Abstract
Saccharomyces cerevisiae has been used for millennia in winemaking, but little is known about the selective forces acting on the wine yeast genome. We sequenced the complete genome of the diploid commercial wine yeast EC1118, resulting in an assembly of 31 scaffolds covering 97% of the S288c reference genome. The wine yeast differed strikingly from the other S. cerevisiae isolates in possessing 3 unique large regions, 2 of which were subtelomeric, the other being inserted within an EC1118 chromosome. These regions encompass 34 genes involved in key wine fermentation functions. Phylogeny and synteny analyses showed that 1 of these regions originated from a species closely related to the Saccharomyces genus, whereas the 2 other regions were of non-Saccharomyces origin. We identified Zygosaccharomyces bailii, a major contaminant of wine fermentations, as the donor species for 1 of these 2 regions. Although natural hybridization between Saccharomyces strains has been described, this report provides evidence that gene transfer may occur between Saccharomyces and non-Saccharomyces species. We show that the regions identified are frequent and differentially distributed among S. cerevisiae clades, being found almost exclusively in wine strains, suggesting acquisition through recent transfer events. Overall, these data show that the wine yeast genome is subject to constant remodeling through the contribution of exogenous genes. Our results suggest that these processes are favored by ecologic proximity and are involved in the molecular adaptation of wine yeasts to conditions of high sugar, low nitrogen, and high ethanol concentrations.
Keywords: adaptive evolution, comparative genomics, horizontal gene transfer, introgression, Zygosaccharomyces bailii
The yeast Saccharomyces cerevisiae has been associated with human activity for thousands of years. The earliest evidence of winemaking has been dated to ≈7,000 years ago (1). The fermentation of grape juice exposes yeast cells to harsh environmental conditions (high sugar concentration, increasing alcohol concentration, acidity, presence of sulfites, anaerobiosis, and progressive depletion of essential nutrients, such as nitrogen, vitamins, and lipids). These conditions, as well as unwitting selection by man for optimal winemaking traits (fermentation performance, alcohol tolerance, and good flavor production) have generated hundred of strains that are currently used in the wine industry. As a result, wine yeast isolates belong to a well-defined lineage (2–6).
Deciphering the mechanisms that participate to these evolutionary processes and identifying the variations contributing to the properties of wine yeast remain challenging issues. Wine yeasts are often diploid, heterozygous, and homothallic (4, 7, 8). They have a large capacity for genome reorganization through chromosome rearrangements (9–11), promoting rapid adaptation to environmental changes. Comparative genomics is a suitable approach for cataloguing multiple types of sequence variation between yeast strains. Comparative genome hybridization analysis of several S. cerevisiae genomes has resulted in the identification of gene deletions and amplifications common to most wine yeast strains (7, 12). Analyses of the genome sequences of yeast strains of various origins have shown that nucleotide polymorphism may be the main source of phenotypic variation (5, 6, 13−15).
With a view to deciphering the genetic basis of winemaking traits, we determined the complete genome sequence of the diploid commercial wine yeast strain EC1118. The full gene repertoire of EC1118 was established and provided evidence for several gene transfer events, which were analyzed in detail. These findings provide unprecedented insight into the molecular mechanisms contributing to the adaptation of yeast to winemaking.
Results
Genome Sequence and Analysis.
The diploid EC1118 genome was sequenced and assembled using a Sanger/pyrosequencing hybrid approach [supporting information (SI) Table S1 and SI Materials and Methods]. Early in the assembly process it became clear that distinct haplotypes could not be obtained in most cases, because heterozygosity levels were very low (approximately 0.2%). An 11.7-Mb high-quality “pseudohaploid” assembly with 31 scaffolds was obtained (Table S1), corresponding to 96.7% of the S288c nuclear genome, as determined from genome alignments. Nucleotide alignments with other S. cerevisiae strains (Table S2) revealed a similar level of nucleotide polymorphism between EC1118 and S288c or the clinical isolate derivative YJM789 (46,825 and 47,253, respectively) and, as expected, a much lower level of nucleotide variation compared with the wine yeast derivatives RM11–1a and AWRI1631 (19,142 and 18,315, respectively).
A total of 5,728 ORFs (except dubious and Ty-associated genes) has been predicted for the nuclear genome of EC1118 (Table S3), of which 5,685 are common to EC1118 and S288c. Several of these ORFs are predicted to be affected by frameshifts (303 ORFs), in-frame stop codons (25 ORFs), or the absence of start or stop codons due to the presence of SNPs or indels (15 ORFs). We identified 11 Ty elements in the assembly (2 Ty1, 7 Ty2, 1 Ty4, and 1 Ty5), whereas 50 such elements have been identified in the S288c genome. We detected no Ty3 elements. This depletion of Ty elements is consistent with the results of comparative genome hybridization for the EC1118 strain (12). This overall picture was further supported by a direct estimate of the overall Ty abundance from sequencing reads (1.8%), much lower than that in S288c (3.4%), which was found to have the highest Ty abundance in a previous population study (5). This analysis also confirmed a clear inversion in the proportions of Ty1 and Ty2 in EC1118 compared with S288c.
Genes Present in S288c but Missing from EC1118.
In total, 111 of the genes present in S288c were not found in the EC1118 genome (Table S4). Most of these genes are repeated and located in subtelomeric regions, which have not been accurately assembled, making it difficult to estimate copy number precisely. However, several of these genes (e.g., HXT16, PAU21, and SOR1) are known to vary in copy number between strains (7, 12, 14). A large 17-kb telomeric region on chromosome VI encompassing YFL052W to YFL058W was absent in EC1118. Nontelomeric genes (21 genes) were also found absent from EC1118. They consist mainly of genes that are present in tandem duplicated arrays (ENA2/5, MST27, PRM8, ASP3, and FCY22) or in a 20.5-kb region of chromosome XII adjacent to the rDNA array, including 4 copies of ASP3. Most of the missing nontelomeric genes were found frequently deleted in other S. cerevisiae strains (Table S4). Two missing genes, MST27 and PRM8, belonging to the DUP240 family, have been found depleted in other wine yeasts (12, 16).
Genes Present in EC1118 but Missing from S288c.
We identified 34 ORFs in EC1118, encoding proteins of 50 to 150 aa, that were absent from S288c. Only 6 of these ORFs were kept in EC1118 annotation (Table S3), thanks to the presence of identified orthologs in S. cerevisiae strains YJM789, RM11–1a, and AWRI1631 and conserved genomic sequences in Saccharomyces sensu stricto: EC1118_1J19_0562g, present in most of these strains and species, EC1118_1G1_0023g, highly conserved in S. mikatae, the duplicated EC1118_1M36_0034g and EC1118_1M36_0045g, present in a single copy in S. mikatae and in AWRI1631 and in 2 copies in Schizosaccharomyces japonicus, and two other genes with a defined function. The first gene, KHR1 (EC1118_1I12_1684g), which encodes a heat-resistant killer toxin, is located in a 1.6-kb fragment inserted into EC1118 chromosome IX and flanked by 2 LTR elements. KHR1 was also found at the same location in the genome of YJM789. The second gene, EC1118_1O30_0012g, is predicted to encode Mpr1, a protein with N-acetyltransferase activity conferring resistance to oxidative stress and ethanol tolerance (17). This ORF has been identified in the Σ1278b strain and, interestingly, also in other wine yeasts (RM11–1a and AWRI1631).
We also found another 34 genes and 5 pseudogenes to be present in EC1118 but missing from S288c. Unlike the genes described above, these genes were organized into 3 large clusters that have been analyzed in detail (see below).
Identification and Localization of Large Chromosomal Regions Unique to EC1118.
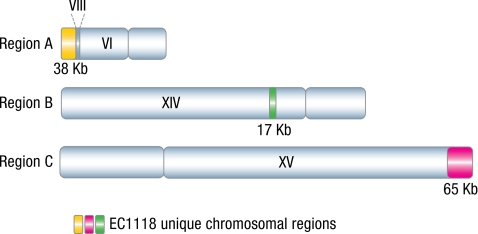
Three large regions of the EC1118 genome, a total of 120 kb in length, which could not be aligned with the S288c reference genome, were identified (Fig. 1).
Fig. 1.
Chromosomal distribution of the 3 unique EC1118 regions. The alignment of EC1118 contigs with S288c chromosomes led to the identification of 3 genomic regions unique to EC1118. The localization and length of these 3 regions are indicated by colored chromosomal segments. The insertion into chromosome VI of a 12-kb fragment from chromosome VIII is also shown.
The first of these regions was 38 kb long (region A) and was located in the subtelomeric region of the left arm of chromosome VI. The extremity of this chromosome displays a high degree of rearrangement (Fig. 1). A 23-kb fragment in the left arm of chromosome VI (including YFL052W to YFL062W in S288c) is absent. An internal part of this region encompassing the genes from YFL059W to YFL062W (5 kb) was found inserted into the right telomeric end of chromosome X. Second, a 12-kb fragment originating from chromosome VIII (including YHR211W to YHR217C) was found in the 3′ region of YFL051C (Fig. 1) resulting in YFL051C being fused to YHR211W (gene EC1118_1F14_0155g). The sequences of YFL051C and of YHR211W are highly similar, suggesting that the translocation was mediated by homologous recombination. Similar translocations to chromosome X were also found in strains YJM789 and RM11–1a. PCR, sequencing, and Southern blot analysis on EC1118 chromosomes confirmed these rearrangements.
We identified a second unique region (region B) as a 17-kb insertion into chromosome XIV, between genes YNL037C and YNL038W. Interestingly, a sequence similar to region B was detected in the RM11–1a genome, but the sequence is slightly rearranged compared with EC1118 and located between genes YNL248C and YNL249C. We confirmed the localization of region B in EC1118 by PCR amplification of the breakpoints.
A third region, 65 kb in length (region C), was identified in the subtelomeric region of the right arm of chromosome XV, replacing the last 9.7 kb of this chromosome. Southern blot analysis confirmed the location of region C on chromosome XV.
Function of the EC1118 ORFs Encompassed by the Unique Regions.
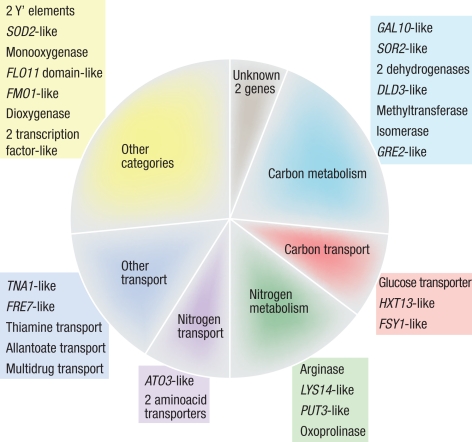
Within the three unique EC1118 regions, 34 ORFs predicted to code for proteins of >150 aa in length and with homologs in other species were identified (Table S5). These genes were classified according to the Munich Information Center for Protein Sequences (MIPS) functional catalog and were found to be involved mostly in key functions of the winemaking process, such as carbon and nitrogen metabolism, cellular transport, and the stress response (Fig. 2).
Fig. 2.
Functional classification of the unique genes of EC1118. The potential functions of the 34 unique genes of EC1118 were deduced from their S. cerevisiae orthologs. EC1118 genes were clustered according to the MIPS functional catalog. Each category is represented in the chart by a color and a description of function.
During wine fermentation, yeast cells must convert large amounts of glucose and fructose into alcohol. This process is also limited by nitrogen. Twenty of the 34 newly identified genes were found to encode proteins potentially involved in the metabolism and transport of sugar or nitrogen. These genes included genes similar to those encoding a Kluyveromyces thermotolerans glucose transporter, the S. cerevisiae glucose high-affinity transporter HXT13, and the S. pastorianus–specific fructose symporter FSY1. Several of these genes have homologs with known functions in amino acid metabolism, such as a transcription factor involved in proline utilization (PUT3), a S. cerevisiae permease potentially involved in the export of ammonia (ATO3), and 2 tandem-repeated genes encoding permeases of neutral amino acids. Another example of genes encoding proteins with nitrogen-related functions is provided by the gene encoding 5-oxo-L-prolinase, which catalyzes the ATP-dependent cleavage of 5-oxoproline to give L-glutamate.
We also identified 5 pseudogenes in subtelomeric regions A and C. In region A, we found a highly degenerate relic displaying sequence similarity to S. cerevisiae BIO3 and an intriguing pseudogene, EC1118_1F14_0067g, very similar to the AGL264W gene of Eremothecium gossypi encoding a bacterial transposase. These 2 genes do not encode hAT-like transposases, whereas gene 10980010 of AWRI1631 (15) and various Kluyveromyces genes encode proteins from this family (18). Three pseudogenes were identified in region C and shown to display similarity to S. cerevisiae ARB1, SOR2, and NFT1. Rapid changes in coding sequences leading to gene inactivation are more frequent at the telomeres in Saccharomyces (19), resulting in relics being largely concentrated in the subtelomeric regions (20), as observed here.
Origin of the Unique Genes of EC1118.
The existence of genes unique to EC1118 suggests the loss of these genes from other S. cerevisiae strains or their acquisition from non–S. cerevisiae donors. Blastp analysis supported the second of these hypotheses, because the closest relatives were found in species belonging to a clade containing the Lachancea, Zygosaccharomyces, Kluyveromyces, Saccharomyces, and Eremothecium genera (21) (clade I) and species belonging to a large, recently reassessed clade (22) containing Debaryomyces, some Pichia, and a number of medically important Candida species (clade II) (Table S5). For accurate identification of the hypothetical donor species for these genes, we carried out a combined phylogeny and synteny analysis. From these analyses, we observed different situations for each region.
Region A shows 2 different syntenic blocks: the first block has genes most closely related to Zygosaccharomyces rouxii genes, and the second block, whose synteny is conserved in species from both clade I and clade II, carries genes with their closest relatives belonging to clade II species (Fig. S1).
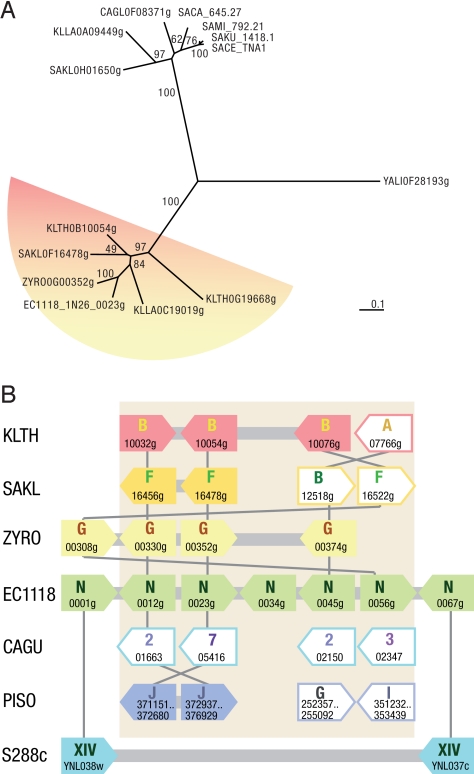
Genes of region B were systematically grouped with Z. rouxii in phylogenetic analysis, consistent with Z. rouxii genes being the best hits in blastp analysis (Fig. 3). In agreement with this observation, region B gene organization was rather well conserved with the related Zygosaccharomyces and Kluyveromyces species (Fig. 3). The exception is EC1118_1N26_0034g, which only shows a good match to RM11–1a strain.
Fig. 3.
Analysis of the phylogeny and synteny of the genes in EC1118 region B and in various yeast species. (A) Phylogram of the EC1118_1N26_0023g homologs. Primary protein sequence alignment of EC1118_1N26_0023g and its homologs, searched using blastp in the National Center for Biotechnology Information and Génolevures databases, was performed with ClustalX (47). Alignments were manually curated with GeneDoc (48). The unrooted bootstrapped neighbor-joining tree was built with ClustalX and visualized with Treeview (49). Bootstrap values (percentages) based on 1,000 replicates are indicated at the nodes. A very similar tree was obtained using Phyml. (Scale bar, 0.1 substitutions per site.) (B) The genomic localization and orientation of orthologs of region B–specific genes are represented for the following species: Z. rouxii (ZYRO, pale yellow arrows), Lachancea kluyveri (SAKL, gold arrows), K. thermotolerans (KLTH, pink arrows), Candida guilliermondii (CAGU, turquoise arrows), and Pichia sorbitophila (PISO, blue arrows). S288c orthologs for the EC1118 genes flanking the region B are shown in light blue. Arrows represent ORFs and their orientation. Genes are identified by their name, and the chromosome or the scaffold to which they belong is shown by a letter or a number within the arrow. In EC1118, N refers to the scaffold N26, and in P. sorbitophila the numbers refer to the gene coordinates on the chromosome. Genes syntenic with those of EC1118 are shown as fully colored arrows. Gene order was analyzed with a genome browser for all species except P. sorbitophila, for which tblastn was used, because this genome is not yet annotated.
Finally, genes in region C displayed some synteny with the genes of species closely related to the Saccharomyces clade, consistent with the observed phylogenetic relationships (Fig. S2).
Donor Species of the Unique Regions.
All natural hybrids discovered to date in Saccharomyces have involved species from the same genus (23–26). The phylogenetic analysis described above identified at least 1 potential donor of genetic material not closely related to Saccharomyces. We tried to identify the origin of the foreign genes found in EC1118, by carrying out PCR amplification with primers based on the sequences of genes from regions A, B, and C on genomic DNA isolated from strains (mostly type strains) belonging to 77 species from clade I or clade II (Table S6).
Only the type strain of Zygosaccharomyces bailii CBS 680T gave positive results with primers based on region B. All of the primer pairs amplified specific fragments of the expected size. We therefore checked by PCR whether the organization of the various foreign genes detected in EC1118 was similar to that in Z. bailii. The organization of genes in region B was found similar in EC1118 and Z. bailii CBS 680T, with the exception of gene EC1118_1N26_0056g, which was located upstream from EC1118_1N26_0012g in Z. bailii, in an arrangement similar to that found in Z. rouxii (Fig. 3). The gene organization in Z. bailii was confirmed by sequencing a 14-kb nucleotide sequence (European Molecular Biology Laboratory accession no. FN295481) that was found to be 99.7% identical to that of EC1118, confirming the identification of Z. bailii as a donor of unique EC1118 genes. In addition, we also detected the presence of this region in 7 other strains of Z. bailii of various origins (Table S6).
Analyses of phylogeny and synteny suggested that regions A and B might have a common origin (Figs. 3 and S1). However, no amplification was obtained when primers based on region A genes were used with DNA from Z. bailii. Similarly, no positive results were obtained for any of the other 46 species tested. The contributor of region A must therefore be an unidentified species related to clade I or II, as suggested by the 2 synteny blocks detected (Fig. S1). The presence, in region A, of a gene encoding a protein resembling a bacterial DNA transposase found only in the clade I species Eremothecium gossypii and the higher level of synteny with clade I than with clade II species strongly suggest that the contributor of region A belongs to clade I.
No amplification was observed with primers based on the 16 genes of region C, with any of the 44 species tested (Table S6), including 26 species either found in the wine microflora or belonging to the group previously known as Saccharomyces sensu lato (21). The contributor of this region may be a non-described species very closely related to the Saccharomyces genus, as suggested by synteny and phylogeny analysis. Consistent with this hypothesis, we have shown that EC1118_1O4_6645g and EC1118_1O4_6656g (right end of region C), also found in strain AWRI1631, display some similarity to S288c telomeric Y' element-encoded DNA helicases (Table S5). Y' elements are only found in S. cerevisiae and in its closest relative, S. paradoxus (27).
Thus, EC1118 contains gene clusters from Z. bailii and from 2 other species that we have not identified or that have not yet been described, one belonging to clade I and the other to the Saccharomyces genus.
Distribution of the Unique Genes and Regions Among S. cerevisiae Strains of Different Origins.
In a previous study of yeast diversity based on multilocus microsatellite typing, we showed that wine yeast strains clustered in a distinct phylogenetic group (4). We investigated the distribution of unique EC1118 regions among yeast populations, and particularly among wine yeasts, by carrying out PCR analysis on strains representative of the established clades (4).
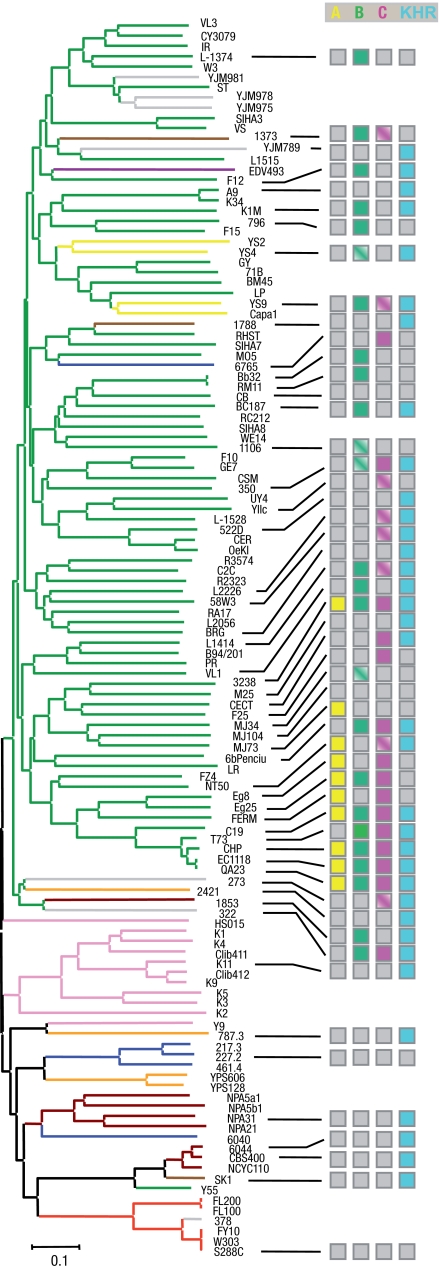
A phylogenic tree based on 120 strains was obtained (Fig. 4), including 66 wine strains and 19 isolates from various other origins (e.g., laboratory, palm wine, bakery, distillery, clinic, and sake). The 35 strains recently sequenced by Liti et al. (5) were also included in the analysis.
Fig. 4.
Dendrogram showing the presence of newly characterized genes among a set of 120 strains isolated from different sources. The 120 strains shown here include the 35 strains of the S. cerevisiae Genome Resequencing Project (5). The full name of each strain is available in Table S6. The neighbor-joining tree was constructed from the Dc chord distance between strains based on polymorphism at 11 loci, and is rooted according to the midpoint method. Source or geographic origin of the strains is denoted by colored branches: green for wine, pink for sake, gold for oak and opuntia isolates (America), blue for fermented fruits (Malaysia and Netherlands), brown for palm and bili wine (Africa), violet for rum (French Indies), yellow for bread, pale brown for soil, pearl gray for clinical, and orange for laboratory. The distribution of unique EC1118 regions is represented by colored squares: yellow for region A, green for region B, pink for region C, and blue for the KHR1 gene. Half-filled squares indicate that at least 1 gene of the corresponding region is absent.
Unique genes were searched in 53 strains, with a set of probes used for each unique region. Region A was found in only 2 groups—the Champagne group, containing EC1118-related strains, and a closely related group containing flor yeasts—suggesting that this region was acquired recently. Region B was found in the same groups and in more distantly related strains. Region C was found to be as widespread as region B among S. cerevisiae strains. Regions B and C were found to be incomplete in several strains, the missing genes differing between strains (Fig. 4). These data suggest that these regions are unstable in S. cerevisiae.
Most strains carrying the unique regions were closely related to the wine yeast group. Overall, regions B and C were found to be present in almost half the 53 strains tested. The 3 regions are exclusively (region A) or mostly (regions B and C) found in wine strains (29 of 35 wine strains contain at least 1 of the 3 unique regions). The differential presence of the genes from regions A, B, and C in a number of wine yeast isolates may be accounted for the progressive diffusion of these events by outcrossing inside the wine yeast population. The transfers of regions B and C seem to be older than that of region A, but the timing of these events cannot be determined, because the subtelomeric location of regions A and C is a source of instability. The KHR1 gene was found to be widespread among strains, suggesting an ancient acquisition event that has subsequently been lost from many strains. The presence of LTR flanking KHR1 might account for this instability.
To obtain a broader view of the distribution of regions A, B, and C within the S. cerevisiae species, we performed a blastn survey of the unique genes in the genome of YJM789 (13), RM11–1a, AWRI1631 (15), and the 36 S. cerevisiae strains sequenced by Liti et al. (5) (Fig. S3). The region A was absent from all strains, consistent with the local distribution of this region in the “Champagne/Flor yeasts” cluster (Fig. 4). Region B was found in the wine yeast derivative RM11–1a, in strains of the clusters called “Wine/European” and “mosaic genomes” (5). Region C was also found in the latter strains, in only 1 strain belonging to the “Wine/European” cluster, and in strain AWRI1631. The fact that this region was largely found in wine yeasts in our PCR survey (Fig. 4) suggests that the “Wine/European” group of Liti et al. (5) is not fully representative of the wine yeast community. It is also possible that because of heterozygosity, some regions are present in the parental strains but not in the derivative strains whose genome was sequenced. The different distribution of regions B and C suggests that these regions have a different history. Interestingly, region C was almost exclusively found in strains of European origin.
Discussion
Various mechanisms are known to be involved in the adaptive evolution of yeasts to the fermentation process, such as gene duplication, polyploidy, chromosomal rearrangements, interspecific hybridization, and introgression (28). Recent analyses have shown that yeast hybrids may be more abundant in both natural and industrial environments than previously thought. Indeed, almost 10% of Saccharomyces strains previously classified as sensu stricto seem to be hybrids between different species (29). Lager brewing yeasts are natural hybrids generated by interspecific hybridization between S. cerevisiae, S. bayanus, and an as-yet non-described species (30–32). Double and triple hybrids of S. cerevisiae with S. uvarum, S. kudriavzevii, or both were recently identified in yeast populations isolated from grape and cider fermentations [for a review see Sipiczki (33)]. However, most of the hybrids described to date have either the genome of each parental species or chimeric genomes, and all of the donor species belong to the Saccharomyces genus.
Horizontal gene transfers have rarely been described in yeast, and all previous examples have involved bacterial single genes (13, 34, 35). The introgression of 23 kb from S. cerevisiae into S. paradoxus has also been described (24) and resembled our findings, in particular for region C. The most likely explanation for this region is that a cross has occurred between S. cerevisiae and a Saccharomyces species to yield the present hybrid. It is generally thought that such hybrids are resolved by the gradual loss of one of the contributing genomes (33).
The situation is clearly different for regions A and B, which represent the first example of gene transfer between S. cerevisiae and non-Saccharomyces species. The phylogenetic topologies for genes of regions A and B indicate that the non-Saccharomyces species are the donors and S. cerevisiae the recipient. The presence of region B in Z. bailii strains from various origins further supports this hypothesis. Z. bailii is a major yeast contaminant of wine. It tolerates common food preservatives, high concentrations of sugar and ethanol, and low pH. These properties confer on this species an outstanding capacity to survive during wine fermentations. With S. cerevisiae, it is one of the rare yeasts able to persist until the end of the fermentation process (36). It is therefore found in close contact with S. cerevisiae in many natural fermentations. This proximity may have favored genetic transfer, either in a direct lateral transfer or through introgression after hybridization. Although Z. bailii has been described as a diploid that does not undergo meiosis but produces tetrads with mitotic spores (37) neither of the 2 hypotheses can be excluded.
This strategy of evolution by gene transfer is an important aspect of yeast diversification and may play a major role in adaptation to the wine fermentation ecosystem. Two of the unique regions are located in subtelomeric regions, which are known to be enriched in genes involved in adaptation (29). In some situations, hybrids between species show increased fitness and acquire unique properties compared with the parental species. For example, hybrids between different Saccharomyces species have been shown to grow over a broader range of temperatures or to produce larger amounts of glycerol or aroma compounds than the parental strains (25, 38, 39). The potential functions associated with the transferred genes, such as those related to fructose utilization, oxidative stress, or nitrogen metabolism, may contribute to the adaptation to the fermentation of high-sugar, low-nitrogen grape musts and may confer a selective advantage during wine fermentation.
Materials and Methods
Strains and Media.
Lalvin EC1118 (EC1118), also known as “Prise de mousse,” is a S. cerevisiae wine strain isolated in Champagne (France) and manufactured by Lallemand Inc. EC1118 has been deposited in the Collection Nationale de Cultures de Microorganismes (Institut Pasteur, France) as strain I-4215. This strain is one of the most frequently used fermentation starters worldwide and has been extensively studied as a model wine yeast (40–42). The other yeast isolates used are detailed in Table S6. The non-Saccharomyces isolates were obtained from the Centre International de Ressources Microbiennes-Levures in France, and from the Centraalbureau voor Schimmelcultures in the Netherlands. Cells were routinely grown in YPD medium (1% yeast extract, 1% peptone, and 1% glucose) at 28 °C, with shaking.
Gene Prediction and Annotation.
Genome annotation was based on a combination of methods including ORF calling (minimum size, 150 bp), gene prediction with GlimmerHMM (43), and direct mapping of S288c ORFs from the Saccharomyces Genome Database. The detailed annotation procedure and a complete annotation file are available in SI Materials and Methods and Table S3.
Microsatellite Analysis.
The 120 strains were characterized for allelic variation at 11 microsatellite loci, as described by Legras et al. (4). The chord distance Dc between strains was calculated, as described by Cavalli-Sforza and Edwards (44). The neighbor-joining tree was constructed with the PHYLIP 3.67 package (45) and drawn with MEGA software version 4.0 (46). The tree was rooted by the midpoint method.
Additional Materials and Methods.
Further details are available in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Lallemand Inc. for providing the strain EC1118, Noemie Jacques for providing strains from the Centre International de Ressources Microbiennes–Levures, and Giani Liti and Justin Fay for providing S. cerevisiae strains; the Génolevures Consortium for providing free access to the genome of Pichia sorbitophila; Claude Gaillardin for helpful discussions and for providing Lebanese strains; Philippe Abbal for bioinformatics assistance; and Jean-Jacques Gonod for drawing figures. This work was supported by grants from the Consortium National de Recherche en Génomique-Genoscope (no. 2005/74 to S.D.) and the Bureau des Ressources Génétiques (no. 347 “Exploration de la diversité fongique” to S.C.). M.N. and E.B. hold postdoctoral fellowships from the Generalitat de Catalunya and Institut National de la Recherche Agronomique (INRA), respectively. This work was supported by INRA and Centre National de la Recherche Scientifique.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the European Molecular Biology Laboratory database (accession nos. FN393058–FN393060, FN393062–FN393087, FN394216, and FN394217).
This article contains supporting information online at www.pnas.org/cgi/content/full/0904673106/DCSupplemental.
References
- 1.McGovern PE, et al. Fermented beverages of pre- and proto-historic China. Proc Natl Acad Sci USA. 2004;101:17593–17598. doi: 10.1073/pnas.0407921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aa E, Townsend JP, Adams RI, Nielsen KM, Taylor JW. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:702–715. doi: 10.1111/j.1567-1364.2006.00059.x. [DOI] [PubMed] [Google Scholar]
- 3.Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legras JL, Merdinoglu D, Cornuet JM, Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 5.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn B, Levine RP, Sherlock G. Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genomics. 2005;6:53. doi: 10.1186/1471-2164-6-53. doi: 10.1186/1471-2164-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradbury JE, et al. A homozygous diploid subset of commercial wine yeast strains. Antonie Van Leeuwenhoek. 2006;89:27–37. doi: 10.1007/s10482-005-9006-1. [DOI] [PubMed] [Google Scholar]
- 9.Bidenne C, Blondin B, Dequin S, Vezinhet F. Analysis of the chromosomal DNA polymorphism of wine strains of Saccharomyces cerevisiae. Curr Genet. 1992;22:1–7. doi: 10.1007/BF00351734. [DOI] [PubMed] [Google Scholar]
- 10.Rachidi N, Barre P, Blondin B. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:841–850. doi: 10.1007/s004380050028. [DOI] [PubMed] [Google Scholar]
- 11.Puig S, Querol A, Barrio E, Perez-Ortin JE. Mitotic recombination and genetic changes in Saccharomyces cerevisiae during wine fermentation. Appl Environ Microbiol. 2000;66:2057–2061. doi: 10.1128/aem.66.5.2057-2061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carreto L, et al. Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics. 2008;9:524. doi: 10.1186/1471-2164-9-524. doi: 10.1186/1471-2164-9-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei W, et al. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc Natl Acad Sci USA. 2007;104:12825–12830. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doniger SW, et al. A catalog of neutral and deleterious polymorphisms in yeast. PLoS Genet. 2008;(8):e1000183. doi: 10.1371/journal.pgen.1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borneman AR, Forgan AH, Pretorius IS, Chambers PJ. Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res. 2008;8:1185–1195. doi: 10.1111/j.1567-1364.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 16.Leh-Louis V, Wirth B, Potier S, Souciet JL, Despons L. Expansion and contraction of the DUP240 multigene family in Saccharomyces cerevisiae populations. Genetics. 2004;167:1611–1619. doi: 10.1534/genetics.104.028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du X, Takagi H. N-Acetyltransferase Mpr1 confers ethanol tolerance on Saccharomyces cerevisiae by reducing reactive oxygen species. Appl Microbiol Biotechnol. 2007;75:1343–1351. doi: 10.1007/s00253-007-0940-x. [DOI] [PubMed] [Google Scholar]
- 18.Souciet JL, et al. Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 2009 doi: 10.1101/gr.091546.109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 20.Lafontaine I, Fischer G, Talla E, Dujon B. Gene relics in the genome of the yeast Saccharomyces cerevisiae. Gene. 2004;335:1–17. doi: 10.1016/j.gene.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 21.Kurtzman CP, Robnett CJ. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex' determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 22.Tsui CK, Daniel HM, Robert V, Meyer W. Re-examining the phylogeny of clinically relevant Candida species and allied genera based on multigene analyses. FEMS Yeast Res. 2008;8:651–659. doi: 10.1111/j.1567-1364.2007.00342.x. [DOI] [PubMed] [Google Scholar]
- 23.Masneuf I, Hansen J, Groth C, Piskur J, Dubourdieu D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl Environ Microbiol. 1998;64:3887–3892. doi: 10.1128/aem.64.10.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liti G, Barton DB, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez SS, Gallo L, Climent MA, Barrio E, Querol A. Enological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int J Food Microbiol. 2007;116:11–18. doi: 10.1016/j.ijfoodmicro.2006.10.047. [DOI] [PubMed] [Google Scholar]
- 26.Groth C, Hansen J, Piskur J. A natural chimeric yeast containing genetic material from three species. Int J Syst Bacteriol. 1999;49:1933–1938. doi: 10.1099/00207713-49-4-1933. [DOI] [PubMed] [Google Scholar]
- 27.Naumov GI, Naumova ES, Lantto RA, Louis EJ, Korhola M. Genetic homology between Saccharomyces cerevisiae and its sibling species S. paradoxus and S. bayanus: Electrophoretic karyotypes. Yeast. 1992;8:599–612. doi: 10.1002/yea.320080804. [DOI] [PubMed] [Google Scholar]
- 28.Barrio E, González SS, Arias A, Belloch C, Querol A. In: The Yeast Handbook, Vol. 2: Yeast in Food and Beverages. Querol A, Fleet GH, editors. Berlin: Springer; 2006. pp. 153–174. [Google Scholar]
- 29.Liti G, Louis EJ. Yeast evolution and comparative genomics. Annu Rev Microbiol. 2005;59:135–153. doi: 10.1146/annurev.micro.59.030804.121400. [DOI] [PubMed] [Google Scholar]
- 30.Vaughan Martini A, Martini A. Three newly delimited species of Saccharomyces sensu stricto. Antonie van Leeuwenhoek. 1987;53:77–84. doi: 10.1007/BF00419503. [DOI] [PubMed] [Google Scholar]
- 31.Casaregola S, Nguyen HV, Lapathitis G, Kotyk A, Gaillardin C. Analysis of the constitution of the beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization. Int J Syst Evol Microbiol. 2001;51:1607–1618. doi: 10.1099/00207713-51-4-1607. [DOI] [PubMed] [Google Scholar]
- 32.Rainieri S, et al. Pure and mixed genetic lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl Environ Microbiol. 2006;72:3968–3974. doi: 10.1128/AEM.02769-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sipiczki M. Interspecies hybridization and recombination in Saccharomyces wine yeasts. FEMS Yeast Res. 2008;8:996–1007. doi: 10.1111/j.1567-1364.2008.00369.x. [DOI] [PubMed] [Google Scholar]
- 34.Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 35.Hall C, Brachat S, Dietrich FS. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James SA, Stratford M. In: Yeasts in Food: Beneficial and Detrimental Aspects. Boekhout T, Robert V, editors. Hamburg: Behr's; 2003. pp. 171–191. [Google Scholar]
- 37.Rodrigues F, et al. The spoilage yeast Zygosaccharomyces bailii forms mitotic spores: A screening method for haploidization. Appl Environ Microbiol. 2003;69:649–653. doi: 10.1128/AEM.69.1.649-653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zambonelli C, et al. Technological properties and temperature response of interspecific Saccharomyces hybrids. J Sci Food Agric. 1997;74:7–12. [Google Scholar]
- 39.Greig D, Louis EJ, Borts RH, Travisano M. Hybrid speciation in experimental populations of yeast. Science. 2002;298:1773–1775. doi: 10.1126/science.1076374. [DOI] [PubMed] [Google Scholar]
- 40.Rossignol T, Dulau L, Julien A, Blondin B. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast. 2003;20:1369–1385. doi: 10.1002/yea.1046. [DOI] [PubMed] [Google Scholar]
- 41.Varela C, Cardenas J, Melo F, Agosin E. Quantitative analysis of wine yeast gene expression profiles under winemaking conditions. Yeast. 2005;22:369–383. doi: 10.1002/yea.1217. [DOI] [PubMed] [Google Scholar]
- 42.Pizarro FJ, Jewett MC, Nielsen J, Agosin E. Growth temperature exerts differential physiological and transcriptional responses in laboratory and wine strains of Saccharomyces cerevisiae. Appl Environ Microbiol. 2008;74:6358–6368. doi: 10.1128/AEM.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 44.Cavalli-Sforza LL, Edwards AW. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- 45.Felsenstein J. Using the quantitative genetic threshold model for inferences between and within species. Philos Trans R Soc Lond B Biol Sci. 2005;360:1427–1434. doi: 10.1098/rstb.2005.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 47.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 48.Nicholas KB, Nicholas HB, Deerfield DW., II GeneDoc: Analysis and visualization of genetic variation. EMBnet News. 1997;4:1–4. [Google Scholar]
- 49.Page RD. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.