Abstract
Background
Genetic variation contributes to risk and outcomes of sepsis. We sought to determine if variation in inflammation related genes is associated with severity of sepsis in trauma patients.
Methods
A cohort of severely injured Caucasian patients was studied and genotyped for candidate single nucleotide polymorphisms (SNPs). These were toll-like receptor 4 (TLR4) A896G, tumor necrosis factor-α G-308A, interleukin-6 G-174C, interleukin-1β C-31T, and cluster of differentiation marker-14 C-159T. SNP genotypes among patients with sepsis and complicated sepsis were analyzed by chi-square and logistic regression. Six haplotype-tagging SNPs in the TLR4 gene were subsequently examined to determine their influence on TLR4 A896G SNP’s relationship to sepsis severity.
Results
We enrolled 598 patients. Complicated sepsis developed in 147 (25%). Adjusting for independent risk factors, carriage of the variant TLR4 896 G allele was associated with decreased risk of complicated sepsis (OR = 0.3, 95%CI = 0.1–0.7, p = 0.008). Furthermore, two haplotypes seemed to better characterize this risk than the variant TLR4 896 G allele. The variant TLR4 896G allele is linked to one common haplotype, which seems to confer a considerably reduced risk of complicated sepsis. (aOR = 0.2 95% CI = 0.05–0.7, p = 0.01)
Conclusions
Variation within TLR4 gene is associated with severity of post-traumatic sepsis. This risk may not be solely related to TLR4 A896G SNP. It is likely that other, uncharacterized variations in the TLR4 gene contribute to sepsis severity. A thorough evaluation of variability within the TLR4 gene is needed to characterize sepsis risk.
Keywords: Polymorphism, SNP, htSNP, Allelic association
Introduction
The outcomes from trauma have improved due to advances in prehospital care, improved resuscitation, and organ system support. However, severely injured patients who survive the initial physiological derangements frequently develop sepsis and organ failure and, despite optimal care, some still die.1 Although clinical factors identify many patients likely to develop complications after severe injury, marked and seemingly unpredictable variation in infectious outcomes is observed among injured patients.
The notion that death from infectious disease has a heritable component has been evident for many years, but exact genetic factors are unknown.2,3 It is therefore possible that inherited differences may contribute to the variation that is observed after traumatic injury and other severe, acute illnesses.
Single nucleotide polymorphisms (SNPs) are single base pair positions in genomic DNA for which sequence alternatives exist, and are the most common human genetic variation.4 While some SNPs may exhibit functionality, such as changing the sequence or the amount of protein product, many more are silent. Nevertheless, these silent SNPs can be useful in characterizing common genetic variation when they are a part of a haplotype. A haplotype is a set of alleles that tend to be inherited together within a block of an individual chromosome. Haplotype tagging uses a relatively small subset of SNPs – called haplotype-tagging SNPs (htSNPs) to capture the overall variation in a gene or region based upon the phenomenon of linkage disequilibrium.5
SNPs in genes important in the innate immune response such as toll-like receptor 4 (TLR4) A896G, tumor necrosis factor alpha (TNF-α) G-308A, interleukin-6 (IL-6) G-174C, interleukin-1β (IL-1β) C-31T, and cluster of differentiation marker 14 (CD14) C-159T have been reported to contribute to the risk for sepsis following major trauma. 6–12 We have previously reported sepsis risk associated with candidate SNPs in these genes. Specifically, we observed an increased risk of severe sepsis associated with the TNF-α-308 A-allele and a reduced risk with the TLR4 896 G-allele.7,10,13
In this study, we sought to determine the effect of known innate immune gene polymorphisms on sepsis related organ failure and shock in Caucasian patients with severe injuries. Based upon the observed association between the TLR4 A896G SNP and complicated sepsis, we then examined TLR4 haplotypes to gain a better understanding of how TLR4 variation contributes to sepsis severity.
METHODS
Patients
With approval from the University of Washington Institutional Review Board, blunt and penetrating trauma patients admitted to the intensive care unit at Harborview Medical Center (Seattle, WA) for 48 hours or longer were prospectively enrolled between August 2003 and August 2004. Individuals were excluded if they were expected to die from the severity of their injuries, had an isolated traumatic brain injury, or suffered a burn injury. Individuals were excluded if they were expected to die from the severity of their injuries, had an isolated traumatic brain injury, or suffered a burn injury. Although our trauma population is multi-ethnic, there were relatively few non-Caucasian subjects. This report is limited to Caucasian subjects.
Clinical data were obtained from two different sources. The injury severity score (ISS), and the abbreviated injury score (AIS) were obtained from a prospectively acquired trauma registry. Admission data, hospital course, and complications were obtained from the electronic medical record. These data were de-identified once the clinical information were linked to the genetic information.
Patients were followed until hospital discharge or death. The primary outcome was complicated sepsis, i.e. sepsis with organ failure and/or septic shock in accordance with the definition set by the American College of Chest Physicians and the Society for Critical Care Medicine consensus statement.7,14 Lower respiratory tract infections diagnosis required quantitative protected specimen demonstrating at least 105 colony forming units/ml. Bacteremia was defined as bacterial growth in a blood culture. Urinary tract infection (UTI) was diagnosed by a positive urine culture of greater than 105 organisms per high power field. Wound infections were diagnosed by direct inspection and culture confirmation when available.
Individual organ failure was defined as a Marshall organ dysfunction score of ≥ 3 in the corresponding organ system, while severe multiple organ dysfunction is defined as a cumulative Marshall score ≥ 6.15 We excluded the central nervous system assessment due to its subjectivity and because of the possible influence of traumatic brain injury on this measurement.16
DNA Isolation and Genotyping
Discarded venous blood samples collected in EDTA were obtained. DNA was isolated via the Qiagen QIAamp DNA Blood Midi Kit. DNA samples were then genotyped for the presence of mutations in TLR4 (A896G), TNF-α (G-308A), IL-6 (G-174C), IL-1β (C-31T), and CD14 (C-159T). The primer and probe sequences are provided in Table 1.
Table 1.
Primer and probe sequences used in SNP detection
| SNP | ref SNP ID# | Major allele | Minor allele | Location | Sequence**(5′ to 3′) |
|---|---|---|---|---|---|
| Forward primer | |||||
| Reverse primer | |||||
| Probe (wild-type) | |||||
| Probe (variant) | |||||
| TLR4 (A896G) | rs4986790 | A | G | Exon 3 | TGACCATTGAAGAATTCCGATTAGCA |
| ACACTCACCAGGGAAAATGAAGAA | |||||
| FAM-TACCTCGATGATATTATT-MGB | |||||
| VIC-CCTCGATGGTATTATT-MGB | |||||
|
| |||||
| TNF-α (G-308A) | 1800629 | G | A | Promoter | CCAAAAGAAATGGAGGCAATAGGTT |
| GGACCCTGGAGGCTGAAC | |||||
| FAM-CCCGTCCTCATGCC-MGB | |||||
| VIC-CCCGTCCCCATGCC-MGB | |||||
|
| |||||
| IL-6 (G-174C) | rs1800795 | G | C | Promoter | GACGACCTAAGCTGCACTTTTC |
| GGGCTGATTGGAAACCTTATTAAGATTG | |||||
| FAM-CTTTAGCATCGCAAGAC-MGB | |||||
| VIC-CCTTTAGCATGGCAAGAC-MGB | |||||
|
| |||||
| IL-1β(T-31C) | 1143627 | T | C | Promoter | CAGCCTCCTACTTCTGCTTTTGA |
| AGGTTTGGTATCTGCCAGTTTCTC | |||||
| VIC-TCGCTGTTTTTATAGCTT-MGB | |||||
| FAM-CGCTGTTTTTATGGCTT-MGB | |||||
|
| |||||
| CD-14 (C-159T) | 2569190 | C | T | Promoter | CTAGATGCCCTGCAGAATCCTT |
| CCCTTCCTTTCCTGGAAATATTGCA | |||||
| VIC-CTGTTACGGCCCCCCT-MGB | |||||
| FAM-CTGTTACGGTCCCCCT-MGB | |||||
TLR4, Toll-like receptor 4; TNF-α, tumor necrosis factor alpha; IL, interleukin; CD-14, cluster of differentiation marker 14.
MGB, minor groove binder
Polymorphic nucleotide is underlined.
All genotyping assays were designed by Applied Biosystems (Foster City, CA, USA). An investigator blinded to the patients’ clinical status assigned the genotypes. The genotyping reaction utilizes two dual-labeled Taqman probes, which specifically target the alternate alleles. The two probes are labeled with a fluorescent reporter dye (VIC or FAM) and a non-fluorescing quencher/minor groove binder (MGB). When a probe specifically binds to the SNP site, the 5′ nuclease activity of Taq polymerase during PCR allows for the cleaving and subsequent fluorescence of the reporter dye. At the conclusion of PCR, samples are genotyped via analysis of the fluorescence of the two dyes. Each 5.0 ul PCR contains the following: TaqMan® Universal PCR Master Mix, No AmpErase® UNG (2X); Assays-on-Demand™ (20X) or Assays-by-Design™ (40X) SNP Genotyping Assay Mix, and 1 ng of genomic DNA. Assays were conducted in 384-well format on the ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems). Reaction conditions were the following: initial denaturation at 95°C for 10 min, followed by 40 cycles each of denaturation (95°C for 15 s) and annealing/extension (60°C for 60 s).
Subsequently, all subjects were genotyped for haplotype-tagging SNPs (htSNPs) in the TLR4 gene (figure 1) and genotyping for TLR4 A896G was repeated, verifying the accuracy of the original assay. The primer and probe sequences for the TLR4 htSNP genotyping assays are provided in Table 2. The selected htSNPs were derived from TLR4 genotypes, and the resultant PHASE and HaploBlockFinder output for 23 European American individuals available at InnateImmunity.net.17,18
Figure 1.
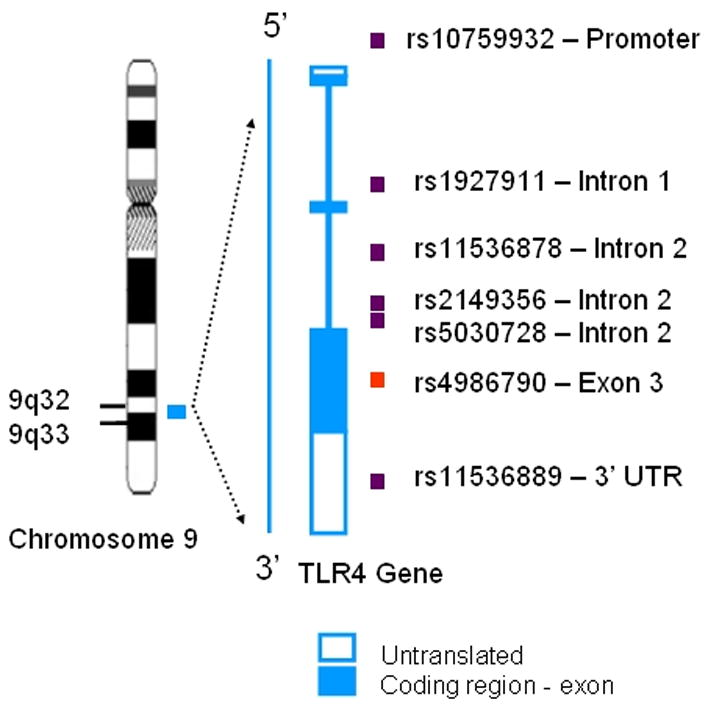
Location of htSNPs on the TLR4 gene. The TLR4 gene has 3 exons and 2 introns
Table 2.
Primer and probe sequences used in htSNP detection
| htSNP* | ref SNP I1D#) | IIPGA# | Major allele | Minor allele | Location | Sequence**(5′ to 3′) |
|---|---|---|---|---|---|---|
| Forward primer | ||||||
| Reverse primer | ||||||
| Probe (wild-type) | ||||||
| Probe (variant) | ||||||
| 1 | 10759932 | 2856 | T | C | Promoter | GCAAGCTTCTGCTATGATTAAAAGTGA |
| CACAAATGGTGTACAGGAGTTCTCA | ||||||
| FAM-TTCACCAACACTTATT-MGB | ||||||
| VIC-CTTCACCAACGCTTATT–MGB | ||||||
|
| ||||||
| 2 | 1927911 | 7764 | C | T | Intron 1 | Applied Biosystems Assay on Demand-proprietary |
| N/A*** | ||||||
| N/A | ||||||
| N/A | ||||||
|
| ||||||
| 3 | 11536878 | 9263 | C | A | Intron 2 | CTGGAAACTGATATAAAGATAGCGACATATAACA |
| CTTGACTACCCACCACAGAGAAG | ||||||
| FAM- AAACTAAAGGTAACTAATTG-MGB | ||||||
| VIC- TTTAAACTAAAGGTAAATAATTG–MGB | ||||||
|
| ||||||
| 4 | 2149356 | 1191 | G | T | Intron 2 | GCTGTCATGTAAGCACTTTTCATAAACA |
| 2 | GTTGGTAGCCAAGATAAATGACTGGTA | |||||
| VIC-ACTTATGTGTAATGTTTCG-MGB | ||||||
| FAM-TTATGTGTAATTTTTCG –MGB | ||||||
|
| ||||||
| 5 | 5030728 | 1199 | G | A | Intron 2 | CCAGTCATTTATCTTGGCTACCAACT |
| 5 | CAAGTTTAGCCATTTTCTGTCACACA | |||||
| FAM- CTGTACCAATCAGATGTAT-MGB | ||||||
| VIC- CTGTACCAATCAAATGTAT–MGB | ||||||
|
| ||||||
| 6 | 11536889 | 1584 | C | G | 3′UTR | GTTGGGCAATGCTCCTTGAC |
| 4 | ACCCCATTAATTCCAGACACATTGT | |||||
| FAM-ATAACATCCACTCTTCCCA-MGB | ||||||
| VIC-ATAACATCCACTGTTCCCA–MGB | ||||||
htSNP, Haplotype tagging SNP
PGA# Innate Immunity PGA number
MGB, minor groove binder
For ease of discussion, each SNP was assigned a number from 1 to 6.
Polymorphic nucleotide is underlined.
Not available
Data Analysis
Data were analyzed with SPSS 14.0 statistical software (SPSS, Chicago, IL). Continuous data are presented as medians and interquartile range (25th to 75th percentile) and were analyzed using Mann-Whitney U test, or in case of multiple categories, ANOVA. Categorical variables were compared using Pearson’s chi-square test. For each comparison, the actual p values are reported. Homozygous carriers of variant-type alleles at TNF-α, TLR4, IL-6, IL-1β, and CD-14 were grouped with heterozygotes for analysis. Hardy-Weinberg Equilibrium analysis was performed for each SNP by comparing the detected genotype distribution with the theoretical distribution estimated on the basis of the SNP allele frequencies. P> 0.05 (Chi-square) was considered to indicate equilibrium.
Multiple logistic regression analysis was used to evaluate independent risk factors for developing complicated sepsis. Variables were examined for their effect as risk factors (considered significant if p < 0.05) and as confounders (altered the effect of other risk factors). Variables that were risk factors or confounders were included in the final model.
RESULTS
Between August 1st 2003 and August 31st 2004, 598 Caucasian trauma patients were admitted to the intensive care unit for ≥ 48 hours. Sepsis developed in 278 patients and complicated sepsis developed in 147 (figure 2). Table 3 summarizes demographic variables, pertinent injury characteristics, and clinical outcomes of these 598 patients. Patients with complicated sepsis had a higher mortality and longer ICU length of stay compared to patients with SIRS or uncomplicated sepsis alone (Table 3 and Figure 3). The type of sepsis was similar between the two groups with 39% of sepsis occurring due to Gram positive organisms and the remainder due to Gram negative organisms or polymicrobial in etiology. Respiratory failure (ARDS or ALI) was the most common organ dysfunction, and occurred among almost all patients (132, 90%) with complicated sepsis.
Figure 2.
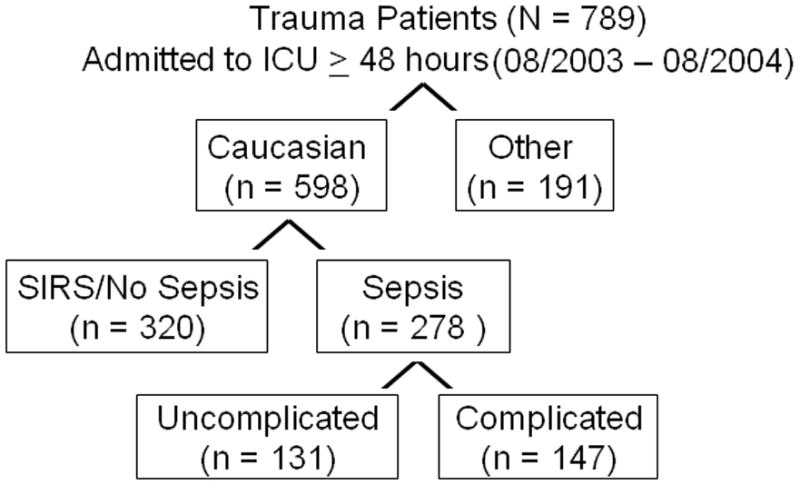
Patient enrollment into the study. Sepsis complicated by organ failure or shock (complicated sepsis), occurred in 147 patients. Of those, 43 had septic shock.
Table 3.
Demographics, Injury Characteristics and Outcomes According to Sepsis Severity
| SIRS or uncomplicated sepsis (N = 451) | Complicated sepsis (n = 147) | p-value | |
|---|---|---|---|
| Age* (years) | 39 (21–58) | 42 (26–57) | 0.054 |
| Male | 306 (68) | 110 (75) | 0.110 |
| Diabetes Mellitus | 19 (4) | 17 (12) | < 0.01 |
| ISS* | 22 (16–29) | 29 (20–36) | < 0.01 |
| Severe thoracic injury | 175 (39) | 93 (64) | < 0.01 |
| Initial Base Deficit* | 3.0 (0.2–5.8) | 5.5 (3–9) | < 0.01 |
| Blood transfusion | 223 (49) | 130 (88) | < 0.01 |
| Died | 31 (7) | 26 (18) | < 0.01 |
| Cumulative Marshall Score* | 4 (2–6) | 9 (7–11) | < 0.01 |
| ARDS or ALI | 124 (27) | 132 (90) | < 0.01 |
| ICU Length of stay* (days) | 3 (2–6) | 15 (9–22) | < 0.01 |
| Hospital length of stay* (days) | 10 (7–16) | 22 (13–35) | < 0.01 |
Median (25th–75th percentile)
ISS: Injury Severity Score
ARDS: Acute Respiratory Distress Syndrome
ALI: Acute lung injury
Figure 3.
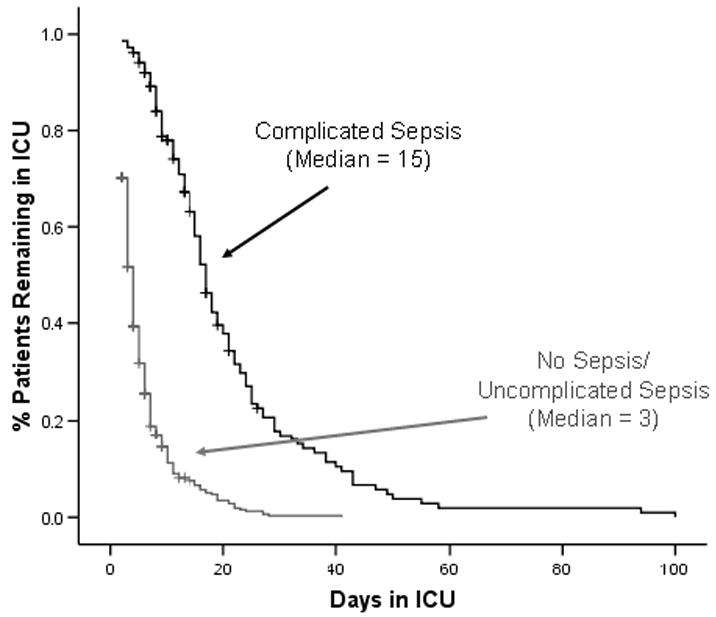
Patients with complicated sepsis had a longer ICU length of stay than those with uncomplicated sepsis
Examination of the association between candidate SNPs and complicated sepsis
Allele frequencies are in Table 4. All SNPs were in Hardy-Weinberg equilibrium (p>0.20). After adjusting for age, ISS, and clinical risk factors for complicated sepsis (sex, initial base deficit, blood transfusions, and severe thoracic trauma), carriage of the TLR4 896 G-allele was associated with a decreased risk of complicated sepsis (OR 0.29, 95% CI 0.12–0.69, p = 0.005, Table 5). This observation is similar to our previous finding in a smaller, independent cohort of trauma patients13, but does conflict with other published reports. Others have found the +896 A-allele to increase the risk for sepsis.6,11 Contrary to our previous finding regarding the TNF-α G-308A SNP7, we observed that none of the other promoter SNPs studied were associated with sepsis severity in this cohort.
Table 4.
Allele frequencies of candidate SNPs among patients according to complicated sepsis
| Polymorphism | SIRS or uncomplicated sepsis (n = 451) | Complicated sepsis (n = 147) | p-value |
|---|---|---|---|
| TLR4 A896G | |||
| Wild type - A allele | 850 (94) | 286 (97) | 0.038 |
| Variant-G allele | 52 (6) | 8 (3) | |
| TNF-αG-308A | |||
| Wild type-G allele | 779 (86) | 252 (86) | 0.779 |
| Variant-A allele | 123 (14) | 42 (14) | |
| IL-6 G-174C | |||
| Wild type-G allele | 528 (59) | 180 (61) | 0.415 |
| Variant-C allele | 374 (41) | 114 (39) | |
| IL-1βT-31C | |||
| Wild type-T allele | 620 (70) | 202 (69) | 0.992 |
| Variant-C allele | 282 (31) | 92 (31) | |
| CD-14 C-159T | |||
| Wild type-C allele | 148 (57) | 152 (52) | 0.258 |
| Variant-T allele | 114 (44) | 142 (48) | |
Values are expressed as number of alleles (%). There are two alleles per patient. Exact p-values are presented for the comparison of allele frequencies between the two groups by chi-square analysis
Table 5.
Risk factors for developing complicated sepsis
| Factor | OR (95% CI) | p-value |
|---|---|---|
| Age | 1.01 (1.00–1.02) | 0.095 |
| Male Sex | 2.01 (1.27–3.31) | 0.003 |
| Injury Severity Score | 1.03 (1.00–1.05) | 0.030 |
| Severe Thoracic Injury | 1.86 (1.15–3.00) | 0.011 |
| Initial Base Deficit | 1.09 (1.03–1.15) | 0.002 |
| Blood Transfusion (yes/no) | 5.48 (3.01–9.99) | 0.000 |
| History of diabetes mellitus | 3.01 (1.32–7.11) | 0.009 |
| Variant G allele TLR4 A896G SNP | 0.29 (0.12–0.69) | 0.005 |
The data presented are the result of stepwise logistic regression analysis. Clinical factors found to be important in determining the risk for complicated sepsis were male sex, injury severity score, severe thoracic injury as determined by an abbreviated injury severity score (AIS) greater than or equal to 3, initial base deficit measured in the emergency department, receiving a transfusion at any point during the hospital stay, and history of diabetes mellitus. Clinical variables that were included in the initial model but were found not to be important in determining the risk for complicated sepsis were age, severe head injury, and a history of cardiovascular or pulmonary disease.
TLR4 htSNPs are associated with risk of developing complicated sepsis
We sought to further characterize how variation in the TLR4 gene might influence the risk of complicated sepsis. In order to do so, DNA samples were genotyped for six htSNPs (figure 1), for which the overall allele frequencies are shown in Table 6. These additional markers of TLR4 variation were evaluated in the context of the previously described risk factors by including them in the multivariate logistic regression analysis. Including these htSNPs markedly changed the results of our initial analysis and demonstrated that the risk of complicated sepsis associated with TLR4 genetic variation was associated with two particular htSNPs shown in Table 7. Furthermore, after adjusting for the htSNPs, the +896 variant was no longer associated with complicated sepsis. Additional examination of the TLR4 haplotypes demonstrated that the +896 SNP, exists in association with (is linked to) the haplotype tagged by htSNP4. In this large cohort of severely injured trauma patients, the decreased risk for complicated sepsis seems more strongly associated with carriage of htSNP4 than carrier status of the specific TLR4 G896A variant.
Table 6.
Allele frequencies of TLR4 htSNPs among study cohort (n=598)
| Genotypes | Allele Frequencies (N = 1196) |
|---|---|
| TLR4 htSNP1 | |
| Wild type – T allele | 1025 (86) |
| Variant – C allele | 169 (14) |
| Undetermined | 2 (0.2) |
| TLR4 htSNP2 | |
| Wild type – C allele | 867 (72) |
| Variant – T allele | 323 (27) |
| Undetermined | 6 (0.5) |
| TLR4 htSNP3 | |
| Wild type – C allele | 1033 (86) |
| Variant – A allele | 157 (13) |
| Undetermined | 6 (0.5) |
| TLR4 htSNP4 | |
| Wild type – G allele | 808 (68) |
| Variant – T allele | 384 (32) |
| Undetermined | 4 (0.3) |
| TLR4 htSNP5 | |
| Wild type – G allele | 862 (72) |
| Variant – A allele | 326 (27) |
| Undetermined | 8 (0.6) |
| TLR4 htSNP6 | |
| Wild type – C allele | 1005 (84) |
| Variant – G allele | 185 (16) |
| Undetermined | 6 (0.5) |
Values are expressed as number of alleles (%). There are two alleles per patient.
Table 7.
Risk factors for developing complicated sepsis among Caucasian patients with post-traumatic sepsis (N = 598)
| Factor | OR (95% CI) | p-value |
|---|---|---|
| Age | 1.01 (1.00–1.02) | 0.125 |
| Male Sex | 2.1 (1.27–3.31) | 0.003 |
| Injury Severity Score | 1.03 (1.00–1.05) | 0.027 |
| Severe Thoracic Injury | 1.9 (1.17–3.04) | 0.009 |
| Initial Base Deficit | 1.0 (1.03–1.15) | 0.002 |
| Blood Transfusion (yes/no) | 5.5 (2.98–9.95) | < 0.001 |
| History of diabetes mellitus | 2.9 (1.27–6.88) | 0.012 |
| Variant T allele of TLR4 htSNP2 (rs1927911) | 6.1 (2.18–17.16) | 0.001 |
| Variant T allele of TLR4 htSNP4 (rs2149356) | 0.2 (0.06–0.50) | 0.001 |
The data presented are the result of stepwise logistic regression analysis. Carriage of the TLR4 896 variant was no longer associated with complicated sepsis after adjusting for TLR4 htSNP2 and htSNP4.
Assessing the contribution of genetic influences to the outcome of post-traumatic sepsis
Clinical risk factors for post-traumatic sepsis and organ failure are well characterized in the literature. The value of genetic variability in improving risk stratification is uncertain. Therefore, we generated receiver-operating characteristic curves (ROC) using the predicted probabilities generated from two models and compared the area under the curve (AUC) from each model. The first model included only clinical factors and the second incorporated TLR4 htSNP2 and TLR4 htSNP4 in the prediction model. The predicted probabilities were obtained from the logistic regression models presented in Tables 5 & 7 and the results are shown in Figure 4. While the model incorporating the TLR4 genotypes does provide an improved prediction (i.e. greater AUC), this improvement is marginal.
Figure 4.
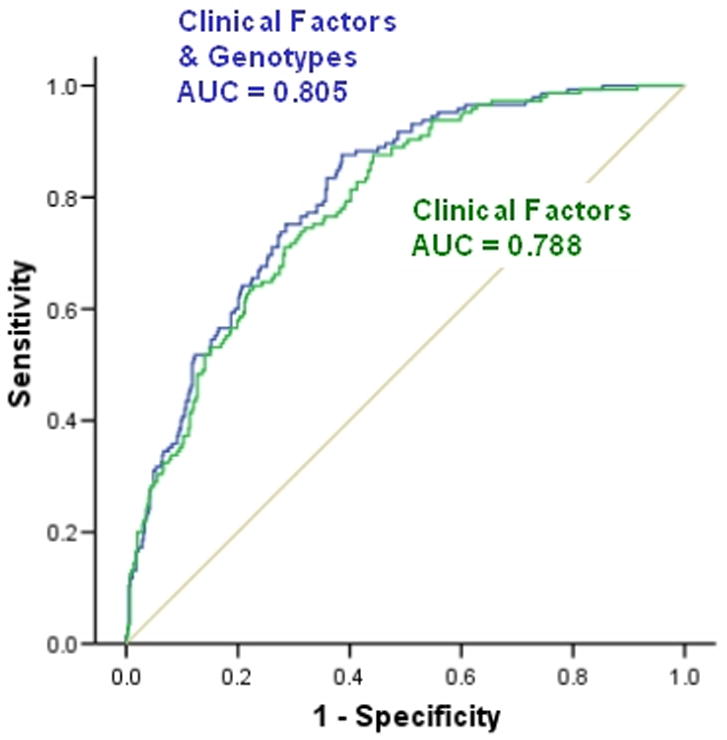
Comparative Receiver Operating Characteristic (ROC) Curves of the model used to predict the development of complicated sepsis among Caucasian patients with post-traumatic sepsis. Including patients TLR4 genotypes (TLR4 htSNP2 and TLR4 htSNP4) in the model improves the area under the curve (AUC). (The greater the area under the curve the more predictive the test is of the outcome of interest)
DISCUSSION
Many factors including age, sex, and genetic variation undoubtedly influence the host response to injury and infection and therefore influence outcomes in important ways. 6–11 Furthermore, the severity of sepsis, as indicated by associated organ failure or shock has considerable influence on the outcome after traumatic injury. We have therefore chosen to examine whether genetic variation influences the risk for the most severe forms of sepsis in injury victims. Our study and our observations reflect both the promise and the limitations of genetic association studies for common conditions such as post-traumatic sepsis. First, associations were not observed between SNPs in a number of genes considered important to innate immunity and outcomes of post-traumatic sepsis. 6–12 Second, we confirmed and expanded upon a genotype-phenotype association that our previous data suggested, yet this observation contradicts the findings of other, albeit smaller, cohorts. Finally, we have characterized that even where there is a fairly strong genetic association with disease risk, having that genetic information adds marginal predictive value over well-described clinical risk factors.
Why did we not observe an association between the TNF-α G-308A SNP and complicated sepsis? There are many explanations for the failure to confirm initial findings, including small sample sizes, population stratification (dissimilar genetic background between cases and controls, such as that associated with race), and the generally weak associations that likely exist between SNPs and complex diseases, such as sepsis. 19,20 Taken together, these factors have limited the reproducibility of other genotype-phenotype associations and are likely responsible for our failure to confirm the previously observed association between complicated sepsis and TNF-α genetic variation.7
Our main observation was that variation in the TLR4 gene is associated and potentially causally related to the development of complicated sepsis. This confirmed an observation we have previously made in a similar cohort13 but contradicts other reports. Prior studies have documented that TLR4 variant allele carriers have an impaired response to bacterial endotoxin 21,22 and an increased incidence of SIRS, gram-negative infections, severe sepsis, and septic shock.6,11,23–25 We and others have previously demonstrated that in-vitro response to LPS is similar between carriers of the wild type and heterozygote variants as measured by cytokine production.26–29 These findings are supported by observations in clinical studies demonstrating no effect of this polymorphism in patients with meningococcal disease and invasive pneumococcal infection.30,31 We sought to examine TLR4 variation in more detail and thus studied TLR4 haplotypes in addition to the G896A variant. Of the six htSNPs evaluated TLR4 htSNP2 and htSNP4 were associated with complicated sepsis. Furthermore, carriage of the TLR4 +896 variant was no longer associated with complicated sepsis after adjusting for TLR4 htSNP4. It appears that the variant TLR4 +896 arises on the background of the haplotype marked by htSNP4 and it is this haplotype that appears to be associated with a lower risk of serious infection. We are uncertain of the basis for this observed association, but given that the human TLR4 gene is highly polymorphic (> 20 reported SNPs change the amino acid sequence of TLR4), studying how TLR4 genotype influences phenotype must extend beyond one or even a few SNPs. In addition to more broadly characterizing how TLR4 variation influences functional responses, it will be important to understand how altered (decreased LPS binding affinity, for example) function might influence clinical outcomes. It is perhaps too simplistic to consider that a decreased (or increased) responsiveness to LPS is detrimental under all conditions. Post-traumatic sepsis, whether due to gram negative or gram positive organisms is a complex phenomenon, and we simply know too little about how alterations in innate immune responses due to variation in TLR4 or other genes directly or indirectly affect outcomes. Our observations that the severity of both gram positive and gram negative sepsis is reduced may reflect a more global influence of TLR4 variation rather than an immediate effect on LPS signaling. Alternatively, the lower risk of complicated sepsis due to gram positive organisms may simply be a spurious association. Finally, it has been observed that a variety of relatively rare coding SNPs in the TLR4 gene are responsible for altered LPS signaling and affect clinical outcomes.32,33 It is therefore possible that detrimental but rare SNPs are more likely to exist on haplotypes other than that tagged by htSNP4. This would explain why studying a single or at most a few polymorphisms located in one gene has yielded results that are difficult to reproduce. More complete characterization of variation in a gene, rather than simply studying a few “candidate SNPs” is necessary. As we have shown, this should involve htSNPs and will likely also require follow-up sequencing.
The question remains regarding the clinical usefulness of SNPs in predicting prognosis in injured patients. Our observations indicate that the improved predictive ability is small. Comparing receiver operating characteristic curves, the clinical model for predicting complicated sepsis can be improved by adding the results of patients’ htSNP genotypes, however this resulted in only marginal improvement. Nevertheless, in this study, we have only incorporated a single variant in our model. Perhaps, once clear associations with other genes are identified, better and clinically useful predictive models can be developed.
In summary, haplotypes in TLR4 seem to be related to disease severity, and possibly play a causative role in post-traumatic sepsis. More detailed investigation of the nucleotide sequence in and around the TLR4 gene of those persons carrying the TLR4 htSNP rs1927911 (htSNP2) should provide information regarding how altered TLR4 function influences sepsis risk.
Acknowledgments
This work has been supported by the following NIGMS grants: R01 GM066946-01, and T32 GM007037-31
Definition of Terms
- Allele
Variants of the same gene. Each person carries two alleles (maternal and paternal copies)
- Exon
A region of a gene that codes for a protein
- Haplotype
Variant alleles located together along portions of individual chromosomes that appear to be inherited together in blocks
- Haplotype Tagging SNP (htSNP)
A representative non-redundant single nucleotide polymorphism in a region of the genome with high linkage disequilibrium essentially capturing a major fraction of the “variation” present within a population
- Heterozygous
Having two different alleles at a specific gene locus
- Homozygous
Having two identical alleles at a specific gene locus
- Intron
A region of a gene that does not code for a protein
- Linkage Disequilibrium (LD)
A phenomenon whereby genetic variants are associated, i.e. people who have one variant tend to have a second variant as well
- Phenotype
The observable properties of genes and environmental factors
- Single nucleotide polymorphism (SNP)
A DNA sequence variation occuring when a single nucleotide (A, T, C or G) in the genome differes between memebers of a species
Footnotes
This work was presented at the 66th Annual Meeting of the American Association for the Surgery of Trauma (September 2007)
References
- 1.O’Keefe GE, Maier RV, Diehr P, et al. The complications of trauma and their associated costs in a level I trauma center. Arch Surg. 1997;132:920–924. doi: 10.1001/archsurg.1997.01430320122021. [DOI] [PubMed] [Google Scholar]
- 2.Sorensen TI, Nielsen GG, Andersen PK, et al. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 3.Westendorp RG, Langermans JA, Huizinga TW, et al. Genetic influence on cytokine production and fatal meningococcal disease. Lancet. 1997;349:170–173. doi: 10.1016/s0140-6736(96)06413-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Z, Zhang F. Sequence context analysis of 8.2 million single nucleotide polymorphisms in the human genome. Gene. 2006;366:316–324. doi: 10.1016/j.gene.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 5.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 6.Agnese DM, Calvano JE, Hahm SJ, et al. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J Infect Dis. 2002;186:1522–1525. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 7.O’Keefe GE, Hybki DL, Munford RS. The G-->A single nucleotide polymorphism at the -308 position in the tumor necrosis factor-alpha promoter increases the risk for severe sepsis after trauma. J Trauma. 2002;52:817–825. doi: 10.1097/00005373-200205000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Arnalich F, Lopez-Maderuelo D, Codoceo R, et al. Interleukin-1 receptor antagonist gene polymorphism and mortality in patients with severe sepsis. Clin Exp Immunol. 2002;127:331–336. doi: 10.1046/j.1365-2249.2002.01743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivera-Chavez FA, Peters-Hybki DL, Barber RC, et al. Interleukin-6 promoter haplotypes and interleukin-6 cytokine responses. Shock. 2003;20:218–223. doi: 10.1097/01.shk.0000079425.52617.db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera-Chavez FA, Peters-Hybki DL, Barber RC, et al. Innate immunity genes influence the severity of acute appendicitis. Ann Surg. 2004;240:269–277. doi: 10.1097/01.sla.0000133184.10676.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barber RC, Chang LY, Arnoldo BD, et al. Innate immunity SNPs are associated with risk for severe sepsis after burn injury. Clin Med Res. 2006;4:250–255. doi: 10.3121/cmr.4.4.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menges T, Konig IR, Hossain H, et al. Sepsis syndrome and death in trauma patients are associated with variation in the gene encoding tumor necrosis factor. Crit Care Med. 2008;36:1456. doi: 10.1097/CCM.0B013E318170ABB6. [DOI] [PubMed] [Google Scholar]
- 13.Staudenmayer K, Barber RC, O’Keefe GE. The TLR4 +896 AtoG Polymorphism in Sepsis. Presented at the Society of University of Surgeons; 2003. [Google Scholar]
- 14.Levy MM, Fink MP, Marshall JC, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31:1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 15.Marshall JC, Cook DJ, Christou NV, et al. Multiple organ dysfunction score: a reliable descriptor of a complex clinical outcome. Crit Care Med. 1995;23:1638–1652. doi: 10.1097/00003246-199510000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Cumming J, Purdue GF, Hunt JL, et al. Objective estimates of the incidence and consequences of multiple organ dysfunction and sepsis after burn trauma. J Trauma. 2001;50:510–515. doi: 10.1097/00005373-200103000-00016. [DOI] [PubMed] [Google Scholar]
- 17.Zhang K. HaploBlockFinder V0.7. Online Source. 2004.
- 18.Innate Immunity Net. Online Source. 2007.
- 19.Hirschhorn JN, Lohmueller K, Byrne E, et al. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Wacholder S, Chanock S, Garcia-Closas M, et al. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arbour NC, Lorenz E, Schutte BC, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 22.Michel O, LeVan TD, Stern D, et al. Systemic responsiveness to lipopolysaccharide and polymorphisms in the toll-like receptor 4 gene in human beings. J Allergy Clin Immunol. 2003;112:923–929. doi: 10.1016/j.jaci.2003.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Lorenz E, Mira JP, Frees KL, et al. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 24.Child NJ, Yang IA, Pulletz MC, et al. Polymorphisms in Toll-like receptor 4 and the systemic inflammatory response syndrome. Biochem Soc Trans. 2003;31:652–653. doi: 10.1042/bst0310652. [DOI] [PubMed] [Google Scholar]
- 25.Montes AH, Asensi V, Alvarez V, et al. The Toll-like receptor 4 (Asp299Gly) polymorphism is a risk factor for Gram-negative and haematogenous osteomyelitis. Clin Exp Immunol. 2006;143:404–413. doi: 10.1111/j.1365-2249.2005.03002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heesen M, Bloemeke B, Kunz D. The cytokine synthesis by heterozygous carriers of the Toll-like receptor 4 Asp299Gly polymorphism does not differ from that of wild type homozygotes. Eur Cytokine Netw. 2003;14:234–237. [PubMed] [Google Scholar]
- 27.Imahara SD, Jelacic S, Junker CE, et al. The TLR4 +896 polymorphism is not associated with lipopolysaccharide hypo-responsiveness in leukocytes. Genes Immun. 2005;6:37–43. doi: 10.1038/sj.gene.6364147. [DOI] [PubMed] [Google Scholar]
- 28.Erridge C, Stewart J, Poxton IR. Monocytes heterozygous for the Asp299Gly and Thr399Ile mutations in the Toll-like receptor 4 gene show no deficit in lipopolysaccharide signalling. J Exp Med. 2003;197:1787–1791. doi: 10.1084/jem.20022078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Von AS, Schroder NW, Gueinzius K, et al. Heterozygous toll-like receptor 4 polymorphism does not influence lipopolysaccharide-induced cytokine release in human whole blood. J Infect Dis. 2003;188:938–943. doi: 10.1086/378095. [DOI] [PubMed] [Google Scholar]
- 30.Read RC, Pullin J, Gregory S, et al. A functional polymorphism of toll-like receptor 4 is not associated with likelihood or severity of meningococcal disease. J Infect Dis. 2001;184:640–642. doi: 10.1086/322798. [DOI] [PubMed] [Google Scholar]
- 31.Moens L, Verhaegen J, Pierik M, et al. Toll-like receptor 2 and Toll-like receptor 4 polymorphisms in invasive pneumococcal disease. Microbes Infect. 2007;9:15–20. doi: 10.1016/j.micinf.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Smirnova I, Hamblin MT, McBride C, et al. Excess of rare amino acid polymorphisms in the Toll-like receptor 4 in humans. Genetics. 2001;158:1657–1664. doi: 10.1093/genetics/158.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smirnova I, Mann N, Dols A, et al. Assay of locus-specific genetic load implicates rare Toll-like receptor 4 mutations in meningococcal susceptibility. Proc Natl Acad Sci U S A. 2003;100:6075–6080. doi: 10.1073/pnas.1031605100. [DOI] [PMC free article] [PubMed] [Google Scholar]


