Abstract
Non‐syndromic cleft lip with or without palate (CL/P) is one of the most common malformations among live births, but most of the genetic components and environmental factors involved remain to be identified. Among the different causes, MYH9, the gene encoding for the heavy chain of non‐muscle myosin IIA, was considered a potential candidate, because it was found to be abundantly and specifically expressed in epithelial cells of palatal shelves before fusion. After fusion, its expression level was shown to decrease and to become limited to epithelial triangles before disappearing, as fusion is completed.
To determine whether MYH9 plays a role in CL/P aetiology, a family‐based association analysis was performed in 218 case/parent triads using single‐nucleotide polymorphism (SNP) markers. Pairwise and multilocus haplotype analyses identified linkage disequilibrium between polymorphism alleles at the MYH9 locus and the disease. The strongest deviation from a null hypothesis of random sharing was obtained with two adjacent SNPs, rs3752462 and rs2009930 (global p value = 0.001), indicating that MYH9 might be a predisposing factor for CL/P, although its pathogenetic role needs to be investigated more accurately.
Non‐syndromic cleft lip with or without cleft palate (CL/P, MIM 119530) is a common newborn malformation caused by complex and still unknown pathogenetic mechanisms determined by genetic and environmental factors.1,2 The lip and palatal regions develop from the convergence of different growing processes. Nasal processes and maxillary prominences contribute to the lip, the anterior tooth‐bearing alveolus and the anterior palate up to the incisal foramen. As revealed mainly in the mouse, the secondary palate develops as an outgrowth of the maxillary prominences starting from embryo day 11.5. The palatal shelves initially grow vertically down the side of the tongue and then elevate above the tongue as it drops in the oral cavity. With continued horizontal growth, the shelves appose in the midline and fuse by embryo day 15.5. On meeting at the midline of the oropharyngeal cavity, the medial edge epithelia (MEE) of shelves adhere to each other, leading to the medial epithelial seam (MES). The MES initially consists of a multilayered epithelium, and later becomes a single epithelial layer that, through different mechanisms, including epithelial–mesenchymal transformation, apoptosis and cell migration, seals shelves and forms the palate.
Candidate gene studies or genomewide mapping searches for novel loci have led to the identification of genetic factors involved in clefting, such as transcription factors MSX1, IRF6 and TBX22, as well as growth factors transforming growth factor α and transforming growth factor β3 (TGFβ3), or the cell adhesion molecule poliovirus receptor‐related 1.2 Consistent with their role in palatogenesis, they are highly transcribed during palate formation. For instance, TGFβ3 is abundantly and specifically expressed in the MEE cells of pre‐fusion shelves, and its expression ceases shortly after the MES is formed, whereas IRF6—responsible for van der Woude syndrome, an autosomal dominant form of cleft lip and palate—is highly expressed in the MEE of the paired palatal shelves immediately before and during their fusion.3,4
To improve our understanding of the physiopathological mechanisms responsible for CL/P, we regarded the MYH9 gene as a novel potential candidate because we found that it was highly expressed in palatal shelves.5 MYH9 encodes for the heavy chain of non‐muscle myosin IIA (NMMHCIIA, MIM 160775), one of the three myosins of class II expressed in non‐muscle cells to exert contractile force, which is needed for many often unknown cellular functions.6 We first characterised more deeply its pattern of expression during palatal development in the mouse, and then explored whether the gene was in linkage disequilibrium (LD) with CL/P, using a family‐based LD approach.6 Pairwise and multilocus haplotype analyses identified an LD between MYH9 alleles and the disease, suggesting that determinants of CL/P susceptibility that are not yet identified are localised within the MYH9 locus.
Materials and methods
Myh9 and TGFβ3 in situ hybridisation and immunohistochemistry
A mouse Myh9 antisense probe was obtained by linearisation of clone AA575507 (IMAGE) with SalI and transcription with SP6 RNA polymerase, while digestion of the same plasmid with NotI and transcription with T7 RNA polymerase generated a sense control probe. The mouse Tgfβ3 probe was obtained by reverse transcription‐PCR from the RNA of mouse palate at embryo day 14.5 using the primers Tgfβ3IF (5′‐GAGCCCCTGACCATCTTGTAC‐3′) and Tgfβ3IR (5′‐CCTCTGCTTTTGAGTCCAGC‐3′) for amplification. The PCR product was subcloned into the pCRII‐TOPO vector (TOPO TA Cloning, Invitrogen, Milan, Italy). The plasmid was linearised either with BamHI (antisense probe) or with NotI (sense probe), and transcribed with T7 and SP6 RNA polymerase, respectively.
Mouse embryos at day 14.5 were harvested from CD1 pregnant females and fixed with 4% paraformaldehyde in phosphate‐buffered saline (PBS) overnight at 4°C. For in situ hybridisation, sections were analysed as described previously.5 We used adjacent sections for hybridisation of the two Myh9 and Tgfβ3 genes. Slides were coverslipped with 70% glycerol in PBS and photographed (AxioCam digital camera, Zeiss, CarlZeiss, Milan, Italy) using a microscope with Nomarski optics (Axioplan, Zeiss).
For immunohistochemical analysis, gelatin tissues were sectioned (15 μm) and mounted on Superfrost‐pretreated glass slides (Fisher Scientific, Pittsburgh, Pennsylvania, USA). The mouse sections were fixed in 4% paraformaldehyde in PBS for 15 min and then incubated with a blocking reagent, 10% goat serum in PBS for 1 h. The blocked sections were incubated in the presence and absence of a polyclonal antibody against NMMHCIIA (Covance, Princeton, New Jersey, USA) at dilution 1:200. The sections were then washed with PBS and incubated with a biotin‐conjugated antirabbit IgG secondary antibody as described previously.7
Sets of families
A sample consisting of 218 patients from Italy with non‐syndromic CL/P and their parents was used in this study. Specifically, 65 patients had only cleft lip, whereas 153 had both lip and palate fusion defects. Patients with only cleft palate were excluded from the study. Since it is not clear whether cleft lip and lip and palate fusion defects are separate clinical entities or are different expressions of the same pathology, the two groups were pooled, thus increasing the power of statistical analyses.8,9 To classify the CL/P as non‐syndromic and to exclude potential teratogenic influences, a careful anamnesis was carried out to evaluate the presence of any other somatic or neurological disorders in the family and the use of clefting substances, such as phenytoin, warfarin and ethanol, during pregnancy. Although 131 cases were considered sporadic or non‐familial, as no other relatives manifested the malformation, 87 were familiar cases, because of the recurrence of CL/P within each unrelated pedigree. After obtaining informed consent, DNA was extracted from peripheral blood samples as described previously.10
Markers
Eight SNPs within the MYH9 locus were analysed for LD (table 1). SNP rs5995288 was selected because it maps in a regulative region, whereas SNP rs2269529 introduces the Ile1626Val protein variant in the NMMHCIIA and might be regarded as an actual susceptibility factor.11 These two polymorphisms were typed by restriction enzyme digestion of PCR products (primers and protocols are available on request). The other six SNPs were selected from among validated assays, using the Applied Biosystems SNPbrowser Software. Selection was made considering the exon position in the MYH9 locus, and preference was accorded to SNPs with low inter marker LD and minor allele frequency >0.2. Genotypes were obtained using an ABI PRISM 7700 Sequence Detection System and the TaqMan chemistry according to Applied Biosystems (Milan, Italy) protocols.
Table 1 Single‐nucleotide polymorphisms selected from within the MYH9 locus and their relative transmission disequilibrium test results.
| n | dbSNP id | Genome position* | bp to next | Gene position | Alleles† | MAF‡ | T/NT§ | p Value¶ |
|---|---|---|---|---|---|---|---|---|
| 1 | rs5995288 | chr22:35087134 | 16555 | Intron 1 | G/A | 0.36 | 91/88 | 0.82 |
| 2 | rs739097 | chr22:35070579 | 27221 | Intron 1 | T/C | 0.49 | 85/94 | 0.50 |
| 3 | rs2071731 | chr22:35043358 | 4199 | Intron 6 | C/T | 0.41 | 93/95 | 0.88 |
| 4 | rs1002246 | chr22:35039159 | 4476 | Intron 10 | C/T | 0.29 | 81/81 | 1.00 |
| 5 | rs3752462 | chr22:35034683 | 10877 | Intron 13 | G/A | 0.41 | 84/116 | 0.02 |
| 6 | rs2009930 | chr22:35023806 | 14952 | Intron 19 | T/C | 0.40 | 95/107 | 0.40 |
| 7 | rs2269529 | chr22:35008854 | 6440 | Exon 33 | A/G | 0.26 | 78/83 | 0.69 |
| 8 | rs7078 | chr22:35002414 | — | Exon.40‐3′UTR | T/C | 0.29 | 72/84 | 0.34 |
MAF, minor allele frequency; SNP, single‐nucleotide polymorphism; T/NT, transmission/non‐transmission; UTR, untranslated.
*UCSC Genome Browser on Human May 2004 Assembly.
†SNP alleles in coding frame, major allele first.
‡MAF calculated from all parental chromosomes.
§T/NT counts from heterozygous parents are given for the major allele.
¶p value for transmission disequilibrium test.
Statistical analysis
LD between SNP alleles and disease was tested by the transmission disequilibrium test (TDT), which examines the transmission of alleles from heterozygous parents to affected offspring.12 Pairwise and multilocus haplotype analyses, in a sliding window up to five markers, were performed using the program TDTPHASE, as part of the UNPHASED package.13 This program provides both global p values, which assess the significance of distortion in transmission for all the test haplotypes, and p values that assess the significance of distortion in transmission for specific haplotypes. The analysis was restricted to phase‐certain haplotypes only, in a conditional logistic regression model. This is equivalent to the extended transmission disequilibrium test that ensures a valid test for either genetic linkage or allelic association.14 A rare haplotype frequency threshold of 0.03 was adopted, because likelihood ratio statistics may be sensitive to rare haplotypes.
LD between markers was also calculated using the D′ and r2 statistics from parental haplotypes, using the program ldmax of the GOLD package.15
Results
MYH9 is highly expressed in the mouse during palate fusion
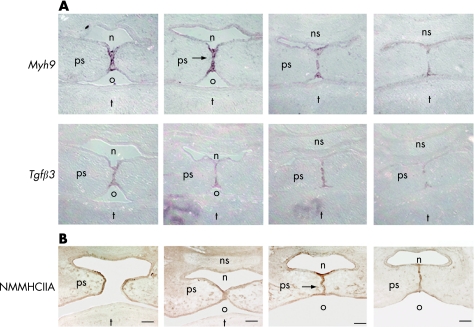
Our previous studies on Myh9 in mice during development showed a high expression of the gene in the palate.5 We looked more accurately into its expression by RNA in situ hybridisation on adjacent sections of the palate at embryo day 14.5, and revealed that the gene was abundantly and specifically expressed in MEE cells of shelves before fusion (fig 1A). Its expression gradually decreases and becomes limited to epithelial triangles after the MES is formed. This pattern of expression overlaps with that of the TGFβ3 gene in both temporal and spatial expression. We also analysed the expression level of the Myh9 product by immunohistochemical analysis. Similar to the mRNA, the protein is expressed at high level, confirming the presence of NMMHCIIA in the MEE cells of paired palate shelves immediately before and during their fusion (fig 1B).
Figure 1 Expression studies during palate fusion in mouse. (A) RNA in situ hybridisation of the Myh9 and transforming growth factorβ3 (Tgfβ3) genes in adjacent sections of embryonic mouse at day 14.5 showing an overlapping pattern of expression. (B) Immunohistochemistry using antibodies against heavy chain of non‐muscle myosin IIA (NMMHCIIA) during palate fusion on mouse sections at embryo day 14.5. The arrows indicate the medial epithelial seam (MES) with the highest expression levels of Myh9 mRNA and protein. n, nasal cavity; ns, nasal septum; o, oral cavity; ps, palate shelf; t, tongue. Scale bars represent 100 μm.
SNPs within MYH9 are in LD with CL/P
Eight MYH9 intragenic SNPs were used to investigate allele and haplotype transmission in a CL/P sample of 218 patient/parent triads. LD between each SNP allele and disease was analysed by standard TDT (table 1).
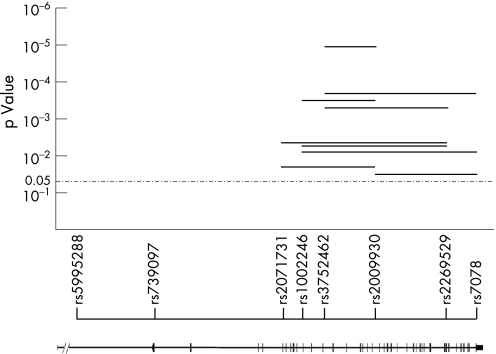
None of the SNPs tested showed evidence of deviation from the Hardy–Weinberg equilibrium in either affected or unaffected individuals. Significant distortion in allele transmission was detected at the rs3752462 locus, where the nucleotide A allele was found to be transmitted and not transmitted in 116 and 84 cases, respectively (p = 0.02). Haplotype analysis of multilocus data is potentially a powerful means to detect association in multifactorial diseases.16 Sliding windows including two to five SNPs were examined. Significant distortion in transmission was observed with different combinations of markers (fig 2). The highest departure from random sharing was observed with two adjacent markers, rs3752462 and rs2009930 (nominal p value <0.001, Bonferroni‐adjusted p value <0.001). Combining single‐marker and multipoint analyses, we performed 30 association tests, 10 of which produced p values lower than the nominal level of significance. The conservative Bonferroni correction for multiple testing produced an adjusted α level of 0.0017. At this level, we obtained four significant tests in multipoint analysis.
Figure 2 Global p values obtained with multilocus haplotype analyses. p Values are plotted on a log10 scale corresponding to the physical map of the single‐nucleotide polymorphisms (SNPs) and exon–intron map of the MYH9 gene.
In order to identify any conserved blocks carrying putative CL/P susceptibility mutation(s), we also defined the haplotypes that were significantly overtransmitted to patients with CL/P from heterozygous parents (table 2).
Table 2 Single‐nucleotide polymorphism alleles and haplotypes overtransmitted to patients with cleft lip with or without cleft palate from heterozygous parents.
| SNPs | T* | NT* | T† (%) | p Value‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||||
| A | 116 | 84 | 58 | 0.014 | |||||||
| C | A | 64 | 38 | 63 | 0.014 | ||||||
| A | C | 80 | 48 | 63 | 0.015 | ||||||
| T | C | A | 36 | 19 | 65 | 0.023 | |||||
| C | A | C | 55 | 28 | 66 | 0.004 | |||||
| C | G | C | 39 | 20 | 66 | 0.017 | |||||
| T | C | A | C | 31 | 15 | 67 | 0.019 | ||||
| A | C | G | C | 39 | 16 | 71 | 0.003 | ||||
| T | C | A | C | G | 25 | 8 | 76 | 0.002 | |||
| C | A | C | A | T | 16 | 6 | 73 | 0.030 | |||
| C | A | C | G | C | 20 | 9 | 69 | 0.039 | |||
NT, non‐transmitted; SNP, single‐nucleotide polymorphism; T, transmitted.
*Counts of T and NT haplotypes from heterozygous parents.
†Transmission percentage.
‡ p Value for transmission disequilibrium test.
All the overtransmitted haplotypes are characterised by the presence of either allele A at marker 5 (rs3752462) or allele C at marker 6 (rs2009930), or both A and C. Interestingly, the longer the haplotype, the higher its percentage of transmission. LD between alleles at different loci was examined by means of D′ and r2 estimation (table 3). A considerable amount of LD—that is, D′>0.9 and r2>0.45 (typed bold in table 3), was observed between markers 2 and 3, and between markers 5, 6 and 7.
Table 3 Linkage disequilibrium (D′ and r2) between single‐nucleotide polymorphisms within the MYH9 locus.
| SNPs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.21 | 0.18 | 0.04 | 0.13 | 0.13 | 0.19 | 0.19 | |
| 2 | 0.03 | 0.92 | 0.29 | 0.61 | 0.62 | 0.72 | 0.21 | |
| 3 | 0.03 | 0.61 | 0.30 | 0.40 | 0.40 | 0.58 | 0.29 | |
| 4 | 0.00 | 0.04 | 0.05 | 0.48 | 0.46 | 0.26 | 0.20 | |
| 5 | 0.02 | 0.26 | 0.15 | 0.14 | 0.90 | 0.94 | 0.27 | |
| 6 | 0.01 | 0.26 | 0.15 | 0.13 | 0.80 | 0.95 | 0.30 | |
| 7 | 0.02 | 0.19 | 0.17 | 0.06 | 0.46 | 0.48 | 0.30 | |
| 8 | 0.01 | 0.02 | 0.05 | 0.04 | 0.04 | 0.06 | 0.08 |
SNP, single‐nucleotide polymorphism.
D′ values are shown above the diagonal, whereas r2 values are shown below the diagonal; the highest are in bold.
Discussion
Non‐syndromic CL/P is a complex trait with no obvious mode of inheritance, and numerous studies have failed to identify genes with any major influence on the disease.2 The MYH9 gene was analysed in the present study because, as previously observed, it was highly expressed in shelves during palate fusion.5 Accurate studies revealed that both gene and protein were abundantly expressed in the MEE before fusion; after the MES was formed, their expression remained high but was limited to epithelial triangles and, as fusion was completed, it was no longer detectable. This pattern of expression overlaps with that of the TGFβ3 gene in both temporal and spatial expression, suggesting that MYH9 might play an important role during palate development. Since we supposed that the molecular mechanisms involved in epithelial fusion and epithelial–mesenchymal differentiation during lip or palate development at least partially overlap, we undertook a family‐based association study using a CL/P sample, enrolled previously,17 to test the hypothesis of MYH9 involvement in CL/P. The analysis of allele and haplotype transmission in 218 patient–parent triads strongly supports an association between CL/P and MYH9. Using both pairwise and multipoint analyses, we performed 30 significance tests to verify the hypothesis. Of note, in our study, 10 out of 30 association tests obtained p values <0.05, when it is accepted that 1 out of 20 is significant by chance alone. Such a result should be indicative of a true association. Methods to correct the significance threshold for multiple testing in genetic association studies with linked markers and in multipoint analysis are still a matter of debate.18 Bonferroni correction by the number of combinations considered is one method to deal with multiple‐testing problems. This method is appropriate for independent tests, but may be judged excessively conservative in the present case. Nevertheless, four tests still proved significant using a crude and conservative Bonferroni‐corrected threshold of 0.0017. A maximum value of LD was obtained via multipoint analysis with SNPs rs3752462 and rs2009930 (nominal p value ⩽ = 0.001, Bonferroni‐adjusted p value <0.001), which are localised in the MYH9 genomic region spanning from exon 13 to exon 20.
As in other complex multifactorial diseases, the goal is to identify susceptibility alleles. This is itself a difficult task in complex diseases, and is still more complicated in our case owing to the complexity of MYH9, being a large gene with 40 coding exons spread >100 kb over the genome. Independent LD studies should thus be undertaken to confirm the involvement of MYH9, before proceeding to any fine mutational analysis.
Mutations in the MYH9 gene cause MYH9‐related disease (MYH9RD), an autosomal dominant disease characterised by congenital macrothrombocytopenia, NMMHCIIA inclusions in the neutrophils, hearing impairment, cataract and renal failure.19 At least to the best of our knowledge, no patient with MYH9RD has been reported as having palate defects. However, it is to be noted that there is a very limited spectrum of MYH9 mutations associated with the disease. They consist mainly of missense mutations, occurring to a few amino acids—out of the possible 1961 residues—that are well conserved in evolution but are located in regions of poorly defined functional significance. They also include two in‐frame deletions, as well as a few nonsense and frameshift mutations, all localised within a small region at the extreme COOH‐terminus (http://www.hgmd.cf.ac.uk/ac/gene.php?gene = MYH9). In particular, four of these residues, 702, 1424, 1881 and 1933, are mutated in >50% of families.
Moreover, whether these mutations determine haploinsufficiency or act through a dominant‐negative mechanism is still unclear.20,21,22,23 Specifically, no gross deletions at the MYH9 locus have been identified in patients, which prevents us from confirming haploinsufficiency as the pathogenetic mechanism. In the mouse, by contrast, homozygous destruction of MYH9 is lethal early during embryogenesis, while one knockout allele is not associated with any pathological phenotype, suggesting that the mouse is not a good model for the disease, nor is haploinsufficiency the pathogenetic mechanism of MYH9RD.24,25 Accordingly, we cannot exclude that MYH9 alleles different from those identified so far might be associated with or predisposed to allelic disorders such as CL/P, although patients with MYH9RD do not manifest palate or lip defects.26
Besides the well‐characterised role of class II myosins in contraction and force production in muscles, little is known about their specific functional role in non‐muscle cells.6 Some evidence indicates that the non‐muscle isoforms have different roles in cytokinesis, phagocytosis, maintenance of shape, organelle/particle trafficking, as well as cell mobility. NMMHCIIA is likely to play a role during the diverse cellular processes implicated in palate fusion, including epithelial–mesenchymal transformation, migration and cell death. Although epithelial–mesenchymal transformation is considered relevant for the degeneration of MEE,27,28 migration of MEE cells towards the nasal and oral regions has also been proposed as contributing to shelf fusion.29 Recent data indicate that migration mainly involves periderm cells, ordered migration of which from the basal MEE is a prerequisite for normal fusion.30 Another mechanism involved in shelf fusion is MEE cell death, which seems to lead to basal lamina degradation and fusion.31
Turning an epithelial cell into a mesenchymal cell requires alterations in morphology, cellular architecture, adhesion and migration capacity, all mechanisms in which NMMHCIIA is expected to play a fundamental role.6 Although it is well known that motility is based on actin and myosin, NMMHCIIA function in cell death has not been investigated accurately. Apoptosis leads to morphological changes including cell contraction, dynamic membrane blebbing and nuclear disintegration, the contractile force required for which is generated by actin–myosin cytoskeletal structures.32
Key points
We considered MYH9, the gene encoding for the heavy chain of non‐muscle myosin IIA, as a potential candidate for non‐syndromic cleft lip with or without cleft palate (CL/P) because it was found to be abundantly and specifically expressed in epithelial cells of palatal shelves before fusion.
Family‐based association analysis identified linkage disequilibrium between polymorphism alleles at the MYH9 locus and the disease. The strongest deviation from a null hypothesis of random sharing was obtained with two adjacent single‐nucleotide polymorphisms, rs3752462 and rs2009930 (global p value <0.001).
Expression and genetic data indicate that MYH9 might be a predisposing factor for CL/P, although its pathogenetic role needs to be investigated more accurately.
The absence of the TGFβ3 gene, the high expression level of which overlaps with that of MYH9 during palate fusion, leads to alterations in the development of MEE, with the absence of the filopodia and lamellipodia needed for adhesion and for triggering the fusion event.33 Consistent with a role of NMMHCIIA in the formation of these structures, it colocalises with the actin bundles and is detected in the rear part of the lamellipodia and in the lamellipodia–cell body transition zone, where it presumably provides force for the cell body to adapt the advancing lamellipodium.34
In conclusion, on the basis of the expression data, we conjectured that MYH9 was a candidate gene for CL/P and carried out LD analyses. We found a statistically significant correlation between two SNPs and the disease, suggesting that further analysis of this gene is of fundamental importance for unravelling the pathogenetic mechanisms involved in palate defects.
Electronic database information
Accession numbers and the URL for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
Human genome draft, http://genome.ucsc.edu/cgi‐bin/hgGateway
SNP database, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = snp
Acknowledgements
This study was supported by the Italian Telethon Foundation with grants E. 1147 (PC) and GGP05147 (PC and AS), the Italian Ministry of University and Scientific Research (FC and LS), and the Fondazione Cassa di Risparmio di Bologna (PC).
Abbreviations
CL/P - cleft lip with or without cleft palate
LD - linkage disequilibrium
MEE - medial edge epithelia
MES - medial epithelial seam
NMMHCIIA - heavy chain of non‐muscle myosin IIA
PBS - phosphate‐buffered saline
SNP - single‐nucleotide polymorphism
TDT - transmission disequilibrium test
TGFβ3 - transforming growth factor β3
Footnotes
Competing interests: None declared.
References
- 1.Carinci F, Pezzetti F, Scapoli L, Martinelli M, Avantaggiato A, Carinci P, Padula E, Baciliero U, Gombos F, Laino G, Rullo R, Cenzi R, Carls F, Tognon M. Recent developments in orofacial cleft genetics. J Craniofac Surg 200314130–143. [DOI] [PubMed] [Google Scholar]
- 2.Jugessur A, Murray J C. Orofacial clefting: recent insights into a complex trait. Curr Opin Genet Dev 200515270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Proetzel G, Pawlowski S A, Wiles M V, Yin M, Boivin G P, Howles P N, Ding J, Ferguson M W, Doetschman T. Transforming growth factor‐beta 3 is required for secondary palate fusion. Nat Genet 199511409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondo S, Schutte B C, Richardson R J.et al Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet 200232285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marigo V, Nigro A, Pecci A, Montanaro D, Di Stazio M, Balduini C L, Savoia A. Correlation between the clinical phenotype of MYH9‐related disease and tissue distribution of class II nonmuscle myosin heavy chains. Genomics 2004831125–1133. [DOI] [PubMed] [Google Scholar]
- 6.Sellers J R. Myosins: a diverse superfamily. Biochim Biophys Acta 200014963–22. [DOI] [PubMed] [Google Scholar]
- 7.Donaudy F, Snoeckx R, Pfister M, Zenner H P, Blin N, Di Stazio M, Ferrara A, Lanzara C, Ficarella R, Declau F, Pusch C M, Nurnberg P, Melchionda S, Zelante L, Ballana E, Estivill X, Van Camp G, Gasparini P, Savoia A. Nonmuscle myosin heavy‐chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4). Am J Hum Genet 200474770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harville E W, Wilcox A J, Lie R T, Vindenes H, Abyholm F. Cleft lip and palate versus cleft lip only: are they distinct defects? Am J Epidemiol 2005162448–453. [DOI] [PubMed] [Google Scholar]
- 9.Fraser F C. The genetics of cleft lip and cleft palate. Am J Hum Genet 197022336–352. [PMC free article] [PubMed] [Google Scholar]
- 10.Carinci F, Pezzetti F, Scapoli L, Padula E, Baciliero U, Curioni C, Tognon M. Nonsyndromic cleft lip and palate: evidence of linkage to a microsatellite marker on 6p23. Am J Hum Genet 199556337–339. [PMC free article] [PubMed] [Google Scholar]
- 11.Beohar N, Kawamoto S. Transcriptional regulation of the human nonmuscle myosin II heavy chain‐A gene. Identification of three clustered cis‐elements in intron‐1 which modulate transcription in a cell type‐ and differentiation state‐dependent manner. J Biol Chem 19982739168–9178. [DOI] [PubMed] [Google Scholar]
- 12.Spielman R S, McGinnis R E, Ewens W J. Transmission test for linkage disequilibrium: the insulin gene region and insulin‐dependent diabetes mellitus (IDDM). Am J Hum Genet 199352506–516. [PMC free article] [PubMed] [Google Scholar]
- 13.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 200325115–121. [DOI] [PubMed] [Google Scholar]
- 14.Sham P C, Curtis D. An extended transmission/disequilibrium test (TDT) for multi‐allele marker loci. Ann Hum Genet 199559(Pt 3)323–336. [DOI] [PubMed] [Google Scholar]
- 15.Abecasis G R, Cookson W O. GOLD—graphical overview of linkage disequilibrium. Bioinformatics 200016182–183. [DOI] [PubMed] [Google Scholar]
- 16.Merriman T R, Eaves I A, Twells R C, Merriman M E, Danoy P A, Muxworthy C E, Hunter K M, Cox R D, Cucca F, McKinney P A, Shield J P, Baum J D, Tuomilehto J, Tuomilehto‐Wolf E, Ionesco‐Tirgoviste C, Joner G, Thorsby E, Undlien D E, Pociot F, Nerup J, Ronningen K S, Bain S C, Todd J A. Transmission of haplotypes of microsatellite markers rather than single marker alleles in the mapping of a putative type 1 diabetes susceptibility gene (IDDM6). Hum Mol Genet 19987517–524. [DOI] [PubMed] [Google Scholar]
- 17.Martinelli M, Scapoli L, Palmieri A, Pezzetti F, Baciliero U, Padula E, Carinci P, Morselli P G, Carinci F. Study of four genes belonging to the folate pathway: transcobalamin 2 is involved in the onset of non‐syndromic cleft lip with or without cleft palate. Hum Mutat 200627294. [DOI] [PubMed] [Google Scholar]
- 18.Nyholt D R. A simple correction for multiple testing for single‐nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 200474765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seri M, Pecci A, Di Bari F, Cusano R, Savino M, Panza E, Nigro A, Noris P, Gangarossa S, Rocca B, Gresele P, Bizzaro N, Malatesta P, Koivisto P A, Longo I, Musso R, Pecoraro C, Iolascon A, Magrini U, Rodriguez Soriano J, Renieri A, Ghiggeri G M, Ravazzolo R, Balduini C L, Savoia A. MYH9‐related disease: May‐Hegglin anomaly, Sebastian syndrome, Fechtner syndrome, and Epstein syndrome are not distinct entities but represent a variable expression of a single illness. Medicine (Baltimore) 200382203–215. [DOI] [PubMed] [Google Scholar]
- 20.Kunishima S, Matsushita T, Kojima T, Sako M, Kimura F, Jo E K, Inoue C, Kamiya T, Saito H. Immunofluorescence analysis of neutrophil nonmuscle myosin heavy chain‐A in MYH9 disorders: association of subcellular localization with MYH9 mutations. Lab Invest 200383115–122. [DOI] [PubMed] [Google Scholar]
- 21.Deutsch S, Rideau A, Bochaton‐Piallat M L, Merla G, Geinoz A, Gabbiani G, Schwede T, Matthes T, Antonarakis S E, Beris P. Asp1424Asn MYH9 mutation results in an unstable protein responsible for the phenotypes in May‐Hegglin anomaly/Fechtner syndrome. Blood 2003102529–534. [DOI] [PubMed] [Google Scholar]
- 22.Pecci A, Canobbio I, Balduini A, Stefanini L, Cisterna B, Marseglia C, Noris P, Savoia A, Balduini C L, Torti M. Pathogenetic mechanisms of hematological abnormalities of patients with MYH9 mutations. Hum Mol Genet 2005143169–3178. [DOI] [PubMed] [Google Scholar]
- 23.Franke J D, Dong F, Rickoll W L, Kelley M J, Kiehart D P. Rod mutations associated with MYH9‐related disorders disrupt nonmuscle myosin‐IIA assembly. Blood 2005105161–169. [DOI] [PubMed] [Google Scholar]
- 24.Conti M A, Even‐Ram S, Liu C, Yamada K M, Adelstein R S. Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II‐A in mice. J Biol Chem 200427941263–41266. [DOI] [PubMed] [Google Scholar]
- 25.Matsushita T, Hayashi H, Kunishima S, Hayashi M, Ikejiri M, Takeshita K, Yuzawa Y, Adachi T, Hirashima K, Sone M, Yamamoto K, Takagi A, Katsumi A, Kawai K, Nezu T, Takahashi M, Nakashima T, Naoe T, Kojima T, Saito H. Targeted disruption of mouse ortholog of the human MYH9 responsible for macrothrombocytopenia with different organ involvement: hematological, nephrological, and otological studies of heterozygous KO mice. Biochem Biophys Res Commun 20043251163–1171. [DOI] [PubMed] [Google Scholar]
- 26.Romeo G, McKusick V A. Phenotypic diversity, allelic series and modifier genes. Nat Genet 19947451–453. [DOI] [PubMed] [Google Scholar]
- 27. Fitchett JE, Hay ED. Medial edge epithelium transforms to mesenchyme after embryonic palatal shelves fuse. Dev Biol 1989131455–474. [DOI] [PubMed] [Google Scholar]
- 28.Martinez‐Alvarez C, Tudela C, Perez‐Miguelsanz J, O'Kane S, Puerta J, Ferguson M W. Medial edge epithelial cell fate during palatal fusion. Dev Biol 2000220343–357. [DOI] [PubMed] [Google Scholar]
- 29.Carette M J, Ferguson M W. Mouse embryonic palatal epithelial sheets in culture: an immunocytochemical study of proliferative activity using bromodeoxyuridine. Epithelial Cell Biol 19921119–127. [PubMed] [Google Scholar]
- 30.Cuervo R, Covarrubias L. Death is the major fate of medial edge epithelial cells and the cause of basal lamina degradation during palatogenesis. Development 200413115–24. [DOI] [PubMed] [Google Scholar]
- 31.Cuervo R, Valencia C, Chandraratna R A, Covarrubias L. Programmed cell death is required for palate shelf fusion and is regulated by retinoic acid. Dev Biol 2002245145–156. [DOI] [PubMed] [Google Scholar]
- 32.Croft D R, Coleman M L, Li S, Robertson D, Sullivan T, Stewart C L, Olson M F. Actin‐myosin‐based contraction is responsible for apoptotic nuclear disintegration. J Cell Biol 2005168245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lidral A C, Romitti P A, Basart A M, Doetschman T, Leysens N J, Daack‐Hirsch S, Semina E V, Johnson L R, Machida J, Burds A, Parnell T J, Rubenstein J L, Murray J C. Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. Am J Hum Genet 199863557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson K I, Wang Y L, Small J V. Coordination of protrusion and translocation of the keratocyte involves rolling of the cell body. J Cell Biol 19961341209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]