Summary
Although it is well established that some protein tyrosine kinases have a prognostic value in breast cancer, the involvement of protein tyrosine phosphatases (PTPs) is poorly substantiated for breast tumours. Three of these enzymes (PTP-gamma, LAR and PTPL1) are already known to be regulated by estrogens or their antagonists in human breast cancer cells. We used a real-time reverse-transcriptase-polymerase-chain-reaction method to test the expression levels of PTP-gamma, LAR and its neuronal isoform, and PTPL1 in a training set of RNA from 59 breast tumours. We sought correlations between levels of these molecular markers, current tumour markers, and survival. We then quantified the expression level of the selected phosphatase in 232 additional samples, resulting in a testing set of 291 breast tumour RNAs from patients with a median follow-up of 6.4 years. The Spearman nonparametric test revealed correlations between PTPL1 expression and differentiation markers. Cox univariate analysis of the overall survival studies demonstrated that PTPL1 is a prognostic factor (risk ratio = 0.45), together with the progesterone receptor (PR) (risk ratio = 0.52) and node involvement (risk ratio = 1.58). In multivariate analyses, PTPL1 and PR retained their prognostic value (risk ratios of 0.48 and 0.55, respectively). This study demonstrates for the first time that PTPL1 expression level is an independent prognostic indicator of favourable outcome for patients with breast cancer. In conjunction with our mechanistic studies, this finding identifies PTPL1 as an important regulatory element of human breast tumour aggressiveness and sensitivity to treatments such as anti-estrogens and anti-aromatase.
Keywords: Adult; Aged; Aged, 80 and over; Breast Neoplasms; genetics; mortality; pathology; Female; Gene Expression; Genes, Tumor Suppressor; Humans; Kaplan-Meiers Estimate; Middle Aged; Pilot Projects; Prognosis; Protein Tyrosine Phosphatase, Non-Receptor Type 13; biosynthesis; RNA, Messenger; analysis; Receptors, Progesterone; metabolism; Reverse Transcriptase Polymerase Chain Reaction; Tumor Markers, Biological; genetics
Keywords: breast cancer, PTPL1/ PTPN13, phosphatase, prognosis
Introduction
In breast cancer, the clinical and biological variables commonly used to predict the outcome of primary chirurgical treatments include regional lymph node invasion, histological grade, and hormone receptor expression. All of these parameters are well-recognized prognostic and predictive factors. Additionally, the expression of new markers associated with proliferation (Ki-67) and cell cycle (cyclin E, cyclin D1) (1), with mitogenic and survival pathways (HER tyrosine kinase receptor family) (2) and with invasion processes (urokinase-type Plasminogen Activator, Cathepsin D) (3;4), has also been linked to the survival of breast cancer patients or to their response to hormonal or cytotoxic therapies.
Though it is now well established that some protein tyrosine kinases have a prognostic value in breast cancer, the involvement of protein tyrosine phosphatases (PTPs) is poorly substantiated for breast tumours (5). Initially, we showed that, in a breast cancer cell line model, PTP activity was involved in anti-estrogen inhibition of growth factor-stimulated proliferation (6). Furthermore, through mutational analysis of the tyrosine phosphatase gene superfamily in human cancers, a recent study identified six PTPs that are quite commonly affected (7). Three of these enzymes, consisting of two transmembrane subtypes (PTP gamma and LAR) and one intracellular subtype (PTPL1), are already known to be regulated by estrogens (8) or their antagonists (9) in human breast cancer cells, and they are known to play a role in the growth of these tumours in in vitro models.
PTP gamma, which has been regarded as a potential tumour suppressor gene in kidney and lung adenocarcinoma (10), is more highly expressed in normal breast tissue than in breast tumours or breast cancer cell lines (8;11). Moreover, Liu et al. (12) have recently demonstrated that PTP gamma is able to inhibit anchorage-independent growth of breast cancer cells in soft agar and to reduce the proliferative response of MCF-7 cells to oestradiol, thus suggesting that PTP gamma may be a potential estrogen-regulated tumour suppressor gene in human breast cancer. However, Lamprianou et al. did not describe mammary gland phenotypic effects in PTP gamma knockout mice (13). Thus, in order to verify the tumour suppressor properties of PTP gamma, the susceptibility of these mice to various carcinogens should be tested.
LAR gene deletion in mice suggests an important role for LAR-mediated signalling in mammary gland development and final differentiation (14). Moreover, the inhibitory effect of LAR ectopic expression on the growth of neu-transformed human breast carcinoma cells (15) implies a negative role of LAR on the growth or survival of breast cancer cells. On the other hand, Yang et al. (16) showed increased expression of a LAR isoform in malignant breast tissues. This LAR isoform, generated by neuronal-type alternative splicing (17), contains an insertion in the extracellular domain and could have potential clinical relevance as a tumour marker.
We have demonstrated increased PTPL1 mRNA levels after anti-estrogen treatment (9) and have demonstrated by using an antisense strategy that PTPL1 expression and resulting regulation are crucial for mediation of 4-hydroxytamoxifen inhibitory effects on growth factor activity (18). In addition, we have shown that PTPL1 induces apoptosis by inhibiting the PI3-kinase/Akt survival pathway through IRS-1 dephosphorylation (19). It is interesting to note that the PTPL1/PTPN13 gene presents the characteristics of a tumour suppressor gene (20;21). It is located on chromosome 4q21, a region frequently deleted in ovarian and liver cancers (22), and its expression is frequently down-regulated or silenced through promoter hypermethylation in several tumour types (23;24).
In the present study, we compared the expression level and prognostic value of three PTPs (PTP gamma, PTPL1, and the two LAR splicing variants) in a training set of 59 breast tumours. We confirmed the expression of the LAR neuronal variant in 58 of 59 tumours, and we demonstrated that the level of PTPL1 expression is a prognostic indicator of favourable outcome for breast cancer patients. In the testing set of 291 patients that included 232 complementary tumours and had a median follow-up of 6.4 years, we confirmed that the level of PTPL1 expression is an independent prognostic marker of increased overall survival (OS) for breast cancer patients.
Patients and methods
Patients
The pilot study involved 59 breast tumour samples; the mean age of the patients was 59.7 years (range 35–81). The median follow-up duration in living patients was 7.5 years. The number of deaths was 21, and the number of relapses was 27.
To confirm the results in a larger sample set, 232 complementary breast tumour samples were added, resulting in a testing set of 291 patients (Table I). All these patients underwent surgery for locoregional disease in the Centre Oscar Lambret, the Anticancer Centre of the North of France, between May 1989 and December 1991. The mean age of the patients was 57.9 years (range 26–90).
TABLE I.
PATIENTS AND TUMOUR CHARACTERISTICS (n=291)
| Characteristics | No of cases |
|---|---|
| Age | |
| <50 years | 69 |
| ≥50 years | 222 |
| Node involvement | |
| negative | 134 |
| positive | 155 |
| unknown | 2 |
| HPG | |
| I | 42 |
| II | 111 |
| III | 109 |
| unkown | 29 |
| Tumor type | |
| Ductular | 212 |
| Lobular | 22 |
| Others | 57 |
| Tumor diameter | |
| < 2cm | 21 |
| 2 to 5cm | 188 |
| > 5cm | 68 |
| unknown | 14 |
| ER | |
| < 10 fmol/mg protein | 88 |
| ≥ 10 fmol/mg protein | 201 |
| unknown | 2 |
| PgR | |
| <10 fmol/mg protein | 88 |
| ≥ 10 fmol/mg protein | 200 |
| unknown | 3 |
Patients were treated by segmentectomy when tumours were smaller than 3 cm in width and by total mastectomy if tumours were larger or centrally located. Surgery was followed by radiation therapy. Node-positive premenopausal patients, estrogen receptor (ER)-negative and progesterone receptor (PR)-negative postmenopausal patients received adjuvant treatment consisting of six cycles of chemotherapy. The node-positive, ER-positive, and PR-positive postmenopausal patients received tamoxifen for two years. Node-negative patients received no adjuvant treatment. The median follow-up duration in living patients was 6.4 years. The number of deaths was 83, and the number of relapses was 108. The biopsies were obtained with the agreement of the Institutional Review Board.
Tumour samples
All tumours were adenocarcinomas. At the time of collection, fat and necrotic tissues were discarded, and two adjacent tumour pieces were removed. The first piece was submitted to histological studies. The second was immediately frozen in liquid nitrogen for hormone receptor (ER, PR) and RT-PCR assays. Both ER and PR levels were determined by the dextran-coated charcoal method as described previously (25). The Laboratoire d’Oncologie Moléculaire Humaine is affiliated with the European Organisation for Research and Treatment of Cancer Receptor Study Group, which undertook quality control of the assays (26).
PTP mRNA expression
Total RNA was isolated (RNeasy Mini Kit, Qiagen, France) from 40 mg of each tumour sample. Disruption and homogenisation of the tumour samples were performed using a Rotor-Stator Homogeniser (Ribolyzer, Hybaid). The amount of extracted RNA was quantified by measuring the absorbance at 260 nm. The quality of the RNA was checked by assessing the ratio between the absorbance values at 260 nm and 280 nm, and it was then confirmed by electrophoresis of the RNA on a 1.5% agarose gel containing ethidium bromide. Total RNA (2 μg) was reverse transcribed into cDNA using 5 μM of random hexamers (Roche) and Superscript II RNAse H- reverse transcriptase according to the manufacturer’s instructions (Invitrogen).
Real time Polymerase Chain Reaction (PCR) quantification of the expression of each gene was carried out on cDNA corresponding to 12.5 ng of total RNA using a LightCycler 3 device (Roche) with LightCycler FastStart DNA MasterPLUS SYBR GreenI Kit (Roche) according to the manufacturer’s instructions. Primer sequences, amplification position, product length, time and temperature used for each gene are indicated in Table II.
TABLE II.
PCR CONDITIONS AND PRIMERS
| Gene | Primer sequence | Exon | Product size | Annealing temperature |
|---|---|---|---|---|
| PTP gamma | Upper 5′ GCA ACT CGA TGG CTT CGA CAA 3′ | 3 | 84 bp | 61 °C |
| Lower 5′ TCT TTC AGA AGG ATG GCG ACT GTT 3′ | 3–4 | |||
| LAR-Neu | Upper 5′ TGG ACT CCC CAT CAT CCA AGA 3′ | 13 | 85 bp | 66 °C |
| Lower 5′ CCG CTG ATA GTG GTT TCA TAG TCC T 3′ | 14–15 | |||
| LAR | Upper 5′ TAG CCG AGG CCC AGG AAA C 3′ | 13–15 | 87 bp | 61 °C |
| Lower 5′ CCC TTG GTG GTA TAG GCA GCA 3′ | 15 | |||
| PTPL1 | Upper 5′ CAA AGG TGA TCG CGT CCT A 3′ | 26–27 | 148 bp | 61 °C |
| Lower 5′ CGG GAC ATG TTC TTT AGA TGT T 3′ | 28 | |||
| HPRT | Upper 5′ CTG ACC TGC TGG ATT ACA 3′ | 3 | 256 bp | 55 °C |
| Lower 5′ GCG ACC TTG ACC ATC TTT 3′ | 5 | |||
To confirm the specificity of the sequences chosen for the primers, we performed nucleotide-nucleotide BLASTn against a database of expressed sequence tags and nr (the nonredundant set of the GenBank, EMBL, and DDBJ database sequences). To avoid amplification of contaminating genomic DNA, one of the two primers consisted of sequences derived from two adjacent exons.
The relative quantification of PTP gene expression was performed using the comparative cycle threshold (CT) method where the CT parameter is defined as the cycle number at which the fluorescent signal is first detectable. This method is based on the use of a calibrator sample and an endogenous RNA control, which permits the quantification of unknown samples. The human breast cancer cell line MCF7, which is known to express the three PTPs, was chosen as the calibrator sample (PTP expression = 1), and hypoxanthine phosphoribosyltransferase 1 (HPRT) mRNA was used as the endogenous RNA internal control. Relative PTP expression was given by the 2−ΔΔCT, where ΔΔCT = ΔCT patient sample − ΔCT calibrator sample; with Δ CT = CT PTP − CT HPRT.
Statistical analyses
All statistical analyses were performed using SPSS software (version 11.0). Correlations between parameters were assessed using the Spearman nonparametric test. Overall survival and relapse-free survival were analyzed using the Kaplan Meier method. Comparison between curves was carried out using the log rank test. The proportional hazard regression method of Cox (27) was used to assess the prognostic significance of parameters taken in association.
Results
Training set
The distribution of the expression levels of the three PTPs in breast cancer samples was not Gaussian (Fig. 1). PTPL1, PTP gamma and LAR were expressed in all tumours, with median values that were 2.46 (ranging from 0.31 to 19.56)-fold, 5.16 (ranging from 0.21 to 34.06)-fold and 4.01 (ranging from 0.99 to 22.32)-fold greater than the expression levels in MCF7 cells, respectively (Fig. 1). The LAR neuronal isoform, which comprised from 3 to 4% of the total LAR mRNA in MCF7 cells, was detected in 58 out of 59 tumours (median 0.40, ranging from 0.01 to 17.91) (Fig. 1). Its expression in 13 tumours was higher than in MCF7 cells but very low (at least 10-fold lower than in MCF7 cells) in 17 tumours.
Figure 1.
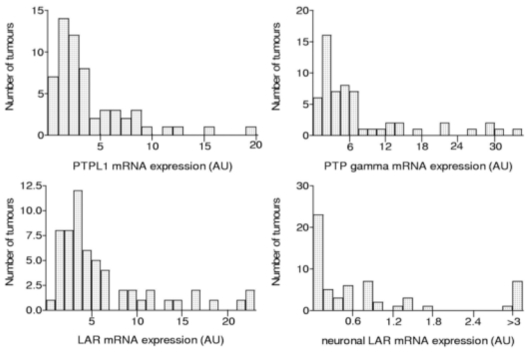
Distribution of breast cancer samples (n = 59) as a function of their PTP transcript levels.
Classical correlations observed in breast cancer were found between ER and PR (p<0.001, r = 0.761), the histoprognostic grade and ER (p = 0.05, r = − 0.256) or PR (p = 0.045, r = − 0.274), and tumour size and ER (p = 0.05, r = − 0.271).
Among the PTPs, we observed several positive correlations. The expression of PTP gamma was correlated to that of the three other PTPs (p<0.001, r= 0.705 for LAR; p<0.001, r= 0.644 for PTPL1; and p=0.04, r= 0.266 for LAR neuronal isoform), and LAR expression was correlated with those of both its neuronal isoform and PTPL1 (p = 0.029, r = 0.284 and p<0.001, r = 0.418, respectively). No correlation was observed between PTP expression and bio-clinical parameters except that of PTPL1, which was positively correlated with hormonal receptor status (p=0.02, r=0.307 between PTPL1 and PR).
We observed that expression of the two LAR isoforms and PTP gamma did not have any prognostic value. On the other hand, the overall survival (OS) was significantly longer among patients with a high level of PTPL1 expression when compared to patients whose tumours had low levels of this protein (P=0.04 by the log-rank test).
Testing set
Based on the results of the training set, the PTPL1 expression level was next determined in a complementary cohort of 232 breast cancer patients. In the total cohort (291 patients), classical correlations observed in breast cancer were found between ER and PR (p<0.001, r=0.626), ER and age (p<0.001, r=0.268) and between PR and age (p=0.01, r=0.141). Expression of hormone receptors was negatively correlated with histoprognostic grade (p<0.001, r=−0.366 and r=−0.342 for ER and PR respectively) and tumour size (p<0.001, r=−0.229 and p=0.037, r=−0.126 for ER and PR respectively). Node involvement was positively correlated with tumour size (p=0.0015, r=0.19). PTPL1 expression was positively correlated with hormonal receptor status and negatively correlated with the histoprognostic grade and node invasion (Table III).
TABLE III.
CORRELATION BETWEEN PTPL1 AND CLINICAL, HISTOLOGICAL, OR BIOLOGICAL PARAMETERS
| P | r | |
|---|---|---|
| Age | NS | |
| Histoprognostic Grade | <0.001 | −0.333 |
| Node involvement | 0.016 | −0.142 |
| Tumour size | NS | |
| ER | <0.001 | 0.246 |
| PR | <0.001 | 0.262 |
OS was found to be significantly longer among patients with a high level of PTPL1 expression (P=0.01 by the log rank test) (Fig. 2).
Figure 2.
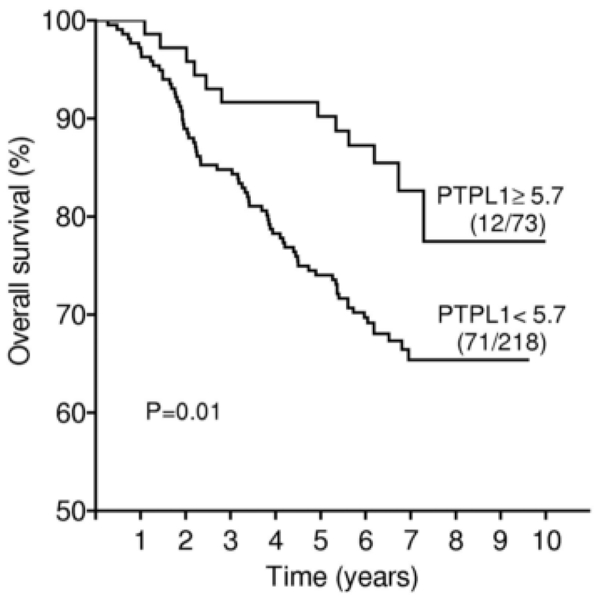
Overall survival curve based on relative PTPL1 expression levels (clinical cut-off, upper quartile) in the whole population of patients. Numbers in parentheses indicate failures/total number of patients in each group.
Cox univariate analyses revealed that, similarly to PTPL1 expression, ER and PR expression, tumour size and node invasion were prognostic factors for global survival (Table IV). Multivariate analyses identified PTPL1 as an independent prognostic factor for OS; PR and tumour size, but not node invasion, also preserved their prognostic value. However, in univariate and multivariate analyses, only node involvement and tumour size were prognostic factors for relapse-free survival (Table IV).
TABLE IV.
PROGNOSTIC FACTORS IN COX UNIVARIATE AND MULTIVARIATE ANALYSES
| Overall survival | Relapse-free survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | multivariate | |||||
| P | RR | P | RR | P | RR | P | RR | |
| Histoprognostic grade (I, II, III) | 0.23 | 0.24 | ||||||
| Node involvement (0; >0) | 0.043 | 1.58 | NS | 0.004 | 1.78 | 0.020 | 1.62 | |
| Tumour size (≤ 2; 2–5; >5 cm) | 0.003 | 1.85 | 0.024 | 1.62 | 0.005 | 1.67 | 0.012 | 1.59 |
| ER (< 10; ≥ 10fmol/mg protein) | 0.047 | 0.63 | NS | 0.13 | ||||
| PR (< 10; ≥ 10fmol/mg protein) | 0.004 | 0.52 | 0.013 | 0.55 | 0.086 | |||
| PTPL1 (<5.7; ≥5.7) | 0.012 | 0.45 | 0.035 | 0.48 | 0.075 | |||
Interestingly, for ER+ (n=201) but not ER− patients, OS was significantly longer among patients with high levels of PTPL1 expression (P=0.034 by the log rank test for ER+, P=0.3 for ER−) (Fig. 3).
Figure 3.
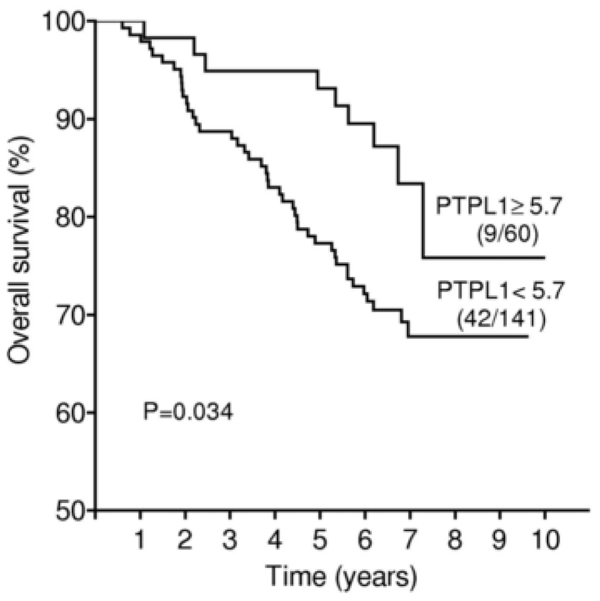
Overall survival curve based on relative PTPL1 expression levels (clinical cut-off, upper quartile) in the ER+ subgroup of patients. Numbers in parentheses indicate failures/total number of patients in each group.
Discussion
In this study, we demonstrated that transcripts of PTP gamma, LAR and its neuronal isoform, and PTPL1 are expressed in almost all human breast cancers. These results confirm previous studies demonstrating the expression of phosphatases in breast cancer (8;16). Using the Spearman test, we showed that PTPL1 expression was positively correlated with that of ER and PR. These results are in agreement with our previous observations (28).
PTP gamma and LAR are differently expressed in breast tumour and normal tissue (8;16), and they have been shown to influence the growth of breast cancer cell lines after ectopic surexpression (12;15). The absence of correlations between their expression and classical clinico-pathological features or survival did not support an effect of these PTPs in tumour growth or invasiveness; rather, it suggests the possible importance of these enzymes in the early steps of tumour development. However, translational and post-translational modifications in addition to mutations and differential splicing can also regulate the expression and activity of these two PTPs and may have caused the divergence between the findings of the previous in vitro study and our present results.
We have demonstrated that PTPL1 has a pro-apoptotic role in breast cancer cell lines. Indeed, studying anti-growth factor activity of the anti-estrogens, we found that PTPL1 mRNA levels were increased by non-steroidal partial antagonist (4-hydroxytamoxifen) or steroidal pure antagonist (ICI 182, 780) (9), as well as by benzothiophenes (29) without regulation by estrogens (9). PTPL1 suppression using an antisense strategy completely abrogated the antagonistic effect of 4-hydroxytamoxifen on growth factor activity, thus demonstrating that PTPL1 and its resulting regulation are crucial for the mediation of this inhibitory effect (9). In addition, PTPL1 affected apoptosis by inhibition of the IRS-1/PI3K survival pathway (18); this inhibition was sufficient to induce apoptosis and necessary for UV-induced cell death in MCF7, HEK 293 and HeLa cells (19).
PTPL1 has also been implicated in the regulation of biological phenomena associated with the cytoskeleton such as cell motility and cellular adhesion (30;31); these processes play a fundamental role in invasion and metastasis. Furthermore, PTPL1 has been implicated in the regulation of cytokinesis in HeLa cells (32), and in the control of the meiotic cell cycle (33), clearly supporting its importance in cell growth regulation. More recently, Zhu et al (34) demonstrated that PTPL1 can inhibit HER2/Neu, a signalling pathway that is frequently deregulated in breast cancer. Published studies using mutant mice that lack PTPN13 protein product or phosphatase activity did not report any effect on tumour susceptibility. Indeed none phenotypic consequences have been reported for PTPN13 KO mice (35) and studies of mice that lack PTPN13 phosphatase activity have focused on haematopoietic cell lineages and the peripheral nervous system (36), which were previously shown to express this phosphatase (37;38). Thus, crossbreeding of these mice with mammary tumour model mice could be used to evaluate the role of PTPL1 in tumour progression and susceptibility.
Considering its links with classical clinico-pathological features, we observed that PTPL1 expression was negatively correlated with node involvement and histoprognostic grade. This indicates that elevated expression of PTPL1 may be a molecular marker of a more differentiated phenotype.
It is well established that mRNA expression does not necessarily reflect protein expression. Indeed, gene expression is regulated at many levels, including post-transcriptionnal downregulation by microRNAs (39). Mammary epithelial tumour cells are the major tissue component in primary breast cancer, which also contains stromal cells and endothelial cells. It should be noted that PTP-BL, the PTPL1 mouse orthologue, is predominantly expressed in epithelial and neuronal cells (40). In addition, our in vitro studies have demonstrated that human breast cancer cell lines express and produce PTPL1 (18). Furthermore, in the Human Protein Atlas program, immunochemical studies of breast cancer using a specific antibody against PTPN13 showed specific staining of tumour cells with little or no signal in the stromal cells (www.proteinatlas.org). Taken together, these observations support the hypothesis that the PTPL1 transcripts that we quantified by real time RT-PCR were produced by the tumour cells.
Univariate and multivariate Cox analyses of our results revealed that PTPL1 mRNA expression is a favourable prognostic indicator of OS with a median duration of follow-up of 6.4 years. It is not unexpected that tumours containing high levels of PTPL1, which induces apoptosis of breast cancer cells, have a better prognosis than tumours without this phosphatase. This observation is in line with the positive links observed between PTPL1 and steroid hormone receptors or low histoprognostic grading, which are parameters associated with a better prognosis.
It is interesting that PTPL1 retains its prognostic value for OS in patients with ER-positive tumours in spite the strong correlation of its expression with that of ER. The presence of ER/PR is typically used as a rational basis for hormonal treatment. Our results suggest that PTPL1 could provide an additional criterion for implementation of such therapies.
In conclusion, this study demonstrates for the first time that PTPL1 expression is an independent prognostic factor of favourable outcome for patients with breast cancer. In conjunction with our mechanistic studies, this finding suggests that PTPL1 is an important regulatory element of human breast tumour aggressiveness and sensitivity to treatments such as anti-estrogens and anti-aromatase.
Acknowledgments
We are particularly grateful to Françoise Vignon who played an essential role in the initiation of this work. This work was supported by Inserm, by the Ligue Régionale Contre le Cancer Languedoc Roussillon and by INCa (Institut National du Cancer) grant N°0611-3D1019-34/Valo and N°0610-3D1616-118/PL2006.
Abbreviations
- ER
estrogen receptors
- HPRT
hypoxanthine phosphoribosyltransferase 1
- OS
overall survival
- PCR
Polymerase-Chain-Reaction
- PR
progesterone receptors
- PTP
protein tyrosine phosphatase
- RFS
relapse-free survival
Footnotes
2 brief statements describing the novelty and impact of our paper:
- PTPL1 phosphatase has a positive prognostic value in breast cancer
- In spite of the links between estradiol receptors and PTPL1, this phosphatase has additional prognostic value.
References
- 1.Esteva FJ, Hortobagyi GN. Prognostic molecular markers in early breast cancer. Breast Cancer Res. 2004;6:109–18. doi: 10.1186/bcr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pawlowski V, Revillion F, Hebbar M, Hornez L, Peyrat JP. Prognostic value of the type I growth factor receptors in a large series of human primary breast cancers quantified with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res. 2000;6:4217–25. [PubMed] [Google Scholar]
- 3.Peyrat JP, Vanlemmens L, Fournier J, Huet G, Revillion F, Bonneterre J. Prognostic value of p53 and urokinase-type plasminogen activator in node-negative human breast cancers. Clin Cancer Res. 1998;4:189–96. [PubMed] [Google Scholar]
- 4.Rochefort H, Garcia M, Glondu M, Laurent V, Liaudet E, Rey JM, Roger P. Cathepsin D in breast cancer: mechanisms and clinical applications, a 1999 overview. Clin Chim Acta. 2000;291:157–70. doi: 10.1016/s0009-8981(99)00226-0. [DOI] [PubMed] [Google Scholar]
- 5.Freiss G, Vignon F. Protein tyrosine phosphatases and breast cancer. Crit Rev Oncol Hematol. 2004;52:9–17. doi: 10.1016/j.critrevonc.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Freiss G, Vignon F. Antiestrogens increase protein tyrosine phosphatase activity in human breast cancer cells. Mol Endocrinol. 1994;8:1389–96. doi: 10.1210/mend.8.10.7854356. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Silliman N, Peters BA, van der Heijden MS, Parmigiani G, Yan H, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–6. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 8.Zheng J, Kulp SK, Zhang Y, Sugimoto Y, Dayton MA, Govindan MV, Brueggemeier RW, Lin YC. 17 beta-estradiol-regulated expression of protein tyrosine phosphatase gamma gene in cultured human normal breast and breast cancer cells. Anticancer Res. 2000;20:11–9. [PubMed] [Google Scholar]
- 9.Freiss G, Puech C, Vignon F. Extinction of insulin-like growth factor-I mitogenic signaling by antiestrogen-stimulated Fas-associated protein tyrosine phosphatase-1 in human breast cancer cells. Mol Endocrinol. 1998;12:568–79. doi: 10.1210/mend.12.4.0088. [DOI] [PubMed] [Google Scholar]
- 10.LaForgia S, Morse B, Levy J, Barnea G, Cannizzaro LA, Li F, Nowell PC, Boghosian-Sell L, Glick J, Weston A. Receptor protein-tyrosine phosphatase gamma is a candidate tumor suppressor gene at human chromosome region 3p21. Proc Natl Acad Sci U S A. 1991;88:5036–40. doi: 10.1073/pnas.88.11.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vezzalini M, Mombello A, Menestrina F, Mafficini A, Della PM, van Niekerk C, Barbareschi M, Scarpa A, Sorio C. Expression of transmembrane protein tyrosine phosphatase gamma (PTPgamma) in normal and neoplastic human tissues. Histopathology. 2007;50:615–28. doi: 10.1111/j.1365-2559.2007.02661.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu S, Sugimoto Y, Sorio C, Tecchio C, Lin YC. Function analysis of estrogenically regulated protein tyrosine phosphatase gamma (PTPgamma) in human breast cancer cell line MCF-7. Oncogene. 2003 doi: 10.1038/sj.onc.1207235. [DOI] [PubMed] [Google Scholar]
- 13.Lamprianou S, Vacaresse N, Suzuki Y, Meziane H, Buxbaum JD, Schlessinger J, Harroch S. Receptor protein tyrosine phosphatase gamma is a marker for pyramidal cells and sensory neurons in the nervous system and is not necessary for normal development. Mol Cell Biol. 2006;26:5106–19. doi: 10.1128/MCB.00101-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaapveld RQ, Schepens JT, Robinson GW, Attema J, Oerlemans FT, Fransen JA, Streuli M, Wieringa B, Hennighausen L, Hendriks WJ. Impaired mammary gland development and function in mice lacking LAR receptor-like tyrosine phosphatase activity. Dev Biol. 1997;188:134–46. doi: 10.1006/dbio.1997.8630. [DOI] [PubMed] [Google Scholar]
- 15.Zhai Y, Wirth J, Kang S, Welsch CW, Esselman WJ. LAR-PTPase cDNA transfection suppression of tumor growth of neu oncogene-transformed human breast carcinoma cells. Mol Carcinog. 1995;14:103–10. doi: 10.1002/mc.2940140206. [DOI] [PubMed] [Google Scholar]
- 16.Yang T, Zhang JS, Massa SM, Han X, Longo FM. Leukocyte common antigen-related tyrosine phosphatase receptor: increased expression and neuronal-type splicing in breast cancer cells and tissue. Mol Carcinog. 1999;25:139–49. [PubMed] [Google Scholar]
- 17.Zhang JS, Longo FM. LAR tyrosine phosphatase receptor: alternative splicing is preferential to the nervous system, coordinated with cell growth and generates novel isoforms containing extensive CAG repeats. J Cell Biol. 1995;128:415–31. doi: 10.1083/jcb.128.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bompard G, Puech C, Prebois C, Vignon F, Freiss G. Protein-tyrosine phosphatase PTPL1/FAP-1 triggers apoptosis in human breast cancer cells. J Biol Chem. 2002;277:47861–9. doi: 10.1074/jbc.M208950200. [DOI] [PubMed] [Google Scholar]
- 19.Dromard M, Bompard G, Glondu-Lassis M, Puech C, Chalbos D, Freiss G. The Putative Tumor Suppressor Gene PTPN13/PTPL1 Induces Apoptosis through Insulin Receptor Substrate-1 Dephosphorylation. Cancer Res. 2007;67:6806–13. doi: 10.1158/0008-5472.CAN-07-0513. [DOI] [PubMed] [Google Scholar]
- 20.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–46. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 21.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–20. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 22.Inazawa J, Ariyama T, Abe T, Druck T, Ohta M, Huebner K, Yanagisawa J, Reed JC, Sato T. PTPN13, a fas-associated protein tyrosine phosphatase, is located on the long arm of chromosome 4 at band q21.3. Genomics. 1996;31:240–2. doi: 10.1006/geno.1996.0039. [DOI] [PubMed] [Google Scholar]
- 23.Yeh SH, Wu DC, Tsai CY, Kuo TJ, Yu WC, Chang YS, Chen CL, Chang CF, Chen DS, Chen PJ. Genetic characterization of fas-associated phosphatase-1 as a putative tumor suppressor gene on chromosome 4q21.3 in hepatocellular carcinoma. Clin Cancer Res. 2006;12:1097–108. doi: 10.1158/1078-0432.CCR-05-1383. [DOI] [PubMed] [Google Scholar]
- 24.Ying J, Li H, Cui Y, Wong AH, Langford C, Tao Q. Epigenetic disruption of two proapoptotic genes MAPK10/JNK3 and PTPN13/FAP-1 in multiple lymphomas and carcinomas through hypermethylation of a common bidirectional promoter. Leukemia. 2006;20:1173–5. doi: 10.1038/sj.leu.2404193. [DOI] [PubMed] [Google Scholar]
- 25.Peyrat JP, Bonneterre J, Beuscart R, Djiane J, Demaille A. Insulin-like growth factor 1 receptors in human breast cancer and their relation to estradiol and progesterone receptors. Cancer Res. 1988;48:6429–33. [PubMed] [Google Scholar]
- 26.Koenders T, Benraad TJ. Quality control of estrogen receptor assays in The Netherlands. Breast Cancer Res Treat. 1983;3:255–66. doi: 10.1007/BF01806699. [DOI] [PubMed] [Google Scholar]
- 27.Cox DR. Regression models and lifetables. J R Stat Soc. 1972;34:187–200. [Google Scholar]
- 28.Rey JM, Pujol P, Callier P, Cavailles V, Freiss G, Maudelonde T, Brouillet JP. Semiquantitative reverse transcription-polymerase chain reaction to evaluate the expression patterns of genes involved in the oestrogen pathway. J Mol Endocrinol. 2000;24:433–40. doi: 10.1677/jme.0.0240433. [DOI] [PubMed] [Google Scholar]
- 29.Freiss G, Galtier F, Puech C, Aknin C, Maudelonde T, Chalbos D, Vignon F. Anti-growth factor activities of benzothiophenes in human breast cancer cells. J Steroid Biochem Mol Biol. 2005:94. doi: 10.1016/j.jsbmb.2004.12.043. [DOI] [PubMed] [Google Scholar]
- 30.Bompard G, Martin M, Roy C, Vignon F, Freiss G. Membrane targeting of protein tyrosine phosphatase PTPL1 through its FERM domain via binding to phosphatidylinositol 4,5-biphosphate. J Cell Sci. 2003;116:2519–30. doi: 10.1242/jcs.00448. [DOI] [PubMed] [Google Scholar]
- 31.Kimber WA, Deak M, Prescott AR, Alessi DR. Interaction of the protein tyrosine phosphatase PTPL1 with the PtdIns (3,4)P 2 binding adaptor protein TAPP1. Biochem J. 2003 doi: 10.1042/BJ20031154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrmann L, Dittmar T, Erdmann KS. The protein tyrosine phosphatase PTP-BL associates with the midbody and is involved in the regulation of cytokinesis. Mol Biol Cell. 2003;14:230–40. doi: 10.1091/mbc.E02-04-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nedachi T, Conti M. Potential role of protein tyrosine phosphatase nonreceptor type 13 in the control of oocyte meiotic maturation. Development. 2004;131:4987–98. doi: 10.1242/dev.01368. [DOI] [PubMed] [Google Scholar]
- 34.Zhu JH, Chen R, Yi W, Cantin GT, Fearns C, Yang Y, Yates JR, III, Lee JD. Protein tyrosine phosphatase PTPN13 negatively regulates Her2/ErbB2 malignant signaling. Oncogene. 2007 doi: 10.1038/sj.onc.1210922. [DOI] [PubMed] [Google Scholar]
- 35.Nakahira M, Tanaka T, Robson BE, Mizgerd JP, Grusby MJ. Regulation of signal transducer and activator of transcription signaling by the tyrosine phosphatase PTP-BL. Immunity. 2007;26:163–76. doi: 10.1016/j.immuni.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 36.Wansink DG, Peters W, Schaafsma I, Sutmuller RP, Oerlemans F, Adema GJ, Wieringa B, van der Zee CE, Hendriks W. Mild impairment of motor nerve repair in mice lacking PTP-BL tyrosine phosphatase activity. Physiol Genomics. 2004;19:50–60. doi: 10.1152/physiolgenomics.00079.2004. [DOI] [PubMed] [Google Scholar]
- 37.Gjorloff-Wingren A, Saxena M, Han S, Wang X, Alonso A, Renedo M, Oh P, Williams S, Schnitzer J, Mustelin T. Subcellular localization of intracellular protein tyrosine phosphatases in T cells. Eur J Immunol. 2000;30:2412–21. doi: 10.1002/1521-4141(2000)30:8<2412::AID-IMMU2412>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 38.Thomas T, Voss AK, Gruss P. Distribution of a murine protein tyrosine phosphatase BL-beta-galactosidase fusion protein suggests a role in neurite outgrowth. Dev Dyn. 1998;212:250–7. doi: 10.1002/(SICI)1097-0177(199806)212:2<250::AID-AJA9>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 39.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 40.van Ham M, Croes H, Schepens J, Fransen J, Wieringa B, Hendriks W. Cloning and characterization of mCRIP2, a mouse LIM-only protein that interacts with PDZ domain IV of PTP-BL. Genes Cells. 2003;8:631–44. doi: 10.1046/j.1365-2443.2003.00660.x. [DOI] [PubMed] [Google Scholar]


