Abstract
The FYVE domain is a small zinc binding module that recognizes phosphatidylinositol 3-phosphate [PtdIns(3)P], a phospholipid enriched in membranes of early endosomes and other endocytic vesicles. It is usually present as a single module or rarely as a tandem repeat in eukaryotic proteins involved in a variety of biological processes including endo- and exocytosis, membrane trafficking and phosphoinositide metabolism. A number of FYVE domain-containing proteins are recruited to endocytic membranes through the specific interaction of their FYVE domains with PtdIns(3)P. Structures and PtdIns(3)P binding modes of several FYVE domains have recently been characterized, shedding light on the molecular basis underlying multiple cellular functions of these proteins. Here, structural and functional aspects and the current mechanism of the multivalent membrane anchoring by monomeric or dimeric FYVE domain are reviewed. This mechanism involves stereospecific recognition of PtdIns(3)P that is facilitated by non-specific electrostatic contacts and modulated by the histidine switch, and is accompanied by hydrophobic insertion. Contributions of each component to the FYVE domain specificity and affinity for PtdIns(3)P-containing membranes are discussed.
Keywords: FYVE domain, Phosphoinositide, Phosphatidylinositol 3-phosphate, Membrane, Endosome
The FYVE domain was identified in 1996 as a zinc binding finger that is present in a range of eukaryotic proteins involved in membrane trafficking and phosphoinositide metabolism [1]. This conserved ∼70-residue module specifically recognizes phosphatidylinositol 3-phosphate [PtdIns(3)P] and targets many FYVE domain-containing proteins to PtdIns(3)P-enriched endocytic membranes [2–4]. Named after the four proteins, Fab1, YOTB, Vac1 and EEA1, it can be found as a single finger or as a tandem repeat in modular proteins that often contain other phosphoinositide binding domains, protein interaction modules, and catalytic units. The FYVE domain is defined by the three conserved sequences: the N-terminal WxxD, the central RR/KHHCR, and the C-terminal RVC motifs that form a compact PtdIns(3)P binding site and distinguish FYVE from other structurally related RING fingers. Structures of five typical FYVE domains [5–8] and molecular mechanisms of the PtdIns (3)P recognition and membrane targeting have recently been elucidated, providing mechanistic insights into multiple cellular functions of these proteins.
1. Biological role of FYVE proteins
The family of FYVE domain-containing proteins is extensive, particularly in higher eukaryotes, and comprises over three hundred members according to the UniGen NCBI database. These proteins usually localize to specific subcellular compartments, such as early endosomes, internal vesicles of multi-vesicular bodies and phagosomes, where the bulk of intracellular PtdIns(3)P is found. PtdIns(3)P comprises nearly 0.25% of the inositol-containing lipids in mammalian cells, yet its concentration is maintained at a relatively high local level of ∼200 μM [9]. The endosomal PtdIns(3)P pool is targeted by the majority of FYVE domains and this interaction is disrupted by the phosphatidylinositol 3-kinase (PI3K) inhibitor wortmannin [10,11]. One of the best-characterized FYVE proteins, early endosome antigen 1 (EEA1), is recruited to early endosomes through binding to PtdIns(3)P and the small GTPase Rab5 [12] and is required for homo- and heterotypic fusion of endosomes and for endocytic transport [12–16]. Owing to the exclusive localization to early endosomes, EEA1 is commonly used as a marker of these organelles. Its relative and also a Rab5 effector, Rabenosyn-5, is targeted to early endosomes by the FYVE domain. Like EEA1, Rabenosin-5 is necessary for the regulated fusion of endocytic membranes and is involved in transport of lysosomal enzymes from the TGN to endosomes [17]. Similarly, FYVE protein Rabip4 translocates to early endosomes through the interactions with PtdIns(3)P and Rab4-GTP and is implicated in recycling from early endosomes [18]. Fusion of vesicles derived from the plasma membrane and Golgi with the yeast vacuole is mediated by FYVE domain-containing Vac1p [19]. Another FYVE protein, hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) [11,20–22] and its yeast homolog Vps27p [23] are involved in trafficking from endosomes to lysosomes or vacuoles. Thus, FYVE domains serve as a driving force that brings host proteins to PtdIns(3)P-enriched endosomal membranes.
A small pool of PtdIns(3)P has been identified in the nucleus and mitochondria and a fraction of this PI can be found in Golgi and plasma membrane [24], yet several FYVE proteins are recruited to these organelles. For example, DFCP1, which contains a pair of FYVE domains and binds PtdIns(3)P, localizes to the Golgi apparatus and endoplasmic reticulum [25,26]. The Lz-fyve protein is found in the nucleus during embryogenic development [27]. Fgd1, implicated in the human disease faciogenital dysplasia [28], associates with actin filaments, Golgi and plasma membrane [29,30]. It contains an atypical FYVE domain that lacks an N-terminal WxxD motif and recognizes both PtdIns(3)P and PtdIns(5)P [31]. The FGD1's close relative, Frabin, is primarily recruited to the plasma membrane, most likely with the help of its PH domain [32]. Autophagy-linked FYVE protein (Alfy) binds PtdIns(3)P in vitro and partially co-localizes with the lipid in vivo; however, it is mainly found in the nuclear envelope [33].
Similar to their non-catalytic relatives, a number of enzymes contain FYVE domain and localize to endosomal membranes. PIKfyve is a phosphoinositide 5-kinase that produces PtdIns 3,5-bisphosphate and regulates membrane invagination [34]. Its N-terminal FYVE domain is absolutely required for targeting of PIKfyve to the vesicles of the late endocytic pathway and for controlling their size [35]. The homologous yeast kinase, Fab1, is involved in regulation of vacuole homeostasis and vesicle formation [36,37]. The FYVE finger containing Myotubularin-related protein 3 (MTMR3) and MTMR4 function as dual specificity phosphatases that can act on PtdIns(3)P [38]. It was long thought that these phosphatases utilize PtdIns(3)P as a ligand and a substrate in competitive binding. However, recent studies reveal that the FYVE domain of MTMR3 does not recognize PtdIns(3)P [39]. Another enzyme, Pib1p ubiquitin ligase, also contains FYVE domain that directs endosomal and vacuolar localization of this protein [40].
Although the majority of the FYVE domain-containing proteins are implicated in membrane trafficking and protein sorting, a number of them, including Hrs and Smad anchor for receptor activation (SARA), play roles in signal transduction. The FYVE domain of SARA is required [41] and sufficient for targeting of this protein to endosomal membranes [42–45] where it interacts with the activated transforming growth factor β receptor. This intracellular complex recruits and phosphorylates Smad proteins, which translocate to the nucleus to control transcription [41,42]. Hrs is shown to bind Smad [46] and STAM, a signal transducing adaptor molecule [47], and appears to be involved in signaling for cytokine-mediated c-fos [48].
2. Structure of the FYVE domain
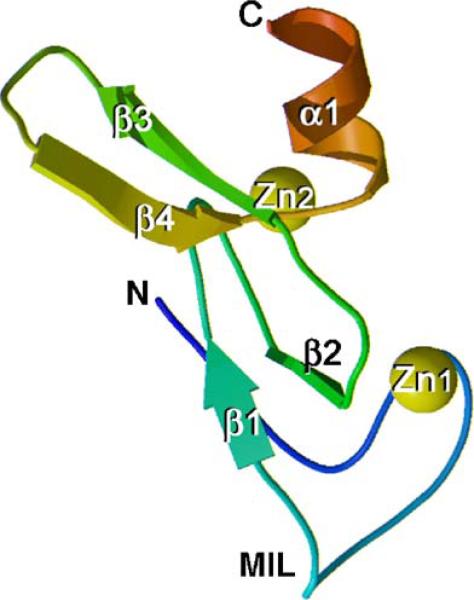
Three-dimensional structures of the S. cerevisiae Vps27p, Drosophila Hrs, and human EEA1 FYVE domains have been determined by X-ray crystallography and NMR spectroscopy [5–8]. Two other FYVE domain solution structures, of Leishmania Major Lm5−1 and of the human FYVE domain-containing 27 isoform B protein, have recently been solved (PDB codes 1Z2Q and 1WFK, respectively, unpublished data). All structures reveal a similar overall fold consisting of two double-stranded antiparallel β sheets and a C-terminal α-helix (Fig. 1). An additional N-terminal α-helical turn is seen in the EEA1 structure, and a short α-helix connecting β2 and β3 is present in the structure of Lm5−1. The functionally critical β1 spans three residues of the RR/KHHCR motif and pairs with the β2 strand, which links the two zinc clusters. The β1 strand is preceded by an exposed hydrophobic protrusion, a so-called membrane insertion loop (MIL), which penetrates into the bilayers upon binding of the FYVE domain to PtdIns(3)P-containing membranes.
Fig. 1.
Crystal structure of the Vps27p FYVE domain [5] (PDB code 1VFY). The backbone ribbon is colored using a graded scheme from the blue N-terminus to the red C-terminus. The zinc ions are shown as yellow spheres. This figure and Fig. 5b are prepared with MOLSCRIPT [92] and Raster3D [93].
The FYVE domain fold is stabilized by tetrahedral coordination of two zinc ions, which are bound by four CxxC motifs in a cross-braced topology (Fig. 1). One zinc ion (Zn1) is coordinated by the first and the third cysteine motifs, whereas Zn2 is bound by the second and the fourth motifs in all human proteins. In yeast Vps27p, the fourth Cys residue is replaced by His (Fig. 2). All residues that coordinate zinc ions are in α helical conformations, except for the second pair of Zn1-binding cysteines, which are in the 3/10 conformations [5]. Zinc coordination is required for structural stability and biological activity of the FYVE domain, as mutation of any of the zinc coordinating residues [2,3,25,40,49] or zinc removal with chelators [3,50] result in loss of structure and function of the domain. Interestingly, the unfolding due to removal of zinc is reversible as evidenced by the restoration of NMR signals corresponding to the folded protein upon the reintroduction of zinc ions [50].
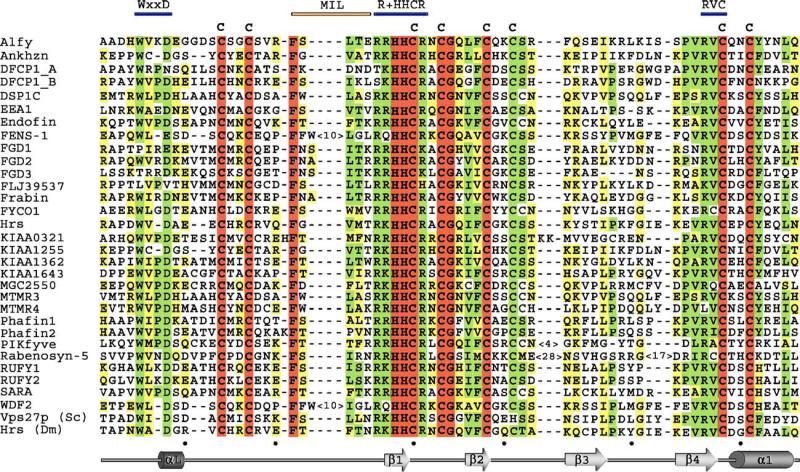
Fig. 2.
Alignment of the FYVE finger sequences of 30 human proteins and Drosophila Hrs and S. cerevisae Vps27p: absolutely, moderately and weakly conserved residues are colored brown, green and yellow, respectively. Three PtdIns(3)P binding elements and the MIL residues are indicated above the alignment by blue and orange bars, respectively. The secondary structure of EEA1 FYVE is shown below with every tenth residue of EEA1 marked with a black dot. The following human protein sequences are aligned: Alfy or WDF3 (aa 3447−3514); Ankhzn (aa 1097−1161); DFCP1_A (aa 591−659); DSP1_B (aa 708−775); DSP1C (aa 1112−1179); EEA1 (aa 1345−1410); Endofin (aa 740−805); FENS-1 or WDF1 (aa 277−352); FGD1 (aa 723−790); FGD2 (aa 451−518); FGD3 (aa 525−588); FLJ39537 (aa 100− 166); Frabin (aa 552−619); FYCO1 or FCCD1 (aa 1166−1231); Hrs (aa 156−220); KIAA0321 (aa 808−875); KIAA1255 (aa 1163−1227); KIAA1362 (aa 547−6134); KIAA1643 (aa 916−983); MGC2550 (aa 37−104); MTMR3 or DSP1A (aa 1075−1142); MTMR4 (aa 1087−1149); Phafin1 (aa 145−212); Phafin2 (aa 145−212); PIKfyve (aa 151−229); Rabenosyn-5 (aa 150−260); RUFY1 or EIP1 (aa 527−592); RUFY2 or Rabip4 (533−598); SARA (aa 692−758); WDF2 (aa 277−352).
The zinc coordination pattern of the FYVE domain is similar to that of the RING finger [51,52], and the bulk of the Vps27p structure superimposes well with rabphilin zinc binding domain [53], with a root mean squared deviation of 1.2 Å. However, the two modules are functionally unrelated and bear no sequence similarities outside the zinc coordinating residues. Unlike other zinc fingers, that typically bind nucleic acids or proteins [51,52], FYVE domains recognize phospholipids with the help of three highly conserved motifs, which became a hallmark of this module.
3. Function of the FYVE domain
FYVE domains bind PtdIns(3)P and direct a wide variety of cytosolic proteins to membranes during key signaling and trafficking events. Although the specific recognition of PtdIns (3)P remains a major distinguishing feature of the FYVE finger [2–4], the overall mechanism of membrane anchoring is found to be multivalent and involves non-specific electrostatic contacts with acidic lipids other than PtdIns(3)P [54–56], activation of the histidine switch [57], hydrophobic insertion into the bilayers [7,55,56,58], and in some cases dimerization [8,59–61]. Each component within the multistep binding mechanism uniquely contributes to the FYVE domain specificity and affinity for PtdIns(3)P embedded in membranes.
4. PtdIns(3)P binding
The ability of FYVE finger to specifically recognize PtdIns (3)P was discovered by in vitro liposome binding experiments and was substantiated by the protein's subcellular localization in vivo [2–4]. The FYVE domain association with PtdIns(3)P-containing liposomes was abolished by removal of zinc or by mutation of the residues that coordinate the lipid or metal [2,3,49,50]. The recruitment of FYVE proteins to endosomes and yeast vacuoles was blocked by elimination of PI3K, mutations of conserved FYVE domain residues, or by the presence of kinase inhibitor wortmannin, as was shown by fluorescence microscopy [2,3,17,24,35,62]. Consequently, the vesicular localization of these proteins was attributed to the recognition of PtdIns(3)P by their FYVE domains.
Following these initial discoveries, a variety of biophysical and biochemical assays have been employed to characterize the binding properties of FYVE fingers. Based on tryptophan fluorescence and NMR spectroscopy studies, the EEA1 FYVE domain binds soluble dibutanoyl form of PtdIns(3)P with a micromolar affinity (∼130 μM) [8,55,61]. A nanomolar-range affinity of various FYVE domains for PtdIns(3)P in bilayers and monolayers was measured using the liposome binding [31,35,49,63], surface plasmon resonance (SPR) [24,25,49,58], and monolayer penetration [56,58] experiments. The results of these studies have established a general mode of PtdIns(3)P recognition shared among all FYVE proteins while providing details on the unique features of individual FYVE fingers.
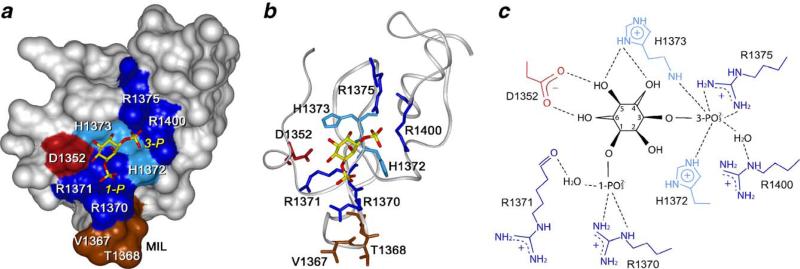
Structural insights of PtdIns(3)P recognition by the FYVE domain are provided by crystal [8] and NMR [7] structures of the EEA1 FYVE domain complexed with inositol 1,3-bisphosphate and dibutanoyl PtdIns(3)P, respectively. Three conserved elements of the FYVE domain, including the N-terminal WxxD, the central basic RR/KHHCR and the C-terminal RVC motifs that together comprise a concave binding pocket, coordinate the PtdIns(3)P head group (Fig. 3). The RR/KHHCR motif, particularly the guanidino moiety of R1375, the imidazole ring of H1372 and the backbone amide of H1373, provides critical hydrogen bonds to the 3-phospate group of PtdIns(3)P [8] (Fig. 3c). The 1-phosphate is recognized by R1370, and forms a water-mediated contact with the R1371's backbone carboxyl group. R1400 of the RVC motif, positioned between H1372 and R1375, forms another water-mediated hydrogen bond to the 3-phosphate group. Recognition of the 4-, 5- and 6-hydroxyl groups of the inositol ring is crucial for stereospecificity and exclusion of alternatively phosphorylated phosphoinositides. This recognition is mediated by D1352 of the N-terminal WxxD motif and H1373. The 4- and 5-hydroxyl groups are hydrogen bonded to the imidazole ring of H1373, whereas the side chain of D1352 contacts the hydroxyls at the 5 and 6 positions. Strong conservation of the binding site elements in the FYVE finger sequences suggests a similar mode of PtdIns(3)P recognition among the majority of these domains (Fig. 2).
Fig. 3.
PtdIns(3)P binding pocket defined from the crystal structure of the EEA1 FYVE domain complexed with 1,3-inositol bisphosphate, a head group of PtdIns(3)P [8] (PDB code 1JOC). (a) Surface of the FYVE domain with the residues involved in PtdIns(3)P binding labeled and colored blue (basic) and red (acidic). MIL is in brown. 1,3-inositol bisphosphate is depicted as a stick model with C/P and O atoms colored yellow and red, respectively. (b) Gray ribbon represents the protein's backbone. Side chains of the binding site residues are shown and colored as in (a). (c) Diagram illustrating 1,3-inositol bisphosphate coordination. Intermolecular hydrogen bonds are shown as dotted lines. The numbering of the FYVE domain residues here and in the text corresponds to the new NCBI deposited sequence of EEA1 (GI: 34222508), which replaced the old sequence (GI:2135066) in 2005. The new sequence matches the numbering in the crystal structure [8] while it differs by one residue from the numbering in solution structure [7] of the EEA1 FYVE domain. This figure and Figs. 5a and 6b are prepared with InsightII.
The critical role of the binding site residues is underscored by mutagenesis. Substitution of the Arg or His residues of the RR/KHHCR motif abolishes PtdIns(3)P interaction in vitro and in vivo, yielding proteins that are diffusely distributed in the cytosol, while mutation of the conserved Trp or Asp residues of the WxxD motif substantially reduces the FYVE domain's affinity for PtdIns(3)P-containing membranes [2,8,49,56].
5. PtdIns(3)P binding is mediated by the histidine switch
Recently we have found that recruitment of the EEA1 FYVE domain to PtdIns(3)P-enriched membranes is strongly pH-dependent and regulated by a histidine switch [57]. This novel mode of regulation differentiates FYVE finger from other phosphoinositide-binding domains and establishes its unique function as a low pH sensor of PtdIns(3)P. As determined by NMR and liposome binding assay, the EEA1 FYVE domain is active and fully PtdIns(3)P-bound in acidic environments, whereas it becomes inactive at pH≥8.0. At low pH (6.0), dibutanoyl PtdIns(3)P is bound by the FYVE domain with a KD of 71 μM. However, the same interaction appears to be much weaker in neutral or basic conditions, that is twice, 20 times or 150 times weaker at pH values of 6.8, 7.4 or 8.0, respectively. Thus, the FYVE domain affinity for PtdIns(3)P varies considerably and is increased in the acidic media.
Similar pH dependence is observed for the in vivo localization of the enhanced green fluorescent protein (EGFP)-fusion EEA1 in mammalian and yeast cells [57]. Lowering the cytosolic pH enhances PtdIns(3)P affinity of the FYVE domain reinforcing the anchoring of EEA1 to endosomal membranes. Reversibly, increasing the pH disrupts the phosphoinositide binding and leads to the cytoplasmic redistribution of EEA1. The most significant changes in the FYVE domain localization occur in the physiological pH range of 6.5 to 7.5. Furthermore, EEA1 translocates to early endosomes during apoptosis-induced acidification of the cytosol with the kinase inhibitor staurosporine (STS). While in the untreated cells, the ECFP-FYVE domain is equally distributed between cytosolic and endosome-bound fractions, the STS treatment substantially shifts the equilibrium toward the membrane-bound state of the protein. These in vivo data corroborate the in vitro experiments and demonstrate the physiological relevance of the pH sensing by EEA1, suggesting that the membrane recruitment of this protein can be regulated by intracellular pH.
The pH dependency is attributed to the histidine switch comprising of a pair of adjacent H1372 and H1373 residues of the RR/KHHCR motif and representing the most conserved amino acids in the FYVE domain sequences [57] (Fig. 2). The EEA1 FYVE domain binds PtdIns(3)P only when both histidines are positively charged and releases the lipid upon their deprotonation. Mutations in the histidine switch completely disrupt PtdIns(3)P binding in vitro and in vivo. Because the two histidine residues are absolutely conserved among PtdIns(3)P-binding FYVE domains, we predict that EEA1 and other FYVE domain-containing proteins exist in a balance between diffuse cytosolic and membrane-anchored populations and are sensitive both to changes in PtdIns(3)P concentrations and acidity within the cell. Based on the estimated affinities and the lipid's physiological concentrations, the EEA1 FYVE domain exists mainly in a bound state at low pH (6.0−6.6). At the cytosolic pH level of 7.3, only half of the protein should be active, while essentially no activity is expected under more basic conditions.
The pH-dependence can influence the function of FYVE proteins in cells with unusual cytosolic pH levels and in normal cells during physiological processes that involve changing of pH. Intracellular pH often fluctuates in response to cell growth, development, and apoptosis, with pH values ranging from 6.3 to 7.5 [64–67], and varies even greater in anomalous cells [68–70]. For example, the cytosol acidification by as much as 0.8 pH units is associated with apoptosis [67], whereas several growth and survival factors induce alkalinization of cytosol by 0.25 pH units [71]. It will be imperative to further investigate the importance of the pH dependency and to test whether the FYVE domain localization is altered in biological processes other than apoptosis that are accompanied by the changes in cytosolic pH.
Intriguingly, the FYVE domains target PtdIns(3)P-containing membranes of primarily low pH compartments, i.e., early endosomes, multivesicular bodies, phagosomes, and Golgi [2–4,41,72]. However the neutral or slightly alkaline organelles with significant PtdIns(3)P concentrations such as the nucleus or mitochondria do not attract FYVE proteins [24,73,74]. Can membrane anchoring of the FYVE domain be mediated by the acidic lumenal pH or local microenvironments and gradients? This hypothesis remains to be tested. Interestingly, EEA1 co-localizes with the endosomal Na+/H+-exchanger around which the formation of acidic microdomains has been proposed [75]. In addition, reduced association of EEA1 to Mycobacterium tuberculosis-containing phagosomes, which are defective in lumenal acidification, has been reported [76].
6. Non-specific electrostatic interactions with acidic lipids
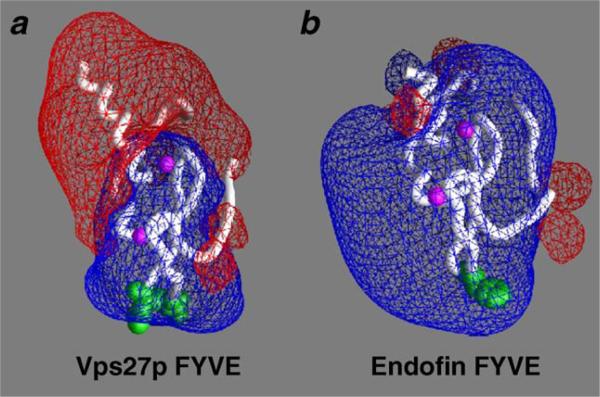
Mammalian early endosomes are enriched in acidic phospholipids other than PtdIns(3)P, such as phosphatidylserine (PS) or phosphatidic acid [77] that contribute to the association of FYVE domains with endosomal membranes. The calculated electrostatic properties of the ligand-free Vps27p, Hrs, EEA1, FENS-1 and Endofin FYVE domains suggest that membrane recruitment of these proteins is facilitated by non-specific electrostatic interactions which occur between basic residues of the FYVE fingers and negatively charged phospholipids in the membrane [54,56]. The calculations show a strong positive potential surrounding the MIL, which may drive the initial membrane association of the proteins and facilitate PtdIns(3)P binding (Fig. 4).
Fig 4.
Electrostatic properties of the Vps27p and Endofin FYVE domains. The FYVE domains are depicted as the white ribbons with zinc ions represented as the magenta balls. The MIL residues are shown in space-filling representation and colored green. The electrostatic potentials of the FYVE domains were calculated using GRASP [94]. Red and blue meshes correspond to the negative and positive potentials, respectively. This figure is kindly provided by D. Murray and R. Stahelin.
Furthermore, non-specific electrostatic interactions continue to play a role after PtdIns(3)P is fully bound. Titration of PS into the EEA1 FYVE domain, which has been pre-bound to PtdIns(3) P and DPC micelles, causes NMR resonance perturbations in residues located in and around the MIL and the site of PtdIns(3)P coordination [55]. The most significant changes are observed in the contiguous polar residues, which form a predominantly basic patch surrounding the MIL, that could easily accommodate a PS headgroup next to a bound PtdIns(3)P molecule. Conservation of the basic residues suggests that the non-specific electrostatic contacts are a common feature among FYVE fingers.
The electrostatic interactions stabilize anchoring of the EEA1 FYVE domain, amplifying its binding affinity by three fold when 10% PS is present in PtdIns(3)P-enriched DPC micelles [55]. Furthermore, these interactions change the domain's orientation with respect to the micelle surface, tilting the FYVE finger by ∼73° and allowing the PS site to interact more extensively with the micelle [55]. PS-stabilization nearly aligns the FYVE domain with the micelle surface in orientation similar to that suggested by the crystal structure of the EEA1 dimer [8], and by computer modeling [54], thus providing a unifying mechanism for the range of previously proposed insertion modes [5,7] (Fig. 5). Interestingly, computational studies predict similar angled membrane orientation for most FYVE domains, with exception of Vps27p, which is positioned to bind in a perpendicular ‘side-on’ orientation [54,56].
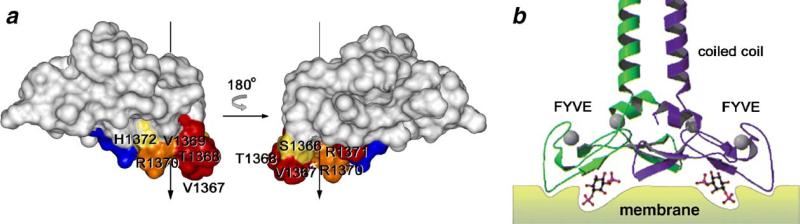
Fig. 5.
Insertion of the MIL into membranes accompanies PtdIns(3)P binding. (a) Depth and angle of the FYVE domain insertion into DPC micelles determined NMR experiments using spin label probes. Inserted residues and residues located near the micelle surface are colored in red and grades of orange, respectively, on FYVE domain surface (NMR structure, PDB code 1HYI). The insertion axis is shown as a vertical vector. (b) Crystal structure of the homodimeric region of containing coiled coil and FYVE domain, shows similar orientation of the protein (PDB code 1JOC).
7. Membrane insertion
In addition to the stereospecific recognition of PtdIns(3)P and non-specific electrostatic contacts with acidic lipids, the exposed hydrophobic residues of MIL (membrane insertion loop) of the FYVE domain penetrate the bilayer. Insertion of the residues at the tip of this loop into membranes upon interaction with PtdIns(3)P was initially suggested based on the crystal structure of Vps27p FYVE domain [5] and on the micelle-induced changes in NMR resonances of the MIL residues of the EEA1 FYVE finger [50]. It was further corroborated by monolayer penetration [56,58], liposome binding [31,56], computational modeling [54] and NMR studies with membrane mimetic micelle systems [7,55].
The multiple anchoring resulting from binding the PtdIns(3) P headgroup, non-specific electrostatic interactions and insertion of a set of aliphatic or aromatic residues provides the strength and selectivity that are necessary for the proper localization and function of the FYVE fingers. Thus, FYVE domains bind PtdIns(3)P-enriched acidic vesicles several orders of magnitude more tightly than soluble lipids or isolated inositol headgroups [8,49]. The FYVE domain of EEA1 exhibits a 130 μM affinity for short chain dibutanoyl PtdIns(3)P [8,55], while a 50 nM affinity is measured for a long chain lipid embedded in acidic liposomes [49]. Other FYVE proteins including FENS-1, Endofin, Drosophila Hrs and yeast Vps27p bind PtdIns(3)P-containing liposomes with comparable affinities of 0.6 nM, 1 nM, 25 nM and 32 nM, respectively [25,56,58]. However, soluble 1,3-inositol bisphosphate is recognized by these FYVE domains three orders of magnitude weaker [58]. Even in the neutral vesicles, human Hrs FYVE domain greatly prefers intact lipid in a bilayer over the isolated headgroup [31]. Likewise, the EEA1 FYVE domain affinity is significantly increased in the presence of dodecylphosphocholine (DPC) micelles [55]. Consequently, the hydrophobic insertion in concert with electrostatic interactions stabilizes anchoring of the FYVE domains to membranes.
Recent computational and monolayer penetration studies have shown that binding of PtdIns(3)P facilitates penetration of the FYVE domains and increases their membrane residence time by decreasing the positive charge surrounding the MIL [54,56]. In agreement, selective broadening of MIL resonances of the EEA1 FYVE domain by paramagnetic probes incorporated into DPC micelles is not observed in the absence of PtdIns (3)P [55], supporting that the deep and stable insertion into membrane requires PtdIns(3)P.
The depth and angle of the insertion into micelles have been determined by NMR experiments using spin label probes incorporated at various positions within micelles and based on intermolecular nuclear Overhauser effects observed between the MIL residues of EEA1 FYVE and the alkyl chain of DPC [55,78] (Fig. 5a). The VT1368 sequence of the EEA1 FYVE domain is immersed into the hydrophobic core of DPC micelles, and is surrounded by polar and hydrophilic residues located at the level of the lipid's headgroups and properly positioned for the non-specific electrostatic contacts. Conformational changes in the MIL accompany the micelle interaction, in which hydrophobic residues of the loop tend to move deeper into the non-polar core of micelles, whereas hydrophilic residues move toward the aqueous interface, hence stretching the MIL [78].
Substitution of the membrane-inserting residues of the FYVE domains abolishes or significantly decreases the membrane association and disrupts the normal biological functions of these proteins [50,56,61]. Thus, the endosomal localization of EEA1 is completely lost when V1367 and T1368 are mutated to Gly or Glu [50]. Similarly, replacement of corresponding hydrophobic residues of the Vps27p or Hrs FYVE domains results in a seven to twenty fold reduction of their affinities for the membrane-bound PtdIns(3)P [56].
While the hydrophobic nature of MIL is conserved among FYVE domains, suggesting a similar mode of insertion, variations occur. In particular, hydrophobic residue immediately preceding the RR/KHHCR motif is often replaced by a basic or a polar residue. Two human FYVE domains, FENS-1 and WDF2 contain an 11-residue insert in their MILs. Furthermore, biophysical measurements and computational studies suggest that minor structural variations in the MIL cause considerable changes in the membrane binding properties and in subcellular localizations of the FYVE domains [58]. The FYVE fingers of EEA1, FENS-1 and Endofin penetrate membranes much better than the FYVE domains of Vps27p and Hrs [56,58]. Although the contribution of MIL penetration to the energetics of membrane targeting varies, it has been noticed that, remarkably, all FYVE proteins, which are able to localize to endosomal membranes, penetrate the bilayer [58].
8. Multiple PtdIns(3)P interactions and dimerization
Membrane localization of a number of FYVE proteins is greatly enhanced by bivalent or multivalent PtdIns(3)P interactions. For example, the synthetic construct containing a pair of covalently linked Hrs FYVE domains shows much higher affinity for PtdIns(3)P-containing membranes and was cleverly used to map the intracellular distribution of PtdIns(3)P [24]. While a singly expressed GFP-fusion FYVE domain of Hrs appears to be cytosolic, it translocates to endosomal membranes when a region adjacent to the FYVE finger is dimerized [61]. The stable dimers and higher oligomers are intrinsically formed by the FYVE-containing SARA and this ability to oligomerize is important for the endosomal localization of this protein [61]. Furthermore, some proteins, such as DFCP1, naturally contain a FYVE domain tandem, which selectively binds PtdIns(3)P [25,26].
The FYVE domain-containing proteins can also dimerize by their coiled coil modules. The coiled coil region of EEA1 forms a parallel homodimer that juxtaposes two C-terminal FYVE domains allowing for simultaneous interactions with two PtdIns (3)P headgroups [8,59] (Fig. 5b). When conjugated with this coiled coil, other cytosolic FYVE domains are able to localize to endosomes [3]. The coiled coils are also predicted next to the FYVE domains of human proteins including FYCO1, Hrs, MTMR3, MTMR4, Rabenosyn-5 and RUFY2. Increased avidity due to the parallel dimerization may represent yet another means to boost the membrane binding by the FYVE fingers.
Dimerization-mediated targeting is suggested from the crystal structure of a long EEA1 construct, which contains a substantial portion of the coiled coil region and an adjacent FYVE domain [8]. The structure identifies a 1200 Å2 interface between the two FYVE domains that was devoid of hydrogen bonding and electrostatic interactions (Fig. 5b). The side chains of the conserved tryptophan of the WxxD motif and a non-conserved proline of the variable β3/β4 hairpin comprise the core of the interface. It contributes weakly to the stability of the EEA1 homodimer and together with more extensive and robust coiled coil interaction juxtaposes the two FYVE domains to concomitantly bind two PtdIns(3)P molecules [8] (Fig. 5b). In this model, the MILs of both FYVE domains are positioned to angularly penetrate the membrane, which is in good agreement with the NMR-derived orientation of a single FYVE domain [7,55], computational modeling [54] and biophysical studies [56,58].
A distinct dimer has been observed in the crystal structure of the Drosophila Hrs FYVE domain [6]. Antiparallel packing between the Hrs FYVE domains, bridged by a citrate ion, buries 1920 Å2 in the interface, including the functionally important N-terminal tryptophan and the second histidine of the RR/KHHCR motif. Due to partial occlusion of the binding site, a different PtdIns(3)P coordination and membrane docking mode have been proposed. The general relevance of the Drosophila Hrs dimer interface remains unclear.
Despite the ability of some FYVE domain-containing proteins to dimerize or oligomerize, it appears that isolated FYVE fingers can strongly bind PtdIns(3)P in vitro and target proteins to endosomes in vivo. For example, both full-length FENS-1 and its isolated FYVE domain bind PtdIns(3)P and localize to endosomes to the same extent [25]. The endosomal localization of Endofin is also comparable to the distribution of its isolated FYVE domain in cytoplasmic vesicles [79]. Ultracentrifugation experiments show that FENS-1, Endofin and Hrs remain monomeric even at high protein concentrations [58]. In addition, localization of Rabenosyn-5 to early endosomes is shown to be similar to that of a truncated version, which contains primarily its FYVE domains [2,17]. Thus, dimerization of FYVE domain-containing proteins is not strictly required. It is, however, important for the membrane targeting of a subset of FYVE domains.
9. Regulation of activity
Function of the FYVE domain can be regulated at several levels. The intracellular concentration of PtdIns(3)P is increased by the activity of phosphatidylinositol 3-kinases [80,81] and decreased by PtdIns(3)P 5-kinases [34,36,82,83] and MTMR phosphatases [38]. The endocytic PtdIns(3)P pool is hydrolyzed in lysosomal compartments, which is a principal pathway of PtdIns(3)P turnover [84]. In addition, local pools of this lipid may be affected and stabilized by proteins that contain the PtdIns(3)P-recognizing FYVE, PX [85–88] and PH [89] domains.
The activity of FYVE domains may also be modulated by protein interactions. For example, the EEA1 FYVE domain's affinity for PtdIns(3)P-enriched membranes is increased by coiled coil dimerization [8]. Furthermore, the membrane recruitment of EEA1 and Rabenosin-5 is found to be regulated by p38 mitogen-activated protein kinase (MAPK) that phosphorylates their FYVE domains on T1392 and S215 residues, respectively [90]. Other proteins may directly bind FYVE fingers, as has been reported for Src homology 3 domain of Etk, which recognizes the FYVE domain of RUFY1 [91].
Recent studies have shown that the FYVE domain of EEA1 binds PtdIns(3)P in a pH dependent manner in vitro and in vivo [57]. However, several questions remain unanswered. Is targeting of the EEA1 FYVE domain to endosomal membranes regulated by the changes in cytosolic pH that occur in a variety of biological processes? Does the EEA1 FYVE domain sense acidic intralumenal pH? Do other FYVE domains exhibit similar pH dependence? Why do most FYVE proteins target endocytic pools of PtdIns(3)P and not its nuclear or mitochondrial accumulations? Furthermore, the mechanisms by which FYVE proteins localize to the Golgi [25,26], plasma membrane [32] or nuclear envelope [27,33] remain to be determined, as do the roles of their FYVE fingers.
10. Conclusion
Remarkable progress has been made since the discovery of FYVE domain 10 years ago. The structural, biophysical and cellular studies offer a detailed molecular mechanism of the multivalent docking and anchoring of the monomeric or dimeric FYVE domain to PtdIns(3)P-containing membranes (Fig. 6). This mechanism involves stereospecific PtdIns(3)P head group recognition, which is facilitated by non-specific electrostatic contacts with acidic lipids and modulated by the histidine switch, and is accompanied by hydrophobic insertion into the bilayers.
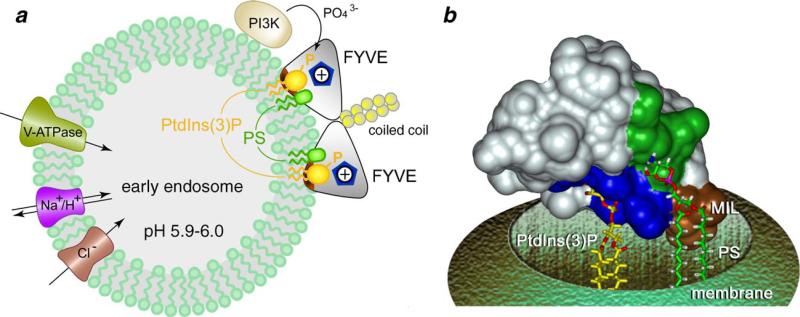
Fig 6.
Multivalent molecular mechanism of the membrane docking by the FYVE domain. (a) Schematics of the dimeric EEA1 anchoring to PtdIns(3)P-enriched endosomal membranes. Dimerized coiled coils juxtapose two FYVE domains for the simultaneous interactions with two head groups of PtdIns(3)P (dark yellow). Binding to PtdIns(3)P is facilitated by non-specific electrostatic contacts with acidic lipids (PS, green) and accompanied by a hydrophobic insertion (brown) into the bilayer. The protonated state of the two adjacent His1371 and His1372 residues (blue pentagon) is required for the binding. A signal is initiated by phosphorylation of PtdIns at the third position by PI3K. Three membrane proteins, V-type ATPase, Cl− channel and Na+/H+-exchanger regulate the acidification of early endosomes. (b) A model of the monomeric FYVE domain anchoring (NMR structure, PDB code 1HYI). Conserved His and Arg residues of the PtdIns(3)P binding pocket, as well as MIL and PS binding site are colored in blue, brown and green, respectively. PtdIns(3)P and PS are shown as stick models.
Acknowledgements
The author thanks C. Burd, S. Emr and A. Sorkin for helpful discussions, D. Murray and R. Stahelin for kindly providing Fig. 4, and J. Kice, S. Lee and H. Witte for critical reading of the manuscript. This work is supported by the NIH, American Cancer Society and University of Colorado Cancer Center.
References
- 1.Stenmark H, Aasland R, Toh BH, D'Arrigo A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J. Biol. Chem. 1996;271:24048–24054. doi: 10.1074/jbc.271.39.24048. [DOI] [PubMed] [Google Scholar]
- 2.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 3.Gaullier JM, Simonsen A, D'Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P [letter; comment] Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 4.Patki V, Lawe DC, Corvera S, Virbasius JV, Chawla A. A functional PtdIns(3)P-binding motif [letter] [see comments] Nature. 1998;394:433–434. doi: 10.1038/28771. [DOI] [PubMed] [Google Scholar]
- 5.Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- 6.Mao Y, Nickitenko A, Duan X, Lloyd TE, Wu MN, Bellen H, Quiocho FA. Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell. 2000;100:447–456. doi: 10.1016/s0092-8674(00)80680-7. [DOI] [PubMed] [Google Scholar]
- 7.Kutateladze TG, Overduin M. Structural mechanism of endosome docking by the FYVE domain. Science. 2001;291:1793–1796. doi: 10.1126/science.291.5509.1793. [DOI] [PubMed] [Google Scholar]
- 8.Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright D. Multivalent endosome targeting by homodimeric EEA1. Mol. Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 9.Stenmark H, Gillooly DJ. Intracellular trafficking and turnover of phosphatidylinositol 3-phosphate. Cell Dev. Biol. 2001;12:193–199. doi: 10.1006/scdb.2000.0236. [DOI] [PubMed] [Google Scholar]
- 10.Patki V, Virbasius J, Lane WS, Toh BH, Shpetner HS, Corvera S. Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7326–7330. doi: 10.1073/pnas.94.14.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raiborg C, Bache KG, Mehlum A, Stang E, Stenmark H. Hrs recruits clathrin to early endosomes. EMBO J. 2001;20:5008–5021. doi: 10.1093/emboj/20.17.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI (3)K function to Rab5 regulation of endosome fusion [see comments] Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 13.Mills IG, Jones AT, Clague MJ. Involvement of the endosomal autoantigen EEA1 in homotypic fusion of early endosomes. Curr. Biol. 1998;8:881–884. doi: 10.1016/s0960-9822(07)00351-x. [DOI] [PubMed] [Google Scholar]
- 14.McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 15.Rubino M, Miaczynska M, Lippe R, Zerial M. Selective membrane recruitment of EEA1 suggests a role in directional transport of clathrin-coated vesicles to early endosomes. J. Biol. Chem. 2000;275:3745–3748. doi: 10.1074/jbc.275.6.3745. [DOI] [PubMed] [Google Scholar]
- 16.Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, Hoflack B, Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cormont M, Mari M, Galmiche A, Hofman P, Le Marchand-Brustel Y. A FYVE-finger-containing protein, Rabip4, is a Rab4 effector involved in early endosomal traffic. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1637–1642. doi: 10.1073/pnas.031586998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson MR, Burd CG, Emr SD. Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr. Biol. 1999;9:159–162. doi: 10.1016/s0960-9822(99)80071-2. [DOI] [PubMed] [Google Scholar]
- 20.Komada M, Soriano P. Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Gen. Dev. 1999;13:1475–1485. doi: 10.1101/gad.13.11.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vieira OV, Harrison RE, Scott CC, Stenmark H, Alexander D, Liu J, Gruenberg J, Schreiber AD, Grinstein S. Acquisition of Hrs, an essential component of phagosomal maturation, is impaired by mycobacteria. Mol. Cell. Biol. 2004;24:4593–4604. doi: 10.1128/MCB.24.10.4593-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morino C, Kato M, Yamamoto A, Mizuno E, Hayakawa A, Komada M, Kitamura N. A role for Hrs in endosomal sorting of ligand-stimulated and unstimulated epidermal growth factor receptor. Exp. Cell Res. 2004;297:380–391. doi: 10.1016/j.yexcr.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 23.Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridley SH, Ktistakis N, Davidson K, Anderson KE, Manifava M, Ellson CD, Lipp P, Bootman M, Coadwell J, Nazarian A, Erdjument-Bromage H, Tempst P, Cooper MA, Thuring JW, Lim ZY, Holmes AB, Stephens LR, Hawkins PT. FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments. J. Cell Sci. 2001;114:3991–4000. doi: 10.1242/jcs.114.22.3991. [DOI] [PubMed] [Google Scholar]
- 26.Cheung PC, Trinkle-Mulcahy L, Cohen P, Lucocq JM. Characterization of a novel phosphatidylinositol 3-phosphate-binding protein containing two FYVE fingers in tandem that is targeted to the Golgi. Biochem. J. 2001;355:113–121. doi: 10.1042/0264-6021:3550113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunkelberg JC, Gutierrez-Hartmann A. LZ-FYVE: a novel developmental stage-specific leucine zipper, FYVE-finger protein. DNA Cell Biol. 2001;20:403–412. doi: 10.1089/104454901750361460. [DOI] [PubMed] [Google Scholar]
- 28.Pasteris NG, Cadle A, Logie LJ, Porteous ME, Schwartz CE, Stevenson RE, Glover TW, Wilroy RS, Gorski JL. Isolation and characterization of the faciogenital dysplasia (Aarskog–Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell. 1994;79:669–678. doi: 10.1016/0092-8674(94)90552-5. [DOI] [PubMed] [Google Scholar]
- 29.Olson MF, Pasteris NG, Gorski JL, Hall A. Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho GTPases. Curr. Biol. 1996;6:1628–1633. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- 30.Nagata K, Driessens M, Lamarche N, Gorski JL, Hall A. Activation of G1 progression, JNK mitogen-activated protein kinase, and actin filament assembly by the exchange factor FGD1. J. Biol. Chem. 1998;273:15453–15457. doi: 10.1074/jbc.273.25.15453. [DOI] [PubMed] [Google Scholar]
- 31.Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA. High affinity binding of a FYVE domain to phosphatidylinositol 3-phosphate requires intact phospholipid but not FYVE domain oligomerization. Biochemistry. 2001;40:8581–8587. doi: 10.1021/bi010425d. [DOI] [PubMed] [Google Scholar]
- 32.Obaishi H, Nakanishi H, Mandai K, Satoh K, Satoh A, Takahashi K, Miyahara M, Nishioka H, Takaishi K, Takai Y. Frabin, a novel FGD1-related actin filament-binding protein capable of changing cell shape and activating c-Jun N-terminal kinase. J. Biol. Chem. 1998;273:18697–18700. doi: 10.1074/jbc.273.30.18697. [DOI] [PubMed] [Google Scholar]
- 33.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 34.Ikonomov OC, Sbrissa D, Shisheva A. Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J. Biol. Chem. 2001;276:26141–26147. doi: 10.1074/jbc.M101722200. [DOI] [PubMed] [Google Scholar]
- 35.Sbrissa D, Ikonomov OC, Shisheva A. Phosphatidylinositol 3-phosphate-interacting domains in PIKfyve. J. Biol. Chem. 2002;277:6073–6079. doi: 10.1074/jbc.M110194200. [DOI] [PubMed] [Google Scholar]
- 36.Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, Hall MN, Michell RH, Parker PJ. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr. Biol. 1998;8:1219–1222. doi: 10.1016/s0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- 37.Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 38.Taylor GS, Maehama T, Dixon JE. Inaugural article: myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorenzo O, Urbe S, Clague MJ. Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J. Cell Sci. 2005;118:2005–2012. doi: 10.1242/jcs.02325. [DOI] [PubMed] [Google Scholar]
- 40.Shin ME, Ogburn KD, Varban OA, Gilbert PM, Burd CG. FYVE domain targets Pib1p ubiquitin ligase to endosome and vacuolar membranes. J. Biol. Chem. 2001;276:41388–41393. doi: 10.1074/jbc.M105665200. [DOI] [PubMed] [Google Scholar]
- 41.Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 42.Runyan CE, Schnaper HW, Poncelet AC. The role of internalization in transforming growth factor beta1-induced Smad2 association with Smad anchor for receptor activation (SARA) and Smad2-dependent signaling in human mesangial cells. J. Biol. Chem. 2005;280:8300–8308. doi: 10.1074/jbc.M407939200. [DOI] [PubMed] [Google Scholar]
- 43.Panopoulou E, Gillooly DJ, Wrana JL, Zerial M, Stenmark H, Murphy C, Fotsis T. Early endosomal regulation of Smad-dependent signaling in endothelial cells. J. Biol. Chem. 2002;277:18046–18052. doi: 10.1074/jbc.M107983200. [DOI] [PubMed] [Google Scholar]
- 44.Hayes S, Chawla A, Corvera S. TGF beta receptor internalization into EEA1-enriched early endosomes: role in signaling to Smad2. J. Cell Biol. 2002;158:1239–1249. doi: 10.1083/jcb.200204088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itoh F, Divecha N, Brocks L, Oomen L, Janssen H, Calafat J, Itoh S, Dijke Pt P. The FYVE domain in Smad anchor for receptor activation (SARA) is sufficient for localization of SARA in early endosomes and regulates TGF-beta/Smad signalling. Genes Cells. 2002;7:321–331. doi: 10.1046/j.1365-2443.2002.00519.x. [DOI] [PubMed] [Google Scholar]
- 46.Miura S, Takeshita T, Asao H, Kimura Y, Murata K, Sasaki Y, Hanai JI, Beppu H, Tsukazaki T, Wrana JL, Miyazono K, Sugamura K. Hgs (Hrs), a FYVE domain protein, is involved in Smad signaling through cooperation with SARA. Mol. Cell. Biol. 2000;20:9346–9355. doi: 10.1128/mcb.20.24.9346-9355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asao H, Sasaki Y, Arita T, Tanaka N, Endo K, Kasai H, Takeshita T, Endo Y, Fujita T, Sugamura K. Hrs is associated with STAM, a signal-transducing adaptor molecule. Its suppressive effect on cytokine-induced cell growth. J. Biol. Chem. 1997;272:32785–32791. doi: 10.1074/jbc.272.52.32785. [DOI] [PubMed] [Google Scholar]
- 48.Sasaki Y, Sugamura K. Involvement of Hgs/Hrs in signaling for cytokine-mediated c-fos induction through interaction with TAK1 and Pak1. J. Biol. Chem. 2001;276:29943–29952. doi: 10.1074/jbc.M104230200. [DOI] [PubMed] [Google Scholar]
- 49.Gaullier JM, Ronning E, Gillooly DJ, Stenmark H. Interaction of the EEA1 FYVE finger with phosphatidylinositol 3-phosphate and early endosomes. Role of conserved residues. J. Biol. Chem. 2000;275:24595–24600. doi: 10.1074/jbc.M906554199. [DOI] [PubMed] [Google Scholar]
- 50.Kutateladze TG, Ogburn KD, Watson WT, de Beer T, Emr SD, Burd CG, Overduin M. Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol. Cell. 1999;3:805–811. doi: 10.1016/s1097-2765(01)80013-7. [DOI] [PubMed] [Google Scholar]
- 51.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struck. Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 52.Brown RS. Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struck. Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 53.Ostermeier C, Brunger AT. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- 54.Diraviyam K, Stahelin RV, Cho W, Murray D. Computer modeling of the membrane interaction of FYVE domains. J. Mol. Biol. 2003;328:721–736. doi: 10.1016/s0022-2836(03)00325-5. [DOI] [PubMed] [Google Scholar]
- 55.Kutateladze TG, Capelluto DGS, Ferguson CG, Cheever ML, Kutateladze AG, Prestwich GD, Overduin M. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 2004;279:3050–3057. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]
- 56.Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J. Biol. Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 57.Lee SA, Eyeson R, Cheever ML, Geng J, Verkhusha VV, Burd C, Overduin M, Kutateladze TG. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13052–13057. doi: 10.1073/pnas.0503900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blatner NR, Stahelin RV, Diraviyam K, Hawkins PT, Hong W, Murray D, Cho W. The molecular basis of the differential subcellular localization of FYVE domains. J. Biol. Chem. 2004;279:53818–53827. doi: 10.1074/jbc.M408408200. [DOI] [PubMed] [Google Scholar]
- 59.Callaghan J, Simonsen A, Gaullier JM, Toh BH, Stenmark H. The endosome fusion regulator early-endosomal autoantigen 1 (EEA1) is a dimer. Biochem. J. 1999;338:539–543. [PMC free article] [PubMed] [Google Scholar]
- 60.Lawe DC, Patki V, Heller-Harrison R, Lambright D, Corvera S. The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J. Biol. Chem. 2000;275:3699–3705. doi: 10.1074/jbc.275.5.3699. [DOI] [PubMed] [Google Scholar]
- 61.Hayakawa A, Hayes SJ, Lawe DC, Sudharshan E, Tuft R, Fogarty K, Lambright D, Corvera S. Structural basis for endosomal targeting by FYVE domains. J. Biol. Chem. 2004;279:5958–5966. doi: 10.1074/jbc.M310503200. [DOI] [PubMed] [Google Scholar]
- 62.Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayakawa A, Kitamura N. Early endosomal localization of hrs requires a sequence within the proline- and glutamine-rich region but not the FYVE finger. J. Biol. Chem. 2000;275:29636–29642. doi: 10.1074/jbc.M002696200. [DOI] [PubMed] [Google Scholar]
- 64.Gottlieb RA, Giesing HA, Zhu JY, Engler RL, Babior BM. Cell acidification in apoptosis: granulocyte colony-stimulating factor delays programmed cell death in neutrophils by up-regulating the vacuolar H(+)-ATPase. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5965–5968. doi: 10.1073/pnas.92.13.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moolenaar WH, Tsien RY, van der Saag PT, de Laat SW. Na+/H+ exchange and cytoplasmic pH in the action of growth factors in human fibroblasts. Nature. 1983;304:645–648. doi: 10.1038/304645a0. [DOI] [PubMed] [Google Scholar]
- 66.Schuldiner S, Rozengurt E. Na+/H+ antiport in Swiss 3T3 cells: mitogenic stimulation leads to cytoplasmic alkalinization. Proc. Natl. Acad. Sci. U. S. A. 1982;79:7778–7782. doi: 10.1073/pnas.79.24.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thangaraju M, Sharma K, Leber B, Andrews DW, Shen SH, Srikant CB. Regulation of acidification and apoptosis by SHP-1 and Bcl-2. J. Biol. Chem. 1999;274:29549–29557. doi: 10.1074/jbc.274.41.29549. [DOI] [PubMed] [Google Scholar]
- 68.LaManna JC. Hypoxia/ischemia and the pH paradox. Adv. Exp. Med. Biol. 1996;388:283–292. doi: 10.1007/978-1-4613-0333-6_36. [DOI] [PubMed] [Google Scholar]
- 69.Punnia-Moorthy A. Evaluation of pH changes in inflammation of the subcutaneous air pouch lining in the rat, induced by carrageenan, dextran and Staphylococcus aureus. J. Oral Pathol. 1987;16:36–44. doi: 10.1111/j.1600-0714.1987.tb00674.x. [DOI] [PubMed] [Google Scholar]
- 70.Gerweck LE, Seetharaman K. Cellular pH gradient in tumor versus normal tissue: potential exploitation for the treatment of cancer. Cancer Res. 1996;56:1194–1198. [PubMed] [Google Scholar]
- 71.Rajotte D, Haddad P, Haman A, Cragoe EJ, Jr., Hoang T. Role of protein kinase C and the Na+/H+ antiporter in suppression of apoptosis by granulocyte macrophage colony-stimulating factor and interleukin-3. J. Biol. Chem. 1992;267:9980–9987. [PubMed] [Google Scholar]
- 72.Derubeis AR, Young MF, Jia L, Robey PG, Fisher LW. Double FYVE-containing protein 1 (DFCP1): isolation, cloning and characterization of a novel FYVE finger protein from a human bone marrow cDNA library. Gene. 2000;255:195–203. doi: 10.1016/s0378-1119(00)00303-6. [DOI] [PubMed] [Google Scholar]
- 73.Matsuyama S, Llopis J, Deveraux QL, Tsien RY, Reed JC. Changes in intramitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat. Cell Biol. 2000;2:318–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- 74.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akhter S, Kovbasnjuk O, Li X, Cavet M, Noel J, Arpin M, Hubbard AL, Donowitz M. Na(+)/H(+) exchanger 3 is in large complexes in the center of the apical surface of proximal tubule-derived OK cells. Am. J. Physiol., Cell Physiol. 2002;283:C927–C940. doi: 10.1152/ajpcell.00613.2001. [DOI] [PubMed] [Google Scholar]
- 76.Fratti RA, Chua J, Deretic V. Induction of p38 mitogen-activated protein kinase reduces early endosome autoantigen 1 (EEA1) recruitment to phagosomal membranes. J. Biol. Chem. 2003;278:46961–46967. doi: 10.1074/jbc.M305225200. [DOI] [PubMed] [Google Scholar]
- 77.Kobayashi T. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- 78.Brunecky R, Lee S, Rzepecki PW, Overduin M, Prestwich GD, Kutateladze AG, Kutateladze TG. Investigation of the binding geometry of a peripheral membrane protein. Biochemistry. 2005;44:16064–16071. doi: 10.1021/bi051127+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seet LF, Hong W. Endofin, an endosomal FYVE domain protein. J. Biol. Chem. 2001;276:42445–42454. doi: 10.1074/jbc.M105917200. [DOI] [PubMed] [Google Scholar]
- 80.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 81.Volinia S, Dhand R, Vanhaesebroeck B, MacDougall LK, Stein R, Zvelebil MJ, Domin J, Panaretou C, Waterfield MD. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sbrissa D, Ikonomov OC, Shisheva A. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides—Effect of insulin. J. Biol. Chem. 1999;274:21589–21597. doi: 10.1074/jbc.274.31.21589. [DOI] [PubMed] [Google Scholar]
- 84.Wurmser AE, Emr SD. Phosphoinositide signaling and turnover: PtdIns (3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 1998;17:4930–4942. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Y, Hortsman H, Seet L, Wong SH, Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- 86.Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, Cantley LC, Yaffe MB. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 87.Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, Tempst P, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Gaffney PR, Coadwell J, Chilvers ER, Hawkins PT, Stephens LR. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox) Nat. Cell Biol. 2001;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- 88.Song X, Xu W, Zhang A, Huang G, Liang X, Virbasius JV, Czech MP, Zhou GW. Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry. 2001;40:8940–8944. doi: 10.1021/bi0155100. [DOI] [PubMed] [Google Scholar]
- 89.Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mace G, Miaczynska M, Zerial M, Nebreda AR. Phosphorylation of EEA1 by p38 MAP kinase regulates mu opioid receptor endocytosis. EMBO J. 2005;24:3235–3246. doi: 10.1038/sj.emboj.7600799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang J, Qiu Y. Interaction between tyrosine kinase Etk and a RUN domain- and FYVE domain-containing protein RUFY1. A possible role of ETK in regulation of vesicle trafficking. J. Biol. Chem. 2002;277:30066–30071. doi: 10.1074/jbc.M111933200. [DOI] [PubMed] [Google Scholar]
- 92.Kraulis P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 93.Merritt EA, Bacon DJ. Raster3D: Photorealistic Molecular Graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 94.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]