Summary
We have shown previously that lack of molybdenum cofactor (MoCo) in Escherichia coli leads to hypersensitivity to the mutagenic and toxic effects of N-hydroxylated base analogs, such as 6-N-hydroxylaminopurine (HAP). However, the nature of the MoCo-dependent mechanism is unknown, as inactivation of all known and putative E. coli molybdoenzymes does not produce any sensitivity. Presently, we report on the isolation and characterization of two novel HAP-hypersensitive mutants carrying defects in the ycbX or yiiM open reading frames. Genetic analysis suggests that the two genes operate within the MoCo-dependent pathway. In the absence of the ycbX- and yiiM-dependent pathways, biotin sulfoxide reductase (BisC) plays also a role in the detoxification pathway. YcbX and YiiM are hypothetical members of the MOSC protein superfamily, which contain the C-terminal domain (MOSC) of the eukaryotic MoCo sulfurases. However, deletion of ycbX or yiiM did not affect the activity of human xanthine dehydrogenase expressed in E. coli, suggesting that the role of YcbX and YiiM proteins is not related to MoCo sulfuration. Instead, YcbX and YiiM may represent novel MoCo-dependent enzymatic activities. We also demonstrate that the MoCo/YcbX/YiiM-dependent detoxification of HAP proceeds by reduction to adenine.
Introduction
Modified nucleobases that can participate in cellular processes alongside the natural substrates but are toxic and/or mutagenic are traditionally referred to as base analogs (Freese, 1959). In vivo, such analogs may be produced from normal cellular metabolism or by the action of chemical and physical factors, such as alkylating agents or ionizing radiation. In addition, synthetic base analogs with mutagenic or inhibitory properties have been used as tools for study of fundamental cellular processes, such as the metabolism of nucleic acid precursors or the mechanisms of DNA replication and repair. An important group of toxic and mutagenic analogs are the N-hydroxylated derivatives of purines and pyrimidines, such as 6-N-hydroxylaminopurine (HAP), 2-amino-HAP (AHAP) and N4-hydroxycytidine (see Khromov-Borisov, 1997). In particular, HAP has been shown to possess highly potent mutagenic effects in bacteria, yeast, and mammalian cells (Pavlov et al., 1991; Kozmin et al., 1998a; Barrett, 1981). HAP is believed to be incorporated like adenine or hypoxanthine via the purine salvage pathways and to be converted to a deoxynucleoside triphosphate (dHAPTP), serving as a precursor for enzymatic DNA synthesis, and inducing mutations due to its ambivalent coding capacity (Kozmin et al., 1998a).
Studies of the genetic control of HAP-induced mutagenesis in Saccharomyces cerevisiae and Escherichia coli has led to identification of HAP-hypersensitive mutants. In yeast, drastically enhanced sensitivity to the mutagenic and toxic effects of HAP was observed in ham1 mutants (Pavlov, 1986; Noskov et al., 1996). Ham1p has been shown to be a specific pyrophosphatase that acts to hydrolyze dHAPTP to the corresponding mononucleotide, hence preempting incorporation into the DNA (Kozmin et al., 1998a; Hwang et al., 1999; Burgis and Cunningham, 2007). While hydrolysis of dHAPTP is a major HAP-protective mechanism in yeast, it does not play a significant role in bacteria because inactivation of RdgB, the E. coli Ham1p homolog, does not confer HAP-sensitivity (Burgis et al., 2003).
In E. coli, HAP-hypersensitivity was observed in strains carrying a deletion of the uvrB-bio chromosomal region (Pavlov et al., 1996). The Δ(uvrB-bio) strains were also AHAP-hypermutable and sensitive to growth inhibition or killing by hydroxylamine (Pavlov et al., 1996; Kozmin et al., 1998b). Likewise, Salmonella strains carrying a corresponding deletion of the uvrB-bio region proved hypermutable by AHAP and N4-hydroxycytidine (Janion, 1978; Janion, 1979; Janion and Myszkowska, 1981). Our laboratory established that the base-analog hypersensitivity of E. coli Δ(uvrB-bio) mutants is due to a defect in biosynthesis of the Molybdenum Cofactor (MoCo), an essential cofactor for a variety of oxidoreductases (Kozmin et al., 2000). Based on this result, we proposed that E. coli contains an as yet unidentified enzymatic activity that requires MoCo and is capable of inactivating HAP, presumably by some oxidation/reduction reaction.
MoCo is a pterin derivative, called molybdopterin (MPT), carrying a unique dithiolene group that coordinates a molybdenum atom acting as a catalytic redox center (for review, see Rajagopalan, 1996; Mendel and Bittner, 2006). At least four families of molybdoenzymes have been recognized based on sequence homology and the chemical structure of the Mo-containing redox center (Kisker et al., 1999; Hille, 2002). The predominant family in E. coli and most other prokaryotes (the DMSO reductase family) utilizes a guanylated version of MoCo, named molybdopterin guanine dinucleotide (MGD). In contrast, enzymes of the xanthine oxidase and sulfite oxidase families, common to eukaryotes, utilize the guanylate-free MPT form of MoCo. The xanthine oxidase family is further characterized by the need for a MoCo maturation step, in which the molybdenum atom is additionally coordinated to an extra sulfur atom (MoCo sulfuration) (Mendel and Bittner, 2006). Interestingly, we recently showed that the base-analog protective mechanism in E. coli does not depend on MGD; instead, the MPT form of MoCo is sufficient (Kozmin and Schaaper, 2007). We also showed that deletion of all the known and putative molybdoenzymes of E. coli did not lead to base-analog sensitivity (Kozmin and Schaaper, 2007). Thus, it was proposed that perhaps an additional, as yet unrecognized, family of MoCo-dependent activities might exist in E. coli that is responsible for base-analog detoxification.
In the present work, we describe the isolation and characterization of novel HAP-sensitive mutants carrying defects in the ycbX and yiiM open reading frames. Genetic analysis of these mutants indicates that they control different sub-pathways within the overall MoCo-dependent pathway. The corresponding YcbX and YiiM proteins are characterized by a so-called MOSC domain, which has been found in eukaryotic MoCo sulfurases (Anantharaman and Aravind, 2002) and which may represent a novel MoCo-binding domain (Havemeyer et al., 2006). From these and other data we propose that YcbX and YiiM represent novel MPT-containing molybdoenzymes in E. coli. Using E. coli cell-free extracts, we show that the main ycbX/yiiM-dependent HAP-inactivation pathways proceed by reduction to adenine.
Results
YcbX, a novel HAP-resistance determinant
As part of our efforts to better understand the mechanisms for the MoCo-dependent resistance of E. coli to N-hydroxylated base analogs, we searched for additional HAP-sensitive mutants beyond those affected in MoCo biosynthesis such as moaE or moeA mutants (Kozmin et al., 2000; Kozmin and Schaaper, 2007). Several genome-wide searches using random transposon mutagenesis of the E. coli chromosome (see Experimental procedures) were performed, which yielded a number of HAP-sensitive mutants carrying a transposon insert in the ycbX open-reading frame. Some tests demonstrating the sensitivity of these mutants are shown in Figs. 1 and 2. In simple spot-tests (Fig. 1), a small aliquot of chemical is placed in the center of a plate containing the strain of interest and the plates are inspected for a zone of inhibition (or killing) after overnight growth. If desired, these plates can then be replicated on rifampicin-containing plates to obtain an estimate of the chemical-induced frequency of rifampicin-resistant mutants. In liquid incubation tests, growing cells are exposed to various concentrations of the chemical and the fraction of surviving cells is determined as a function of dose (Fig. 2). As seen in both Fig. 1 and 2, the ycbX defect leads to strongly increased sensitivity to both the toxic and mutagenic effects of HAP. On the other hand, the sensitivity of the new isolates was not as severe as that of the moaE or moeA mutants defective in MoCo biosynthesis. For example, in Fig. 1, the diameter of the inhibition zone is 50 mm and 35 mm in the moaE and ycbX strains, respectively. We also noticed certain specificity differences between the ycbX and MoCo defects. The ycbX mutant did not show any clear sensitivity to two other hydroxylated compounds, 2-amino-HAP (AHAP) and hydroxylamine (HA) to which MoCo-deficient strains are clearly sensitized (Figs. 3 and 4).
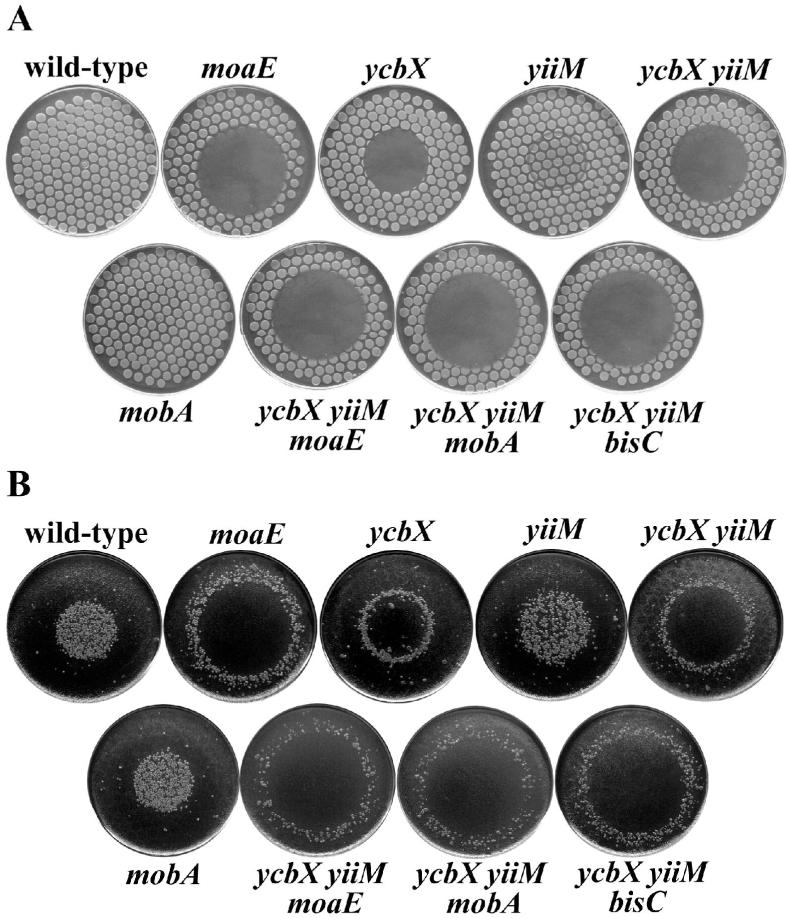
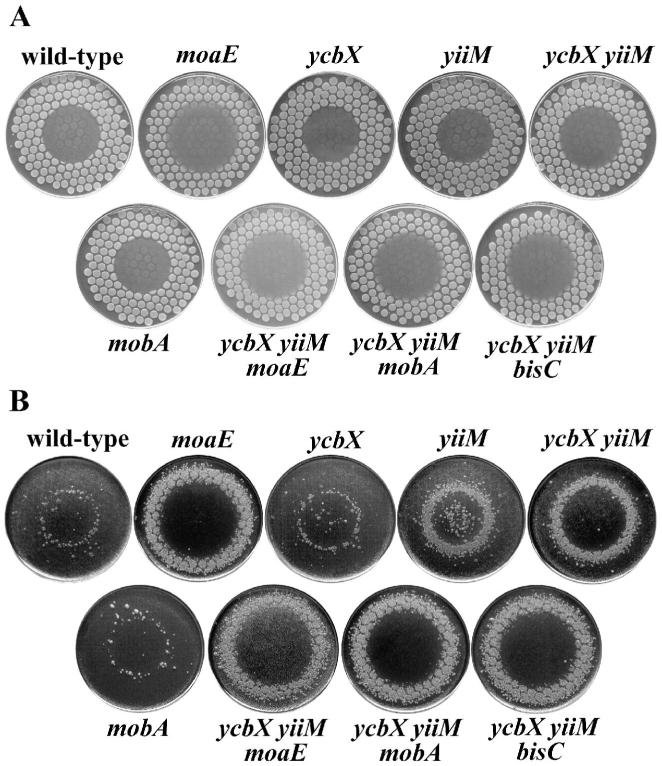
Fig. 1.
HAP-induced killing (A) and mutagenesis (B) for various E. coli mutants. The strains used were wild-type (NR10836), moaE (NR15996), ycbX (NR15873), yiiM (NR15871), ycbX yiiM (NR15876), mobA (NR15994), ycbX yiiM moaE (NR16023), ycbX yiiM mobA (NR16028), and ycbX yiiM bisC (NR16047). Cells were spotted on a minimal medium plate using a multi-prong replicator device, and 50 μg of HAP was spotted onto the center of each plate (see Experimental procedures). The plates, after overnight incubation, are shown in panel A. These plates were replica-plated onto LB plates containing rifampicin and incubated overnight (panel B).
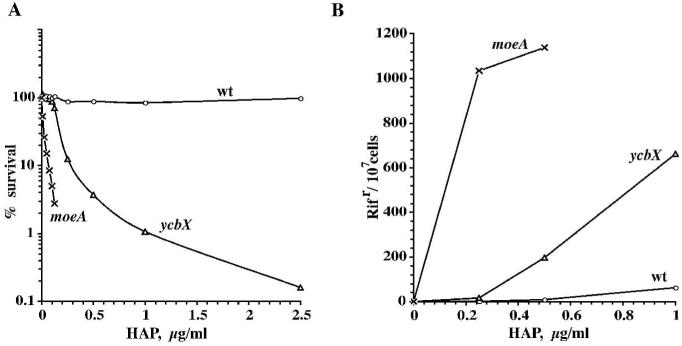
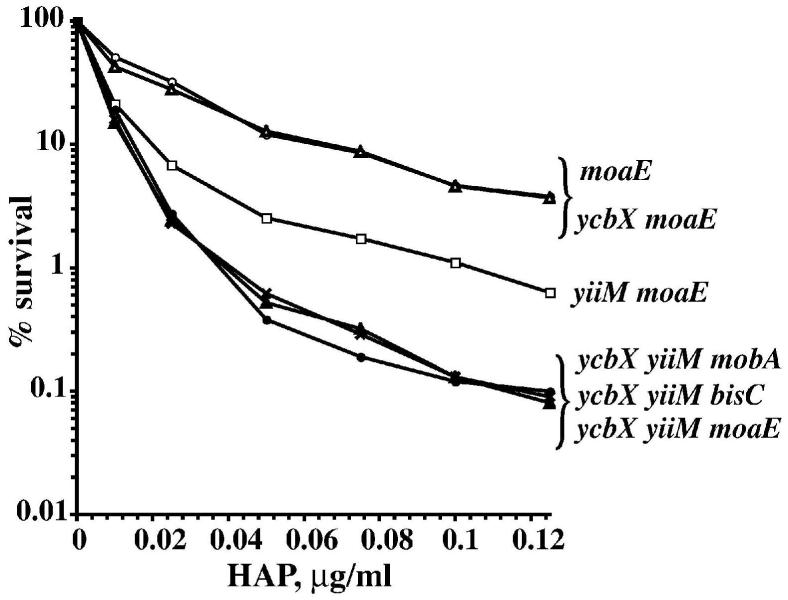
Fig. 2.
Survival (A) and mutagenesis (B) of ycbX and moeA E. coli exposed to HAP for 2 h in liquid medium. Strains used were NR10836 (wild-type) (○), NR15873 (ycbX::mini-Tn10cam) (Δ), and NR12383 (moeA121::mini-Tn10cam) (×). See Experimental procedures for details.
Fig. 3.
Spot-test for determination of 2-amino-HAP (AHAP)-induced killing (A) and mutagenesis (B). All strains and procedures were as indicated in the legend to Fig. 1, except that AHAP was used instead of HAP.
Fig. 4.
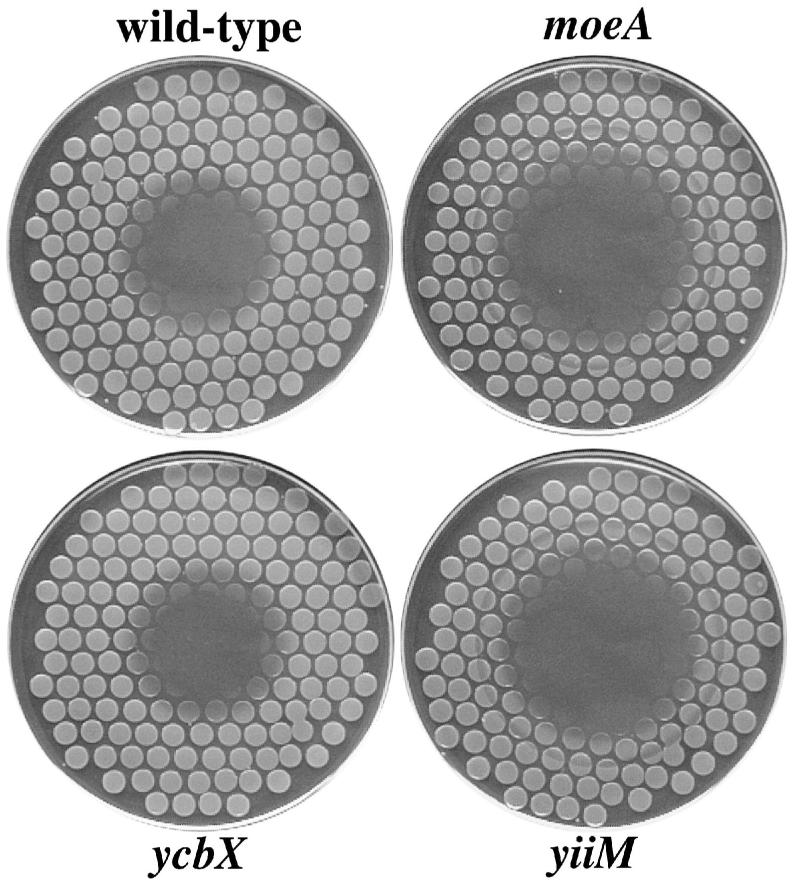
Sensitivity of E. coli strains to hydroxylamine. Cells were plated on minimal medium plates using a multi-prong replicator, and 150 μg of hydroxylamine (in water) was spotted onto the center of each plate. Plates were incubated overnight. See Experimental procedures for details. Strains used were NR10836 (wt), NR12383 (moeA), NR15873 (ycbX), and NR15871 (yiiM). When replicated to LB-Rif plates, no induction of Rifr mutants was observed for any of the strains (not shown).
To investigate the genetic relationship between the ycbX and MoCo mutants we tested ycbX moaE double mutants. The result of the quantitative liquid incubation test showed that the sensitivity of the ycbX moaE double mutant was essentially indistinguishable from that of the single moaE mutant (Fig. 5). The ycbX moaE double mutant also proved as sensitive as the single moaE mutant when tested for AHAP and hydroxylamine using direct spot tests (data not shown). Consequently, it appears that ycbX likely controls a subpathway within the MoCo-dependent pathway, with a specificity that is geared towards HAP.
Fig. 5.
Role of BisC (biotin sulfoxide reductase) in HAP detoxification. The liquid culture tests were performed as in Fig. 2. The strains used were moaE (NR15996) (○), ycbX moaE (NR16021) (Δ), yiiM moaE (NR16019) (□), yiiM ycbX mobA (NR16028) (×), yiiM ycbX bisC (NR16047) (σ), and yiiM ycbX moaE (NR16023) (λ).
YcbX protein contains three essential conserved domains: β-barrel, MOSC and ferredoxin
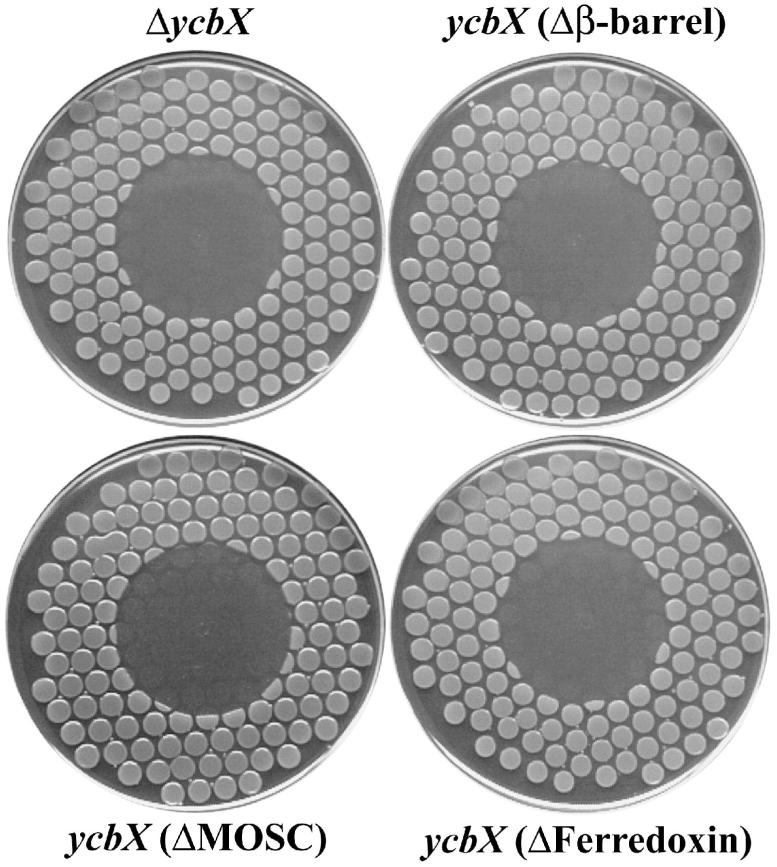
Analysis of YcbX protein using the NCBI Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=cdd) (Marchler-Bauer et al., 2005) indicates the presence of three conserved domains: an N-terminal β-barrel domain, a central MOSC domain, and a C-terminal [2Fe-2S]-ferredoxin-like domain (Fig. 6). The first two domains, β-barrel and MOSC, were first found in eukaryotic MoCo sulfurases (the abbreviation MOSC was introduced by Anantharaman and Aravind [2002] and refers to MoCo sulfurase C-terminal domain). MoCo sulfurases transfer an inorganic sulfur from cysteine to the molybdenum center of MoCo as part of a final MoCo maturation (sulfuration) step. Such a sulfurated version of MoCo is utilized specifically by enzymes of the xanthine oxidase family, which includes xanthine dehydrogenase and aldehyde oxidase (Kisker et al., 1999; Hille, 2002; Mendel and Bittner, 2006). Additional MOSC-containing proteins are distributed widely in pro- and eukaryotes; however, except for the MoCo sulfurases, all other members of the ‘MOSC superfamily’ are proteins without any confirmed function (Anantharaman and Aravind, 2002). Finally, the third domain of YcbX, ferredoxin, belongs to a widely distributed class of small iron-sulfur proteins serving as an electron carriers in various cellular processes (Sticht and Rösch, 1998). To check if all three YcbX domains are essential for YcbX function, we constructed a set of strains carrying in-frame deletions of each of the domains (see Fig. 6 and Experimental procedures). When testing for HAP-sensitivity we found that deletion of any YcbX domain led to HAP-hypersensitivity, in a manner indistinguishable from a complete ycbX deletion (ΔycbX) in strain NR16262 (Fig. 7).
Fig. 6.
Sequence and predicted domain architecture of YcbX protein. The predicted domains (indicated by colored boxes) are β-barrel (M1-E117) (blue), MOSC (T128-I263) (red), and Ferredoxin (V291-T363) (green). The Figure also indicates (by small arrows) the endpoints of the domain-specific ycbX deletions created in this study. In strain NR16261 (ΔycbX[Δ(β-barrel)]) β-barrel amino acids A2 through E117 (endpoints marked by blue arrows) are replaced by the stuffer peptide IPGIRRPAVRSSYSLESIGTSKQLQPTQ. In strain NR16259 (ΔycbX[Δ(MOSC)]) MOSC-domain amino acids K132 to I263 (red arrows) are replaced by the polypeptide IPGIRRPAVRSSYSLESIGTSKQLQPTL. In strain NR16258 (ΔycbX[Δ(Ferredoxin)]) Ferredoxin-domain amino acids D292 through A362 (green arrows) are replaced by polypeptide IPGIRRPAVRSSYSLESIGTSKQLQPT. In strain NR16262 (ΔycbX), carrying a complete deletion of the ycbX gene, the polypeptide MIPGIRRPAVRSSYSLESIGTSKQLQPTRR replaces the YcbX protein. See text and Experimental procedures for further details.
Fig. 7.
HAP-sensitivity of strains carrying in-frame deletions of the ycbX gene. The ΔycbX strain (NR16262) carries a complete deletion of the ycbX gene, whereas NR16261, NR16259, and NR16258 encode truncated versions of the YcbX protein lacking, respectively, the β-barrel, MOSC or ferredoxin domain as indicated, and as more fully described in Fig. 6. 50 μg of HAP was spotted onto the center of each plate (see Experimental procedures for more details).
YiiM, a second E. coli MOSC-containing protein, is also involved in protection against N-hydroxylated analogs
Homology searches have predicted the existence of a second MOSC-containing protein in E. coli, YiiM (Anantharaman and Aravind, 2002). YiiM is a predicted 224-amino acid protein composed of a MOSC domain and a C-terminal domain with an assumed trihelical structure (Anantharaman and Aravind, 2002). To check for its possible role in base-analog detoxification we created a yiiM deletion strain (see Experimental procedures) and found that it displayed moderately increased sensitivity to HAP (Fig. 1A) as well as significantly enhanced AHAP-induced mutagenesis (Fig. 3B). Moreover, the yiiM mutant, in contrast to the ycbX mutant, was sensitized to hydroxylamine, resembling, in fact, the MoCo-deficient strain (Fig. 4). A double ycbX yiiM mutant showed a significant increase in HAP- and AHAP-sensitivity relative to each of the single mutants (Figs. 1 and 3). This observation, as well as the noted differences between ycbX and yiiM with respect to HAP, AHAP, and hydroxylamine (compare Figs. 1, 3, and 4), leads us to propose that the two genes likely participate in two different pathways within the base-analog protective mechanisms (see Fig. 9).
Fig. 9.
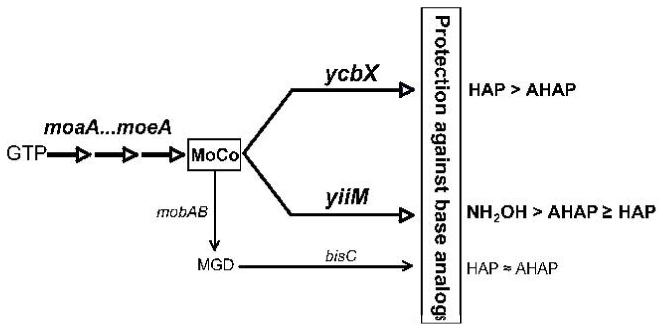
Genetic framework for pathways of detoxification of N-hydroxylated base analogs in E. coli based on the current findings. See text for more details.
Interestingly, we noted that the combination of yiiM and moaE increased HAP-sensitivity relative to the single moaE strain (Fig. 5), suggesting that yiiM may also work outside the MoCo pathway; moreover, a triple ycbX yiiM moaE mutant was more HAP-sensitive than the yiiM moaE strain (Fig. 5), suggesting that at least in the yiiM moaE background ycbX may also have an additional function. These results suggest that while ycbX, and presumably also yiiM, are clearly involved in the MoCo-dependent pathway, both genes also may function to some extent in a MoCo-independent manner.
YcbX and YiiM are not involved in MoCo biosynthesis
The possibility needed to be considered that the ycbX and yiiM genes are involved indirectly in base-analog detoxification through involvement in an as yet unrecognized aspect of MoCo biosynthesis, an area still subject of continuing investigation (ee Mendel and Bittner, 2006). A common phenotype of E. coli mutants defective in the MoCo biosynthesis is resistance to chlorate under anaerobic conditions (Shanmugam et al., 1992). In wild-type strains, but not MoCo-deficient strains, chlorate is converted to toxic chlorite due to the action of several molybdoenzymes of the DMSO reductase family, such as nitrate reductase and trimethylamine N-oxide (TMAO) reductase (Stewart and MacGregor, 1982). We found that the ycbX (NR15873), yiiM (NR15871), and double ycbX yiiM (NR15876) strains were fully sensitive to chlorate similar to a wild-type strain (NR10836), even at a very low chlorate concentration, 0.5 mM (data not shown). We also tested for the activity of nitrate and TMAO reductases using a simple colony pigmentation test using MacConkey (MC) indicator plates supplemented with nitrate or TMAO (Stewart and MacGregor, 1982; and Experimental procedures). On these media, usage of the substrates (nitrate or TMAO) leads to acidification of the colonies and, hence, altered coloration of the colonies (Stewart and MacGregor, 1982). We observed that on MC plates containing nitrate or TMAO, the MoCo-deficient strain moeA formed deep-red colonies, whereas wild-type, ycbX, yiiM, and ycbX yiiM strains formed light salmon-colored colonies (data not shown). In addition, TMAO reduction in the wild-type, ycbX, yiiM, and ycbX yiiM strains was easily detectable by formation of the odorous product trimethylamine, which was not detectable in the MoCo-deficient mutant. Taken together, these experiments indicate that neither ycbX nor yiiM mutants have any significant defects in MoCo (MGD) biosynthesis.
A limited involvement of Biotin Sulfoxide Reductase in base-analog detoxification
As seen from Figs. 1 and 3, the double ycbX yiiM mutant was less sensitive to HAP and AHAP than a MoCo-deficient strain. This suggested that there might exist one or more additional MoCo-dependent pathways in addition to those controlled by ycbX and yiiM. In previous work (Kozmin and Schaaper, 2007) we demonstrated that the major base-analog protective mechanism utilizes the GMP-free molybdopterin (MPT) version of MoCo rather than the common molybdopterin-guanine-dinucleotide (MGD). This can been seen, for example, in Figs. 1 and 3 where the mobA strain (defective in conversion of MPT to MGD) is shown to be resistant to HAP and normally sensitive to AHAP. However, in the ycbX yiiM background, the mobA defect clearly further sensitizes the strain such that it is now indistinguishable from the ycbX yiiM moaE mutant (Figs. 1, 3, 5). Thus, the additional pathway(s) are mobA-dependent and are predicted to utilize the MGD version of MoCo. To find this responsible MGD-utilizing activity, we systematically combined the ycbX yiiM double deficiency with deletions of the genes encoding the 13 known and putative MGD-requiring enzymes (strains NR16047 through NR16060) (see Table1, and Kozmin and Schaaper, 2007). This experiment showed that only the combination of yiiM ycbX with bisC (encoding Biotin Sulfoxide Reductase) (del Campillo-Campbell and Campbell, 1982; Pierson and Campbell, 1990) increased HAP- and AHAP-sensitivity. Moreover, the yiiM ycbX bisC strain is indistinguishable from the ycbX yiiM mobA or ycbX yiiM moaE strains (see Figs. 1, 3, 5), indicating that no additional factors are required to account for the sensitivity of the MoCo mutants. Note that no contribution of the bisC pathway to HAP resistance was detected in a MoCo-proficient background (Kozmin and Schaaper, 2007). Therefore, the bisC pathway must be considered a minor pathway. The overall scheme of HAP detoxification based on our results is presented in Fig. 9
TABLE 1.
E. coli strains used in this study
| Strain | Genotype | Source or derivation |
|---|---|---|
| BL21 | ompT hsdSB(rB- mB-) gal dcm (Chlr) | Novagen |
| BW15113 | lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | Datsenko and Wanner, 2000 |
| KA796 | ara thi Δ(pro-lac) | Schaaper et al., 1985 |
| NR10836 | ara thi Δ(pro-lac) F’CC106 | Kozmin et al., 2000 |
| NR12383 | NR10836, but moeA121::mini-Tn10cam | Kozmin et al., 2000 |
| NR15870 | NR10836, but ΔyiiM::kan | This work |
| NR15871 | NR10836, but ΔyiiM | This work |
| NR15873 | NR10836, but ycbX::mini-Tn10cam | This work |
| NR15876 | NR10836, but ΔyiiM ycbX::mini-Tn10cam | NR15871 × P1/NR15873 |
| NR15967 | BL21, but gal+, Chls | BL21 × P1/NR10836 |
| NR15994 | NR10836, but ΔmobA::kan | Kozmin and Schaaper, 2007 |
| NR15996 | NR10836, but ΔmoaE::kan | Kozmin and Schaaper, 2007 |
| NR16019 | NR10836, but ΔyiiM ΔmoaE::kan | NR15871 × P1/NR15996 |
| NR16021 | NR10836, but ycbX::mini-Tn10cam ΔmoaE::kan | NR15873 × P1/NR15996 |
| NR16023 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔmoaE::kan | NR15876 × P1/NR15996 |
| NR16028 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔmobA::kan | NR15876 × P1/NR15994 |
| NR16047 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔbisC::kan | This work |
| NR16048 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔdmsA::kan | This work |
| NR16049 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔtorA::tet | This work |
| NR16050 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔtorZ::kan | This work |
| NR16051 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔnarG::tet | This work |
| NR16052 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔnarZ::kan | This work |
| NR16053 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔnapA::kan | This work |
| NR16054 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔfdnG::kan | This work |
| NR16055 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔfdhF::kan | This work |
| NR16056 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔfdoG::tet | This work |
| NR16057 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔynfE::kan | This work |
| NR16058 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔynfF::kan | This work |
| NR16060 | NR10836, but ΔyiiM ycbX::mini-Tn10cam ΔydeP::kan | This work |
| NR16258 | NR10836, but ΔycbX[Δ(Ferredoxin)] | This work |
| NR16259 | NR10836, but ΔycbX[Δ(MOSC)] | This work |
| NR16261 | NR10836, but ΔycbX[Δ(β-barrel)] | This work |
| NR16262 | NR10836, but ΔycbX[Δ(β-barrel, MOSC, Ferredoxin)] | This work |
| NR17070 | NR15967, but λ(DE3) ΔmobA ΔguaB::tet | This work |
| NR17072 | NR17070, but ΔmoaE::kan | NR17070 × P1/NR15996 |
| NR17073 | NR17070, but ycbX::mini-Tn10cam | NR17070 × P1/NR15873 |
| NR17074 | NR17070, but ΔyiiM::kan | NR17070 × P1/NR15870 |
| NR17075 | NR17070, but ΔyiiM::kan ycbX::mini-Tn10cam | NR17074 × P1/NR15873 |
| NR17315 | BW25113, but ΔycbX::kan | This work |
| NR17326 | NR10836, but Δadd Δade::cam | This work |
| NR17330 | NR17326, but ΔyiiM | NR17070 × P1/NR15870 and elimination of kan |
| NR17485 | NR17326, but ΔmoaE::kan | NR17326 × P1/NR15996 |
| NR17486 | NR17326, but ΔycbX::kan | NR17326 × P1/NR17315 |
| NR17487 | NR17326, but ΔyiiM ΔycbX::kan | NR17330 × P1/NR17315 |
Details about the various strain constructions can be found in Experimental procedures.
Chlr and Chls indicates chlorate resistance and sensitivity, respectively. The ΔycbX::kan present in NR17315 is the complete deletion ΔycbXΔ(β-barrel, MOSC, Ferredoxin)::kan.
Base-analog sensitivity of ycbX and yiiM strains is not due to lack of MoCo sulfuration
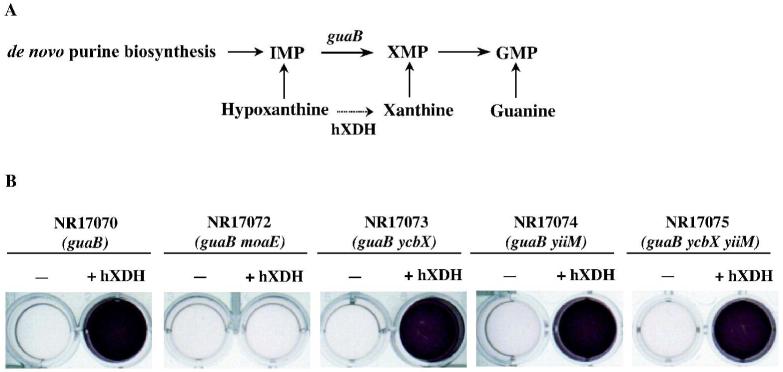
While ycbX and yiiM are not required for MoCo biosynthesis, it cannot be excluded that the genes are involved in some form of MoCo modification specific for the base-analog detoxification. For example, since ycbX and yiiM share the MOSC domain with the eukaryotic MoCo sulfurases involved in maturation (sulfuration) of MoCo used by members of the xanthine oxidase family (Kisker et al., 1999; Hille, 2002), the possibility of a role of YcbX and YiiM in MoCo sulfuration in E. coli was considered. To test this hypothesis, we developed an in vivo assay for the activity of human xanthine dehydrogenase (hXDH) expressed in E. coli. A cDNA clone of hXDH was prepared as described in Experimental procedures. The test for activity was based on the inability of E. coli guaB (guanine or xanthine) auxotrophs to utilize hypoxanthine as a GMP source (see Fig. 8A), consistent with the reported absence of physiologically significant XDH activity in E. coli (Xi et al., 2000). However, conversion of hypoxanthine to xanthine by hXDH may overcome this deficiency. The results in Fig. 8B show that expression of hXDH indeed permitted growth of the guaB strain on hypoxanthine (purple color indicative of active respiration; see Legend for details). Allopurinol, a specific inhibitor of xanthine dehydrogenase (Elion, 1989) prevented the use of hypoxanthine (data not shown). In the MoCo-deficient moaE strain no hypoxanthine utilization was detected, confirming the dependence of XDH on MoCo (Fig. 8B). Importantly, neither the single ycbX or yiiM deficiencies nor the combined ycbX yiiM deficiency inhibited this activity (Fig. 8B). In view of the well-documented dependence of xanthine dehydrogenase on MoCo sulfuration (see Mendel and Bittner, 2006), we conclude that, while sulfuration appears to occur in E. coli, the ycbX and yiiM genes do not play a significant role in this maturation process.
Fig. 8.
In vivo assay for human xanthine dehydrogenase (hXDH) activity in E. coli. The cDNA encoding xanthine dehydrogenase was cloned on a pET3a vector, as described in Experimental procedures and tested for its activity in E. coli cells. Panel A shows the purine salvage pathways from hypoxanthine, xanthine, and guanine, leading to production of GMP. It also indicates the blockage for usage of hypoxanthine by the guaB mutation. The guaB block may be relieved by conversion of hypoxanthine to xanthine by hXDH. Panel B shows the results of the experiment. Purple color indicates usage of and growth (respiration) on hypoxanthine as purine source. See text and Experimental procedures for more details.
HAP-detoxification by conversion to adenine
As part of our efforts to elucidate the HAP-detoxification pathways, we further addressed the reaction by which HAP might be converted to less harmful compounds. One plausible reaction for this conversion is reduction to adenine. Reduction of HAP to adenine was previously demonstrated in vitro by mammalian xanthine oxidase (Clement and Kunze, 1992). While our experiments show that xanthine dehydrogenase is not involved in this reaction in E. coli, it is reasonable to assume that other MoCo-dependent activities could perform this reaction as well. Secondly, studies in our laboratory with E. coli purine auxotrophs (pur strains) have indicated that these strains can use HAP as a purine source in a manner suggestive of prior conversion to adenine (data not shown). We tested this hypothesis by following the fate of HAP (and adenine) in E. coli cell free extracts (see Experimental procedures). Initial experiments indicated in these extracts that both HAP and adenine were efficiently deaminated to yield hypoxanthine. We therefore constructed strains lacking the genes for both adenosine deaminase (add) and (cryptic) adenine deaminase (ade, previously called yicP) (Zalkin and Nygaard, 1996; Matsui et al., 2001). The outcome of the reactions was monitored by thin-layer chromatography (TLC) or UV spectroscopy (exploiting the differential λmax values for HAP, adenine, and hypoxanthine, 268, 260, and 250 nm, respectively) (See experimental procedures). An example of the UV experiments is presented in Fig. 10. The Δadd Δade cell extracts are capable of converting 1 mM HAP completely to adenine within 40 min of incubation at 30° C (Fig. 10B, E). Importantly, under the same conditions, no HAP conversion was observed in the corresponding moa or ycbX yiiM cell extracts (Fig. 10C, D, E), while the single ycbX and yiiM extracts provided reduced activities (Fig. 10E). Thin-layer chromatography results fully confirmed these findings (not shown). Thus, these results (i) support the contention that the major detoxification reaction for HAP is reduction to adenine, and (ii) that this reaction requires participation of MoCo, YcbX, and YiiM, as suggested by our genetic experiments.
Fig. 10.
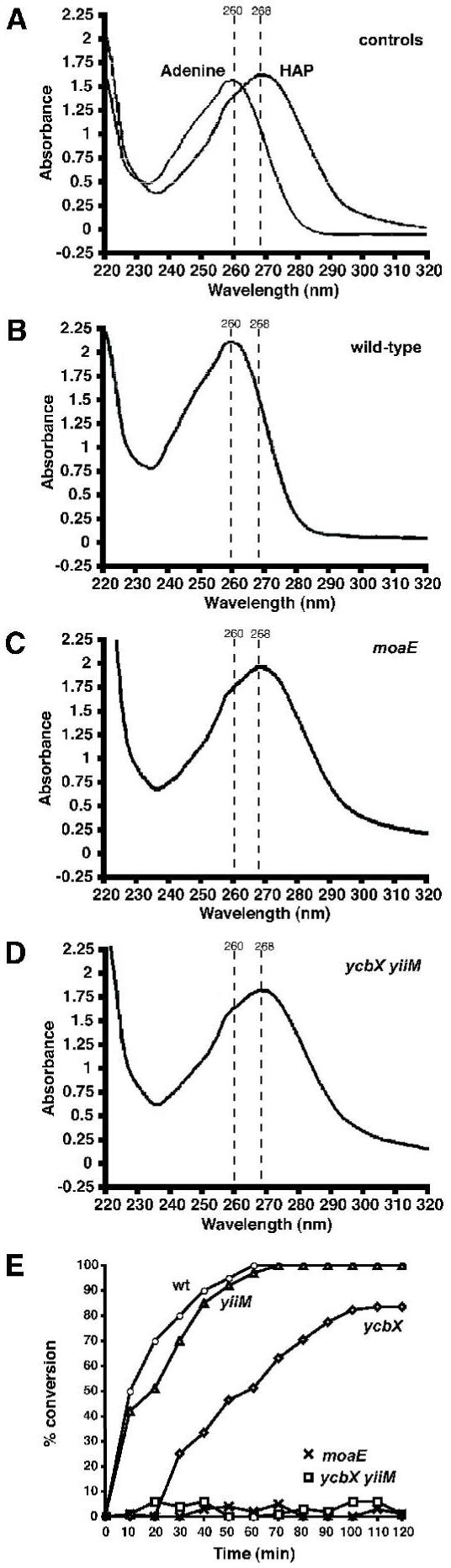
HAP conversion to adenine by E. coli cell-free extracts. HAP (1 mM) was incubated in extracts prepared from various deamination-deficient (add ade) E. coli, and the products were analyzed by UV spectra. Panel A shows zero-time controls for either added HAP or adenine, indicating the different peak wavelengths at which the compounds can be detected. Panels B, C, and D show the obtained products after 2 hr incubation of HAP in extracts derived from wt, moaE, and ycbX yiiM strains, respectively. Panel E shows the time course of conversion of HAP to adenine in the various extracts. See Experimental procedures for details regarding details of extract preparation and sample analysis.
Discussion
N-hydroxylated derivatives of the normal purine or pyrimidine bases, such as 6-N-hydroxyaminopurine (HAP) or N4-hydroxycytidine, are potent mutagens in organisms from bacteria to man. At the same time organisms possess, at least as revealed in yeast and bacteria, efficient mechanisms to prevent the toxic and mutagenic effects of these agents. In E. coli, this protective system requires the molybdenum cofactor (MoCo). The system is remarkably effective as can be estimated based on observed survival and mutagenesis data: when growing overnight in minimal media, moa or moe strains are effectively killed by concentrations of around 0.01 μg/ml HAP, while a wild-type strain readily sustains growth in media containing up to 100 μg/ml (data not shown). In the same experiments, high levels of mutations are obtained for MoCo-deficient strains at 0.001 μg/ml of HAP, while wild-type strains require at least 1 μg/ml for similar mutagenesis levels. Thus, the MoCo-dependent system is capable of reducing the sensitivity of E. coli by 1,000- to 10,000-fold. Our present study establishes a genetic framework delineating the factors involved in this detoxification mechanism and, secondly, provides evidence that the newly discovered YcbX/YiiM pathways operate through an enzymatic reduction of HAP to adenine.
The genetic framework as outlined in Fig. 9 emphasizes as particular feature that the two main MoCo-dependent pathways are dependent on the MPT form of MoCo rather than the common MGD form found in E. coli and related bacteria. This dependence on MPT, combined with our observation that none of the known and putative molybdoenzymes of E. coli significantly affect base-analog resistance (Kozmin and Schaaper 2007), suggests that resistance is mediated by a novel group of smolybdoenzymes. In the simplest case, the YcbX and YiiM proteins themselves represent these novel molybdoactivities. The two proteins are characterized by possession of a MOSC domain (Amrani et al., 2000; Anantharaman and Aravind, 2002) first recognized in eukaryotic MoCo sulfurases. While the MOSC domain appears to be essential for in vivo MoCo sulfuration (Dai et al., 2005; Sagi et al., 2002; Peretz et al., 2007), its precise function is not known. Both a sulfur-carrier and a MoCo hosting function have been considered (Amrani et al., 2000; Anatharaman and Aravind, 2002). Assuming that YcbX and YiiM represent the novel molybdoenzymes, the MOSC domain is a likely site for hosting MoCo and acting as catalytic center.
The above model is fully consistent with important recent findings by Havemeyer et al. (2006) in a mitochondrial system. These investigators studied a mitochondrial prodrug converting system and discovered a novel activity, operating in conjunction with cytochrome b5 and NADH:cytochrome b5 reductase, capable of converting the prodrug benzamidoxime to its active form benzamidine. This conversion is analogous to the conversion of HAP to adenine, as it entails the reduction of an N-hydroxyl compound (benzamidoxime) to the corresponding amino form (benzamidine). Most interestingly, the newly isolated 35 kDa protein was shown to possess a MOSC domain and to be able to bind MoCo. Thus, it appears that a hitherto undescribed but apparently broadly-distributed class of molybdoenzymes exists, as judged from their discovery in both bacteria and mammalian cells. The precise function(s) of these enzymes remain to be determined, and may well extend beyond the examples provided by these current cases.
The scheme of Fig. 9 also shows the detoxification step mediated by the MGD-requiring enzyme biotin sulfoxide reductase (BisC). The function described so far for E. coli BisC is the reduction of biotin sulfoxide (del Campillo-Campbell and Campbell, 1982; Pierson and Campbell, 1990) or methionine sulfoxide (Ezraty et al., 2005). Our present result suggests that its specificity may be broader, as also reported for the biotin sulfoxide reductase from Rhodobacter (Pollock and Barber, 1997). Nevertheless, the action of BisC in the HAP detoxification is only minor, as its effect was only discerned in the ycbX yiiM background and, also, its activity was not observed in the in vitro experiments of Fig. 10.
Of interest with regard to the scheme of Fig. 9 are the different specificities that appear associated with each of the subpathways. While moa or moe strains display increased sensitivity to all three agents (HAP, AHAP, and hydroxylamine), the ycbX pathway is most effective towards HAP, while the yiiM pathway is most effective for AHAP and hydroxylamine (Figs. 1, 3, 4). Hydroxylamine, while not a base analog, represents the simplest N-hydroxylated compound and may be a useful model compound for further studies. Reduction of hydroxylamine in E. coli has been reported mediated by the hybrid cluster protein (Wolfe et al., 2002) or by cytoplasmic cytochrome c nitrite reductase (Angove et al., 2002), neither of which are molybdoenzymes. Thus, a novel pathway for dissimilation of hydroxylamine has been discovered. The specificity differences of Fig. 9 may reflect differences in substrate preference for, say, YcbX vs. YiiM protein, but may also reflect different stages in nucleotide metabolism at which the proteins may act. This will be an important area for further studies.
As noted in the Results, our data leave open the possibility that the ycbX and yiiM genes play additional roles in base-analog detoxification outside the MoCo-dependent pathways. For example, in Fig. 5, the moaE yiiM strain was more HAP-sensitive than the single moaE mutant, while the triple moaE yiiM ycbX was more sensitive than the double moaE yiiM, suggesting that both ycbX and yiiM might have additional function(s). On the other hand, the moaE ycbX double was as sensitive as the single moaE mutant, arguing against an additional function for ycbX. These issues will require further study, and both direct and indirect effects may be operative. However, from the present perspective the most important finding is that the triple ycbX yiiM mobA or ycbX yiiM bisC mutants are as sensitive as the ycbX yiiM moaE strain (as seen in Figs. 1, 3, and 5). This result indicates that after inactivation of BisC (via bisC or mobA) the yiiM and ycbX deficiencies completely eliminate the moa-dependent base-analog resistance.
The conversion of HAP to adenine as demonstrated in cell-free extracts (Fig. 10) is consistent with other genetic data from our laboratory on the usage of HAP as a purine source by E. coli purine auxotrophs (data to be published elsewhere). In brief, pur add strains defective in both de novo purine biosynthesis (pur) and adenosine deaminase (add) are capable of using adenine as a purine source based on conversion of adenine to AMP by Apt activity (adenine phosphoribosyl transferase) (Zalkin and Nygaard, 1996). This strain can also use HAP as purine source, and this usage, like that of adenine, is blocked by an additional apt defect. The simplest interpretation is that, in this kind of experiment, HAP is first converted to adenine by the YcbX/YiiM system prior to its usage in purine salvage. Note that it is not possible to demonstrate the absence of this type of HAP usage in MoCo (or ycbX/yiiM) deficient cells, because these cells are effectively killed by the purine concentrations necessary for bacterial growth (10-50 μg/ml). An interesting parallel to this reductive usage of HAP are the early studies by Popowska and Janion (1975, 1977). Using toluenized E. coli cells these investigators concluded that the base analog N4-hydroxycytidine could be used as pyrimidine source via reduction to cytidine by an unknown system. Thus, it appears that an N-reductive reaction is a current hallmark of the activity of the MOSC systems in both bacteria and higher organisms.
It is an interesting question why E. coli would possess a defensive system against N-hydroxylated base analogs. It has been suggested that HAP can be produced occasionally as a byproduct of normal cellular metabolism (Lieberman, 1956; Budowski, 1976; Clement and Kunze, 1990). If so, its efficient removal from the cell would be desirable. On the other hand, MoCo-deficient strains are not spontaneous mutators, at least under the conditions that we have tested. It is possible that increased amounts of N-hydroxylated compounds will be formed under more stressful situations, such as oxidative stress conditions; a mutator effect of MoCo-deficient strains might be observed under those conditions. We note that exposure of DNA and nucleobases to peroxyl radicals was reported to yielded HAP as the major product (Simandan et al., 1998). Alternatively, as the MoCo-dependent system converts the hydroxylated bases to the normal bases, the system might primarily have a salvage or scavenging function. Also, as inferred perhaps from the mitochondrial data of Havemeyer et al. (2006), the bacterial system may be geared towards a broader range of N-hydroxylated compounds. N-hydroxylated compounds are intermediates in both the oxidation of aromatic amines (by cytochrome P450s) and the reduction of aromatic nitro compounds (by nitroreductases) These N-hydroxylated derivatives are unstable and highly reactive and can produce DNA adducts (Rosenkranz and Mermelstein, 1983).
The present study and that of Havemeyer et al. (2006) suggest that MOSC-containing proteins represent a novel class of molybdoenzymes with similar biochemical properties in bacteria and mammals. The precise biological function of these molybdoenzymes is not yet known. However, an indispensable role of these proteins in base-analog detoxification in bacteria is clear. It is possible that these molybdoenzymes play a role in protection against mutagenic base analogs and related carcinogens in humans as well, providing an incentive for further research in this area.
Experimental procedures
Bacterial strains and constructions
The E. coli strains used in this study are listed in Table 1 along with their source or derivation. P1 transductions were performed using P1virA. All mutagenesis and base-analog sensitivity tests were performed using strain NR10836 (Kozmin et al., 2000) and its mutant derivatives. The ycbX::mini-Tn10cam allele was obtained initially in strain KA796 upon random transposon insertion mutagenesis, as described in a section below, and then transferred to NR10836 by P1 transduction, yielding strain NR15873. Construction of the ycbX in-frame deletions (NR16258, NR16259, NR16261, and NR16262) is described in a separate section below. A deletion of yiiM was initially generated in strain BW25113 (Datsenko and Wanner, 2000) by the PCR-based gene replacement method, using the Kanr module of plasmid pKD13 as a template (Datsenko and Wanner, 2000). Primers for the PCR were: yiiM-p1: 5′-ttg atg tat aca cag gca aga ttc agg ctt atc ccg aag gca aac cca gcg tgt agg ctg gag ctg ctt c-3′ and yiiM-p4: 5′-taa ccc ggc agc gga gag taa acg gtg ata ttg atc gtc atc aaa cgg cac tgt caa aca tga gaa tta a-3′. The resulting ΔyiiM::kan deletion-insertion was transferred into strain NR10836 by P1 transduction using P1virA, yielding strain NR15870. To obtain strain NR15871 (ΔyiiM), the Kanr insert in yiiM was eliminated by induction of recombination between repeats flanking the kan gene, using plasmid pCP20 (Datsenko and Wanner, 2000). NR10836 derivatives carrying deletions of known and putative molybdoenzymes (ΔbisC::kan, ΔdmsA::kan, ΔtorA::tet, ΔtorZ::kan, ΔnarG::tet, ΔnarZ::kan, ΔnapA::kan, ΔfdnG::kan, ΔfdhF::kan, ΔfdoG::tet, ΔynfE::kan, ΔynfF::kan, ΔydeP::kan) have been described (Kozmin and Schaaper, 2007). These deletions were transferred into strain NR15876 (ycbX::mini-Tn10cam, ΔyiiM) by P1 transduction yielding strains NR16047 through NR16060 (Table 1). Activity of the human xanthine dehydrogenase (see below) was tested in a modified version of strain BL21(DE3) (Table 1) constructed as follows. First, we noted that strain BL21(DE3) (Studier and Moffat, 1986) is chlorate-resistant and HAP-sensitive, suggesting a defect in MoCo biosynthesis (Kozmin et al., 2000). On the assumption that this phenotype might be associated with the gal defect in BL21 (noting that gal is located very close to the modABC and modEF operons (Berlyn, 1998), which are important for molybdenum transport and cofactor biosynthesis) (Shanmugam et al., 1992; Kozmin et al., 2000), we created a gal+ derivative of BL21 by P1 transduction. The gal+ derivative NR15967 proved chlorate-sensitive and HAP-resistant, consistent with our hypothesis. NR15967 was then lysogenized with bacteriophage λDE3 using the lysogenization kit from Novagen, followed by introduction of the ΔmobA::kan allele from strain NR15994 and elimination of the kan insert using plasmid pCP20 (Datsenko and Wanner, 2000). Finally, to yield the strain NR17070, we introduced the ΔguaB::tet allele by P1 transduction. The ΔguaB::tet allele was created in strain BW25113 by the PCR-based gene replacement method of Datsenko and Wanner (2000) using primers guaB-p1T 5′-aaa cag ata ata taa atc gcc cga cat gaa gtc ggg cga aga gaa tca ggA AGA GGG TCA TTA TAT TTC G-3′ and guaB-p4T 5′-taa tgc cgc ggc aat att tat taa cca ctc tgg tcg aga tat tgc cca tgA CTC GAC ATC TTG GTT ACC G-3′ (upper case letters indicate the sequences used to amplify the 2-kb Tetr module containing the tetR and tetA genes from transposon Tn10). The Δadd::kan and ΔyicP::cam (Δade::cam) deletion/insertions were created in strain BW25113 by the method of Datsenko and Wanner (2000) using primers add-p1 5′-ttg atg gca aca ttc gtc ccc aga cca ttc ttg aac ttg gcc gcc agt atg tgt agg ctg gag ctg ctt c-3′ plus add-p4 5′-gtt aat gct ggc acg aat gcc atg ctc aag gaa cgt ttt cag cgg atg tgc tgt caa aca tga gaa tta a-3′ (using pKD13 as a template), and yicP-Cm1 5′-ttc ttt atg ata aat aat aat caa att gat aaa atc aaa atg aga aaa atgtg tag gct gga gct gct tcg-3′ plus yicP-Cm2 5′-tgg cat aaa cgt aac tgg tga ctt ttg ccc ggc atg acg ccg ggc ttt ttcat atg aat atc ctc ctt ag-3′ (using pKD3 as a template), respectively. The Δadd::kan and ΔyicP::cam alleles were then transferred into the NR10836 background by P1 transduction (see Table 1).
Media and chemicals
Bacteria were cultivated in LB broth (Miller, 1972) or minimal Vogel-Bonner medium (VB) (Vogel and Bonner, 1956) containing 0.2% glucose as carbon source. Solid media contained 1.5% agar. For selection of antibiotic-resistant clones, LB medium was used containing 35 μg/ml of kanamycin, 20 μg/ml of chloramphenicol, 15 μg/ml of tetracycline, or 100 μg/ml of rifampicin. MacConkey (MC) indicator plates for monitoring nitrate reduction or trimethylamine N-oxide reduction were prepared according to Stewart and MacGregor (1982). HAP was purchased from ICN Biochemicals; 2-amino-HAP was synthesized from 2-amino-6-chloropurine according to the method of Giner-Sorolla and Bendich (1958) by Ilya Kuchuk, Indiana University. HAP and AHAP were used as the free base. Hydroxylamine hydrochloride and other chemicals were purchased from Sigma-Aldrich.
Isolation and identification of HAP-hypersensitive mutants by random transposon insertion mutagenesis
A library of random transposon insertions was obtained in strain KA796 using the mini-Tn10cam transposon from phage λNK1324 as described by Kleckner et al. (1991). Chloramphenicol-resistant clones were restreaked on minimal VB plates containing 0 or 10 μg/ml of HAP to search for HAP-sensitive mutants. Genomic DNA from HAP-sensitive clones was isolated using Easy-DNA kit (Invitrogen). To determine the location of transposon insertions we used arbitrary primed PCR (Caetano-Anolles, 1993). Briefly, a first round of PCR was performed on genomic DNA of each mutant using three primers: two primers with random 3′ ends, ARB1 (5′-ggc cac gcg tcg act agt acN NNN NNN NNN gat at-3′) and ARB6 (5′-ggc cac gcg tcg act agt acN NNN NNN NNN acg cc-3′), and a third primer complementary to the mini-Tn10cam sequence, CmExt (5′-cag gct ctc ccc gtg gag g-3′). A second round of PCR was then performed with primers ARB2 (5′-ggc cac gcg tcg act agt ac-3′) and CmInt (5′-ctg cct ccc aga gcc tg-3′). The final PCR product was purified with a QIAquick spin PCR purification kit (Qiagen) and sequenced using ARB2 primer and the dRhodamine terminator cycle DNA sequencing kit (PE Biosystems, Warrington, UK).
Construction of the ycbX in-frame deletion alleles
YcbX protein contains three predicted domain, β-barrel, MOSC, and ferredoxin (see Text). To create individual in-frame deletions of each of these domains, we used the PCR-based gene replacement method of Datsenko and Wanner (2000). First, we generated kanamycin-resistant deletion-insertions for each domain in strain BW25113. These kanamycin-resistant alleles were transferred into strain NR10836 by P1 transduction. The kan modules were then eliminated by induction of recombination between the repeats flanking the kan gene using plasmid pCP20 (Datsenko and Wanner, 2000). The resulting in-frame deletions were verified by DNA sequencing. As shown in Fig. 6, after elimination of the Kanr module, the in-frame deletion alleles encode reduced versions of the YcbX protein containing small (27- to 30-residue) polypeptide in place of the deleted domain (see Legend to Fig. 6). To generate the initial deletion-insertion alleles, plasmid pKD13 was used as a template (Datsenko and Wanner, 2000) and the following primer pairs (upper case letters indicate pKD13 sequence): for ΔycbX[Δβ-barrel]: YcbX-ΔN-p1 (5′-gcg ttt cac gcg ccg ggt cat ttg tgg ccc cac cca gcg taa ttg cac tt GTG TAG GCT GGA GCT GCT TCG-3′) and YcbX-ΔN-p4 (5′-cgt cat gtc ata aag ttg agg gct tat ttt cat ttg agg acc gca ccg tg ATT CCG GGG ATC CGT CGA CC-3′); for ΔycbX[ΔMOSC]: YcbX-ΔMOSC-p1 (5′-atc atc agc ggc agc tgc gcc gta aat ttt tgc cgg agc cgt tgc cag aa GTG TAG GCT GGA GCT GCT TCG-3′) and YcbX-MOSC-p4 (5′-ccc gcg aag tgc aat tac gct ggg tgg ggc cac aaa tga ccc ggc gcg tg ATT CCG GGG ATC CGT CGA CC-3′); and for ΔycbX[ΔFerredoxin]: YcbX-ΔFer-p1 (5′-gaa acc gca ggt taa tgt tga cag ctt cag cct cga aca ggc agt cta ac GTG TAG GCT GGA GCT GCT TCG-3′) and YcbX-Fer-p4 (5′-ctg ccg ctg atg ata ccg cca aca tca cgc aac aac cgg acg caa atg ta ATT CCG GGG ATC CGT CGA CC-3′). To obtain a complete deletion of ycbX (denoted as ΔycbX in Table 1), we used primers YcbX-ΔN-p1 and YcbX-Fer-p4. After elimination of the Kanr modules using plasmid pCP20 (Datsenko and Wanner, 2000), the deletion alleles were confirmed by DNA sequencing via PCR amplification using primers ycbX-seqA (5′-aac cct gcg ttg tta ttg cg-3′) and ycbX-seqB (5′-tga gga ttt cga gca taa cc-3′). Sequencing was done both directions using the same primers, ycbX-seqA and ycbX-seqB.
Spot-tests for base-analog sensitivity
Stationary E. coli cultures grown in LB were diluted 30-fold in 10 ml of 0.9% NaCl in Petri dishes and transferred to VB minimal plates using a multi-prong replicator device (approximately 0.1 ml total per plate). After the spots had dried, a few microliters of a solution of HAP or AHAP (50 μg in DMSO) or hydroxylamine (150 μg in water) were spotted onto the center of the plate. The pH of the hydroxylamine·HCl solution was ∼3.4. Identical results were obtained with either this solution or solutions neutralized to pH 7.0. The plates were incubated overnight at 37° C and inspected the next day for zones of inhibition. In some experiments, the plates were further replicated to LB plates containing rifampicin, incubated overnight at 37° C, and inspected for the induction of Rifr colonies surrounding the place of mutagen placement.
Tests for growth inhibition by base analogs in liquid cultures
A single colony of each strain to be tested was inoculated into 5 ml of liquid LB and grown overnight at 37° C. 80 μl of each culture was inoculated into 40 ml of minimal VB medium (supplemented with 1 μg/ml of thiamine) in 125-ml flasks, and incubated for 6-7 hrs at 37° C at 200 rpm with shaking to reach mid-log-phase (OD600 = 0.2, ∼ 108 cells/ml). At this point, for each strain and each concentration of HAP to be tested, three 1-ml aliquots were transferred into 13×100 mm glass culture tubes. An appropriate amount of HAP water solution (0.1 mg/ml) was added to each tube. Cultures were incubated for 2 hrs at 37° C on a rotating drum at 56 rpm and then placed on ice. The cultures were diluted 104-fold in 0.9% NaCl and appropriate volumes of the dilution were plated on LB plates in triplicate to count survivors. Plates were incubated 20-24 h at 37°C. The fraction of surviving cells was determined by dividing the average number of cells in HAP-exposed cultures divided by the average number in the non-exposed cultures. For each strain, this test was repeated 2-3 times, and average survival values were used to obtain survival curves (Fig. 2).
Mutant frequency determinations
For each strain and each HAP concentration, 10 independent 1-ml LB cultures were started from 104 cells. The cultures were grown overnight at 37° C with agitation. Mutant frequencies were determined by plating appropriate culture volumes on LB-rifampicin plates (to obtain the number of Rifr cells per culture) and 100 μl of 10-6 dilution on LB plates (to obtain the total number of cells per culture).
Tests for chlorate resistance and nitrate or TMAO reduction
To test for chlorate resistance, approximately 103 cells were plated on LB plates containing either 0.5 mM or 25 mM KClO3. The plates were incubated overnight at 37° C under anaerobic conditions using a Becton Dickinson BBL gas pack anaerobic system, after which they were incubated aerobically for an additional 6-10 hrs. Under these conditions, chlorate-sensitive strains do not form colonies, whereas chlorate-resistant strains plate with essentially 100% efficiency. To monitor the ability of cells to reduce nitrate or trimethylamine-N-oxide (TMAO), we plated approximately 102 cells on MC indicator plates supplemented with either 0.1 M KNO3 or 0.1 M TMAO. The plates were incubated aerobically overnight at 37° C and inspected for red pigmentation of the colonies indicative of nitrate reductase or TMAO reductase activities.
Cloning of the human xanthine dehydrogenase (hXDH)
‘PCR Ready First Strand cDNA’ (BioChain, cat# C1234226-10) from human normal adult small intestine was used as source of the hXDH-encoding cDNA. Amplification was performed in three consecutive rounds using PfuTurbo® DNA polymerase (Stratagene) using conditions recommended by the supplier. In round I, we amplified separately the left and right arms (2 kb each) of the hXDH-encoding cDNA using two primers pairs: hXDH-NdeI (5′-atc cta cca tat gac agc aga caa att gg-3′) plus hXDH-int2 (5′-tat gcc caa cac aag taa cc-3′), and hXDH-BglII (5′-ata aag atc ttt aga ccc tca cag acc agg-3′) plus hXDH-int1 (5′-atg atg aga cag tct ttg cg-3′). The int1 and int2 primers were chosen such as to generate an internal 3′ overlap between the two arms. In round II, the entire amounts of purified PCR products from round I were mixed together and further amplified in the presence of the two external primers, hXDH-NdeI and hXDH-BglII. Finally, to obtain sufficiently large amounts of the final full-length (4-kb) product, in round III 1/50 volume of reaction II was used as a template for amplification with primers hXDH-NdeI and hXDH-BglII. The resulting 4-kb PCR product corresponding to the hXDH-encoding DNA was gel-purified and restricted with NdeI and BglII restriction endonucleases. The resulting fragment was cloned into the BamHI and NdeI sites of the pET3a expression vector (Novagen) to yield plasmid pET3a-hXDH, in which the hXDH coding sequence is placed under control of the T7 promoter. The final construct was verified by DNA sequencing.
In vivo test for the hXDH activity using tetrazolium violet indicator
In this test we monitored the ability of guaB strains expressing hXDH to utilize hypoxanthine as a GMP source (see Fig. 8A) in the presence of the tetrazolium violet (TV) indicator dye. In growing actively respiring cultures TV (a colorless substance) is efficiently reduced to a deep-purple insoluble formazan derivative, whereas in starving cultures this reduction does not take place (Bochner and Savageau, 1977; Bochner et al., 2001). Thus, strains NR17070 and NR17072 through NR17075 (Table 1) were transformed with plasmids pET3a (control) and pET3a-hXDH. A single colony of each transformant to be tested was inoculated into 6 ml of LB supplemented with 200 μg/ml of ampicillin, and incubated overnight at 37°C with agitation. Cells from these cultures were collected by centrifugation and resuspended in 20 ml of minimal VB media containing 100 μg/ml of ampicillin, 40 μg/ml of guanosine, 1 mM sodium molybdate, 0.02 mM IPTG, and incubated for 24 hrs at 16° C with agitation. After that, cells were washed and resuspended in 2 ml of indicator media (minimal VB medium containing 0.01% TV, 0.02 mM IPTG, and 40 μg/ml of either hypoxanthine or xanthine) at a density OD600 = 0.1, and incubated for 2-3 days at room temperature without shaking and inspected for purple pigment precipitation.
Conversion of HAP to adenine in cell-free extracts
Cell-free extracts were prepared according to Lu et al. (1983). 500-ml LB cultures were grown to OD600 = 1.0-1.2. Cells were collected by centrifugation, and each gram of cells was resuspended in 1.5 ml of 50 mM Tris-HCl (pH 7.6) containing 10% sucrose. Cell suspensions were quickly frozen in a dry ice/ethanol bath. For thawing, the frozen suspensions were placed at 0°C for 20 min and then in a 20° C waterbath until just melted. DTT, KCl, and lysozyme were added to final concentrations of 1.2 mM, 150 mM, and 0.23 mg/ml, respectively. incubation was continued for 1 h at 0° C followed by placement in a 37° C bath until the temperature of the solution reached 20° C. Cell debris was removed by centrifugation for 1 h at 37,000 rpm at 4° C. Supernatants (100 μl-aliquots) were stored at -80° C. Total protein concentration in the extracts was determined using the Non-Interfering Protein Assay Kit from Calbiochem. Extracts prepared by this procedure contained 6-9 mg of protein per ml. To assay for activity, 2 μl of a 100 mM HAP solution in DMSO or 2 μl of DMSO (control reaction) was added to 200 μl of cell-free extract, and the reaction mixtures were incubated at 30° C. Every ten minutes, 20-μl samples were removed and heat-inactivated for 2 min at 80° C. Samples were chilled on ice, and centrifuged for 5 minutes at 14,000 rpm in a microcentrifuge. 15-μl supernatant samples were removed and used for further analysis. The reaction products were monitored by UV spectrometry and thin-layer chromatography (TLC). The UV spectrometric assay was based on the differences in UV spectra of HAP (λmax = 268 nm), adenine (λmax = 260 nm), and hypoxanthine (λmax = 250 nm). Samples were diluted tenfold in 10 mM Tris-HCl (pH 8.0), and UV spectra were recorded on a ND-1000 spectrometer (NanoDrop Technologies) using a ten-fold diluted control sample (no HAP) as a reference. The percent of conversion of HAP to adenine was calculated as (A260/A268 - 0.8545)/0.0042, the constant values determined with pure solutions containing mixtures of HAP and adenine in various ratios. For TLC, 5 μl of undiluted samples were loaded on Cellulose F plates (Merck). Distilled water was used as a solvent. The spots were visualized with a UV lamp. In these conditions, Rf values for adenine and HAP were 0.23 and 0.29, respectively.
Acknowledgements
We thank Drs. W. Copeland and J. Wang of the NIEHS for their critical reading of the manuscript for this paper. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
References
- Amrani L, Primus J, Glatigny A, Arcangeli L, Scazzocchio C, Finnerty V. Comparison of the sequences of the Aspergillus nidulans hxB and Drosophila melanogaster ma-l genes with nifS from Azotobacter vinelandii suggests a mechanism for the insertion of the terminal sulphur atom in the molybdopterin cofactor. Mol Microbiol. 2000;38:114–125. doi: 10.1046/j.1365-2958.2000.02119.x. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. MOSC domains: ancient, predicted sulfur-carrier domains, present in diverse metal-sulfur cluster biosynthesis proteins including Molybdenum cofactor sulfurases. FEMS Microbiol Lett. 2002;207:55–61. doi: 10.1111/j.1574-6968.2002.tb11028.x. [DOI] [PubMed] [Google Scholar]
- Angove HC, Cole JA, Richardson DJ, Butt JN. Protein film voltammetry reveals distinctive fingerprints of nitrite and hydroxylamine reduction by a cytochrome c nitrite reductase. J Biol Chem. 2002;277:23374–23381. doi: 10.1074/jbc.M200495200. [DOI] [PubMed] [Google Scholar]
- Barrett JC. Induction of gene mutation in and cell transformation of mammalian cells by modified purines: 2-aminopurine and 6-N-hydroxylaminopurine. Proc Natl Acad Sci USA. 1981;78:5685–5689. doi: 10.1073/pnas.78.9.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn MKB. Linkage map of Escherichia coli K-12, Edition 10: the traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Savageau MA. Generalized indicator plate for genetic, metabolic, and taxonomic studies with microorganisms. Appl Environ Microbiol. 1977;33:434–444. doi: 10.1128/aem.33.2.434-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budowski EI. The mechanisms of the mutagenic action of hydroxylamine. Prog Nucleic Acid Res Mol Biol. 1976;16:125–182. doi: 10.1016/s0079-6603(08)60757-6. [DOI] [PubMed] [Google Scholar]
- Burgis NE, Brucker JJ, Cunningham RP. Repair system for noncanonical purines in Escherichia coli. J Bacteriol. 2003;185:3101–3110. doi: 10.1128/JB.185.10.3101-3110.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgiss NE, Cunningham RP. Substrate specificity of RdgB protein, a deoxyribonucleoside triphosphate pyrophosphohydrolase. J Biol Chem. 2007;282:3531–3538. doi: 10.1074/jbc.M608708200. [DOI] [PubMed] [Google Scholar]
- Caetano-Anolles G. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 1993;3:85–94. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- Clement B, Kunze T. Hepatic microsomal N-hydroxylation of adenine to 6-N-hydroxylaminopurine. Biochem Pharmacol. 1990;39:925–933. doi: 10.1016/0006-2952(90)90209-4. [DOI] [PubMed] [Google Scholar]
- Clement B, Kunze T. The reduction of 6-N-hydroxylaminopurine to adenine by xanthine oxidase. Biochem Pharmacol. 1992;44:1501–1509. doi: 10.1016/0006-2952(92)90464-t. [DOI] [PubMed] [Google Scholar]
- Dai X, Hayashi K, Nozaki H, Cheng Y, Zhao Y. Genetic and chemical analyses of the action mechanisms of sirtinol in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:3129–3134. doi: 10.1073/pnas.0500185102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo-Campbell A, Campbell A. Molybdenum cofactor requirement for biotin sulfoxide reduction in Escherichia coli. J Bacteriol. 1982;149:469–478. doi: 10.1128/jb.149.2.469-478.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion GB. The purine path to chemotherapy. Science. 1989;244:41–47. doi: 10.1126/science.2649979. [DOI] [PubMed] [Google Scholar]
- Ezraty B, Bos J, Barras F, Aussel L. Methionine sulfoxide reduction and assimilation in Escherichia coli: new role for the biotin sulfoxide reductase BisC. J Bacteriol. 2005;187:231–237. doi: 10.1128/JB.187.1.231-237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese E. The specific mutagenic effect of base analogs on phage T4. J Mol Biol. 1959;1:87–105. [Google Scholar]
- Giner-Sorolla A, Bendich A. Synthesis and properties of some 6-substituted purines. J Am Chem Soc. 1958;80:3932–3937. [Google Scholar]
- Havemeyer A, Bittner F, Wollers S, Mendel R, Kunze T, Clement B. Identification of the missing component in the mitochondrial benzamidoxime prodrug converting system as a novel molybdenum enzyme. J Biol Chem. 2006;281:34796–34802. doi: 10.1074/jbc.M607697200. [DOI] [PubMed] [Google Scholar]
- Hille R. Molybdenum and tungsten in biology. Trends Biochem Sci. 2002;27:360–367. doi: 10.1016/s0968-0004(02)02107-2. [DOI] [PubMed] [Google Scholar]
- Hwang KY, Chung JH, Kim SH, Han YS, Cho Y. Structure-based identification of a novel NTPase from Methanococcus jannaschii. Nat Struct Biol. 1999;6:691–696. doi: 10.1038/10745. [DOI] [PubMed] [Google Scholar]
- Janion C. The efficiency and extent of mutagenic activity of some new mutagens of base-analogue type. Mutat Res. 1978;56:225–234. doi: 10.1016/0027-5107(78)90189-6. [DOI] [PubMed] [Google Scholar]
- Janion C. On the different response of Salmonella typhimurium hisG46 and TA1530 to mutagenic action of base analogues. Acta Biochim Pol. 1979;26:171–177. [PubMed] [Google Scholar]
- Janion C, Myszkowska K. Mutagenic and inhibitory properties of some new purine analogs on Salmonella typhimurium TA1530. Mutat Res. 1981;91:193–197. doi: 10.1016/0165-7992(81)90030-0. [DOI] [PubMed] [Google Scholar]
- Khromov-Borisov NN. Naming the mutagenic nucleic acid base analogs: the Galatea syndrome. Mutat Res. 1997;379:95–103. doi: 10.1016/s0027-5107(97)00112-7. [DOI] [PubMed] [Google Scholar]
- Kisker C, Schindelin H, Baas D, Retey J, Meckenstock RU, Kroneck PM. A structural comparison of molybdenum cofactor-containing enzymes. FEMS Microbiol Rev. 1999;22:503–521. doi: 10.1111/j.1574-6976.1998.tb00384.x. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- Kozmin SG, Schaaper RM, Shcherbakova PV, Kulikov VN, Noskov VN, Guetsova ML, Alenin VV, Rogozin IB, Makarova KS, Pavlov YI. Multiple antimutagenesis mechanisms affect mutagenic activity and specificity of the base analog 6-N-hydroxylaminopurine in bacteria and yeast. Mutat Res. 1998a;402:41–50. doi: 10.1016/s0027-5107(97)00280-7. [DOI] [PubMed] [Google Scholar]
- Kozmin SG, Leroy P, Pavlov YI. Overexpression of the yeast HAM1 gene prevents 6-N-hydroxylaminopurine mutagenesis in Escherichia coli. Acta Biochim Pol. 1998b;45:645–652. [PubMed] [Google Scholar]
- Kozmin SG, Pavlov YI, Dunn RL, Schaaper RM. Hypersensitivity of Escherichia coli Δ(uvrB-bio) mutants to 6-hydroxylaminopurine and other base analogs is due to a defect in molybdenum cofactor biosynthesis. J Bacteriol. 2000;182:3361–3367. doi: 10.1128/jb.182.12.3361-3367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin SG, Schaaper RM. Molybdenum cofactor-dependent resistance to N-hydroxylated base analogs in E. coli is independent of MobA function. Mutat Res. 2007;619:9–15. doi: 10.1016/j.mrfmmm.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman I. Enzymatic synthesis of adenosine-5′-phosphate from inosine-5′-phosphate. J Biol Chem. 1956;223:327–339. [PubMed] [Google Scholar]
- Lu AL, Clark S, Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci USA. 1983;80:4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Shimaoka M, Kawaski H, Takenaka Y, Kurahashi O. Adenine deaminase activity of the yicP gene product of Escherichia coli. Biosci Biotechnol Biochem. 2001;65:1112–1118. doi: 10.1271/bbb.65.1112. [DOI] [PubMed] [Google Scholar]
- Mendel RR, Bittner F. Cell biology of molybdenum. Biochim Biophys Acta. 2006;1763:621–635. doi: 10.1016/j.bbamcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1972. [Google Scholar]
- Noskov VN, Staak K, Shcherbakova PV, Kozmin SG, Negishi K, Ono B-C, Hayatsu H, Pavlov YI. HAM1, the gene controlling 6-N-hydroxylaminopurine sensitivity and mutagenesis in the yeast Saccharomyces cerevisiae. Yeast. 1996;12:17–29. doi: 10.1002/(SICI)1097-0061(199601)12:1%3C17::AID-YEA875%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Pavlov YI. Saccharomyces cerevisiae mutants highly sensitive to the mutagenic action of 6-N-hydroxylaminopurine. Genetika. 1986;22:1099–1107. [PubMed] [Google Scholar]
- Pavlov YI, Noskov VN, Lange EK, Moiseeva EV, Pshenichnov MR, Khromov-Borisov NN. The genetic activity of N6-hydroxyadenine and 2-amino-N6-hydroxyadenine in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae. Mutat Res. 1991;253:33–46. doi: 10.1016/0165-1161(91)90343-7. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Suslov VV, Shcherbakova PV, Kunkel TA, Ono A, Matsuda A, Schaaper RM. Base analog N6-hydroxylaminopurine mutagenesis in Escherichia coli: genetic control and molecular specificity. Mutat Res. 1996;357:1–15. doi: 10.1016/0027-5107(96)00060-7. [DOI] [PubMed] [Google Scholar]
- Peretz H, Naamati MS, Levartovsky D, Lagziel A, Shani E, Horn I, Shalev H, Landau D. Identification and characterization of the first mutation (Arg776Cys) in the C-terminal domain of the human molybdenum cofactor sulfurase (HMCS) associated with type II classical xanthinuria. Mol Genet Metab. 2007;91:23–29. doi: 10.1016/j.ymgme.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Pierson DE, Campbell A. Cloning and nucleotide sequence of bisC, the structural gene for biotin sulfoxide reductase in Escherichia coli. J Bacteriol. 1990;172:2194–2198. doi: 10.1128/jb.172.4.2194-2198.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock VV, Barber MJ. Biotin sulfoxide reductase. Heterologous expression and characterization of a functional molybdopterin guanine dinucleotide-containing enzyme. J Biol Chem. 1997;272:3355–3362. doi: 10.1074/jbc.272.6.3355. [DOI] [PubMed] [Google Scholar]
- Popowska E, Janion C. The metabolism of N4-hydroxycytidine - a mutagen for Salmonella typhimurium. Nucleic Acids Res. 1975;2:1143–1152. doi: 10.1093/nar/2.7.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowska E, Janion C. The N4-hydroxycytidine reduction system in toluenized cells of Salmonella typhimurium. Acta Biochim Pol. 1977;24:197–205. [PubMed] [Google Scholar]
- Rajagopalan KV. Biosynthesis of the molybdenum cofactor. In: Neidhardt FC, editor. Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology Press; Washington, DC: 1996. pp. 674–679. [Google Scholar]
- Rosenkranz HS, Mermelstein R. Mutagenicity and genotoxicity of nitroarenes. All nitro-containing chemicals were not created equal. Mutat. Res. 1983;114:217–267. doi: 10.1016/0165-1110(83)90034-9. [DOI] [PubMed] [Google Scholar]
- Sagi M, Scazzocchio C, Fluhr R. The absence of molybdenum cofactor sulfuration is the primary cause of the flacca phenotype in tomato plants. Plant J. 2002;31:305–317. doi: 10.1046/j.1365-313x.2002.01363.x. [DOI] [PubMed] [Google Scholar]
- Shanmugam KT, Stewart V, Gunsalus RP, Boxer DH, Cole JA, Chippaux M, DeMoss JA, Giordano G, Lin EC, Rajagopalan KV. Proposed nomenclature for the genes involved in molybdenum metabolism in Escherichia coli and Salmonella typhimurium. Mol Microbiol. 1992;6:3452–3454. doi: 10.1111/j.1365-2958.1992.tb02215.x. [DOI] [PubMed] [Google Scholar]
- Simandan T, Sun J, Dix TA. Oxidation of DNA bases, deoxyribonucleosides and homopolymerase by peroxyl radicals. Biochem J. 1998;335:233–240. doi: 10.1042/bj3350233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V, MacGregor CH. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982;151:788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticht H, Rösch P. The structure of iron-sulfur proteins. Prog Biophys Mol Biol. 1998;70:95–136. doi: 10.1016/s0079-6107(98)00027-3. [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Vogel HJ, Bonner DM. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- Wolfe MT, Heo J, Garavelli JS, Ludden PW. Hydroxylamine reductase activity of the hybrid cluster protein from Escherichia coli. J Bacteriol. 2002;184:5898–5902. doi: 10.1128/JB.184.21.5898-5902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Schneider BL, Reitzer L. Purine catabolism in Escherichia coli and function of xanthine dehydrogenase in purine salvage. J Bacteriol. 2000;182:5332–5341. doi: 10.1128/jb.182.19.5332-5341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt FC, editor. Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology Press; Washington, DC: 1996. pp. 561–579. [Google Scholar]