Abstract
Purpose
To systematically compare two techniques for measuring brain atrophy rates from serial magnetic resonance imaging (MRI) studies.
Materials and Methods
Using the separation in atrophy rate between cohorts of cognitively normal elderly subjects and patients with Alzheimer's disease (AD) as the gold standard, we evaluated 1) different methods of computing volume change; 2) different methods for steps in image preprocessing - intensity normalization, alignment mask used, and bias field correction; 3) the effect of MRI acquisition hardware changes; and 4) the sensitivity of the method to variations in initial manual volume editing. For each of the preceding evaluations, measurements of whole-brain and ventricular atrophy rates were calculated.
Results
In general, greater separation between the clinical groups was seen with ventricular rather than whole-brain measures. Surprisingly, neither the use of bias field correction nor a major hardware change between the scan pairs affected group separation.
Conclusion
Atrophy rate measurements from serial MRI are candidates for use as surrogate markers of disease progression in AD and other dementing neurodegenerative disorders. The final method has excellent precision and accurately captures the expected biology of AD - arguably the two most important features if this technique is to be used as a biomarker of disease progression.
Keywords: serial MRI, Alzheimer's Disease, Brain Atrophy
Introduction
ALZHEIMER'S DISEASE (AD) IS THE MOST common cause of dementia in the elderly. Although there are currently no approved therapeutic agents that conclusively arrest or retard pathologic progression in AD, trials for potentially disease-modifying agents are in the planning phase. Therapeutic efficacy of candidate drugs for the treatment of AD is assessed through serial clinical/psychometric tests in double-blind, placebo-controlled trials. The high test/retest variability of clinical/psychometric measures has led many to consider rates of change in imaging measures as a surrogate endpoint in AD trials. The imaging biomarkers of disease progression in AD that have received the most attention are magnetic resonance imaging (MRI) measures of the rate of change in whole-brain volume or the volume of the hippocampus.
The techniques that have been applied most frequently to track neurodegenerative disease are based on translating intensity changes between time-separated scans into volume change estimates. Consider two image volumes acquired about a year apart. Differences between volume 1 and volume 2 at the brain-cerebrospinal fluid (CSF) boundary are used to obtain an estimate of volume change. Determinants of algorithm performance can be divided into three categories: 1) the specific method employed to compute change in tissue volume, 2) image data preprocessing steps, and 3) technical characteristics of the MR image volumes that were used as input. Variations in any of these categories produce different estimates of rate of change. There is no objective right or wrong answer - it is impossible to measure the “true” rate of volume change in actual subjects.
To test hypotheses and optimize algorithms in this area, an independent measure of algorithm performance is needed. One reasonable metric used to optimize algorithm parameters is the separation in atrophy rates between cognitively normal elderly subjects and patients with AD. On average, the rates of brain atrophy in cognitively normal elderly subjects will be less than those in patients with clinically probable AD.
The purpose of this project was to optimize a semiautomated method of measuring volume change from serial MRIs. Using the maximum separation in atrophy rates between cognitively stable normal elderly subjects and patients with AD as the gold standard, we performed the following evaluations: 1) compared two different methods of computing volume change; 2) compared two different methods for each of several steps in image preprocessing - intensity normalization, alignment mask used, bias field correction; 3) evaluated the effect of MRI acquisition nonuniformity over time; and 4) measured the uncertainty of the final method due to manual volume editing.
For each of the preceding evaluations, two sets or measurements were made. One set integrated change over all brain-CSF interfaces. The other set integrated change only over the surfaces of the third and lateral ventricles.
Materials and Methods
Subjects
Elderly Cognitively Normal Subjects and Alzheimer's Patients (N = 61)
We identified 30 individuals from the Mayo Alzheimer's Patient Registry (ADPR) and Alzheimer's Disease Research Center (ADRC) who met criteria for cognitively stable normal subjects and who had serial MRI studies. The mean age was 78.8 ± 6.13 years, and 22 were women. Thirty-eight subjects were identified from the ADPR/ADRC who carried the diagnosis of clinically probable AD. The diagnosis of probable AD was made according to the Diagnostic and Statistical Manual for Mental Disorders III Revised (DSM-III-R) Criteria for Dementia, and National Institute of Neurological and Communicative Disorders and Stroke/Alzheimer's Disease and Related Disorders Association Criteria (NINCDS/ADRDA) for AD ([1][2]). The mean age of these subjects was 76.2 ± 10.2 years, and 20 were women. The ADPR and the ADRC are Institution Review Board (IRB)-approved longitudinal population-based studies of aging and dementia. As part of the ongoing activities of these projects, the volunteers undergo serial MRI studies at roughly yearly intervals. Additionally, it was required that both scans in each pair of scans were acquired on similar hardware. As discussed later, after interscan compatibility criteria were evaluated, the number of subjects remaining was 29 normals and 32 ADs. Scan pairs from these 61 elderly subjects were used for all subsequent analyses except those involving hardware change and those in normal young and middle-aged volunteers.
Elderly Normal and AD Subjects With Hardware Change (N = 30)
A separate cohort of 15 AD and 15 normal subjects was identified from the ADPR/ADRC in order to evaluate the effects of MR hardware change over time. These subjects were selected using the same clinical criteria as the previous 68 subjects; however, the gradient hardware had been upgraded in the time between scans.
Normal Young and Middle-Aged Volunteers (N = 10)
Ten normal, young volunteer subjects ages 23-45 years underwent MR studies of the brain at two different points in time. The mean age was 32.6 years (range = 24-50 years). Four were women. The scans were performed on the same scanner over a 5-month period. The average interscan interval across the 10 volunteers was 6 weeks. Since negligible neurologic changes are expected in the scans from the young volunteers, these data were used to investigate variability inherent in the measurement process.
MRI Acquisition
All MRI data were acquired at 1.5 T using three-dimensional radio-frequency (RF) spoiled gradient echo (SPGR) acquisition with the following parameters: TR = 27 msec, TE = 9 msec, field of view (FOV) = 22 × 16.6 cm, matrix = 256 × 192, partitions = 124 with 1.6-mm partition thickness, repetition = 1, flip angle = 45°, and bandwidth = ±10.4 kHz.
Image Preprocessing
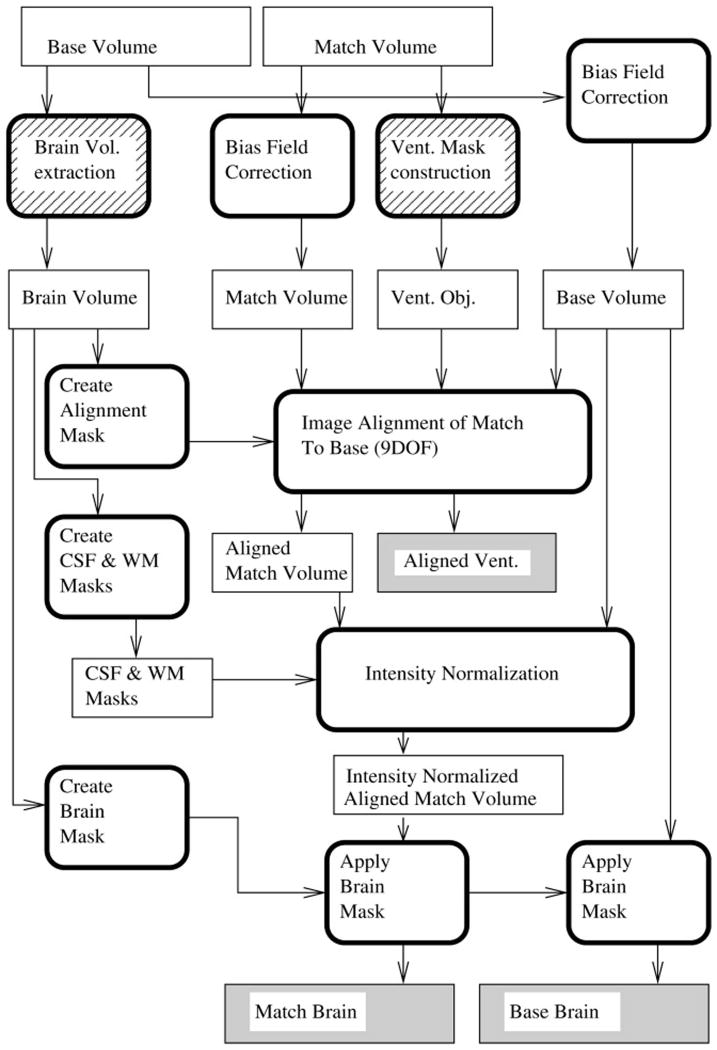
For each subject, we defined the base volume to be the MR volume acquired earlier in time, and the match volume to be the MRI volume acquired later in time. We used our best guess at the optimum set of image preprocessing parameters as the default method. A flowchart is given in Figure 1. The various preprocessing steps are outlined in chronological order below.
Figure 1.
Flowchart for the image preprocessing. Square-cornered boxes represent image volumes. Round-cornered boxes represent processing tasks. Hatched boxes represent steps requiring manual intervention. The shaded boxes are used as input to the change measurement steps.
Manual Preparation
The initial steps in the data preprocessing pipeline were two image preparation operations that required manual intervention. Extraction of the brain from the overlying skull and scalp soft tissue was necessary for each pair of MR volumes. As will be described later, this needs only be done manually for the base volume. A volume containing the base MRI brain was created using the brain extraction tools in the Analyze/AVW software package ([3]). This required identification of the internal region of interest (the brain) and several morphologic operations of erosion, dilation, and internal hole filling. To achieve pristine anatomic segmentation of the base brain, however, some degree of manual editing was required on well over half of all the individual brain slices after the automated brain extractor program had been run. Included in the manual editing is the deletion of the choroid plexus inside the ventricles. The second manual preparation step involved creating a binary mask of the third and lateral ventricles on the match volume of each subject. A trained neuroanatomic expert accomplished this using the autotrace subprogram in Analyze/AVW plus manual editing of anatomic outlines.
Bias Field Correction and Image Alignment
Correction of B1 field nonuniformity was performed on each of the input image volumes using the N3 program suite ([4]). Image alignment was then carried out using the automated image registration (AIR) suite ([5]). To make the alignment process robust, it was carried out in four steps. First the match volume was registered to the base volume using a rigid body transformation with six degrees of freedom (6DOF). A mask was created by dilating a binary mask of the segmented brain image to include the skull and scalp and applied to the base image. The inclusion of the temporally invariant skull and overlying scalp served to define outer volume margins that spatially normalize the voxel sizes between the scans. For the next step, both volumes were blurred with a 10-voxel-wide Gaussian kernel. Starting with the previous (6DOF) alignment parameters, the blurred match volume was aligned to the blurred version of the masked base volume using a transformation that allows voxel size scaling (9DOF). The resulting transformation was used as a starting point, and the alignment repeated with slight blurring (a two-voxel-wide Gaussian kernel). A final registration pass with no blurring was made using the slightly blurred result as a starting point. The match volume was transformed using windowed sinc interpolation with a window half-width of nine voxels. The same transformation was applied to the ventricle object map using nearest-neighbor interpolation. The observed alignment failure rate was about 1 in 300 scan pairs.
Intensity Normalization
The intensity normalization method employed in this analysis was based on matching peaks in intensity histograms for subvolumes dominated by CSF and white matter. A linear remapping that brought the peak values in the match volume into correspondence with those in the base volume peak values was used.
The CSF intensity spectrum was constructed for each volume. A CSF mask was constructed from the manually edited brain volume by morphological operations. A hole-filling filter was passed over a mask of the brain. The result contained the brain and interior CSF spaces. An inverted version of the brain mask was then applied to remove brain voxels, leaving a sample of voxels largely dominated by CSF. To determine the peak position of the CSF intensity distribution, a Gaussian distribution of mean μ and width σ was fitted to the spectrum of voxels in the masked region using a least squares minimization. The distribution had a positive skew if there was gray matter contamination in the spectrum. To minimize these effect of the contamination, the fit was carried out only over the range from zero to μ + σ. Since μ and σ were not known before fitting, we proceeded iteratively. The fit was seeded with initial values μ = m and σ = 0.2m where m is the median intensity in the distribution being fitted. The fit was then repeated using the previous estimates of μ and σ as initial values and to set the range. The entire process typically converged after two or three fitting iterations.
The white matter subvolume was selected using a mask derived by eroding the intersection of the brains from the base and match volumes. The brains were extracted using the brain mask, the resulting gray-scale images were thresholded at 30% of the median intensity of nonzero voxels, the intersection of the two thresholded volumes was found, and two passes of a 3 × 3 × 3 erosion kernel were made over the intersection volume.
Means and widths were again determined by fitting Gaussian distributions to the observed spectra. The fits extended only to data in the range of μ - σ to μ + 3σ. Denoting the intensity distribution as f(I), the initial seed values were μ = argmax(f(I)) and σ = the standard deviation of f(I). The process again proceeded iteratively.
Final Brain Extraction
After alignment and intensity normalization, the brain volumes were extracted by applying a mask derived from the manually segmented brain. The brain mask was derived from a dilated (3 × 3 × 3 neighborhood) version of the thresholded base brain volume.
Volume Compatibility Criteria
The intensity distributions for CSF (or white matter) were drawn from exactly the same voxels within each volume. After intensity normalization, the widths of the peak distributions were compared as an indication of the similarity of the contrast in the image volumes. If the widths were strikingly dissimilar, then evaluation of anatomic changes was found to be unreliable. Denote the mean and width measured for each tissue class and image volume by super- and subscripts, that is, and as the mean and width for volume ν∈{base,match} and approximate voxel class c ∈{wm,csf} before intensity normalization. Define as the width from the match volume after the linear intensity normalization. A mismatch value (γ) was defined as
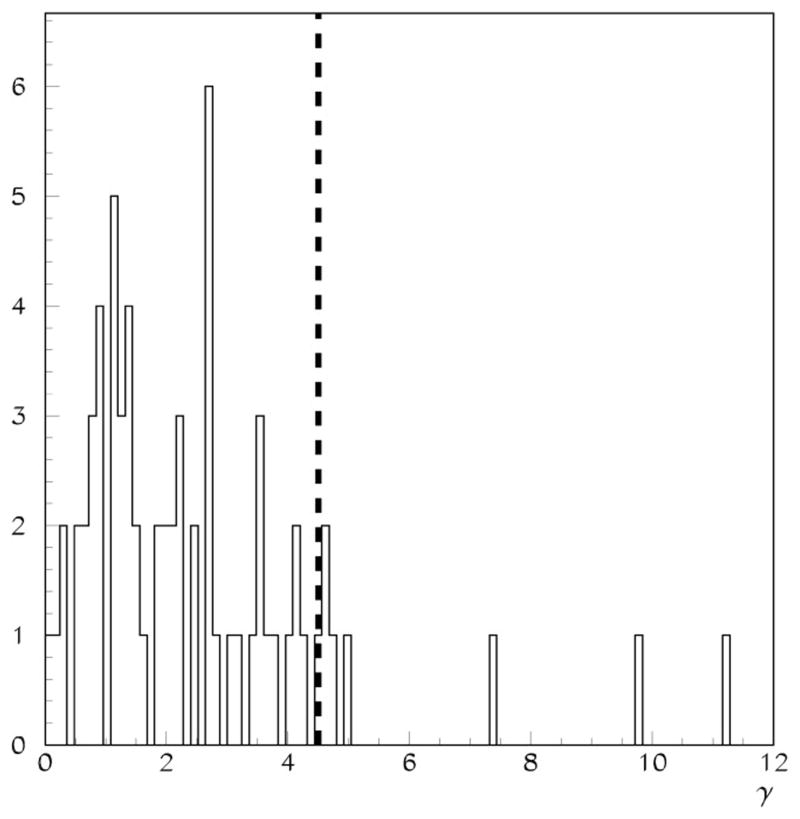
The denominator was chosen to reckon the differences with respect to the intensity range in the base image. The γ distribution for 67 volume pairs is shown in Figure 2. A requirement of γ < 4.5 was used to reject incompatible volume pairs. This requirement rejected less than 10% of the scan pairs in the data sets we have examined. The data presented in the rest of this paper reflect the application of the criteria above for discarding MRI scan pairs. A total of 7 scan pairs (6 ADs and 1 normal) were discarded in order to arrive at the nominal census of 32 normal and 29 AD patients.
Figure 2.
The distribution of γ values for 68 cases used as input for this study. The dashed line is at γ = 4.5, the volume compatibility cutoff used for data presented in this paper.
Method of Computing Volume Change
We compared the boundary shift integral (BSI) method of Freeborough and Fox ([6]) with a variation of a gradient-matching method (GMM) included in the SIENA package of Smith et al ([7]).
BSI Method
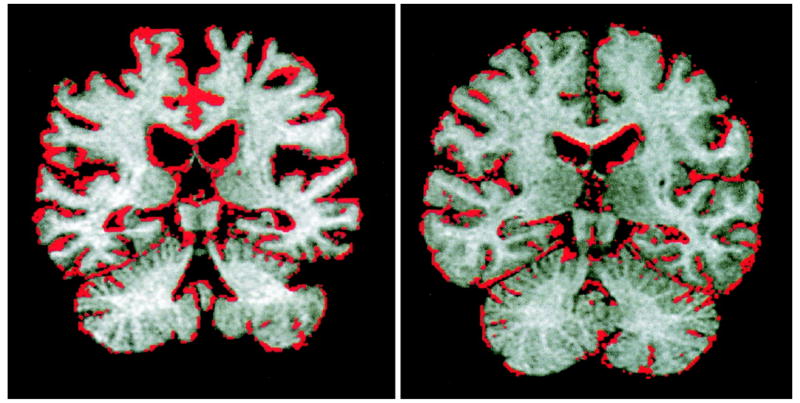
The BSI algorithm was adapted from Freeborough and Fox and is described in detail in reference [6]. In the BSI method, differences in intensity between two spatially co-registered volumes near the brain-CSF interfaces are assumed to be due to a spatial shift in the brain boundary. Consider a line profile perpendicular to the brain surface in the two registered image volumes. Let I1(x) and I2(x) be the voxel intensities along this line in the base and match volumes, respectively. Next, these intensities are clipped at upper and lower bounds Ihigh and Ilow, respectively. Denote the linear shift in the brain boundary between the volumes as Δw, which can be calculated as a discrete approximation of . The integration is performed only over voxels in the brain-CSF boundary region. For ventricular measures, only the voxels also within the ventricle mask are included. When extended to three dimensions, this linear model gives a measure of the shift in the brain boundary between volumes, and hence a measure of volume change. Shown in Figure 3 are single coronal images from two patients, one normal and one AD, with color overlays indicating the level of change as measured by the BSI.
Figure 3.
Color overlays are used to show the change measurements determined by BSI. The coronal image at left is from an AD patient. In contrast, the image at right is for a normal patient of approximately the same age. Markedly more atrophy is apparent in the AD patient image.
The most significant difference in our implementation of the BSI measurement scheme is in the selection of the intensity clipping range. As originally formulated, the BSI included intensities from 25%-75% of the mean intensity of voxels in the brain. In the current formulation with intensities normalized to CSF and white matter, the BSI is calculated over voxel intensities from the peak of the CSF distribution to halfway between the CSF and white matter peaks. Net volume loss is expressed as a percentage of initial brain volume and may be divided by the interscan interval to give the annualized rate of global atrophy.
GMM
The GMM employed here is derived from the SIENA package ([7]). In the gradient-matching algorithm, the white or gray matter voxels determined to be on the CSF-brain interface of the base volume are found using the FAST segmentation tool ([8]). For each brain-CSF interface voxel, intensity profiles perpendicular to the interface surface at that voxel are constructed from intensities in the base and match brain images The profiles are differentiated and weighted. The relative shift necessary to bring the resulting profiles into agreement is then found to subvoxel accuracy. The form of the weight function is W(d; σ) = e-d4/2σ4 with σ = 7 mm and d the distance from along the profile from the edge voxel. This weighting biases the algorithm toward finding small shifts. The shifts are accumulated over all edge volumes and reexpressed as volumetric change.
The chief differences in our implementation of GMM and that in the SIENA package are the use of the preprocessing path discussed previously, the modification of the weighting function to reduce the bias against finding large changes, and the ability to restrict the calculation to the ventricular surfaces. In our formulation, each brain surface voxel is biased by the average motion surface of voxels in the immediate neighborhood. That is, a weight function of the form W(d; d0σ) = e-(d-d0)4/2σ4 is employed. The additional parameter d0 is always zero for the profile from the base volume. For the match volume profile, however, d0 is the average displacement of all surface voxels in the 3 × 3 × 3 voxel neighborhood of the edge voxel under consideration. The values of d0 are initially unknown, so we procede iteratively. Initially all d0 values are set to zero. For subsequent passes, the previous estimates of the match volume displacements are used to calculate new d0 values for the profiles from the match volume. A total of three passes are made over the data. Ventricular measures are obtained by including only brain-CSF interface voxels lying within the ventricle mask.
Gold Standard
The complete algorithm presented has many components and parameters that could affect the final atrophy measurements. Systematic studies varying each of the components of the algorithm were carried out. The measure we used to test the hypothesis that one method was superior to the other was the degree of separation in rates of atrophy of cognitively normal elderly subjects vs. those of AD patients. The metric used was the difference between the group mean rates divided by the pooled variance:
Results
Results from the Baseline Method
The average yearly rate of atrophy for the normal and AD groups using BSI and GMM with calculations over all brain-CSF boundaries and over ventricular surfaces only are given in Table 1. The values given in the Normal and AD rows are the group means and standard deviations. The values given in the bottom row are the group separation metrics with an estimate of the statistical uncertainty in the metrics (±stat.)
Table 1. Summary of Atrophy Rate Results for the Default Algorithm Configurations*.
| Group | BSI WB | GMM WB | BSI vent | GMM vent |
|---|---|---|---|---|
| Normal (N = 29) |
-0.54 (0.74) | -0.27 (0.52) | -2.96 (1.79) | -2.23 (1.55) |
| AD (N = 32) |
-1.09 (0.93) | -1.42 (0.72) | -6.35 (3.77) | -5.62 (3.22) |
| Separation ± stat. | 0.46 ± 0.19 | 1.30 ± 0.22 | 0.81 ± 0.20 | 0.95 ± 0.20 |
BSI and GMM refer to the Boundary Shift Integral and Gradient Matching Method techniques. WB and Vent refer surfaces included in the measurements. WB values are for measurements over the whole brain and vent values are for ventricular surface measurements. Atrophy rates, in percent of the brain or ventricle volume, are given in the form mean (SD) for each group.
The data in Table 1 show that GMM is superior to the BSI method for the whole-brain measurement. GMM is slightly better than the BSI method for the ventricular rate measurement as well; however, this difference is not as striking.
Image Preprocessing Perturbations
For each major step in the image preprocessing tool chain, a pair of different methods was compared using the elderly normal control and AD patients. The patient population (N = 61) and MRI acquisition parameters are as described previously, unless otherwise explicitly noted.
Global Intensity Normalization
We compared the previously described intensity normalization with an alternate approach. The alternate approach simply scaled the intensities such that the white matter peak positions matched. Comparison of the results in Table 2 (single-point white matter normalization) with those in Table 1 (two-point normalization) reveals clear superiority of the two-point method for whole-brain-rate measures with both the BSI method and GMM. A trend toward better clinical group separation of ventricular rates is present for the BSI method and GMM as well.
Table 2. Results With Single Point (WM) Intensity Normalization.
| Group | BSI WB | GMM WB | BSI vent | GMM vent |
|---|---|---|---|---|
| Normal (N = 29) |
-0.12 (0.86) | -0.12 (0.59) | -2.36 (2.28) | -2.05 (1.66) |
| AD (N = 32) |
-0.40 (1.18) | -1.18 (0.72) | -5.26 (4.05) | -5.39 (3.30) |
| Separation ± stat | 0.19 ± 0.18 | 1.12 ± 0.21 | 0.62 ± 0.19 | 0.90 ± 0.20 |
Alignment Mask
In the baseline method, a mask is used to select the region over which the image alignment is to be optimized. The mask is created by morphological operations that “grow” the manually segmented brain to include the skull and scalp. An alternative method of forming the mask is to simply select everything in the image except the brain. The results from using such a mask are presented in Table 3. Note that the AD patient population in Table 3 is reduced to N = 31 because one of the scan pairs blatantly failed to register. Comparison with Table 1 reveals that the group separation for GMM-based calculations is reduced relative to the default alignment mask. The BSI whole-brain measures are largely unchanged, and the BSI ventricular measures are improved.
Table 3. Results With Inverted Brain Mask in Place of the Standard Alignment.
| Group | BSI WB | GMM WB | BSI vent | GMM vent |
|---|---|---|---|---|
| Normal (N = 29) |
-0.47 (0.71) | -0.29 (0.54) | -2.88 (1.77) | -2.31 (1.55) |
| AD (N = 31) |
-1.00 (0.94) | -1.31 (0.90) | -6.79 (3.74) | -5.42 (3.59) |
| Separation ± stat | 0.45 ± 0.19 | 0.98 ± 0.21 | 0.94 ± 0.21 | 0.79 ± 0.20 |
Bias Field Correction
The effect of including bias field correction was investigated by simply removing it from the preprocessing tool chain. Comparison of results in Table 1 (with bias field correction) to those in Table 4 (no bias field correction) reveals no difference in group separation with vs. without this preprocessing step. It should be noted that these scans were acquired using a coil with good spatial uniformity over the volume of the head. For hardware with less uniform response, the inclusion of bias field correction may well be important.
Table 4. Results Without Bias Field Correction.
| Group | BSI WB | GMM WB | BSI vent | GMM vent |
|---|---|---|---|---|
| Normal (N = 29) |
-0.67 (0.74) | -0.27 (0.52) | -2.95 (1.75) | -2.23 (1.56) |
| AD (N = 32) |
-1.17 (0.95) | -1.43 (0.74) | -6.35 (3.76) | -5.63 (3.22) |
| Separation ± stat | 0.42 ± 0.18 | 1.28 ± 0.22 | 0.82 ± 0.20 | 0.95 ± 0.20 |
MRI Acquisition Uniformity: Effect of Hardware Change
A separate cohort of 15 AD and 15 normal subjects was evaluated in whom the gradient hardware had been upgraded in the time between scans. This hardware change was an upgrade in the gradient subsystem from a standard 1.0 gauss/cm gradient set to an “echo speed” gradient set with both faster gradient slew rates and maximum gradient amplitude, 2.2 gauss/cm. The results are summarized in Table 5. Overall, the major changes in hardware had a negligible effect on the observed rates of atrophy in each group for each of the four measurement types (i.e., BSI whole brain, BSI ventricle, GMM whole brain, and GMM ventricle).
Table 5. Gradient Hardware (HW) Effects: Group Separation ± stat.
| BSI WB | GMM WB | BSI Vent | GMM Vent | |
|---|---|---|---|---|
| HW match N = 61 | 0.46 ± 0.19 | 1.30 ± 0.22 | 0.81 ± 0.20 | 0.95 ± 0.20 |
| HW mismatch N = 30 | 0.65 ± 0.27 | 1.03 ± 0.30 | 0.88 ± 0.29 | 0.71 ± 0.28 |
Test/Retest Reproducibility
In this section, we sought to investigate the sensitivity of the algorithm to differences in the semiautomated brain segmentation and definition of ventricular masks. Five normal and five AD subjects were randomly selected from the subjects summarized in Table 1. The change in brain and ventricular volume in these 10 subjects was remeasured starting with the raw unprocessed image files. The manual editing steps were redone by an analyst who was blinded to the results of the first measurement. The coefficient of variation for the pairs of measurements for each subject was computed. The average across all 10 subjects was then computed to assess the sensitivity of each method to differences in manual editing. For each case, the ratio of the test/retest standard deviation to the baseline whole-brain or ventricle volume is used as one estimate of the variability. Alternately, we also calculated the ratio of the standard deviation to the mean of the two volume change measurements for each case. The mean variabilities, expressed in percent, are summarized in Table 6.
Table 6. Manual Editing Effects*.
| BSI WB | GMM WB | BSI vent | GMM vent | |
|---|---|---|---|---|
| SD/base (in %) | 0.0135 | 0.0042 | 0.0101 | 0.0147 |
| SD/mean change (in %) | 1.83 | 1.15 | 1.83 | 0.39 |
Data used were the pooled 61 elderly normal and AD subjects. Values given in the first row are averages of the SD of the test/retest volume change measurements normalized to the respective base brain volumes. The values in the second row are normalized to the mean volume change measurements.
The most direct comparison is the ratio of the standard deviation of the measurements reckoned relative to the base volume with the estimates of atrophy from Table 1. The effects of manual editing are much smaller than the intragroup standard deviations.
Measurements in Young and Middle-Aged Normal Volunteers
The brains of both Alzheimer's patients and cognitively normal elderly subjects atrophy over time, and the rate of atrophy is greater in patients with clinically diagnosed AD. We do not, however, know what the precise rate of atrophy is or should be in either group of subjects. A different way to compare the accuracy and precision of several different measurement methods using realistic models of the human brain (i.e., living subjects) is to perform the measurements in subjects in whom no substantial biologic change over the period of serial MR scanning is expected. Such measurements also provide insight into the sources of error, which can be divided into three categories: 1) variation due to scanner drift, 2) variation due to the actual change in brain/ventricular volume that occurs in normal young subjects over a short time interval, and 3) unknown systematic variation in the image processing tool chain itself when used on a given pair of images. The change in volume between scan 1 and scan 2 in each of these 10 volunteers was computed forward (scan 2 - scan 1) and backward (scan 1 - scan 2) for each of the four major measurement techniques. The standard deviation of the sum of the forward and backward measurements across all 10 volunteers provides an estimate of measurement variability due to image processing alone. Biologic variation and machine variation in any individual that changed in a positive direction from scan 1 to scan 2 were measured in an equal and opposite direction when the measurement was made in the backward direction (i.e., scan 2 to scan 1).
For both the forward and backward measurements, the computed “rate of atrophy” was not significantly different from 0, as would be expected. In addition, the actual rate measured in the forward direction for each of the four measurement techniques had the opposite sign with approximately the same magnitude when the corresponding measurement was done in the backward direction. This implies that on a subject-by-subject basis there were biologic or scanner-related differences between scan 1 and scan 2 embedded in the MR data itself. It also implies that each of the four image processing methods accurately captured this biologic/machine “change,” and reversed it when the measurements were done in the backward direction. For each of the four measurements, the standard deviation of the forward plus backward measures was substantially less than the standard deviation across all 10 volunteers of either the forward or backward measurement alone. The means (standard deviation) of these summed values were as follows: brain_BSI, -0.02 (0.14); ventricle_BSI, -0.34 (0.34); brain_GMM, -0.00 (0.41); and ventricle_GMM, 0.19 (0.18). The standard deviation of the summed values ranged from a high of about 1/2 the forward (or backward) alone standard deviation in the case of the brain_GMM measurement, to a low of about 1/5 in the case of the ventricular_BSI measurement. This implies that the measurement error or variability due to image processing alone accounts for 1/2 to 1/5 of the total measurement error or measurement variability in living subjects (depending on the measurement technique in question).
Discussion
Regardless of perturbations in a variety of technical parameters, the measured rates of whole-brain and ventricular volume change consistently match expectation based on the known differences in clinical behavior between normal elderly subjects and patients with AD. The actual rates for each group and the separation of the group rates, however, vary significantly with technical perturbations. In general, greater separation between the normal and AD groups is seen with ventricular than whole-brain measures. Group separation was better with the GMM than the BSI method of computing volume change; with two-point than single-point intensity normalization; and with the dilated-brain masking method.
Surprisingly, neither the use of bias field correction nor a major hardware change between the scan pairs affected group separation. The upgrade to the higher-performance echo speed gradient subsystem was accompanied by numerous subtle differences in the execution of the imaging pulse sequence itself. For example, the rise times for all of the gradient pulses were subtly changed, and differences in gradient linearity were present between the conventional gradient set and the high-performance system. Therefore, we anticipated dramatically worse separation of the rates of atrophy in the AD vs. normal groups in those MRI volume pairs that were acquired with different hardware systems, due to expected greater variability in the data. Failure to observe a significant degradation of the data due to gradient hardware mismatches has significant implications for longitudinal imaging studies employing serial MRI. As a rule, hardware upgrades improve magnet performance and are viewed as desirable by MRI users. At any individual MRI site it is inconceivable that such an upgrade could be postponed in order to maintain hardware consistency for any single research study. In other words, hardware upgrades will inevitably occur in any longitudinal MRI clinical study if the study lasts long enough. If hardware upgrades had seriously corrupted the calculation of longitudinal rates of change, this would present a major and largely unavoidable problem for the use of this approach to atrophy rate quantitation as a marker of disease progression.
Sources of variation in serial MRI volume quantitation include actual biologic change, change in MRI acquisition factors, positional change of the scanned object with respect to the fixed geometry of the scanner, and variability inherent in the image analysis method. Measurement imprecision due to positional change of the object with respect to the scanner can be pronounced if the structure being measured is relatively small, and the imaging slices are fairly thick. For example, for a small structure like the hippocampus, slice thickness is a major factor in the precision of MRI volume measurements. In the application evaluated here, measurement of whole-brain or ventricular volume, the ratio of the size of the head vs. the most anisotropic voxel dimension (1.6 mm), is on the order of 50 to 1. It seems unlikely, therefore, that change in the spatial orientation of the subject's head with respect to fixed scanner geometry over time should introduce major variation.
The serial MR measurements in 10 young to middle-aged cognitively normal volunteers provide some insight into the relative contribution of the three possible sources of measurement variation - biologic variability, scanner variability over time, and randomness inherent in the image processing method itself. The data show that measurable volume variation was imbedded in the serial MRI data obtained at two different time points. Some of the measured variation may have been due to actual change in volume due to biologic variation, for example, change in hydration status between the scans. It is also possible that measurement variation was due to scanner changes. For example, changes in the quality of shimming between two separate scans might introduce geometric distortions in the difference images, which would be misinterpreted as a volume change. We cannot determine whether biologic change or scanner change over time was the major effect. More importantly, however, by computing volume change forward and backward and then summing these two measurements, we were able to arrive at an estimate of the relative contribution of the image processing method itself to overall measurement stability over time. This contribution is relatively small, representing 1/2 to 1/5 of overall test retest variability.
The data from this study have implications for the use of serial MRI as a surrogate marker of disease progression in both naturalistic and therapeutic trials of neurodegenerative diseases - particularly AD. The final method has excellent precision and seems to capture the expected change in biology well - which are arguably the two most important features of a proposed biomarker of disease progression. Although tracing of specific regions of the brain, for example, the hippocampus, can be performed with a high degree of precision, manual methods are time-consuming and personnel-intensive. Conversely, more automated methods such as those evaluated in this study are better suited to high-throughput, high-efficiency image analyses, which are desirable for large-scale trials.
Acknowledgments
This work was supported in part by the Institute on Aging, National Institutes of Health AG11378, AG16574, AG06786.
References
- 1.DSM-III-R: diagnostic and statistical manual of mental disorders. Washington, D.C.: American Psychiatric Association; 1987. [Google Scholar]
- 2.McKhann G, Drachman D, Folstein M, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of Department of Health and Human Services Task Force on Alzheimer's disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 3.Robb R. The Biomedical Imaging Resource at Mayo Clinic. IEEE Trans Med Imaging. 2001;20:854–867. doi: 10.1109/42.952724. [DOI] [PubMed] [Google Scholar]
- 4.Sled J, Zijdenbos A, Evans A. A non-parametric method for automatic correction of intensity non-uniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 5.Woods R, Grafton S, Holmes C, Cherry S, Mazziotta J. Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998;22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- 6.Freeborough P, Fox N. The boundary shift integral: an accurate and robust measure of cerebral volume changes from registered repeat MRI. IEEE Trans Med Imaging. 1997;16:623–629. doi: 10.1109/42.640753. [DOI] [PubMed] [Google Scholar]
- 7.Smith S, Zhang Y, Jenkinson M, et al. Accurate, robust and automated longitudinal and cross-sectional brain change analysis. Neuroimage. 2002;17:479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]