, was named proHP6I. A mutant at the predicted activation site of proHP6 changed residues 89–92 from LDLH to IEGR by using the mutagenic oligonucleotide primer (5′-CGATACCGCCGATAGAAGGGCGGATACTCGGCGGTG-3′). IEGR is a cleavage site for bovine factor Xa. This construct was named proHP6Xa. The proHP6Xa plasmid was used as template to produce the double mutant proHP6Xa,I, with both the active site Ser → Ala mutation and the factor Xa activation site. After DNA sequence verification the plasmids were used to transfect Drosophila S2 cells to generate stably transformed cell lines for producing recombinant proteins, following the manufacturer's instructions (Invitrogen).
Purification of Recombinant Proteins
The cell cultures were harvested 48 h after induction of expression with CuSO4 at a final concentration of 500 μm, and cells were removed by centrifugation at 5000 × g for 15 min at 4 °C. The cell-free medium (400 ml) was mixed with concanavalin A-Sepharose 4B resin (20 ml) that had been equilibrated with buffer 1 (20 mm Tris-HCl, 0.5 m NaCl, 1 mm CaCl2, 1 mm MgCl2, 1 mm MnCl2, pH 7.5). After rotating overnight at 4 °C, the mixture was packed into a column (2.0-cm inner diameter × 6 cm) by gravity. After washing the column with buffer 1 until A280 was <0.002, bound proteins were eluted with 60 ml of buffer 2 (20 mm Tris-HCl, 0.5 mm NaCl, and 0.5 mm methyl-α-d-mannopyranoside, pH 7.5).
The eluted factions were pooled and dialyzed against 2 liters of buffer 3 (20 mm Tris-HCl, 20 mm NaCl, pH 8.0) (2 liters each time for 8 h, twice). The dialyzed sample was applied to a Q-SepharoseTM Fast Flow column (1.5-cm inner diameter × 9 cm) equilibrated with buffer 3 at a flow rate of 1.0 ml/min. After application, the column was washed with buffer 3 until A280 was lower than 0.002 and then eluted at 1.0 ml/min with a linear gradient of 20–700 mm NaCl in 20 mm Tris-HCl, pH 8.0, for 50 min. Fractions of 1.0 ml were collected and analyzed by immunoblotting. The factions containing recombinant protein were pooled and concentrated on Centricon-10 (Millipore).
After passing through a syringe filter, the concentrated sample (2 ml) was applied to a Sephacryl S-300 HR column (2.5-cm inner diameter × 90 cm) equilibrated with buffer 4 (20 mm Tris-HCl, 150 mm NaCl, pH 8.0). The column was eluted with buffer 4 at a flow rate of 0.25 ml/min, and fractions were collected at 1.0 ml/tube after the first 60 ml. Each faction was analyzed by 10% SDS-PAGE followed by silver staining (38) or immunoblot analysis. After concentration using a Centricon-10 device, protein concentration was determined by using Coomassie PlusTM protein assay reagent (Pierce) with bovine serum albumin as standard. Purified proteins were stored at −80 °C.
Activation of proHP6Xa and proHP6Xa,I by Factor Xa
Purified recombinant protein was incubated with bovine Factor Xa (New England Biolabs) in buffer 4 plus 2 mm CaCl2 at 37 °C for 1 h. Amounts of each protein for a particular experiment are provided in the legends to Figs. 4 and 5.
FIGURE 4.
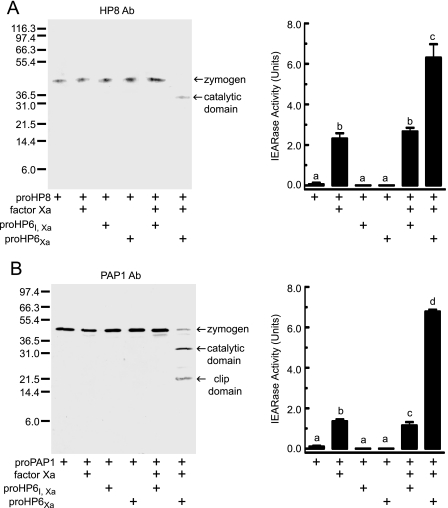
Activation of purified proHP6Xa and proHP6Xa,I by bovine Factor Xa. After incubation of the purified recombinant proHP6 proteins (50 ng/each) with Factor Xa (200 ng), the mixtures were separated by 10% SDS-PAGE followed by immunoblot analysis using HP6 antiserum. The sizes and positions of molecular weight standards are indicated on the left. These were compared with natural HP6 in plasma activated by exposure to M. luteus (far right lane). Bands representing proHP6 zymogen, a cleavage product corresponding to its catalytic domain, and a putative HP6-serpin complex are marked by arrows.
FIGURE 5.
Proteolytic activation of purified proHP8 (A) and proPAP1 (B) by Factor Xa-activated HP6. proHP6Xa (10 ng) or proHP6Xa,I (10 ng) was activated by Factor Xa (40 ng) and then incubated with proHP8 (20 ng) (A) or proPAP1 (20 ng) (B) at 37 °C for 1 h. The mixtures were subjected to 10% SDS-PAGE and immunoblotting using HP8 or PAP1 antibodies (Ab). The sizes and positions of molecular weight standards are indicated on the left. Bands representing proHP8 or proPAP1 zymogen and their catalytic domain are marked by arrows. Catalytic activity of activated HP8 or PAP1 was detected by a spectrophotometric assay using Ile-Glu-Ala-Arg-p-nitroanilide as a substrate, as described under “Experimental Procedures.” The bars represent the mean ± S.D. (n = 3). Bars labeled with different letters (a, b, and c) are significantly different (analysis of variance and Newman-Keuls test, p < 0.05).
ProPO Activation Assays
Recombinant proteins were mixed with plasma from naïve larvae, which had a low basal PO activity and a dramatic increase in PO activity after M. luteus elicitation (28). The mixtures were incubated at room temperature for 10 min in the presence or absence of M. luteus. Details of the reaction mixtures are provided in the figures and corresponding legends. The PO activity in the reaction mixtures was measured as described previously (39) using dopamine as a substrate. One unit of PO activity was defined as the amount of enzyme producing an increase in absorbance (ΔA470) of 0.001/min.
Effects of HP6 and HP8 on AMP Gene Expression
Purified recombinant proteins were injected into day-0 fifth-instar larvae (n = 3 larvae for each treatment). Before injection, larvae were anesthetized on ice for 15 min and then injected with a protein mixture in buffer 4 described above using a 1-ml syringe and a 30-gauge needle. In some experiments larvae were given a second injection 30 min later of dried M. luteus (ATCC 4698) suspended in 50 μl of water. Twenty hours later, fat body and hemolymph samples were collected. Total RNA samples were prepared from fat body, and cDNA was prepared as describe above. Cell-free hemolymph samples were heated at 95 °C for 5 min to remove most high molecular weight proteins and then centrifuged at 10,000 × g for 5 min. The supernatant was stored at −20 °C.
Quantitative real-time PCR was performed using primer pairs (supplemental Table S1) designed with the aid of Beacon Designer 7.0 software. Each 25-μl reaction contained 12.5 μl of 2 × buffer mix (Bio-Rad), 1 μl of forward primer (5 μm), 1 μl of reverse primer (5 μm), and 11.5 μl of diluted cDNA. The thermal cycling conditions were 95 °C for 5 min and 40 cycles of 95 °C for 30 s, 53 °C for 30 s, and 72 °C for 40 s. Amplification was monitored on an iCycler (Bio-Rad) by means of SYBR-Green (Bio-Rad). Thresholds were individually calculated for each target gene. Transcript abundance values (ΔCT = CTtreated − CTcontrol) for each gene were used to calculate the expression level relative to the transcript for rpS3 (2−ΔΔCT, ΔΔCT = ΔCTspecific gene − ΔCTrpS3).
Heat-treated cell-free plasma samples (3 μl each) were treated with 2 × SDS sample buffer containing 0.1 m dithiothreitol at 95 °C for 5 min and separated by electrophoresis on a 15% SDS-polyacrylamide gel and stained with Coomassie Brilliant Blue. The bands of interest were excised and sent to the Nevada Proteomics Center (University of Nevada) for trypsin digestion and analysis with an ABI 4700 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Foster City, CA) using their 4000 Series Explorer software Version 3.6. The eight most intense ions from the MS analysis that were not known trypsin masses were further analyzed by MS/MS. A peak list was created by GPS Explorer software (Applied Biosystems) from the raw data from the ABI 4700. MS/MS peak filtering included a mass range of 60 to 20 Da below each precursor mass, minimum signal-to-noise filter 10, peak density filter of 50 peaks per 200 Da, and a cluster area filter with maximum number of 65 peaks. The resulting MS/MS peaks and intensities were then searched against “other metazoa” sequences in the NCBInr 20081128 data base (356,920 sequences) by Mascot. The data base search parameters included one missed cleavage, fixed carbamidomethylation of cysteines, and variable oxidation of methionines, with a 20-ppm tolerance for precursor ions and a 0.6-Da tolerance for fragment ions. The MS/MS data were also searched against partial sequences for M. sexta cecropin A and B that were obtained from M. sexta EST contigs 5774 and 2488 (40).
Antimicrobial activity assays were performed as described by Hultmark et al. (41) using E. coli strain XL1-Blue and M. luteus strain ATCC 4698. Lysozyme activity was measured as a decrease in the turbidity of the suspension of M. luteus (Sigma) at 450 nm, 25 °C, and pH 7.0 as described by Dunn and Drake (34). An enzyme unit was defined as the amount of enzyme that produced a ΔA450 = 0.001 min−1. For each heat-treated plasma sample, duplicate 10-μl aliquots were assayed.
RESULTS
HP6 and HP8 Sequence Comparisons
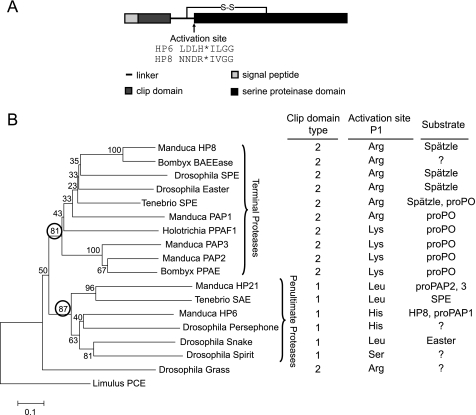
M. sexta HP6 and HP8 are each composed of an amino-terminal clip domain connected by a linker region to a carboxyl-terminal S1 family serine proteinase domain (Fig. 1A) (15). They are each most similar to clip domain proteinases known to function in activation of the Toll pathway (Fig. 1B). HP6 may be an ortholog of Drosophila Persephone (35% identity), whereas HP8 has the highest identity to a group of proteinases whose substrate is the cytokine precursor prospätzle, including Drosophila Easter (42%) (42), SPE from Drosophila (39%) (18), and Tenebrio (45%) (11) and a predicted SPE named BAEEase from the silkworm, B. mori (70%) (18). The predicted proteolytic activation sites (↓) are at 109LDLH↓ILGG in proHP6 and 111NNDR↓IVGG in proHP8 (15) (Fig. 1). After such cleavage, the resulting polypeptide fragments containing the clip domain and the proteinase domain are expected to remain connected by an interchain disulfide bond (Cys98-Cys228 in HP6 and Cys107-Cys241 in HP8). This linkage would be disrupted under reducing conditions to produce two separate chains.
FIGURE 1.
The domain architecture of M. sexta HP6 and HP8 and comparison to other clip domain proteinases. A, domain organization. The positions of cleavage activation sites were predicted based on a sequence alignment of clip-domain proteinases (3, 15). The asterisk indicates the position of the peptide bond cleaved during activation. B, comparison of HP6 and HP8 with insect clip domain proteinases with known function. The phylogenetic tree is based on an alignment of the proteinase domain sequences (see “Experimental Procedures” and supplemental Fig. S1), with horseshoe crab proclotting enzyme (Limulus PCE) as an outgroup. Numbers at the branches indicate bootstrap value, as a percent of 1000 repetitions. A very similar tree was obtained when the clip domains were included in the alignment. The circled bootstrap values indicate two clades that correspond with groups of proteinases that are either the final proteinase in a known pathway (terminal proteinases) or at the next to last proteolytic step of a pathway (penultimate proteinases; known for Manduca HP21, Tenebrio SAE, Manduca HP6, Drosophila Snake). The clip domain type is defined based on the number of residues between the third and fourth Cys residues in the domain (3). The activation site P1 residue is the amino acid residue (determined experimentally or predicted based on sequence alignment) on the amino-terminal side of the peptide bond that is cleaved to activate the proteinase zymogen.
HP6 and HP8 Gene Expression
RT-PCR assay indicated that HP6 and HP8 transcripts were expressed constitutively in fat body and hemocytes and did not increase after larvae were injected with bacteria or the β-1,3-glucan curdlan (supplemental Fig. S2A). Both genes appeared to be expressed at significantly higher levels in fat body than in hemocytes. Immunoblot analysis of plasma samples revealed a specific band of the expected size for proHP6 (39 kDa) and proHP8 (40 kDa), whose intensity did not change significantly after larvae were injected with E. coli, M. luteus, or curdlan (supplemental Fig. S2B).
Purification of Recombinant proHP6, proHP8, and proHP6 Mutants
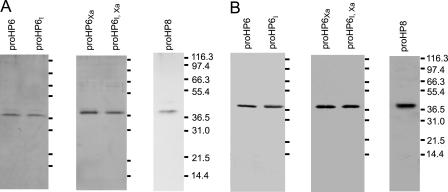
To investigate potential immune functions of HP6 and HP8, we expressed them in their zymogen form as recombinant proteins in Drosophila S2 cells. We also expressed three site-directed mutants of proHP6. To obtain an activated form of HP6, we mutated its predicted activation site from LDLH92 to IEGR92, a sequence preferred for cleavage by bovine Factor Xa (43). This mutant was named proHP6Xa. We also prepared a mutation that changed the catalytic Ser287 residue of proHP6 to Ala287 to produce an inactive form of HP6 (proHP6I) for use as a negative control. A third form of the protein contained both mutations, so that it could be cleaved at the activation site by Factor Xa but lack proteolytic activity (proHP6Xa,I). All of the recombinant proteins were secreted from S2 cells by utilizing their own secretion signal peptides. proHP6 and proHP8 in the cell culture supernatant bound to concanavalin A, indicating that they are glycoproteins. Proteins eluted from a concanavalin A column were further purified by anion exchange chromatography on Q-Sepharose followed by gel permeation chromatography on a Sephacryl S-300 HR column. SDS-PAGE analysis followed by silver staining or immunoblotting indicated that proHP6, proHP8, and proHP6 mutants were of high purity (Fig. 2). proHP6 and its mutants had a similar apparent molecular mass of 39 kDa, whereas proHP8 had an apparent mass of 40 kDa. These masses are slightly larger than those predicted from the cDNA sequences of proHP6 (36.7 kDa) and proHP8 (38.1 kDa), probably because both proteins are glycosylated.
FIGURE 2.
SDS-PAGE and immunoblot analysis of recombinant proHP6, proHP8, and proHP6 mutants. The purified proHP6 (0.1 μg/each) and proHP8 (0.06 μg) were treated with SDS sample buffer containing β-mercaptoethanol and separated by 10% SDS-PAGE followed by silver staining (A) or immunoblotting (B). The sizes and positions of the molecular weight markers are indicated on the right.
Microbial Elicitors Stimulate proHP6 Cleavage and Activation in Plasma
We used immunoblot analysis to investigate whether endogenous or recombinant proHP6 could be activated by factors in M. sexta plasma (Fig. 3). Proteolytic activation is predicted to result in decreased intensity of a band corresponding to the zymogen and the appearance of a band corresponding to the carboxyl-terminal catalytic chain (our antisera to HP6 and HP8 do not recognize the light chain containing the clip domain (15)). The naturally occurring proHP6 zymogen band was detected in plasma, and this band disappeared after plasma was treated with bacteria or curdlan (Fig. 3). The disappearance of the proHP6 zymogen correlated with the appearance of a higher molecular weight band not detected in the control plasma. This band is likely a covalent complex of the HP6 catalytic chain with an inhibitory serpin present in plasma. We previously detected such complexes of HP6 with M. sexta serpins 4 and 5 (28, 44). The appearance of serpin-proteinase complexes can be viewed as evidence that HP6 was indeed activated by plasma factors after exposure to microbial elicitors.
FIGURE 3.
Cleavage of M. sexta proHP6 after exposure to plasma proteins and microbial elicitors. Purified proHP6 (72 ng) was incubated at 37 °C for 30 min with plasma (3 μl) and the microbial elicitors indicated (104 E. coli XL1-Blue cells, 10 μg of M. luteus, 10 μg of lipopolysaccharide from E. coli 0111:B4 (Sigma L-2630), 10 μg of curdlan (Sigma C7821-5G)). The commercial lipopolysaccharide preparation may contain additional bacterial components (54). The reaction mixtures were subjected to 10% SDS-PAGE followed by immunoblot analysis using HP6 antiserum. The sizes and positions of molecular weight standards are indicated on the left. Bands representing proHP6 zymogen, a cleavage product corresponding to its catalytic domain, and a putative HP6-serpin complex are marked by arrows. Ab, antibody.
The addition of recombinant proHP6 to plasma to increase its concentration led to increased intensity of the zymogen band detected by immunoblot analysis. After treating proHP6-supplemented plasma with lipopolysaccharide, E. coli, M. luteus, or curdlan, a putative HP6-serpin complex appeared as well as an additional band at 29 kDa, representing the HP6 catalytic domain (calculated mass, 26.8 kDa). Curdlan and M. luteus had the strongest effect in stimulating proHP6 activation (Fig. 3). Detection of the catalytic domain in these samples supplemented with recombinant proHP6 is probably due to an excess of active HP6 relative to available serpin in the plasma. These results are consistent with a conclusion that microbial elicitors stimulate activation of proHP6 in plasma. Similar results were obtained for proHP8 (data not shown).
HP6 Activates proHP8 and proPAP1
We carried out experiments to identify proteins in M. sexta plasma that might be physiological substrates for HP6 using proHP6Xa activated by bovine Factor Xa (42). Incubation of purified proHP6Xa or proHP6Xa,I with Factor Xa resulted in decreased intensity of the 39-kDa zymogen band and appearance of the expected 29-kDa band corresponding to the catalytic domain (Fig. 4). This band was at the same position as the activated natural HP6 catalytic domain that appeared after plasma was incubated with bacteria (Fig. 4). This result indicates that Factor Xa effectively cleaved proHP6Xa and proHP6Xa,I.
We found that active HP6 can cleave and activate proHP8 and proPAP1 (Fig. 5). When activated proHP6Xa was mixed with proHP8, the proHP8 zymogen disappeared, and a 34-kDa product corresponding to the catalytic domain of HP8 was produced (Fig. 5A). This band did not appear after treatment of proHP8 with Factor Xa alone or with the Factor Xa-treated proHP6Xa,I mutant, indicating that the observed cleavage of proHP8 was a result of HP6 proteolytic activity. Amidase activity of HP8 cleaved by HP6Xa could be detected using a colorimetric substrate, Ile-Glu-Ala-Arg-p-nitroanilide (Fig. 5A). Although Factor Xa had some activity with this substrate, a significant increase in activity was observed in the presence of HP8 activated by HP6Xa but not when proHP8 was treated with the proHP6Xa,I negative control.
Similarly, activated HP6Xa cleaved proPAP1, producing bands corresponding to the catalytic and clip domains (Fig. 5B). Activity of the cleaved PAP1 was detected as hydrolysis of the Ile-Glu-Ala-Arg-p-nitroanilide substrate. The cleavage of proPAP1 and the appearance of elevated Ile-Glu-Ala-Arg-p-nitroanilide amidase activity did not occur in the mixture including the proHP6Xa,I mutant and Factor Xa, indicating that proteolytic activity of HP6Xa was responsible for the processing and activation of proPAP1. Active HP6 did not cleave M. sexta proHP1, proHP21, proPAP2, proPAP3, or proPO.3
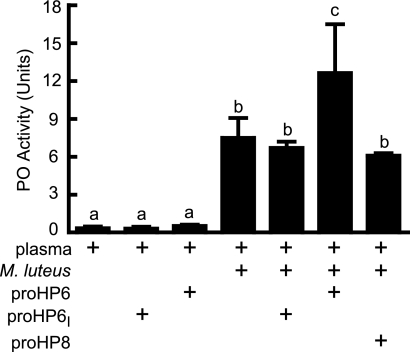
HP6 Functions in Plasma proPO Activation
The finding that HP6 activated proPAP1 led us to test the effect of HP6 in proPO activation in plasma (Fig. 6). Untreated plasma had low basal PO activity that could be activated dramatically by addition of M. luteus. Recombinant proHP6 or proHP6I in the absence of bacteria had no significant effect on plasma proPO activation. However, supplementing bacteria-activated plasma with recombinant proHP6 resulted in an approximate doubling of PO activity. This additional PO activation did not occur in the presence of the inactive proHP6I mutant, indicating that the effect was due to HP6 proteolytic activity. On the other hand, proHP8 did not have a significant effect on proPO activation. These results are consistent with an interpretation that proHP6 is activated in plasma in response to bacteria and then activates proPAP1, which in turn activates proPO.
FIGURE 6.
HP6 stimulates the proPO activation cascade. Samples of plasma (3 μl) were left untreated or mixed with M. luteus (10 μg) or with M. luteus plus proHP6 (40 ng), proHP6I (40 ng), or proHP8 (60 ng). After incubation at room temperature for 10 min, PO activity was assayed using dopamine as a substrate, as described under “Experimental Procedures.” The bars represent the mean ± S.D. (n = 3). Bars labeled with different letters (a, b, and c) are significantly different (analysis of variance and Newman-Keuls test, p < 0.05).
When plasma was treated with Factor Xa-activated proHP6Xa in the absence of bacteria, there was a significant increase in PO activity (supplemental Fig. S3) but of a much smaller magnitude than observed when bacteria were present. This result is consistent with previous results in which active PAP1 alone was not an efficient proPO activator and requires an activated SPH cofactor (26, 45), which may not be produced by treatment of plasma with active HP6. Activated HP8 did not affect PO activity.
HP6 and HP8 Function in Bacteria-induced Antimicrobial Peptide Synthesis
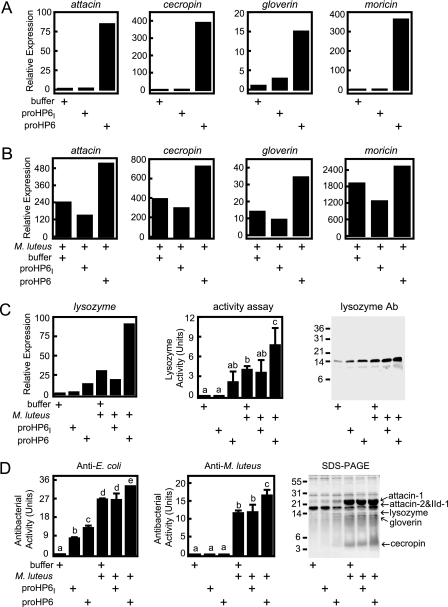
We investigated whether HP8 and its activating enzyme HP6 have a role in stimulating expression of AMP genes. To address this question, we injected zymogen or activated forms of HP6 or HP8 into larvae and measured AMP mRNA levels in fat body and AMP protein levels in hemolymph 20 h later compared with control insects injected with buffer. Transcript levels for moricin, cecropin, attacin, gloverin, and lysozyme increased after larvae were injected with proHP6 but not after injection of proHP6I (Fig. 7). When larvae injected with proHP6 or with proHP6I or buffer alone as controls were injected 30 min later with M. luteus, there was a strong induction of AMP transcript levels. This increase was even greater in larvae that had been first injected with recombinant proHP6 but not in larvae first injected with the inactive proHP6I (Fig. 7B). In fact, the insects injected with bacteria and proHP6I tended to have lower transcript levels than those injected only with bacteria, perhaps indicating a dominant negative effect of the mutant proHP6I.
FIGURE 7.
HP6 stimulates synthesis of antimicrobial plasma proteins in vivo. Fifth-instar day-0 larvae were injected with buffer (180 μl buffer 4), proHP6 (0.4 μg in 180 μl of buffer 4), or the catalytically inactive mutant proHP6I (0.4 μg in 180 μl of buffer 4). For some treatments, the insects were injected again 30 min later with 500 μg of M. luteus. After 20 h, hemolymph was collected from each insect, and RNA samples were prepared from fat body from groups of three insects. mRNA levels for indicated genes was assayed by quantitative RT-PCR (Relative Expression) as described under “Experimental Procedures.” A, antimicrobial peptide mRNA expression stimulated after injection of recombinant proHP6. B, antimicrobial peptide mRNA expression after treatment with M. luteus plus HP6. C, lysozyme mRNA and plasma lysozyme expression (detected by enzyme assay and immunoblotting (1 μl/lane)). D, antimicrobial activity of plasma assayed against E. coli or M. luteus and identification of induced antimicrobial plasma proteins by 15% SDS-PAGE (3 μl/lane) and MALDI-TOF peptide mass fingerprinting. For lysozyme and antibacterial activity, the bars represent mean ± S.D. (n = 3). Bars labeled with different letters (a, b, and c) are significantly different (analysis of variance and Newman-Keuls test, p < 0.05).
With lysozyme, we could also assay the protein in hemolymph by immunoblotting and enzyme activity (Fig. 7C), and these results are consistent with the data for lysozyme transcript level. proHP6 but not proHP6I enhanced expression of lysozyme. Similarly, proHP6 but not proHP6I enhanced the bacterial induction in vivo of plasma antimicrobial activity against E. coli or M. luteus (Fig. 7D). In SDS-PAGE analysis of heat-stable proteins in these plasma samples, several bands increased in intensity after insects were injected with proHP6 or proHP6 followed by M. luteus (Fig. 7D). Analysis of tryptic peptides from these bands by MS/MS and Mascot software identified them as attacin-1, attacin-2, immune-induced protein-1, lysozyme, gloverin, cecropin A, and cecropin B (supplemental Table S2). These results indicate that HP6 proteolytic activity functions in a pathway that leads to bacteria-induced expression in fat body of several genes encoding AMPs.
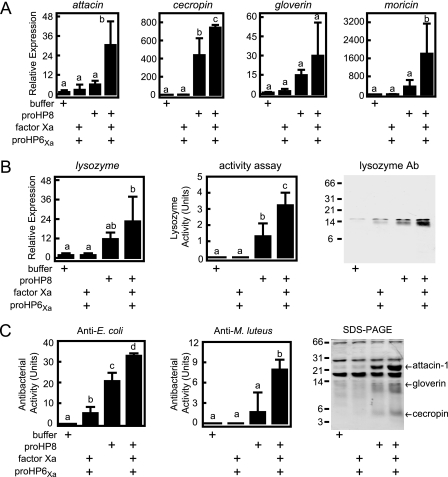
We carried out similar experiments to test the function of HP8 in AMP gene expression. We injected proHP8 or active HP8 (generated by cleavage with activated HP6Xa) into larvae and 20 h later assayed fat body mRNA levels and plasma antimicrobial activity and lysozyme levels (Fig. 8). Note that in these experiments the amount of activated HP6Xa used to activate proHP8 (30 ng) was much less than we used to inject larvae to test HP6 function (400 ng) and did not stimulate a significant increase in antimicrobial peptide mRNA levels. It is possible that this small amount of active HP6 is rapidly inhibited by plasma serpin-4 and serpin-5 (44). Injection of activated HP8 stimulated a significant increase in the transcript levels of attacin, ceropin, moricin, and lysozyme. Gloverin mRNA level was also elevated, but with a large variance in these samples, a statistically significant difference was not observed. Lysozyme enzymatic activity correlated well with the lysozyme mRNA levels and confirmed that injection of HP8 stimulated increased lysozyme concentration in plasma. Activated HP8 also stimulated the appearance of antibacterial activity against E. coli and M. luteus in plasma. SDS-PAGE after injection of larvae with activated HP8 showed increased intensity of multiple bands, three of which were excised and identified by Mascot analysis of MS/MS data as attacin-1, gloverin, and cecropin A and B (supplemental Table S2). Injection of activated HP8 or M. luteus resulted in similar levels of AMP transcripts in fat body and antimicrobial activity in hemolymph, suggesting that HP8 is a component of a pathway leading to bacteria-induced expression of these innate immune response genes.
FIGURE 8.
HP8 stimulates synthesis of antimicrobial plasma proteins in vivo. Fifth-instar day-0 larvae were injected with buffer (116 μl of buffer 4) or recombinant protein mixtures in 116 μl of buffer 4: HP6Xa (30 ng) activated by bovine Factor Xa (120 ng), proHP8 alone (1.6 μg), or HP8 (1.6 μg) activated by HP6Xa (30 ng). After 20 h, hemolymph was collected, and fat body RNA samples were prepared from each insect. mRNA levels for the indicated genes were assayed by quantitative RT-PCR (Relative Expression) as described under “Experimental Procedures.” A, antimicrobial peptide mRNA expression. B, lysozyme mRNA and plasma lysozyme expression (detected by enzyme assay and immunoblotting (1 μl/lane)). Ab, antibody. C, antimicrobial activity of plasma assayed against E. coli or M. luteus and identification of induced antimicrobial plasma proteins by 15% SDS-PAGE and MALDI-TOF peptide mass fingerprinting. The bars represent mean ± S.D. (n = 3). Bars labeled with different letters (a, b, and c) are significantly different (analysis of variance and Newman-Keuls test, p < 0.05).
DISCUSSION
Although proPO activation in insect immunity has been investigated for many years (9), understanding of the proPO activation cascade is still incomplete. To investigate proPO activation and other immune pathways in M. sexta, we have isolated more than 20 serine proteinase cDNAs from fat body and hemocytes (15). Only a handful of these hemolymph proteinases have been studied functionally. ProPAP1, proPAP2, and proPAP3 can cleave and activate proPO (39, 45–47) in a reaction that requires the noncatalytic SPH1 and SPH2 (26, 39, 45, 47). One cascade pathway for proPO activation in M. sexta has been established. proHP14 becomes active in the presence of Gram-positive bacteria or β-1,3-glucan recognition protein and zymosan (21). HP14 can then activate proHP21, which in turn activates proPAP2 or proPAP3 (7, 16), and PAP2 or PAP3 in the presence of active SPHs can activate proPO (Fig. 9). However, the pathways that lead to the cleavage activation of proPAP1 and proSPHs have remained elusive. Likewise, processing of spätzle has recently been found to induce AMP production in lepidopteran insects (48), but the proteinase mediators had not been identified. We have examined the function of two M. sexta serine proteinases, HP6 and HP8, and found that they stimulate expression of AMP genes and that HP6 also promotes proPO activation.
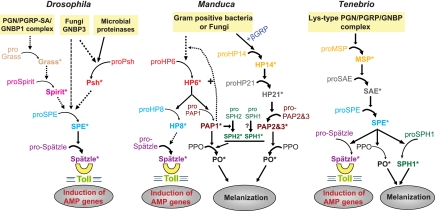
FIGURE 9.
A model for the roles of M. sexta HP6 and HP8 in extracellular immune pathways and comparison with Drosophila (18, 30–32, 50) and Tenebrio (11, 19) innate immune pathways involving serine proteinase cascades. Arrows indicate activation of downstream components or steps. Dashed arrows indicate steps that have not been experimentally verified or in which components of the pathway have not yet been identified. Proteins shown in the same color are putative orthologs. PGN, peptidoglycan; PGRP, peptidoglycan recognition protein; PGRP-SA, peptidoglycan recognition protein-SA; GRP, β-1,3-glucan recognition protein; GNBP, Gram-negative bacteria binding protein; PPO, prophenoloxidase; SAE, SPE activating enzyme; Psh, Persephone.
We produced recombinant proHP6 and proHP8 as well as mutants of proHP6. These purified proteins were used as reagents for in vitro experiments to examine their proteolytic activity and for ex vivo experiments in the complex protein mixture present in plasma, and the proteins were also injected into larvae, permitting assay of their function in vivo. Increasing the proteinase concentration by the addition of recombinant proteins to plasma or injection into larvae provided a means to examine the effects of “overexpression” of the proteinases. Use of a recombinant inactive HP6I mutant provided an important negative control for establishing that observed biological effects were due to the HP6 proteolytic activity.
proHP6 in plasma was cleaved and activated after exposure to bacteria or curdlan. The addition of proHP6 (but not the catalytically inactive proHP6I) to plasma resulted in activation of proPO. This outcome can be explained by our discovery that HP6 cleaves and activates proPAP1, an enzyme that has multiple functions in the proPO activation cascade, including activation of proPO, activation of proSPH2, and functioning in a positive feedback step that promotes proHP6 activation (49). The activator of proHP6 is still unknown; however, activation of proHP6 in plasma can be inhibited by serine proteinase inhibitors such as benzamidine or phenylmethylsulfonyl fluoride, suggesting that activation of proHP6 is mediated by a serine proteinase.4 HP14 does not activate proHP6,3 a finding consistent with our current model in which two different proteinase cascades lead to proPO activation in M. sexta (Fig. 9).
HP6 was also found to activate proHP8. HP8 had no effect on proPO activation in plasma, but we discovered that injection of activated HP8 (or proHP6) into larvae stimulated expression of a set of AMP genes. Injection of proHP6 into larvae with no microbial challenge stimulated expression of several antimicrobial peptide genes. However, the proHP6I mutant did not, indicating that the proteolytic activity of HP6 is required for this process. There may have been a low level of HP6 activation in these insects triggered by the wounding injection, or perhaps there is a low background level of activation of this pathway in the absence of infection that was accentuated by the higher concentration of proHP6 achieved after injection of the recombinant protein. As HP6 and HP8 are similar to Drosophila Persephone and SPE/Easter, respectively, these results suggest that Manduca may have a hemolymph proteinase cascade that leads to induced transcription of innate immune genes via activation of a Toll pathway. We have recently produced a recombinant form of Manduca spätzle and found that it is activated by HP8,5 confirming this prediction. Our results also support the earlier prediction (18) that B. mori BAEEase, which is highly similar to HP8, functions as a spätzle activator. Genetic studies have demonstrated that several Drosophila serine proteinase genes (Persephone, Grass, Spirit) are upstream of SPE in the immune spätzle activation pathway (Fig. 9) (30, 31). Biochemical results presented here support Persephone as a candidate for a proteinase that may directly activate SPE due to its similarity to HP6.
It is notable that HP6 and HP8 were constitutively expressed in naive larvae, and larvae that had been injected with bacteria or curdlan had no detectable increase in mRNA or protein levels for these two enzymes. The presence of HP6 and HP8 in hemolymph of naïve insects fits the idea that proteinase cascade pathways are available for rapid response to infection and do not require synthesis of new proteins. That HP6 and HP8 would not be detected in screens for induced gene expression, such as microarray experiments, should serve as a caution that such screens may miss potentially important innate immune genes.
HP6 occupies a position as the penultimate proteinase in two different immune pathways in M. sexta, leading to activation of proPO and the melanization response and to activation of spätzle and synthesis of AMPs (Fig. 9). In Drosophila, Persephone may serve the same function as HP6 in the Toll immune pathway, activating proSPE, but Persephone has not so far been implicated in the melanization response. Persephone can be activated by fungal and bacterial proteinases, suggesting that it is adapted to stimulate a response to microbial virulence factors (31, 50). It may also be activated by endogenous proteolytic activity in hemolymph downstream of GNBP3 (50), although the identity of such a proteinase is not yet known. M. sexta HP6 can be activated by proteolytic activity in hemolymph in the absence of live microbial infection, indicating the existence of an endogenous activating proteinase. Persphone and HP6 are activated by cleavage after a His residue (Fig. 1), which is unusual for serine proteinases and may point toward an activating proteinase with a unique active site pocket.
A pathway leading to activation of a single proteinase, SPE, which can activate both spätzle and proPO, has been defined in another insect, the beetle T. molitor (11, 19). This pathway is different from the one we have discovered in M. sexta, in which three different proteinases have proPO-activating function, and a different proteinase, HP8, activates spätzle (Fig. 9). A proteinase similar to HP6 and Persephone has not yet been identified in T. molitor, although a related beetle, Tribolium castaneum, has in its genome a predicted gene (XP_974337, SP66) that is a probable ortholog of HP6/Persephone and has a His residue at its predicted activation site (51). The proteinase SAE, which activates T. molitor SPE (11), is more similar to M. sexta HP21 (Fig. 1), which activates proPAP2 and ProPAP3 (7, 16).
We previously found that insect clip domain proteinases fall into two groups based on overall sequence comparison and on some individual sequence features of their clip domains (3, 52). We observe that these two groups of proteinases also can be distinguished based on their position in a cascade pathway (Fig. 1). Phylogenetic analysis based on alignment of the proteinase domains places the clip domain proteinases in two well defined clades. One clade contains the enzymes known to activate spätzle or proPO (terminal proteinases). The other clade contains enzymes upstream in the pathways, with those of known function having terminal proteinases as their substrates (penultimate proteinases). Within the terminal proteinase clade, the enzymes that activate spätzle (HP8, SPEs, Easter) cluster together, as do those that activate proPO (Manduca PAPs, Bombyx proPO-activating enzyme, Holotrichia PPAF1, Tenebrio SPE). The terminal proteinases all have a basic residue, Arg or Lys, at their activation site, whereas the penultimate proteinases instead have Leu (Manduca HP21, Tenebrio SAE, Drosophila snake), His (Drosophila Persephone, Manduca HP6), or Ser (Drosophila Spirit) at this position.
These two groups of enzymes also differ consistently in the length of the sequence between the third and fourth Cys residues of their clip domains, a feature previously used to define two groups of clip domain proteinases (3). Type 2 clip domains have 22–24 residues between Cys-3 and -4, whereas type 1 clip domains typically have 15–17 residues at the same position, with Drosophila Persephone somewhat longer at 20 residues. Drosophila Grass does not fit well in either clade and has an unusually long sequence (29 residues) between Cys-3 and -4 of its clip domain. The region between Cys-3 and -4, forming two antiparallel α-helices in the clip domains of M. sexta PAP2, has been proposed as a potential recognition/binding site (53). A conserved structural difference in this region between the clip domain proteinases that occupy different positions in cascade pathways may be due to binding interactions required for their function to either activate another proteinase or to activate proPO or a cytokine such as spätzle.
Supplementary Material
Acknowledgments
We thank Peter Dunn for antiserum to M. sexta lysozyme and Maureen Gorman for helpful comments on the manuscript. Protein digestion and mass spectrometry was performed by the Nevada Proteomics Center at the University of Nevada, which is supported by P20 RR-016464 from the IDeA Network of Biomedical Research Excellence Program of the National Center for Research Resources, Department of Health and Human Services, National Institutes of Health.
This work was supported, in whole or in part, by National Institutes of Health Grants GM41247 and GM58643. This is Contribution 07-125 from the Kansas Agricultural Experiment Station.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
C. An and M. R. Kanost, unpublished results.
J. Ishibashi, unpublished results.
C. An, H. Jiang, and M. Kanost, unpublished results.
- AMP
- antimicrobial peptide/protein
- PO and proPO
- phenoloxidase and its precursor, respectively
- HP6
- hemolymph proteinase 6
- PAP
- proPO activating proteinase
- SPH
- serine proteinase homolog
- RT
- reverse transcriptase
- contig
- group of overlapping clones
- SPE
- spätzle-processing enzyme
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- MS
- mass spectrometry.
REFERENCES
- 1.Sim R. B., Tsiftsoglou S. A. (2004) Biochem. Soc. Trans. 32, 21–27 [DOI] [PubMed] [Google Scholar]
- 2.Carroll M. C. (2004) Nat. Immunol. 5, 981–986 [DOI] [PubMed] [Google Scholar]
- 3.Jiang H., Kanost M. R. (2000) Insect Biochem. Mol. Biol. 30, 95–105 [DOI] [PubMed] [Google Scholar]
- 4.Osaki T., Kawabata S. (2004) Cell. Mol. Life Sci. 61, 1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theopold U., Schmidt O., Söderhäll K., Dushay M. S. (2004) Trends Immunol. 25, 289–294 [DOI] [PubMed] [Google Scholar]
- 6.Piao S., Song Y. L., Kim J. H., Park S. Y., Park J. W., Lee B. L., Oh B. H., Ha N. C. (2005) EMBO J. 24, 4404–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorman M. J., Wang Y., Jiang H., Kanost M. R. (2007) J. Biol. Chem. 282, 11742–11749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H., Jiravanichpaisal P., Cerenius L., Lee B. L., Söderhäll I., Söderhäll K. (2007) J. Biol. Chem. 282, 33593–33598 [DOI] [PubMed] [Google Scholar]
- 9.Kanost M. R., Gorman M. G. (2008) in Insect Immunology (Beckage N. ed) pp. 69–96, Academic Press, Inc./Elsevier, San Diego, CA [Google Scholar]
- 10.Ferrandon D., Imler J. L., Hetru C., Hoffmann J. A. (2007) Nat. Rev. Immunol. 7, 862–874 [DOI] [PubMed] [Google Scholar]
- 11.Kim C. H., Kim S. J., Kan H., Kwon H. M., Roh K. B., Jiang R., Yang Y., Park J. W., Lee H. H., Ha N. C., Kang H. J., Nonaka M., Söderhäll K., Lee B. L. (2008) J. Biol. Chem. 283, 7599–7607 [DOI] [PubMed] [Google Scholar]
- 12.Kurata S., Ariki S., Kawabata S. (2006) Immunobiology 211, 237–249 [DOI] [PubMed] [Google Scholar]
- 13.Le Saux A., Ng P. M., Koh J. J., Low D. H., Leong G. E., Ho B., Ding J. L. (2008) J. Mol. Biol. 377, 902–913 [DOI] [PubMed] [Google Scholar]
- 14.Belvin M. P., Anderson K. V. (1996) Annu. Rev. Cell Dev. Biol. 12, 393–416 [DOI] [PubMed] [Google Scholar]
- 15.Jiang H., Wang Y., Gu Y., Guo X., Zou Z., Scholz F., Trenczek T. E., Kanost M. R. (2005) Insect Biochem. Mol. Biol. 35, 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Jiang H. (2007) Insect Biochem. Mol. Biol. 37, 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitin A., Whiteway M. (2008) Cell. Microbiol. 10, 1021–1026 [DOI] [PubMed] [Google Scholar]
- 18.Jang I. H., Chosa N., Kim S. H., Nam H. J., Lemaitre B., Ochiai M., Kambris Z., Brun S., Hashimoto C., Ashida M., Brey P. T., Lee W. J. (2006) Dev. Cell 10, 45–55 [DOI] [PubMed] [Google Scholar]
- 19.Kan H., Kim C. H., Kwon H. M., Park J. W., Roh K. B., Lee H., Park B. J., Zhang R., Zhang J., Söderhäll K., Ha N. C., Lee B. L. (2008) J. Biol. Chem. 283, 25316–25323 [DOI] [PubMed] [Google Scholar]
- 20.Yu X. Q., Zhu Y. F., Ma C., Fabrick J. A., Kanost M. R. (2002) Insect Biochem. Mol. Biol. 32, 1287–1293 [DOI] [PubMed] [Google Scholar]
- 21.Wang Y., Jiang H. (2006) J. Biol. Chem. 281, 9271–9278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J. W., Kim C. H., Kim J. H., Je B. R., Roh K. B., Kim S. J., Lee H. H., Ryu J. H., Lim J. H., Oh B. H., Lee W. J., Ha N. C., Lee B. L. (2007) Proc. Natl. Acad. Sci. 104, 6602–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashida M., Brey P. T. (1998) in Molecular Mechanisms of Immune Response in Insects (Brey P. T., Hultmark D. eds) pp. 135–172, Chapman & Hall Ltd., London [Google Scholar]
- 24.Zhao P., Li J., Wang Y., Jiang H. (2007) Insect Biochem. Mol. Biol. 37, 952–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerenius L., Lee B. L., Söderhäll K. (2008) Trends Immunol. 29, 263–271 [DOI] [PubMed] [Google Scholar]
- 26.Yu X. Q., Jiang H., Wang Y., Kanost M. R. (2003) Insect Biochem. Mol. Biol. 33, 197–208 [DOI] [PubMed] [Google Scholar]
- 27.Volz J., Müller H. M., Zdanowicz A., Kafatos F. C., Osta M. A. (2006) Cell. Microbiol. 8, 1392–1405 [DOI] [PubMed] [Google Scholar]
- 28.Tong Y., Kanost M. R. (2005) J. Biol. Chem. 280, 14923–14931 [DOI] [PubMed] [Google Scholar]
- 29.Zhu Y., Wang Y., Gorman M. J., Jiang H., Kanost M. R. (2003) J. Biol. Chem. 278, 46556–46564 [DOI] [PubMed] [Google Scholar]
- 30.Ligoxygakis P., Pelte N., Hoffmann J. A., Reichhart J. M. (2002) Science 297, 114–116 [DOI] [PubMed] [Google Scholar]
- 31.Kambris Z., Brun S., Jang I. H., Nam H. J., Romeo Y., Takahashi K., Lee W. J., Ueda R., Lemaitre B. (2006) Curr. Biol. 16, 808–813 [DOI] [PubMed] [Google Scholar]
- 32.El Chamy L., Leclerc V., Caldelari I., Reichhart J. M. (2008) Nat. Immunol. 9, 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang H., Kambris Z., Lemaitre B., Hashimoto C. (2006) J. Biol. Chem. 281, 28097–28104 [DOI] [PubMed] [Google Scholar]
- 34.Dunn P. E., Drake D. R. (1983) J. Invert. Pathol. 41, 77–85 [Google Scholar]
- 35.Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 36.Yu X. Q., Kanost M. R. (2000) J. Biol. Chem. 275, 37373–37381 [DOI] [PubMed] [Google Scholar]
- 37.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 38.Coligan J. E., Dunn B. M., Ploegh H. L., Speicher D. W., Wingfield P. T. (1995) Current Protocols in Protein Science, pp 10.5.6–10.5.7, John Wiley & Sons, Inc. [Google Scholar]
- 39.Jiang H., Wang Y., Yu X. Q., Kanost M. R. (2003) J. Biol. Chem. 278, 3552–3661 [DOI] [PubMed] [Google Scholar]
- 40.Zou Z., Najar F., Wang Y., Roe B., Jiang H. (2008) Insect Biochem. Mol. Biol. 38, 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hultmark D., Engström A., Bennich H., Kapur R., Boman H. G. (1982) Eur. J. Biochem. 127, 207–217 [DOI] [PubMed] [Google Scholar]
- 42.DeLotto Y., DeLotto R. (1998) Mech. Dev. 72, 141–148 [DOI] [PubMed] [Google Scholar]
- 43.Jenny R. J., Mann K. G., Lundblad R. L. (2003) Protein Expr. Purif. 31, 1–11 [DOI] [PubMed] [Google Scholar]
- 44.Tong Y., Jiang H., Kanost M. R. (2005) J. Biol. Chem. 280, 14932–14942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta S., Wang Y., Jiang H. (2005) Insect Biochem. Mol. Biol. 35, 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang H., Wang Y., Kanost M. R. (1998) Proc. Natl. Acad. Sci. 95, 12220–12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang H., Wang Y., Yu X. Q., Zhu Y., Kanost M.. (2003) Insect Biochem. Mol. Biol. 33, 1049–1060 [DOI] [PubMed] [Google Scholar]
- 48.Wang Y., Cheng T., Rayaprolu S., Zou Z., Xia Q., Xiang Z., Jiang H. (2007) Dev. Comp. Immunol. 31, 1002–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y., Jiang H. (2008) Insect Biochem. Mol. Biol. 38, 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gottar M., Gobert V., Matskevich A. A., Reichhart J. M., Wang C., Butt T. M., Belvin M., Hoffmann J. A., Ferrandon D. (2006) Cell 127, 1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou Z., Evans J. D., Lu Z., Zhao P., Williams M., Sumathipala N., Hetru C., Hultmark D., Jiang H. (2007) Genome Biology 8, R177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ross J., Jiang H., Kanost M. R., Wang Y. (2003) Gene 304, 117–131 [DOI] [PubMed] [Google Scholar]
- 53.Huang R., Lu Z., Dai H., Velde D. V., Prakash O., Jiang H. (2007) Biochemistry 46, 11431–11439 [DOI] [PubMed] [Google Scholar]
- 54.Leulier F., Parquet C., Pili-Floury S., Ryu J. H., Caroff M., Lee W. J., Mengin-Lecreulx D., Lemaitre B. (2003) Nat. Immunol. 4, 478–484 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.