Abstract
Background:
The angiotensin-converting enzyme (ACE) gene contains a common polymorphism based on the insertion (I) or deletion (D) of a 287-bp intronic DNA fragment. The D allele is associated with higher ACE activity, and thus higher angiotensin II levels. Angiotensin II stimulates cardiac fibrosis and conduction heterogeneity.
Objective:
The purpose of this study was to determine whether the ACE I/D polymorphism modulates cardiac electrophysiology.
Methods:
We studied three different cohorts of patients: 69 patients with paroxysmal lone AF, 151 patients with structural heart disease and no history of AF, and 161 healthy subjects without cardiovascular disease or AF. Patients receiving drugs affecting cardiac conduction were excluded. Electrocardiographic parameters during sinus rhythm were compared among the ACE I/D genotypes.
Results:
The ACE I/D polymorphism was associated with the PR interval and heart block in the lone AF cohort. In multivariable linear regression models, the D allele was associated with longer PR interval in the lone AF and heart disease cohorts (12.0 ms and 7.1 ms increase per D allele, respectively). The P-wave duration showed a similar trend with increase in the PR interval across the ACE I/D genotypes in the lone AF and heart disease cohorts.
Conclusion:
The ACE D allele is associated with electrical remodeling in patients with lone AF and those with heart disease, but not in control subjects. ACE activity may play a role in cardiac remodeling after the development of AF and heart disease.
Keywords: atrial fibrillation, angiotensin-converting enzyme, genetics, electrocardiogram, heart block, arrhythmia
Introduction
The renin-angiotensin-aldosterone system (RAAS) plays an important role in regulation of normal cardiovascular function and disorder of the system is associated with cardiovascular diseases.1-6 Genetic variants in angiotensin-converting enzyme (ACE) have been associated with cardiovascular disease including hypertension, myocardial infarction, dilated cardiomyopathy, and left ventricular hypertrophy.1-3, 7 Recent evidence suggests that the ACE gene may also play an important role in the pathogenesis of arrhythmias.4-6
The ACE gene contains a polymorphism based on the presence, insertion (I) or absence, deletion (D) of a 287-bp intronic DNA segment, resulting in three genotypes (DD and II homozygotes, and ID heterozygotes).8 The ACE D allele is associated with higher plasma and cardiac ACE activity resulting in higher angiotensin II levels, and has been reported to increase the risk of arrhythmias including sudden death and atrial fibrillation (AF).4-6, 8, 9 RAAS activation plays an important role in structural and electrophysiological remodeling that is associated with the development of AF.10 It has been recently reported that angiotensin II also provides an electrophysiologic substrate for arrhythmias by modulation of ion channels and gap junctions.11-14 Moreover, we have recently reported that antiarrhythmic drugs are less effective for AF in subjects carrying the D allele, suggesting the hypothesis that the ACE I/D polymorphism modulates cardiac electrophysiology.15 Here, we tested this hypothesis by studying the association of the ACE I/D polymorphism with electrocardiographic parameters in subjects with AF and in those with heart disease.
Methods
Study Population
The study protocol was approved by the Institutional Review Board of Vanderbilt University and Massachusetts General Hospital. All subjects gave written informed consent. This study included three cohorts; 1) patients with lone AF enrolled in the Vanderbilt or Massachusetts AF Registry, which is made up of a clinical and genetic registries;16, 17 2) patients with structural heart disease but no personal or family history of AF;18 and 3) 161 healthy volunteers with no significant medical history, normal physical examinations, and no personal or family history of AF.19 At enrollment into the AF Registries, all subjects complete a symptom questionnaire. This is done to assess symptomatic AF burden, which is measured by using an algorithm for scoring the frequency, duration, and severity of symptoms.15 Although AF symptoms are important when evaluating response to drug therapies, for the purposes of this study, we limited the AF burden score to the frequency and duration of AF.
Lone AF was defined as AF occurring in patients <65 years of age without hypertension or overt structural heart disease by clinical examination, electrocardiogram, and echocardiography. Patients with lone AF who had 12-lead electrocardiograms (ECGs) showing sinus rhythm were included in the lone AF cohort. Patients who received drugs affecting cardiac conduction including antiarrhythmic drugs, β-blockers, calcium channel blockers, and digoxin were excluded from this study.
ECG measurements
All 12 lead ECGs were recorded with the patient resting supine, using an analogue system at 25 mm/second paper speed, 10 mm/mV gain, and 40 Hz low pass filter setting. The P-wave duration and PR intervals were measured on all possible leads by an experienced observer blinded to all clinical details using a semi-automated digitizing program with electronic calipers.20
To determine interobserver variability, a second observer made independent blinded P-wave duration determinations of 20 randomly selected ECGs. Intraobserver variability was evaluated by P-wave duration analysis of 30 randomly assigned ECGs, again in a blinded fashion by a single observer. Interobserver measurement error was avoided by using the measurements of the same experienced observer for statistical comparisons.
Determination of ACE genotype
ACE I/D genotypes were determined by PCR as described previously.15 Briefly, a set of primers was designed to encompass the polymorphic region in intron 16 of the ACE gene (sense primer 5′ CTGGAGACCACTCCCATCCTTTCT 3′, antisense primer 5′ GATGTGGCCATCACATTCGTCAGAT 3′). The products were separated by electrophoresis on 2% agarose gel and identified by ethidium bromide staining. Each sample found to be DD was verified using insertion specific primers (sense primer 5′ TGGGACCACAGCGCCCGCCACTAC3, antisense primer 5′ TCGCCAGCCCTCCCATGCCCATAA 3′).
Statistical Analysis
Statistical analysis was performed using the SPSS statistical package, release 16.0.1 (SPSS Inc, Chicago, IL). All values are expressed as mean ± SD for continuous variables and proportions for categorical variables. Hardy-Weinberg equilibriums were performed using a chi-square test. Statistical tests of parameters by genotypes were analyzed using a Fisher's exact test for categorical variables and ANOVA for continuous variables.
Associations of ECG measurements with ACE genotypes were adjusted for variables affecting cardiac conduction using a linear regression model. All factors adjusted for were determined a priori. Modulating factors for the lone AF cohort included age, length of time with paroxysmal AF (AF burden), heart rate, and left atrial size. For healthy controls, factors consisted of age and heart rate. For patients with cardiovascular disease, factors adjusted for included age, heart rate, a history of hypertension, and congestive heart failure. The PR interval and P-wave durations were checked for normal Gaussian distributions; no transformations were required. A two-sided significance of 0.05 was used for all analyses.
Results
Clinical Characteristics
Among 399 patients with lone AF and 385 heart disease patients without AF in the original cohorts, 69 patients with lone AF and 151 heart disease patients were eligible for this study. The reasons for exclusion from this analysis included concomitant medications that modulate cardiac conduction, and failure to record an ECG during sinus rhythm. Table 1 shows the clinical characteristics of the study subjects. The mean symptomatic AF burden score in the lone AF cohort at enrollment was high, but more importantly, it was similar across the three ACE I/D genotypes.
Table 1.
Clinical characteristics of subjects
| Variables | |||||
|---|---|---|---|---|---|
| Lone AF Patients | All patients, n = 69 |
ACE I/D genotype |
P value | ||
| DD, n = 17 | ID, n = 38 | II, n = 14 | |||
| Age at AF onset, years | 38±14 | 43±13 | 37±14 | 35±13 | 0.12 |
| Duration after AF diagnosis, years | 4±8 | 4±7 | 3±6 | 8±11 | 0.06 |
| AF burden score15 | 12±6 | 11±5 | 12±6 | 11±6 | 0.18 |
| Male sex, n (%) | 33 (72%) | 6 (55%) | 21 (78%) | 6 (75%) | 0.34 |
| Left atrial size, mm | 36±6 | 38±7 | 35±5 | 37±6 | 0.37 |
| Ejection fraction, % | 59±6 | 62±5 | 58±6 | 58±6 | 0.11 |
| Heart disease patients without AF | All patients, n = 151 |
ACE I/D genotype |
P value | ||
| DD, n = 54 | ID, n = 77 | II, n = 20 | |||
| Age, years | 55±16 | 55±16 | 54±16 | 57±14 | 0.69 |
| Male sex, n (%) | 63 (42%) | 25 (46%) | 32 (42%) | 6 (30%) | 0.45 |
| Hypertension, n (%) | 92 (61%) | 37 (70%) | 42 (55%) | 13 (65%) | 0.2 |
| Diabetes, n (%) | 31 (21%) | 14 (26%) | 13 (17%) | 4 (20%) | 0.42 |
| Coronary artery disease, n (%) | 52 (34%) | 16 (30%) | 30 (39%) | 6 (30%) | 0.49 |
| Congestive heart failure, n (%) | 39 (26%) | 14 (26%) | 17 (22%) | 8 (40%) | 0.26 |
| Ejection fraction, % | 44±17 | 42±19 | 44±17 | 44±15 | 0.82 |
| Controls without AF or heart disease | All subjects, n = 118 |
ACE I/D genotype |
P value | ||
| DD, n = 41 | ID, n = 53 | II, n = 24 | |||
| Age, years | 27±6 | 27±6 | 27±6 | 28±5 | 0.82 |
| Male sex, n (%) | 46 (39) | 16 (39) | 21 (40) | 9 (38) | 0.98 |
AF denotes atrial fibrillation; I/D, insertion/deletion polymorphism.
ACE I/D Genotype and ECG parameters
The frequencies of the II, ID and DD genotypes for the lone AF cohort (22%, 53%, and 25%, respectively), the heart disease cohort (16%, 49%, and 35%, respectively), or the control cohort (18%, 48%, and 34%, respectively) did not deviate significantly from those predicted by Hardy-Weinberg criteria (27%, 50%, and 23% respectively; P = NS for each). The intra-class correlation coefficient for the ECG measurement between the two observers was 0.98, indicating excellent agreement and reproducibility of data.
In the lone AF cohort, patients with the DD genotype had longer PR intervals and a higher incidence of heart block (bundle branch block or prolonged PR interval ≥200 ms) (Table 2). To determine whether conduction in the atria is modulated by the ACE I/D polymorphism, P-wave duration was compared among the genotypes. The P-wave duration was not different among the ACE I/D genotypes. The ACE I/D genotypes were not associated with heart rate, QRS interval, QRS axis, or QT interval in any of the cohorts.
Table 2.
Electrocardiographic parameters across ACE I/D genotypes
| Variables | |||||
|---|---|---|---|---|---|
| Patients with lone AF | All patients | ACE I/D genotype |
P value | ||
| DD | ID | II | |||
| Heart rate, beats/min | 65±12 | 64±15 | 66±11 | 65±12 | 0.83 |
| P-wave duration, ms | 109±17 | 110±16 | 111±18 | 101±16 | 0.17 |
| PR Interval, ms | 166±26 | 177±27 | 166±26 | 150±22 | 0.02 |
| Heart block, n (%) | 8 (11) | 6 (33) | 2 (5) | 0 (0) | 0.002 |
| QRS Interval, ms | 89±9 | 92±9 | 89±9 | 89±9 | 0.51 |
| QRS axis, degree | 37±35 | 38±38 | 37±35 | 41±34 | 0.88 |
| QT interval, ms | 402+36 | 400+37 | 400+36 | 410+34 | 0.67 |
| QTc | 408±24 | 408±28 | 411±22 | 398±25 | 0.20 |
| Heart disease patients without AF |
All patients | ACE I/D genotype |
P value | ||
| DD | ID | II | |||
| Heart rate, beats/min | 78±20 | 79±19 | 77±20 | 78±25 | 0.79 |
| P-wave duration, ms | 157±29 | 162±29 | 155±30 | 149±23 | 0.15 |
| PR Interval, ms | 166±30 | 169±32 | 163±28 | 167±33 | 0.52 |
| Heart block, n (%) | 12 (8) | 7 (13) | 5 (7) | 0 (0) | 0.15 |
| QRS Interval, ms | 97±23 | 98±27 | 95±22 | 98±20 | 0.69 |
| QRS axis, degree | 27±51 | 32±61 | 26±45 | 20±42 | 0.72 |
| QT interval, ms | 387+55 | 386+59 | 385+53 | 394+54 | 0.57 |
| QTc | 430±35 | 435±42 | 425±30 | 436±34 | 0.31 |
| Controls without AF and heart disease |
All patients | ACE I/D genotype |
P value | ||
| DD | ID | II | |||
| Heart rate, beats/min | 66±11 | 64±11 | 67±10 | 67±11 | 0.42 |
| P-wave duration, ms | 103±12 | 102±13 | 103±13 | 104±10 | 0.99 |
| PR Interval, ms | 152±23 | 152±23 | 154±24 | 148±19 | 0.56 |
| Heart block, n (%) | 4 (3) | 2 (5) | 2 (4) | 0 (0) | 0.57 |
| QRS Interval, ms | 87±10 | 87±8 | 87±9 | 89±13 | 0.91 |
| QRS axis, degree | 72±20 | 75±15 | 70±22 | 71±19 | 0.32 |
| QT interval, ms | 393+34 | 389+33 | 396+34 | 392+34 | 0.68 |
| QTc | 407±19 | 407±18 | 405±18 | 417±19 | 0.63 |
AF denotes atrial fibrillation; I/D, insertion/deletion polymorphism; QTc, corrected QT interval.
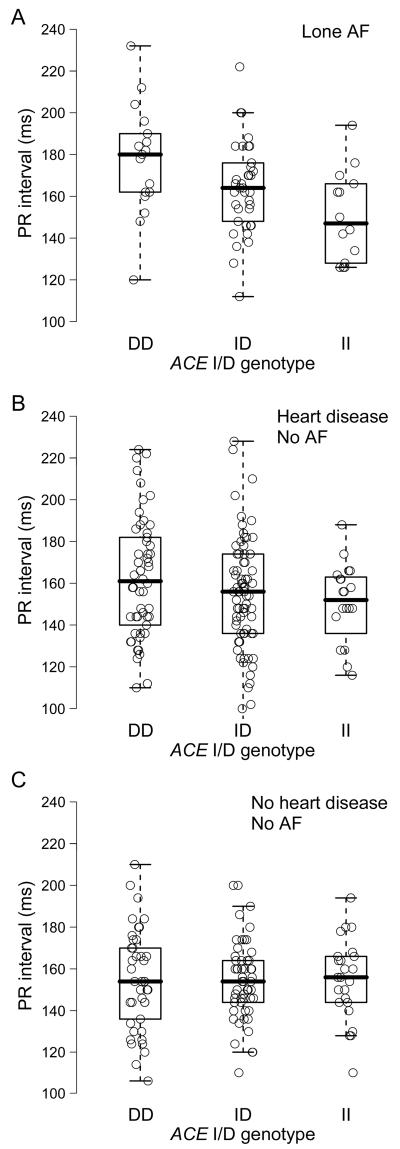
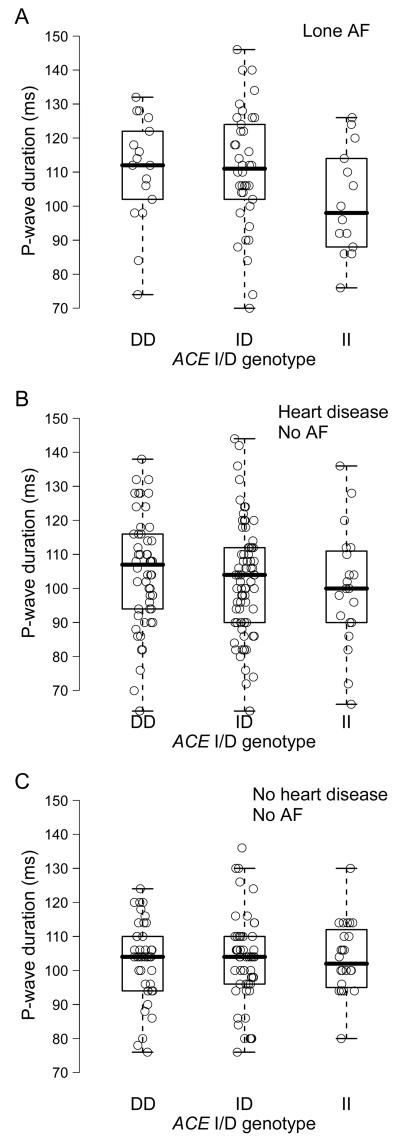
In multivariable linear regression models, the ACE I/D polymorphism was associated with the PR interval in the lone AF and heart disease cohorts (Figure 1). With each D allele, the PR interval prolonged by an average of 12.0 ms and 7.1 ms in the lone AF and heart disease cohorts, respectively. The ACE I/D polymorphism was not significantly related to PR interval in the healthy cohort. The P-wave duration showed a similar trend with increase in the PR interval across the ACE I/D genotypes in the lone AF and heart disease cohorts (Figure 2). P-wave duration was significantly longer in subjects with the D allele than those without the D allele (DD/ID versus II) in the lone AF cohort (P = 0.04), but not in the heart disease or healthy control cohorts (P = NS for each). With each D allele, the P-wave duration prolonged by an average of 4.6 ms and 2.1 ms in the lone AF and heart disease cohorts, respectively. The ACE I/D polymorphism was not related to P-wave duration in the healthy cohort.
Figure 1.
ACE I/D genotype and PR interval in patients with lone AF (A) (P = 0.02), in patients with heart disease and no history of AF (B) (P = 0.04), and in healthy subjects without AF or heart disease (C) (P = 0.84).
Figure 2.
ACE I/D genotype and P-wave duration in patients with lone AF (A) (P = 0.18), in patients with heart disease and no history of AF (B) (P = 0.17), and in healthy subjects without AF or heart disease (C) (P = 0.25).
Discussion
Recent studies strongly support the concept that genetic variation may be important in the pathogenesis of cardiac disorders including arrhythmias and the importance of genetic medicine in clinical practice is increasingly being recognized.21-23 Mutations in genes encoding cardiac ion channels and associated proteins have been identified in inherited arrhythmia syndromes including AF.21, 22 In population association studies, certain polymorphisms (ion channels, ACE, β2 adrenergic receptors) have been associated with increased risk of arrhythmias and sudden cardiac death.5, 6, 21, 24 Furthermore, contribution of genetic variants to electrocardiographic variation in general population has been also reported; polymorphisms in the cardiac potassium channel (KCNH2) and nitric oxide synthase 1 adaptor protein (NOS1AP) genes have been associated with QT interval variation.25, 26 In some cases, the genetic variants are only unmasked when the ‘reserve’ is reduced e.g., exposure to QT-prolonging drugs in patients with formes fruste forms of the congenital long-QT syndrome.27 In this study, we show for the first time that the ACE I/D polymorphism is associated with atrial and atrioventricular conduction in patients with lone AF and those with heart disease, but not in healthy subjects. Our findings suggest that the RAAS plays an important role in electrical remodeling after development of AF and heart disease. The ACE D allele is associated with higher plasma concentration of ACE,14 higher cardiac ACE concentration,21 and increased renal ACE mRNA expression.22 Thus, subjects with the D allele may be exposed to higher angiotensin II levels than those with the I allele. Myocardial fibrosis is strongly correlated with RAAS activation, especially angiotensin II and aldosterone, and chronic exposure to high levels of circulating and/or tissue angiotensin may predispose to both left ventricular hypertrophy and myocardial fibrosis.23
AF is a highly heterogeneous disorder and there is increasing evidence that activation of the RAAS plays a role in the pathophysiology of AF. The ACE I/D polymorphism is associated with serum and cardiac ACE activity, and subjects with the ID or DD genotypes have approximately 25% and 50% higher ACE levels respectively than those with the II genotype.8 It has been reported that genetic variants in ACE are associated with increased AF susceptibility,5, 6 and in this study we found that the polymorphism was also associated with cardiac conduction defects in patients with AF and those with heart disease. Our results are consistent with prior studies, which demonstrated that increased expression of ACE in cardiac tissues of genetically engineered mice results in the development of conduction abnormalities and AF.28, 29 Activation of the RAAS initiates a cascade of processes resulting in hypertrophy, fibroblast proliferation, accumulation of collagen, and apoptosis, all of which predispose to a reduction in conduction velocity.30 Therefore, slow conduction associated with the D allele may result from electrophysiologic remodeling due to higher ACE levels and increased activation of the RAAS. Because blockade of the RAAS attenuates atrial fibrosis,31 ACE inhibitors and angiotensin II receptor blockers may also prevent or reduce conduction defects in AF. Moreover, evidence that angiotensin II affects cardiac electrophysiology by modulation of ion channels and by gap junction remodeling, which in turn impairs cell-to-cell impulse conduction, further supports our findings.11-14
In this study, the ACE I/D polymorphism correlated with cardiac conduction in the atrium and conduction system, but not in the ventricle. The density of angiotensin II receptors is higher in atrial than in ventricular tissue, suggesting that the atrium is more vulnerable to angiotensin effects.32 An alternative explanation is that rapid electrical activity during AF leads to greater electrical remodeling resulting in conduction defects in the atrium and conduction system than in the ventricle as the ventricle is partially protected by atrioventricular block. The ACE I/D polymorphism may have effects on cardiac electrophysiology in the absence of AF or heart disease, but the effects may be subtle and thus it may be difficult to detect on the conventional surface ECG. To address this issue, further studies using more advanced techniques such as signal averaged ECG and intracardiac electrograms, which were not performed in this study, may be important in a large number of subjects.
There are other factors that are important for electrical and structural remodeling. In this study, normal volunteers without a history of AF or cardiac disease were younger than those with lone AF or those with cardiac disease, and aging may also be associated with ACE I/D polymorphism-mediated conduction slowing. However, in multivariate models adjusted for age and other variables, the association remained significant. Although the ACE D allele has been linked with hypertension,7 we found that the ACE I/D polymorphism was associated with cardiac conduction in patients with lone AF who do not have a history of hypertension. Although lone AF patients have no overt structural heart disease, studies have shown subclinical atrial structural abnormalities in some patients. (REF Frustaci et al Circulation 1997;96:1180) Thereby, it is possible that the ACE D allele may merely augment structural remodeling rather than enhance electrical remodeling. Atrial stretch may cause P-wave prolongation and further increases local synthesis of angiotensin II,33 but left atrial size was similar among the ACE I/D genotypes. Our study includes several limitations. We excluded patients taking atrioventricular nodal-modifying drugs and so we could evaluate the exclusive effects of ACE I/D polymorphism on cardiac electrophysiology. Although this could have introduced a selection bias in the study cohorts, this was unlikely because the frequencies of the ACE I/D genotypes were unchanged after the exclusion compared to those in the original populations. Furthermore, the study cohorts were in Hardy-Weinberg equilibrium providing further evidence for the absence of this selection bias. While the DD genotype has been associated with increased risk of AF in a prior study,5 the frequency of ACE I/D genotypes was similar in AF patients and controls in our study similar to another study by Tsai et al.6 They also reported 2-way gene-gene interactions between ACE I/D polymorphism and angiotensinogen gene haplotypes, that may explain this discrepancy.6 Our study only included patients with lone AF, and a further study will be required to evaluate if the ACE I/D polymorphism also modulates electrocardiographic parameters in patients with AF associated with cardiovascular disease.
Conclusion
Our data support the hypothesis that the ACE I/D polymorphism modulates cardiac electrophysiology in patients with lone AF and in those with cardiac disease, consistent with the known effect of the D allele on ACE activity. This study provides further evidence for the role of activation of the RAAS in the pathophysiology of cardiac disorders. Therapies modulating the RAAS may be useful to prevent development of conduction abnormalities in patients with AF, in addition to prevention of AF and other cardiac diseases.34, 35
Acknowledgments
This work was supported by NIH awards HL075266 and HL085690 to DD and HL65962 to DMR.
Abbreviations
- ACE
Angiotensin-converting enzyme
- AF
Atrial fibrillation
- ECG
Electrocardiogram
- RAAS
Renin-angiotensin-aldosterone system
Footnotes
Conflicts of interest: none declared.
References
- 1.Cambien F, Poirier O, Lecerf L, et al. Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature. 1992;359(6396):641–644. doi: 10.1038/359641a0. [DOI] [PubMed] [Google Scholar]
- 2.Raynolds MV, Bristow MR, Bush EW, et al. Angiotensin-converting enzyme DD genotype in patients with ischaemic or idiopathic dilated cardiomyopathy. Lancet. 1993;342(8879):1073–1075. doi: 10.1016/0140-6736(93)92061-w. [DOI] [PubMed] [Google Scholar]
- 3.Schunkert H, Hense HW, Holmer SR, et al. Association between a deletion polymorphism of the angiotensin-converting-enzyme gene and left ventricular hypertrophy. N Engl J Med. 1994;330(23):1634–1638. doi: 10.1056/NEJM199406093302302. [DOI] [PubMed] [Google Scholar]
- 4.Marian AJ, Yu QT, Workman R, et al. Angiotensin-converting enzyme polymorphism in hypertrophic cardiomyopathy and sudden cardiac death. Lancet. 1993;342(8879):1085–1086. doi: 10.1016/0140-6736(93)92064-z. [DOI] [PubMed] [Google Scholar]
- 5.Gensini F, Padeletti L, Fatini C, et al. Angiotensin-converting enzyme and endothelial nitric oxide synthase polymorphisms in patients with atrial fibrillation. Pacing Clin Electrophysiol. 2003;26(1 Pt 2):295–298. doi: 10.1046/j.1460-9592.2003.00036.x. [DOI] [PubMed] [Google Scholar]
- 6.Tsai CT, Hwang JJ, Chiang FT, et al. Renin-angiotensin system gene polymorphisms and atrial fibrillation: a regression approach for the detection of gene-gene interactions in a large hospitalized population. Cardiology. 2008;111(1):1–7. doi: 10.1159/000113419. [DOI] [PubMed] [Google Scholar]
- 7.O'Donnell CJ, Lindpaintner K, Larson MG, et al. Evidence for association and genetic linkage of the angiotensin-converting enzyme locus with hypertension and blood pressure in men but not women in the Framingham Heart Study. Circulation. 1998;97(18):1766–1772. doi: 10.1161/01.cir.97.18.1766. [DOI] [PubMed] [Google Scholar]
- 8.Rigat B, Hubert C, Alhenc-Gelas F, et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86(4):1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danser AH, Schalekamp MA, Bax WA, et al. Angiotensin-converting enzyme in the human heart. Effect of the deletion/insertion polymorphism. Circulation. 1995;92(6):1387–1388. doi: 10.1161/01.cir.92.6.1387. [DOI] [PubMed] [Google Scholar]
- 10.Boldt A, Scholl A, Garbade J, et al. ACE-inhibitor treatment attenuates atrial structural remodeling in patients with lone chronic atrial fibrillation. Basic Res Cardiol. 2006;101(3):261–267. doi: 10.1007/s00395-005-0571-2. [DOI] [PubMed] [Google Scholar]
- 11.Yu H, Gao J, Wang H, et al. Effects of the renin-angiotensin system on the current I(to) in epicardial and endocardial ventricular myocytes from the canine heart. Circ Res. 2000;86(10):1062–1068. doi: 10.1161/01.res.86.10.1062. [DOI] [PubMed] [Google Scholar]
- 12.Zhang TT, Takimoto K, Stewart AF, et al. Independent regulation of cardiac Kv4.3 potassium channel expression by angiotensin II and phenylephrine. Circ Res. 2001;88(5):476–482. doi: 10.1161/01.res.88.5.476. [DOI] [PubMed] [Google Scholar]
- 13.Zankov DP, Omatsu-Kanbe M, Isono T, et al. Angiotensin II potentiates the slow component of delayed rectifier K+ current via the AT1 receptor in guinea pig atrial myocytes. Circulation. 2006;113(10):1278–1286. doi: 10.1161/CIRCULATIONAHA.104.530592. [DOI] [PubMed] [Google Scholar]
- 14.Fischer R, Dechend R, Gapelyuk A, et al. Angiotensin II-induced sudden arrhythmic death and electrical remodeling. Am J Physiol Heart Circ Physiol. 2007;293(2):H1242–1253. doi: 10.1152/ajpheart.01400.2006. [DOI] [PubMed] [Google Scholar]
- 15.Darbar D, Motsinger AA, Ritchie MD, et al. Polymorphism modulates symptomatic response to antiarrhythmic drug therapy in patients with lone atrial fibrillation. Heart Rhythm. 2007;4(6):743–749. doi: 10.1016/j.hrthm.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darbar D, Kucera G, Stubblefield T, et al. Cardiac Sodium Channel (SCN5A) Variants Associated with Atrial Fibrillation. Circulation. 2008;117(15):1927–1935. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellinor PT, Nam EG, Shea MA, et al. Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm. 2008;5(1):99–105. doi: 10.1016/j.hrthm.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Pretorius M, Donahue BS, Yu C, et al. Plasminogen activator inhibitor-1 as a predictor of postoperative atrial fibrillation after cardiopulmonary bypass. Circulation. 2007;116(11 Suppl):I1–7. doi: 10.1161/CIRCULATIONAHA.106.677906. [DOI] [PubMed] [Google Scholar]
- 19.Warsy I, Norris K, Roden D, et al. Ibutilide-induced QT Prolongation Varies Markedly Among Individuals in a Gender and Exercise-independent Fashion. Circulation. 2004;110(17):623. [Google Scholar]
- 20.Choy AM, Darbar D, Dell'Orto S, et al. Exaggerated QT prolongation after cardioversion of atrial fibrillation. J Am Coll Cardiol. 1999;34(2):396–401. doi: 10.1016/s0735-1097(99)00226-0. [DOI] [PubMed] [Google Scholar]
- 21.Priori SG, Napolitano C. Role of genetic analyses in cardiology: part I: mendelian diseases: cardiac channelopathies. Circulation. 2006;113(8):1130–1135. doi: 10.1161/CIRCULATIONAHA.105.563205. [DOI] [PubMed] [Google Scholar]
- 22.Darbar D. Genetics of atrial fibrillation: rare mutations, common polymorphisms, and clinical relevance. Heart Rhythm. 2008;5(3):483–486. doi: 10.1016/j.hrthm.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knollmann BC, Roden DM. A genetic framework for improving arrhythmia therapy. Nature. 2008;451(7181):929–936. doi: 10.1038/nature06799. [DOI] [PubMed] [Google Scholar]
- 24.Sotoodehnia N, Siscovick DS, Vatta M, et al. Beta2-adrenergic receptor genetic variants and risk of sudden cardiac death. Circulation. 2006;113(15):1842–1848. doi: 10.1161/CIRCULATIONAHA.105.582833. [DOI] [PubMed] [Google Scholar]
- 25.Arking DE, Pfeufer A, Post W, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38(6):644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 26.Newton-Cheh C, Guo CY, Larson MG, et al. Common genetic variation in KCNH2 is associated with QT interval duration: the Framingham Heart Study. Circulation. 2007;116(10):1128–1136. doi: 10.1161/CIRCULATIONAHA.107.710780. [DOI] [PubMed] [Google Scholar]
- 27.Fitzgerald PT, Ackerman MJ. Drug-induced torsades de pointes: the evolving role of pharmacogenetics. Heart Rhythm. 2005;2(2 Suppl):S30–37. doi: 10.1016/j.hrthm.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Xiao HD, Fuchs S, Campbell DJ, et al. Mice with cardiac-restricted angiotensin-converting enzyme (ACE) have atrial enlargement, cardiac arrhythmia, and sudden death. Am J Pathol. 2004;165(3):1019–1032. doi: 10.1016/S0002-9440(10)63363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasi VS, Xiao HD, Shang LL, et al. Cardiac Restricted Angiotensin Converting Enzyme Overexpression Causes Conduction Defects and Connexin Dysregulation. Am J Physiol Heart Circ Physiol. 2007;2:2. doi: 10.1152/ajpheart.00684.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goette A, Staack T, Rocken C, et al. Increased expression of extracellular signal-regulated kinase and angiotensin-converting enzyme in human atria during atrial fibrillation. J Am Coll Cardiol. 2000;35(6):1669–1677. doi: 10.1016/s0735-1097(00)00611-2. [DOI] [PubMed] [Google Scholar]
- 31.Kumagai K, Nakashima H, Urata H, et al. Effects of angiotensin II type 1 receptor antagonist on electrical and structural remodeling in atrial fibrillation. J Am Coll Cardiol. 2003;41(12):2197–2204. doi: 10.1016/s0735-1097(03)00464-9. [DOI] [PubMed] [Google Scholar]
- 32.Lenz O, Schmid B, Kilter H, et al. Effects of angiotensin II and angiotensin-converting enzyme inhibitors on human myocardium. Eur J Pharmacol. 1995;294(1):17–27. doi: 10.1016/0014-2999(95)00514-5. [DOI] [PubMed] [Google Scholar]
- 33.Birnie DH, Gollob M, Healey JS. Clinical trials, the renin angiotensin system and atrial fibrillation. Curr Opin Cardiol. 2006;21(4):368–375. doi: 10.1097/01.hco.0000231408.69307.9b. [DOI] [PubMed] [Google Scholar]
- 34.Healey JS, Baranchuk A, Crystal E, et al. Prevention of atrial fibrillation with angiotensin-converting enzyme inhibitors and angiotensin receptor blockers: a meta-analysis. J Am Coll Cardiol. 2005;45(11):1832–1839. doi: 10.1016/j.jacc.2004.11.070. [DOI] [PubMed] [Google Scholar]
- 35.Smith SC, Jr., Allen J, Blair SN, et al. AHA/ACC guidelines for secondary prevention for patients with coronary and other atherosclerotic vascular disease: 2006 update: endorsed by the National Heart, Lung, and Blood Institute. Circulation. 2006;113(19):2363–2372. doi: 10.1161/CIRCULATIONAHA.106.174516. [DOI] [PubMed] [Google Scholar]