Abstract
Background
The Pierre Robin sequence (PRS), consisting of cleft palate, micrognathia and glossoptosis, can be seen as part of the phenotype in other Mendelian syndromes—for instance, campomelic dysplasia (CD) which is caused by SOX9 mutations—but the aetiology of non‐syndromic PRS has not yet been unravelled.
Objective
To gain more insight into the aetiology of PRS by studying patients with PRS using genetic and cytogenetic methods.
Methods
10 unrelated patients with PRS were investigated by chromosome analyses and bacterial artificial chromosome arrays. A balanced translocation was found in one patient, and the breakpoints were mapped with fluorescence in situ hybridisation and Southern blot analysis. All patients were screened for SOX9 and KCNJ2 mutations, and in five of the patients expression analysis of SOX9 and KCNJ2 was carried out by quantitative real‐time PCR.
Results
An abnormal balanced karyotype 46,XX, t(2;17)(q23.3;q24.3) was identified in one patient with PRS and the 17q breakpoint was mapped to 1.13 Mb upstream of the transcription factor SOX9 and 800 kb downstream of the gene KCNJ2. Furthermore, a significantly reduced SOX9 and KCNJ2 mRNA expression was observed in patients with PRS.
Conclusion
Our findings suggest that non‐syndromic PRS may be caused by both SOX9 and KCNJ2 dysregulation.
Cleft lip and/or palate (CL/P) is a common congenital malformation affecting approximately 2 per 1000 newborns worldwide. The aetiology of CL/P is largely unknown; however, recent studies focusing on syndromic forms of CL/P have identified specific genes that may also be involved in non‐syndromic CL/P.1 The Pierre Robin sequence (PRS (MIM 261800)) is a clinically well‐defined subgroup of the CL/P population with an unknown aetiology. PRS is characterised by cleft palate, micrognathia and respiratory difficulties in the early neonatal period (caused by glossoptosis), and is often observed as a part of other Mendelian syndromes, such as Stickler's syndrome, velocardiofacial syndrome and Marshall's syndrome.2 PRS is also seen as a part of campomelic dysplasia (CD (MIM 114290)), a rare skeletal dysplasia, consisting of bowing of the long bones (campomelia), malformation of the pelvis and spine, 11 pairs of ribs, hypoplastic scapulae, club feet, micrognathia, cleft palate and in some patients male‐to‐female sex reversal. When severely affected, the children die in the neonatal period owing to respiratory problems resulting from developmental defects in the respiratory system.3 CD is due to haploinsufficiency of the transcription factor SOX9 on 17q24.3, caused by intragenic mutations, deletions involving the entire gene or chromosomal rearrangements up to about 950 kb upstream of the SOX9 coding region.4,5,6 The phenotypes of the breakpoint cases in CD, believed to be caused by disruption of regulatory regions, are generally less severe than the intragenic mutations.6
The description of patients with skeletal abnormalities, including PRS features caused by breakpoints 900 and 932 kb upstream of the SOX9 coding region7,8 and the finding of a familial translocation t(2;17) cosegregating with PRS,9 suggests SOX9 to be a candidate gene also for non‐syndromic PRS. Animal studies support this hypothesis, as SOX9 mutant mice present with cleft palate and hypoplasia of cartilage‐derived skeletal structures10 and SOX9 is expressed in the fetal mouse mandible.11
To gain more insight into the aetiology of PRS, we performed genetic and cytogenetic analyses in 10 unrelated patients with non‐syndromic PRS.
Methods
The patients included in this study were identified through hospital records and selected if they had non‐syndromic PRS (cleft palate, micrognathia and respiratory difficulties in the early neonatal period). One patient had an additional history of malignant hyperthermia and bilateral inguinal hernias. The patients were aged between 2 and 16 years.
Chromosome analysis was carried out with the G‐banding technique using peripheral blood lymphocytes. Fine mapping of the breakpoints was performed using flourescence in situ hybridisation (FISH), with bacterial artificial chromosome (BAC) and fosmid clones obtained from the Sanger Institute. Genomic DNA was isolated from blood according to standard protocols.
For Southern blot analysis the genomic DNA was digested with the restriction enzymes EcoRI and MscI. Two hybridisation probes (SB817 and SB520) were prepared by PCR using primer sets SB817U/L (5′‐GTAAAGAAACGGTCGCAAATA‐3′/5′‐CAATAGCAGAGCGCAGTAG‐3′) and SB520U/L (5′‐TGGGACAGAAACATTACCTTG‐3′/5′‐TGATTGGAGGAGAAAACGAC‐3′). The SB817 probe hybridised to the 5136 bp MscI and the 7172 bp EcoRI fragments, and SB520 detected the 7102 bp MscI and 3132 bp EcoRI fragments.
Array comparative genome hybridisation (CGH) was carried out using a sub‐megabase resolution whole‐genome tiling path BAC array consisting of the human genome high‐resolution 32 k re‐arrayed clone set (BACPAC Resources), the 1 Mb Sanger set (Wellcome Trust Sanger Institute, Cambridge, UK) and a set of 390 sub‐telomeric clones (assembled by members of the COST B19 initiative: molecular cytogenetics of solid tumours).12
SOX9 and KCNJ2 expression in cultured lymphoblastoid cell lines was analysed by quantitative real‐time PCR (QPCR) using an Opticon DNA engine real‐time PCR machine (MJ Research, Waltham, Massachusetts, USA) using an SYBR green kit (GE Healthcare, Waukesha, Wisconsin, USA). Relative gene expression was calculated using dilution curves and UBC, YWHAZ, GAPDH, G6PD and B2M mRNA as reference genes. Mean values of the housekeeping genes were calculated using the BestKeeper software tool,13 and the experiments were run in duplicate and repeated twice. Genomic PCR and sequencing were performed according to standard protocols (supplementary table showing primer sequences and PCR conditions, mRNA isolation and cDNA preparation is available at http://jmg.bmj.com/supplemental).
The study was approved by the local scientific ethics committee and written informed consent was obtained from the patients and from the parents of children who were aged <12 years.
Results and discussion
We performed chromosome analyses in 10 unrelated patients with features of PRS. All the cases had normal karyotypes except for one, where PRS cosegregated with a balanced translocation t(2;17)(q23.3;q24.3) in a father and a daughter.
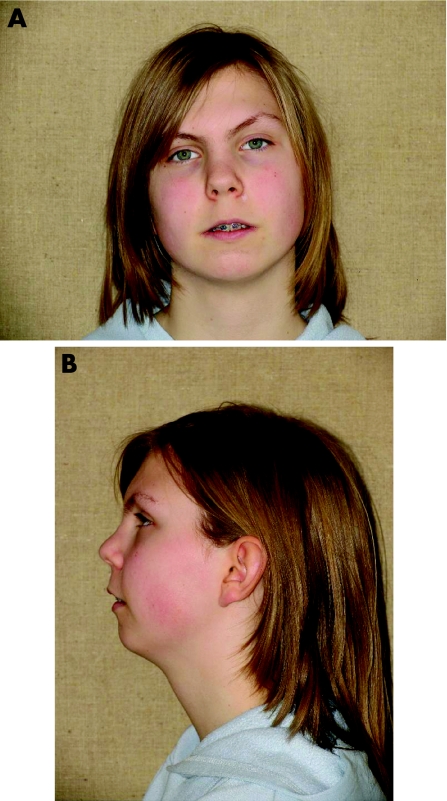
The translocation carrier (fig 1A, B) was a 15‐year‐old girl. The father also had PRS according to hospital records, but he declined to participate in further clinical and genetic studies. The patient had mild facial dysmorphism. In addition to micrognathia, she had a flat face, a broad nasal bridge, low‐set ears and low‐set hairline. She was born as the only child of a 33‐year‐old father and a 41‐year‐old mother with type 2 diabetes. Although the intellectual and psychomotor development was at the lower end of the normal spectrum, she attended a standard school. She had received speech therapy, and a pharyngeal flap operation had been performed to decrease hypernasal speech. No visual or hearing deficits were reported. Growth and sexual maturation were normal. The mother has a healthy daughter from an earlier relationship. No other family member has PRS, cleft palate or other malformations. Material from paternal grandparents was not available, but a paternal uncle and a maternal half‐sister of the father had normal karyotypes.
Figure 1 (A, B) Photographs of the patient with Pierre Robin sequence and the balanced translocation t(2;17)(q23.3;q24.3). Informed consent was obtained for publication of this figure.
Array CGH was performed in seven of the nine non‐translocation patients with PRS, and it showed a 0.4 Mb microdeletion at 7q21.13 (chromosome position 88 473 693–88 909 911, UCSC Genome Browser, March 2006 assembly) in one of the patients. This result was confirmed by FISH. Parental DNA was not available to investigate the origin of this deletion. There is no known copy‐number variation within this region (Database of Genomic Variants and unpublished observations by RU) and there is only one gene, ZNF804B, which encodes a C2H2‐type zinc finger protein. Deletions at 7q21–q22 have been associated previously with PRS, but several other malformations such as ectrodactyly were part of the clinical phenotype.14 The other six patients did not show any significant changes with array CGH.
Translocation breakpoints of the t(2;17) patient were mapped using FISH. The chromosome 2q23.3 breakpoint was within the BAC clone RP11‐373H2 (AC113610) at chromosome position 150 777 641–150 874 211 (UCSC Genome Browser). This breakpoint is located in a gene‐empty region, approximately 200 kb downstream of the RND3 gene and 350 kb upstream of FJL32955 encoding a hypothetical protein (LOC150596). RND3 encodes a plasma membrane‐bound GTPase involved in regulation of cellular response and cytoskeletal dynamics. RND3 could be involved in the aetiology in the present case, but no phenotype is associated with mutations or chromosomal rearrangements in the vicinity of RND3, and a position effect in the present case is speculative. Chromosome abnormalities involving 2q22 and 2q24 have been observed previously in patients with PRS. Jamshidi et al9 reported a familial translocation t(2;17)(q24.1;q24.3) cosegregating with PRS, and mutations of ZFHX1B at 2q22.3 cause Mowat–Wilson syndrome (MIM 235 730), in which submucous cleft palate is a part of the phenotype, although this syndrome does not otherwise resemble CD or PRS. However, these regions are located far from the present breakpoint and do not have an apparent effect on the phenotype of the patient.
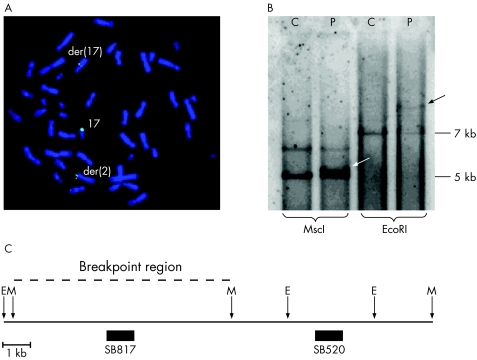
We mapped the 17q24.3 breakpoint within an approximately 15 kb overlapping region of the BAC clone RP11‐7D6 (fig 2A) and two fosmid clones (G248P89352E7 and G248P86963D5). The breakpoint was further mapped with Southern blot hybridisation using two probes, detecting an approximately 12 kb region within the overlapping region of the FISH clones (fig 2B, C). Using the hybridisation probe SB817 and MscI digestion, we detected the 5.1 kb normal fragment and a slightly larger band representing the junction fragment in the patient. This probe also detected the junction fragment using the restriction enzyme EcoRI. Hybridisation probe SB520 showed only the normal band pattern in the patient (data not shown). This suggests that the 17q breakpoint is within an approximately 5 kb region defined by two MscI recognition sites (fig 2C), 1.13 Mb upstream of the SOX9 gene (chromosome position 66 485 382–66 491 517). The total gene‐empty region upstream of SOX9 is approximately 2 Mb where the proximal flanking gene is KCNJ2, which is located about 800 kb from the 17q breakpoint.
Figure 2 The breakpoint region at 17q. (A) FISH analysis showing the breakpoint‐spanning bacterial artificial chromosome (BAC) clone RP11‐7D6. (B) Southern blot hybridisation using probe SB817 and the restriction enzymes MscI and EcoRI. With MscI digests, the 5 kb normal fragment and a larger band representing the junction fragment (white arrow) are detected in the patient. With EcoRI digests, besides the 7 kb normal fragment, the patient has a larger fragment (black arrow). (C) The physical map of the breakpoint region. The 12 kb interval is within the 15 kb overlapping region of the spanning BAC (fig 2A) and the fosmid clones. The breakpoint region (dashed line) is mapped between the two MscI restriction sites (chromosome position 66 485 382–66 491 517). The vertical arrows show the MscI (M) and EcoRI (E) restriction sites. The filled boxes represent the two hybridisation probes SB817 and SB520. C, control; P, patient.
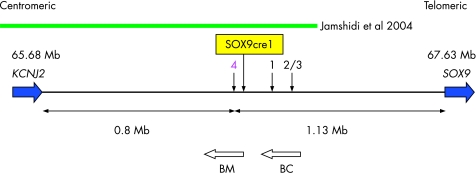
We sequenced the coding region of SOX9 in all the 10 patients with PRS and detected several nucleotide polymorphisms, but no pathogenic mutations. We also screened the 3′ untranslated region of SOX9 for sequence variations, as in silico predictions of microRNA (miRNA) target sites suggest that SOX9 expression is regulated by miRNAs (miRBase Targets V2.0). miRNAs are short non‐coding RNAs involved in downregulation of genes at the translational level by binding to specific target sites in the 3′ untranslated region of the mRNA.15 No sequence variations in the predicted miRNA target sites were identified. Furthermore, we searched for evolutionarily conserved sequences, and hence potentially regulating regions (in human, chimpanzee, mouse, rat, dog, chicken, zebrafish and fugu, UCSC Genome Browser) and identified a conserved region at chromosome position 66 520 880–66 521 660. This region overlaps with the candidate regulatory element “SOX9cre1” (chromosome position 66 520 446–66 521 622, UCSC Genome Browser) proposed by Velagaleti et al8 (fig 3). Screening this conserved region in the 10 patients with PRS revealed two unreported consecutive single‐nucleotide polymorphisms (SNPs) at 66 520 950–66 520 951 (A/T and G/A) in two patients. These SNPs were also detected in two of 18 patients with isolated cleft palate and in 20 of 95 normal controls. We therefore concluded that the two consecutive SNPs were not associated with PRS.
Figure 3 Schematic overview of the 2 Mb gene‐empty region between KCNJ2 and SOX9, showing known breakpoints >900 kb upstream of SOX9. Patients 1 and 2 were reported by Hill‐Harfe et al 7 and patient 3 by Velagaleti et al.8 The patients have breakpoints in the following approximate chromosome positions: patient 1 at 66.7 Mb, and patients 2 and 3 at 66.73 Mb. Patient 4 is the translocation patient presented here with a breakpoint at 66.49 Mb. The green bar represents the breakpoint interval in the three‐generation family with PRS reported by Jamshidi et al.9 Velagaleti et al 8 suggested a potential regulatory region, SOX9cre1, at 66.52 Mb. BM is the spliced human EST BM678241 and BC is the human mRNA BC039327.
Haploinsufficiency of SOX9 as a cause of CD has been shown by deletions involving the entire gene in two unrelated patients with CD,4,5 and we considered whether expression levels of SOX9 were affected in patients with PRS. By QPCR, we detected a significantly reduced SOX9 expression (p = 0.04, two‐tailed Mann–Whitney U test) in cultured lymphoblastoid cell lines from five non‐translocation patients with PRS compared with 11 controls (table 1). Unexpectedly, the expression of SOX9 in the translocation patient was higher than in the group of non‐translocation patients with PRS, although still at the lower end of the normal range. A possible explanation for this is that some upstream regulators (<1 Mb from SOX9 in the present case) are still active in the translocation patient. Likewise, Wirth et al16 found the same expression levels in both SOX9 alleles in a lymphoblastoid cell line from a t(13;17) translocation patient, where the 17q breakpoint was located >130 kb from SOX9.
Table 1 SOX9 expression in lymphoblasts(quantitative real‐time PCR).
| Sample | Gender | Expression values of SOX9 |
|---|---|---|
| PRS 1 | XY | 5.4 |
| PRS 2 | XY | 5.4 |
| PRS 3 | XX | 1.7 |
| PRS 4 | XY | 5.3 |
| PRS 5 | XX | 7.1 |
| PRS mean (range) | 5.0 (1.7–7.1) | |
| PRS 6 t(2;17) | XX | 13.7 |
| C 1 | XY | 10.6 |
| C 2 | XX | 34.6 |
| C 3 | XY | 10.4 |
| C 4 | XY | 15.0 |
| C 5 | XX | 22.4 |
| C 6 | XY | 40.2 |
| C 7 | XX | 22.5 |
| C 8 | XX | 1.0 |
| C 9 | XY | 21.4 |
| C 10 | XX | 4.0 |
| C 11 | XY | 31.9 |
| C mean (range) | 19.5 (1.0–40.2) | |
| p Value | 0.04 |
PRS, Pierre Robin sequence; PRS 1–6, six patients with PRS, PRS 6 is the translocation patient; C 1–11, controls.
*The expression value of SOX9 was normalised to the mean values of the housekeeping genes UBC, YWHAZ, B2M, G6PD and GAPDH. Mean values of the housekeeping genes were calculated using the BestKeeper software tool.13
The experiments were run in duplicate and repeated twice. Values of PRS 1–5 to C 1–11 were compared and p values were calculated using the two‐tailed Mann‐Whitney test.
The expression data should be interpreted with caution, as expression in the patients may not reflect expression during embryonic development. Moreover, expression studies were carried out in lymphoblastoid cell lines, not in chondrocytes, which would have been preferred, but were unavailable.
The breakpoint located most upstream within the SOX9 regulatory landscape reported so far, about 932 kb from the coding region, was associated with hypoplastic scapulae and 11 pairs of ribs and Robin sequence (family F).7 The 17q breakpoint identified in this study was located 1.13 Mb upstream of SOX9, which is 200 kb further upstream than the breakpoint in family F, and the absence of missing ribs in the present patient points to a milder phenotype. Thus, the present case constitutes the mildest phenotype and the most distantly located breakpoint upstream of the SOX9 gene reported to date (fig 3 shows a schematic overview of breakpoints located >900 kb upstream of SOX9). Whether the mild phenotypes may be categorised as CD or represent a distinct clinical entity is a subject of debate.17,18 Considering the growing reports of distantly located chromosomal rearrangements cosegregating with milder types of skeletal dysplasia, it is possible that CD, acampomelic CD, mild acampomelic CD and PRS may represent a continuum of phenotypes caused by dysregulation of SOX9.
The gene most proximal to the 17q breakpoint is KCNJ2 (fig 3), which encodes a potassium channel. In humans, KCNJ2 mutations have been detected in Andersen's syndrome (MIM 170390), which presents with potassium‐sensitive periodic paralysis, variable cardiac arrhythmias and dysmorphic features overlapping with CD to some extent, including low‐set ears, micrognathia, cleft palate and scoliosis. Moreover, mutations in KCNJ2 cause cleft palate in mice.19 To our knowledge, CD with periodic paralysis and cardiac arrhythmias has not been reported, but PRS occurring with cardiac arrhythmias has been described previously20 and a higher prevalence of congenital cardiac anomalies has been found in patients with PRS (13.6%) compared with the general population with CL/P (6.7%).21 We sequenced the coding region of KCNJ2 in the 10 patients with PRS, including the translocation patient, without detecting any mutations. Furthermore, we investigated the KCNJ2 expression using QPCR analyses and detected a significant reduction in the KCNJ2 expression in the non‐translocation patients with PRS compared with 11 controls. The expression levels of KCNJ2 were very low in both controls and patients; consequently, two different KCNJ2 primer sets were tested, and these were in agreement (table 2).
Table 2 KCNJ2 expression in lymphoblasts(QPCR).
| Sample | Gender | Expression values of KCNJ2 A | Expression values of KCNJ2 B |
|---|---|---|---|
| PRS 1 | XY | 2.5 | 1.2 |
| PRS 2 | XY | 3.0 | 1.0 |
| PRS 3 | XX | 2.0 | 1.6 |
| PRS 4 | XY | 2.7 | 3.0 |
| PRS 5 | XX | 1.0 | 2.1 |
| PRS mean (range) | 2.2 (1.0–3.0) | 1.8 (1.0–3.0) | |
| PRS 6 t(2;17) | XX | 3.7 | 2.5 |
| C 1 | XY | 8.2 | 2.1 |
| C 2 | XX | 70.3 | 23.7 |
| C 3 | XY | 2.4 | 1.2 |
| C 4 | XY | 10.8 | 4.3 |
| C 5 | XX | 31.2 | 16.3 |
| C 6 | XY | 17.1 | 5.6 |
| C 7 | XX | 35.2 | 20.0 |
| C 8 | XX | 6.7 | 1.1 |
| C 9 | XY | 27.6 | 30.4 |
| C 10 | XX | 7.6 | 2.3 |
| C 11 | XY | 70.3 | 29.1 |
| C mean (range) | 26.1 (2.4–70.3) | 12.3 (1.1–30.4) | |
| p Value | 0.007 | 0.05 |
PRS, Pierre Robin sequence; PRS 1–6, six patients with PRS, PRS 6 is the translocation patient; C 1–11, controls.
As the expression of KCNJ2 was low, QPCR was performed with two different primer sets A and B (supplementary table). The expression value of KCNJ2 was normalised to the mean values of the housekeeping genes UBC, YWHAZ, B2M, G6PD and GAPDH. Mean values of the housekeeping genes were calculated using the BestKeeper software tool.13
The experiments were run in duplicate and repeated twice. Values of PRS 1–5 and C 1–11 were compared and p values were calculated using the two‐tailed Mann–Whitney U test.
Our expression data therefore suggest that KCNJ2 may play a role in the PRS phenotype, together with SOX9. It has recently been reported that chromosomal aberrations may change the expression of several neighbouring genes,22 thus supporting a role for both KCNJ2 and SOX9 in the aetiology of PRS.
Our sequencing results indicate that mutations in the SOX9 and KCNJ2 coding regions, in SOX9 miRNA target sites and in an evolutionarily conserved upstream region of SOX9 are not a frequent cause of PRS. Identifying and sequencing other candidate cis‐regulatory regions is highly relevant but complicated, time and cost consuming, as regulatory elements may reside even within neighbouring genes.23 Since translocations upstream of SOX9 are causing phenotypes consistent with CD, cis‐regulatory elements have been sought in the conserved neighbouring regions of SOX9 and many have been suggested (reviewed by Hill‐Harfe et al7). A candidate regulatory element SOX9cre1, about 1.1 kb, was suggested using in silico analysis and analysis of chromosomal rearrangements (fig 3).8 The breakpoint in the present case is located even more upstream than the SOX9cre1, thus one or more of the cis‐regulatory regions must be located either in our breakpoint region or even more upstream, or both. Another explanation of the effect of the translocation could be that altered organisation of the chromatin structure causes changes in gene regulation (eg, euchromatic to heterochromatic conformation).23
In the region upstream of SOX9, several non‐coding transcripts, but no protein‐coding transcripts, have been identified.6,7 The 17q breakpoint is located in the intronic region of a 226 kb spliced human EST (BM678241), which has been detected in fetal and adult eyes.24 BM678241 is located proximal to BC039327, a human mRNA exclusively expressed in testis and disrupted by the 17q breakpoint in family F (fig 3). It was suggested that BC039327 was a functional RNA with an unknown role, since there was no testis phenotype in family F.7 Likewise, the role of BM678241 in relation to our breakpoint is unclear, as our translocation carrier does not have a phenotype involving the eyes.
Considering gene regulation, it is also necessary to include genes involved in the SOX9 and KCNJ2 signalling pathways. SOX9 is one of several factors involved in chondrogenesis,25 and mutations and sequence variations in genes encoding collagen subunits (COL2A1, COL11A1 and COL11A2) were found in patients with non‐syndromic PRS,26 supporting the fact that PRS may result from dysregulation of SOX9 signalling pathways in general.
In summary, our data suggest that dysregulation of the genes SOX9 and KCNJ2 may be involved in PRS, evidenced by a familial translocation with a breakpoint located in the gene‐empty region between SOX9 and KCNJ2, and by reduced expression of SOX9 and KCNJ2 in non‐translocated patients with PRS. To determine the extent of the involvement of SOX9 and KCNJ2 in PRS, skeletal x ray surveys and sequencing of SOX9 and KCNJ2 candidate cis‐regulatory regions and signalling partners in a larger group of patients are needed.
Key points
The aetiology of isolated Pierre Robin sequence (PRS) is unknown.
Expression analysis and cytogenetic analysis were performed in patients with isolated PRS.
The data point to a role for SOX9 and KCNJ2 dysregulation in isolated PRS.
Electronic database information
The URLs for data presented herein are as follows:
Database of Genomic Variants, http://www.projects.tcag.ca/variation/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
BACPAC Resources, http://bacpac.chori.org
UCSC Genome Bioinformatics site, March 2006 assembly, http://genome.ucsc.edu/
The Mendelian Cytogenetics Network Database (MCNdb), http://www.mcndb.org/
NCBI dbSNP Build 125, http://www.ncbi.nlm.nih.gov/SNP/
miRBase Targets Version 2.0, http://microrna.sanger.ac.uk/targets/v2/
Sanger Institute, http://sanger.ac.uk/
Supplementary table showing primer sequences and PCR conditions, mRNA isolation and cDNA preparation is available at http://jmg.bmj.com/supplemental
Acknowledgements
We thank the patients and their families for their participation in the study. The study was approved by the local scientific ethics committee, and informed consent was obtained from all the patients and/or their families. Written consent has been obtained from the patient and the mother for the publication of the photographs. We thank David FitzPatrick and Judy Fantes for helpful suggestions and assistance in FISH mapping. We also thank Lillian Rasmussen, Anita Niebuhr, Elisabeth Larsen, Minna Becker and Marei Schubert for skilful technical assistance. DNA for the array CGH set was provided by Pieter de Jong and the clones for the 1 Mb Sanger set were provided by Nigel Carter. The work was partly supported by grants from the Augustinus Foundation, the Vanfoere Foundation, Doctor Johan Boserup and Lise Boserups Foundation, Aase and Ejnar Danielsens Foundation, Jacob Madsen and wife Olga Madsens Foundation and The Research Foundation of the Queen Louise Childrens Hospital. The Wilhelm Johannsen Centre for Functional Genome Research was established by the Danish National Research Foundation.
Abbreviations
BAC - bacterial artificial chromosome
CD - campomelic dysplasia
CGH - comparative genome hybridisation
CL/P cleft lip and/or palate -
FISH - fluorescence in situ hybridisation
MCNdb - Mendelian Cytogenetics Network database
miRNA - microRNA
PRS - Pierre Robin sequence
QPCR - quantitative real‐time PCR
SNP - single‐nucleotide polymorphism
Footnotes
Competing interests: None.
Informed consent was obtained for publication of fig 1.
Supplementary table showing primer sequences and PCR conditions, mRNA isolation and cDNA preparation is available at http://jmg.bmj.com/supplemental
References
- 1.Jugessur A, Murray J C. Orofacial clefting: recent insights into a complex trait. Curr Opin Genet Dev 200515270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holder‐Espinasse M, Abadie V, Cormier‐Daire V, Beyler C, Manach Y, Munnich A, Lyonnet S, Couly G, Amiel J. Pierre Robin sequence: a series of 117 consecutive cases. J Pediatr 2001139588–590. [DOI] [PubMed] [Google Scholar]
- 3.Mansour S, Hall C M, Pembrey M E, Young I D. A clinical and genetic study of campomelic dysplasia. J Med Genet 199532415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pop R, Conz C, Lindenberg K S, Blesson S, Schmalenberger B, Briault S, Pfeifer D, Scherer G. Screening of the 1 Mb SOX9 5′ control region by array CGH identifies a large deletion in a case of campomelic dysplasia with XY sex reversal. J Med Genet 200441e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olney P N, Kean L S, Graham D, Elsas L J, May K M. Campomelic syndrome and deletion of SOX9. Am J Med Genet 19998420–24. [PubMed] [Google Scholar]
- 6.Pfeifer D, Kist R, Dewar K, Devon K, Lander E S, Birren B, Korniszewski L, Back E, Scherer G. Campomelic dysplasia translocation breakpoints are scattered over 1 Mb proximal to SOX9: evidence for an extended control region. Am J Hum Genet 199965111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill‐Harfe K L, Kaplan L, Stalker H J, Zori R T, Pop R, Scherer G, Wallace M R. Fine mapping of chromosome 17 translocation breakpoints > or = 900 kb upstream of SOX9 in acampomelic campomelic dysplasia and a mild, familial skeletal dysplasia. Am J Hum Genet 200576663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velagaleti G V, Bien‐Willner G A, Northup J K, Lockhart L H, Hawkins J C, Jalal S M, Withers M, Lupski J R, Stankiewicz P. Position effects due to chromosome breakpoints that map approximately 900 kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am J Hum Genet 200576652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamshidi N, Macciocca I, Dargaville P A, Thomas P, Kilpatrick N, McKinlay Gardner R J, Farlie P G. Isolated Robin sequence associated with a balanced t(2;17) chromosomal translocation. J Med Genet 200441e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi W, Huang W, Whitworth D J, Deng J M, Zhang Z, Behringer R R, de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci USA 2001986698–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nie X. Sox9 mRNA expression in the developing palate and craniofacial muscles and skeletons. Acta Odontol Scand 20066497–103. [DOI] [PubMed] [Google Scholar]
- 12.Erdogan F, Chen W, Kirchhoff M, Kalscheuer V M, Hultschig C, Müller I, Schulz R, Menzel C, Bryndorf T, Ropers H ‐ H, Ullmann R. Impact of low copy repeats on the generation of balanced and unbalanced chromosomal aberrations in mental retardation. Cytogenet Genome Res 2006115247–253. [DOI] [PubMed] [Google Scholar]
- 13.Pfaffl M W, Tichopad A, Prgomet C, Neuvians T P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper‐Excel‐based tool using pair‐wise correlations. Biotechnol Lett 200426509–515. [DOI] [PubMed] [Google Scholar]
- 14.Nunes M E, Pagon R A, Disteche C J, Evans J P. A contiguous gene deletion syndrome at 7q21‐q22 and implications for a relationship between isolated ectrodactyly and syndromic ectrodactyly. Clin Dysmorphol 19943277–286. [PubMed] [Google Scholar]
- 15.Berezikov E, Plasterk R H. Camels and zebrafish, viruses and cancer: a microRNA update. Hum Mol Genet 200514R183–R190. [DOI] [PubMed] [Google Scholar]
- 16.Wirth J, Wagner T, Meyer J, Pfeiffer R A, Tietze H U, Schempp W, Scherer G. Translocation breakpoints in three patients with campomelic dysplasia and autosomal sex reversal map more than 130 kb from SOX9. Hum Genet 199697186–193. [DOI] [PubMed] [Google Scholar]
- 17.Unger S. The mildest form of campomelic dysplasia. Am J Med Genet A 2005132113. [DOI] [PubMed] [Google Scholar]
- 18.Stalker H J, Zori R T, Wallace M, Hill‐Harfe K L, Kaplan L. Reply to Unger: the mildest form of campomelic dysplasia. Am J Med Genet A 2005132114–115. [DOI] [PubMed] [Google Scholar]
- 19.Zaritsky J J, Eckman D M, Wellman G C, Nelson M T, Schwarz T L. Targeted disruption of Kir2.1 and Kir2.2 genes reveals the essential role of the inwardly rectifying K(+) current in K(+)‐mediated vasodilation. Circ Res 200087160–166. [DOI] [PubMed] [Google Scholar]
- 20.Stoll C, Kieny J R, Dott B, Alembik Y, Finck S. Ventricular extrasystoles with syncopal episodes, perodactyly, and Robin in sequence in three generations: a new inherited MCA syndrome? Am J Med Genet 199242480–486. [DOI] [PubMed] [Google Scholar]
- 21.Geis N, Seto B, Bartoshesky L, Lewis M B, Pashayan H M. The prevalence of congenital heart disease among the population of a metropolitan cleft lip and palate clinic. Cleft Palate J 19811819–23. [PubMed] [Google Scholar]
- 22.Merla G, Howald C, Henrichsen C N, Lyle R, Wyss C, Zabot M T, Antonarakis S E, Reymond A. Submicroscopic deletion in patients with Williams‐Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet 200679332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinjan D A, van Heyningen V. Long‐range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 2005768–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonaldo M F, Lennon G, Soares M B. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res 19966791–806. [DOI] [PubMed] [Google Scholar]
- 25.de Crombrugghe B, Lefebvre V, Behringer R R, Bi W, Murakami S, Huang W. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol 200019389–394. [DOI] [PubMed] [Google Scholar]
- 26.Melkoniemi M, Koillinen H, Mannikko M, Warman M L, Pihlajamaa T, Kaariainen H, Rautio J, Hukki J, Stofko J A, Cisneros G J, Krakow D, Cohn D H, Kere J, Ala‐Kokko L. Collagen XI sequence variations in nonsyndromic cleft palate, Robin sequence and micrognathia. Eur J Hum Genet 200311265–270. [DOI] [PubMed] [Google Scholar]